Abstract
There is an urgent need to develop new pathogenic R5 simian/human immunodeficiency viruses (SHIVs) for the evaluation of candidate anti-HIV vaccines in nonhuman primates. Here, we characterize swarm SHIVAD8 stocks, prepared from three infected rhesus macaques with documented immunodeficiency at the time of euthanasia, for their capacity to establish durable infections in macaques following inoculation by the intravenous (i.v.) or intrarectal (i.r.) route. All three viral stocks (SHIVAD8-CE8J, SHIVAD8-CK15, and SHIVAD8-CL98) exhibited robust replication in vivo and caused marked depletion of CD4+ T cells affecting both memory and naïve CD4+ T lymphocyte subsets following administration by either route. Eleven of 22 macaques inoculated with the new SHIVAD8 stocks were euthanized with clinical symptoms of immunodeficiency and evidence of opportunistic infections (Pneumocystis, Candida, and Mycobacterium). A single but unique founder virus, also present in the SHIVAD8-CE8J swarm stock, was transmitted to two animals following a single i.r. inoculation of approximately 3 50% animal infectious doses, which is close to the threshold required to establish infection in all exposed animals. Because the three new SHIVAD8 viruses are mucosally transmissible, exhibited tier 2 sensitivity to anti-HIV-1 neutralizing antibodies, deplete CD4+ T lymphocytes in vivo, and induce AIDS in macaques, they are eminently suitable as challenge viruses in vaccine experiments.
INTRODUCTION
Nonhuman primate surrogate models of human immunodeficiency virus (HIV)/AIDS have been used for the past 25 years in the quest to develop an effective prophylactic vaccine. In many of these studies, two genetically distinct simian immunodeficiency virus (SIV) lineages (SIVmac239/SIVmac251 and SIVsmE660) have been utilized as challenge viruses in experiments that have focused almost exclusively on immunogens eliciting antivirus cell-mediated immune responses. SIV/HIV-1 chimeric viruses (simian/human immunodeficiency viruses [SHIVs]), which usually carry the HIV-1 env gene inserted into an SIVmac239 genetic background, have also been used to challenge vaccinated macaques. These SHIVs were originally constructed to assess vaccine-directed anti-HIV humoral immune responses. The early pathogenic SHIVs invariably expressed a CXCR4-utilizing (X4) gp120 envelope glycoprotein in infected macaques and induced rapid and complete elimination of CD4+ T lymphocyte subsets and death from immunodeficiency within a few months of virus inoculation (19, 20, 41). Furthermore, most vaccine regimens consistently suppressed the replication of SHIV challenge viruses (1, 2, 43). Because the rapid disease phenotype observed and the uniformly successful vaccine outcomes were discordant with the results obtained following challenge with commonly used (R5) pathogenic SIVs and were inconsistent with the invariable presence of R5-tropic HIV-1 strains in infected individuals following seroconversion (10), attention shifted to the construction of R5-tropic SHIVs.
The effort to develop R5 SHIVs, in fact, received high priority from a HIV Vaccine Summit convened in 2008 at Bethesda, MD, following the announced failure of the phase 2b STEP HIV-1 vaccine trial (9). Although several clade B and clade C R5-tropic SHIVs have been constructed (5, 13, 17, 29, 36), only one, SHIVSF162 (16), has been widely used as a challenge virus in vaccine experiments. Even though SHIVSF162 generates high levels of peak plasma viremia during acute infection, it expresses an HIV-1 envelope glycoprotein with an extremely low tier 1 neutralization phenotype (44), making it exquisitely sensitive to most anti-HIV-1 neutralizing antibodies (NAbs).
We previously reported the construction of the R5 SHIVAD8 and its replicative adaptation to macaques following serial passage in rhesus monkeys (34), but a challenge virus inoculum suitable for vaccine experiments had not been prepared and characterized. In this earlier work, animals were inoculated by transfusing whole blood from SHIVAD8-infected monkeys or with small virus stocks prepared from peripheral blood mononuclear cells (PBMC) and lymphoid tissues collected from macaques with high virus loads. Our goal in the present study was to characterize three new SHIVAD8 stocks prepared from animals that had developed documented immunodeficiency for use in passive-transfer and vaccine experiments. We report that each of these “swarm” virus stocks exhibits tier 2 sensitivity to NAbs and is efficiently transmitted to macaques by intravenous (i.v.) and mucosal routes. Infected animals experience variable levels of set point viremia, sustain depletion of both memory and naïve CD4+ T cell subsets, and develop clinical disease.
MATERIALS AND METHODS
Preparation of SHIVAD8 virus stocks.
PBMC recovered at the time of euthanasia from three animals infected with R5-SHIVAD8 derivatives were cocultured with concanavalin A (ConA)-stimulated PBMC from uninfected rhesus monkeys (Rh PBMC), as previously described (34). Supernatant media were assayed for progeny virus production by measuring 32P-reverse transcriptase (RT) activity (51). SHIVAD8 stocks prepared from each macaque were designated SHIVAD8-CE8J, SHIVAD8-CK15, and SHIVAD8-CL98, corresponding to their animal identifications (IDs). The infectious titers of SHIVAD8-CE8J, SHIVAD8-CK15, and SHIVAD8-CL98 were 1.83 × 104 50% tissue culture infective doses (TCID50)/ml, 4.09 × 104 TCID50/ml, and 3.49 × 105 TCID50/ml, respectively, as determined in Rh PBMC. The replication kinetics of these viral stocks were evaluated by spinoculating (1,200 × g for 1 h) ConA-stimulated PBMC with these virus stocks, following normalization for 32P-RT activity (33). Virus replication was assessed by 32P-RT assay of the culture supernatant.
Animals.
Rhesus macaques (Macaca mulatta) were maintained in accordance with the guidelines of the Committee on Care and Use of Laboratory Animals (30) and were housed in a biosafety level 2 facility. Phlebotomies, euthanasia, and tissue sample collections were performed as previously described (8). Bronchoalveolar lavage (BAL) fluid lymphocytes were prepared from uninfected and infected animals using a pediatric bronchoscope (Olympus BF3C40; Olympus America, Inc., Melville, NY) as previously described (18). All animals were negative for major histocompatibility complex (MHC) class I alleles Mamu-A*01, Mamu-B*08, and Mamu-B*17.
SHIVAD8 inoculation and quantitation of plasma viral RNA.
One milliliter of SHIVAD8-CE8J, SHIVAD8-CK15, or SHIVAD8-CL98 (titers as indicated above) was inoculated i.v. into two macaques. For intrarectal (i.r.) inoculation, 100 TCID50 to 10,000 TCID50 of each virus stock was administered atraumatically in a volume of 1 ml. Macaques that remained uninfected were reinoculated with the initial i.r. challenge dose; animals still uninfected following the two initial inoculations were challenged with a 10-fold-higher dose intrarectally (Table 1).
Table 1.
Number of challenges needed to cause infection
| Animal | No. of challenges needed to cause infection at (TCID50/ml)a: |
||
|---|---|---|---|
| 100 | 1,000 | 10,000 | |
| SHIVAD8-CE8J infected | |||
| CL5E | ● | ||
| DA24 | ○ ○ | ● | |
| DBJE | ● | ||
| DBZL | ● | ||
| CZH | ● | ||
| DBX9 | ● | ||
| SHIVAD8-CL98 infected | |||
| DA55 | ○ ○ | ● | |
| DB18 | ○ ○ | ○ ● | |
| CL5B | ○ ○ | ● | |
| DB0F | ○ ○ | ● | |
| SHIVAD8-CK15 infected | |||
| DC1W | ● | ||
| DC87 | ● | ||
| DBRL | ● | ||
| DC65 | ● | ||
| BIH | ○ | ● | |
| ENF | ○ | ● | |
○, challenge that did not result in detectable SHIVAD8 infection; ●, challenge that did result in detectable SHIVAD8 infection.
Plasma viral-RNA levels were determined by real-time PCR (7900 HT Sequence Detection System; Applied Biosystems, Foster City, CA), using reverse-transcribed viral RNA from plasma samples of SHIVAD8-infected macaques and primer pairs corresponding to the SIVmac239 gag gene, as previously described (8). The cDNA was amplified employing the following thermal-cycling conditions: 50°C for 2 min, 95°C for 10 min, and 45 cycles of 95°C for 15 s and 60°C for 1 min with primer pairs corresponding to SIVmac239 gag gene sequences (forward, nucleotides [nt] 1890 to 1909, and reverse, nt 2010 to 1991) and probe (nt 1963 to 1990). Plasma from infected macaques, previously quantitated by the branched-DNA method (6), served as standards for the RT-PCR assay.
Lymphocyte-immunophenotyping and intracellular-cytokine assays.
EDTA-treated blood samples and BAL fluid lymphocytes were stained for flow cytometric analysis as described previously (12, 31, 32), using combinations of the following fluorochrome-conjugated monoclonal antibodies (MAbs): CD20 fluorescein isothiocyanate (FITC), CD3 phycoerythrin (PE), CD4 peridinin chlorophyll protein-Cy5.5 (PerCP-Cy5.5), CD8 allophycocyanin (APC), CD95 (APC), and CD28 (FITC). All antibodies were obtained from BD Biosciences (San Diego, CA). Flow-cytometric acquisitions were done with a FACSCalibur flow cytometer (Becton, Dickinson, Franklin Lakes, NJ).
For intracellular cytokine assays, immune stimulation using SIVmac239 Gag peptides (New England Peptide, Gardner, MA), 15 amino acids in length, was performed on frozen or freshly prepared lymphocytes as described previously (33).
Measurement of anti-gp120 antibodies.
An enzyme-linked immunosorbent assay (ELISA) was designed to detect antibodies generated against HIV env with specific gp120 protein as an antigen. Ninety-six-well microtiter plates were coated at 4°C overnight with antigen (Advanced Biosciences, Kensington, MD) at 2 μg/ml in phosphate-buffered saline (PBS) solution using 50 μl/well on a rocking platform. The plates were rinsed four times with wash buffer (PBS-0.05% Tween 20), followed by addition of 200 μl/well of blocking buffer (1% bovine serum albumin [BSA], 0.05% Tween 20 in PBS) and incubation for 2 h at room temperature. Plasma samples were diluted in blocking buffer at 1:100 dilution, and 100 μl was added per well. The plates were incubated for 1 h at room temperature, followed by four washes with washing buffer. Secondary-antibody conjugated with horseradish peroxidase (goat anti-rhesus IgG; Southern Biotech, Birmingham, AL) was diluted at 1:8,000 in blocking buffer, and 100 μl was added to each well, followed by incubation for 1 h at room temperature. The plates were washed four times and then developed by addition of 100 μl/well of an o-phenylenediamine (SigmaFast tablets; Sigma, St. Louis, MO) for 30 min at room temperature. The reaction was stopped by adding 100 μl of 1 M H2SO4. Replicate samples were read at 492-nm absorbance with a microplate reader. The background reactivities were subtracted for each time point using prechallenge plasma controls. The results are presented as the optical density (OD) (absorbance value) read at 492 nm using a Multiscan FC microplate reader (Thermo Scientific, Waltham, MA).
Neutralization assays.
Neutralizing activity in plasma samples of SHIVAD8-infected animals was determined as previously described (34). The assay was performed with a 1:20 dilution of plasma samples in TZM-bl target cells using an infectious Env-pseudovirus of SHIVAD8-CK15 generated in 293-T/17 cells. Any sample resulting in a 50% reduction of luciferase activity compared to that obtained with an uninfected control sample was considered positive for NAbs. All experiments were performed in quadruplicate and repeated twice.
The neutralization phenotypes (tiers) of SHIVAD8 virus stocks were determined by TZM-bl assay (27) using plasma samples from a cohort study, which exhibited a wide range of neutralizing activities against subtype B HIV-1 isolates, as previously described (7).
In addition to the phenotype determination, the neutralization sensitivity of SHIVAD8 virus stocks was measured against available monoclonal antibodies (4, 38, 48, 50, 52, 53) using the TZM-bl luciferase reporter gene assay as described previously (27, 44, 45). DEAE-dextran was added to a final concentration of 20 μg/ml to enhance viral entry and to obtain a baseline infection level of ∼200,000 relative light units. Indinavir was added to the medium to a final concentration of 1 μM to limit infection of target cells to a single round of viral replication. Neutralization curves were fitted by nonlinear regression using a 5-parameter hill slope equation as described previously (44). The antibody concentrations required to inhibit infection by 50% or 80% are reported as 50% or 80% inhibitory concentrations (IC50 or IC80), respectively.
Viral-RNA extraction and cDNA synthesis.
From each plasma specimen and inoculum stock, 20,000 viral-RNA copies were extracted using the QIAamp Viral RNA Minikit (Qiagen, Valencia, CA). Reverse transcription of RNA to single-stranded cDNA was performed using SuperScript III reverse transcription according to the manufacturer's recommendations (Invitrogen). In brief, a cDNA reaction mixture of 1× RT buffer, 0.5 mM each deoxynucleoside triphosphate, 5 mM dithiothreitol, 2 U/ml RNaseOut (RNase inhibitor), 10 U/ml of Super-Script III reverse transcriptase, and 0.25 mM antisense primer SIVRR1 (5′-CAC TAG CTT ACT TCT AAA ATG GCA GC-3′) was incubated at 50°C for 60 min and at 55°C for 60 min and then heat inactivated at 70°C for 15 min, followed by treatment with 2 U of RNase H at 37°C for 20 min. The newly synthesized cDNA was used immediately or frozen at −80°C.
SGA of SHIV env.
A 3.5-kb fragment that included the entire env gene was sequenced from two animals (DBJE and DBX9) at peak viremia and the inoculum stock using a limiting-dilution PCR so that only one amplifiable molecule was present in each reaction. Single-genome amplification (SGA) was performed by serially diluting cDNA distributed among independent PCRs to identify a dilution at which amplification occurred in <30% of the total number of reactions. PCR amplification was performed with 1× PCR buffer, 2 mM MgSO4, 0.2 mM each deoxynucleoside triphosphate, 0.2 μM each primer, and 0.025 U/μl of Platinum Taq High Fidelity polymerase (Invitrogen, Carlsbad, CA) in a 20-μl reaction mixture. First-round PCR was performed with sense primer SIVIntF1 (5′-GAA GGG GAG GAA TAG GGG ATA TGA C-3′) and antisense primer SIVRR1 (5′-CAC TAG CTT ACT TCT AAA ATG GCA GC-3′) under the following conditions: 1 cycle of 94°C for 2 min and 35 cycles at 94°C for 15 s, 55°C for 30 s, and 68°C for 5 min, followed by a final extension of 68°C for 10 min. Next, 1 μl of the first-round PCR product was added to a second-round PCR that included the sense primer SIVEnvF1 (5′-CCT CCC CCT CCA GGA CTA GC-3′) and the antisense primer SIVEnvR1 (5′-TGT AAT AAA TCC CTT CCA GTC CCC CC-3′) performed under the same conditions used for first-round PCR but with a total of 45 cycles. Correctly sized amplicons were identified by agarose gel electrophoresis and directly sequenced with second-round PCR primers and 6 HIV-specific primers using BigDye Terminator technology (Applied Biosystems, Foster City, CA). To confirm PCR amplification from a single template, chromatograms were manually examined for multiple peaks, indicative of the presence of amplicons resulting from PCR-generated recombination events, Taq polymerase errors, or multiple variant templates. Sequences containing two or more ambiguous sites were excluded from analysis.
Sequence analysis.
Sequences were aligned using ClustalW and hand edited using MacClade 4.08 to improve alignment quality. Phylogenetic trees were constructed using the neighbor-joining method. In both animals, sequences segregated into distinct homogeneous lineages with at most 2 changes from the consensus sequence of each lineage. Each low-diversity lineage was inferred to be a unique founder lineage.
Nucleotide sequence accession numbers.
All sequences were deposited in GenBank with accession numbers JQ775968 to JQ776025.
RESULTS
Properties of new SHIVAD8 swarm stocks prepared from animals with documented immunodeficiency.
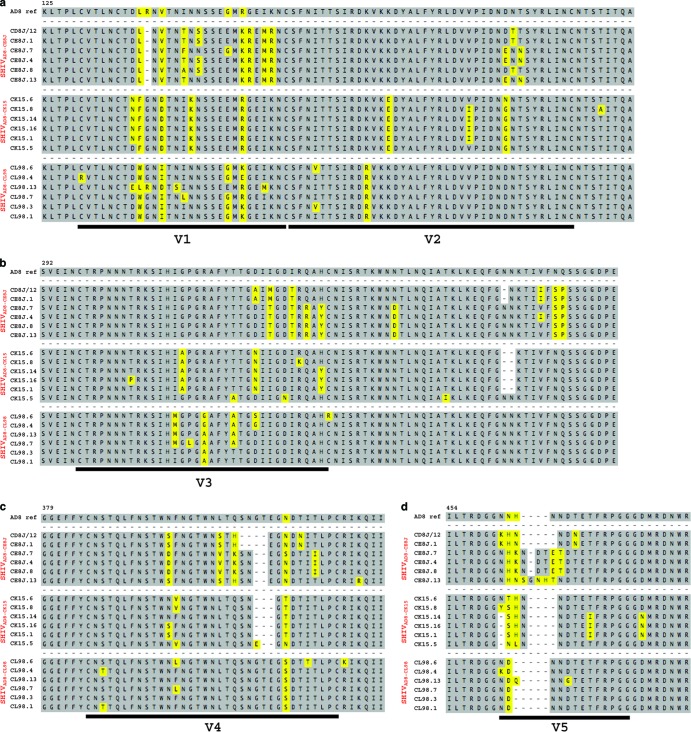
We previously reported the generation of the pathogenic R5-tropic SHIVAD8 following serial passaging in macaques (34). Independent SHIVAD8 stocks were prepared from PBMC, collected at the time of euthanasia, from three monkeys (CE8J, CK15, and CL98) that had developed symptomatic AIDS. The stocks prepared from the macaques were designated SHIVAD8-CE8J, SHIVAD8-CK15, and SHIVAD8-CL98, corresponding to the IDs of the source animals. RT-PCR analyses revealed that each “swarm” stock possessed distinctive env gene sequences reflecting its replication in individual infected monkeys (Fig. 1). All three virus stocks replicated to high levels in ConA-stimulated rhesus macaque PBMC (see Fig. S1 in the supplemental material).
Fig 1.
Gp120 sequence alignments of SHIVAD8 viral stocks (CE8J, CK15, and CL98). Six independent clones amplified from each virus stock by RT-PCR were sequenced, and the deduced amino acid sequences of the V1 and V2 (a), V3 and C3 (b), V4 (c), and V5 (d) regions of gp120 were aligned with the HIVAD8 sequence at the top. Amino acid changes are highlighted in yellow.
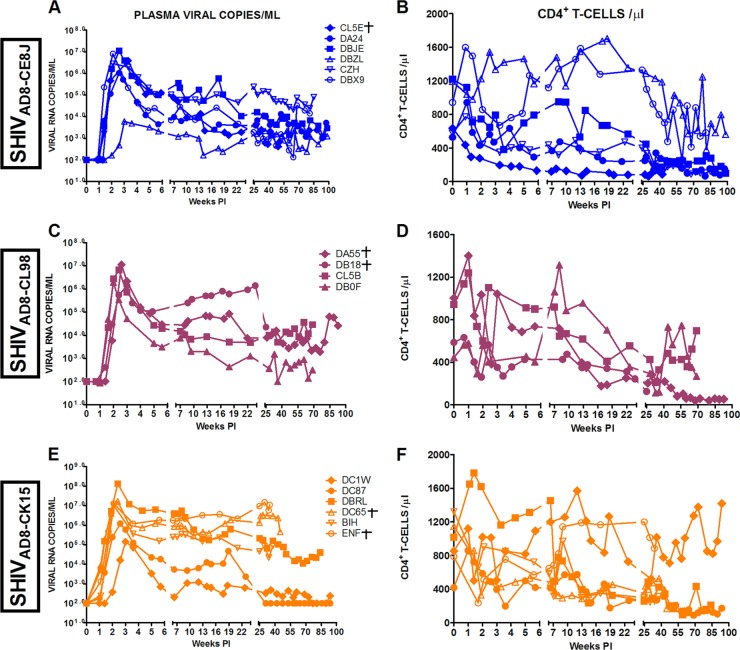
The replication properties of these SHIVAD8 stocks in vivo were assessed following inoculation by parenteral and mucosal routes. In an initial experiment, pairs of macaques were inoculated intravenously with high doses (2 × 104 to 3 × 105 TCID50) of each SHIVAD8 stock. Individual monkeys generated peak plasma viral loads ranging from 1.2 × 105 to 1.9 × 107 viral-RNA copies/ml between days 10 and 14 postinfection (p.i.) (Fig. 2A). Levels of set point viremia ranged from 103 to 106 viral-RNA copies/ml plasma, consistent with values previously reported for “intermediate” SHIVAD8 derivatives (34). All of the macaques sustained a gradual loss of circulating CD4+ T cells, even in the case of one animal (A3E051) with modest levels (∼1 × 103 RNA copies/ml) of plasma viremia (Fig. 2A and B). The depletion of circulating CD4+ T lymphocytes affected both memory and naïve subsets (Fig. 2C and D). At the time of euthanasia, all six of the infected animals (DB93, DB1A, A3E050, CK5H, A3E051, and CJ7D) had sustained significant depletions of circulating memory CD4+ T lymphocytes (144, 55, 9, 3, 4, and 56 circulating memory CD4+ T cells/μl, respectively). As observed previously (34), these SHIVAD8-infected macaques also experienced a decline in circulating naive CD4+ T cells during the chronic phase of infection and then a profound depletion of this T cell subset heralding the onset of clinical AIDS (Fig. 2D). This loss of naïve CD4+ T lymphocytes had also previously been shown not to be associated with altered coreceptor utilization by the contemporaneously circulating SHIVAD8 (34).
Fig 2.
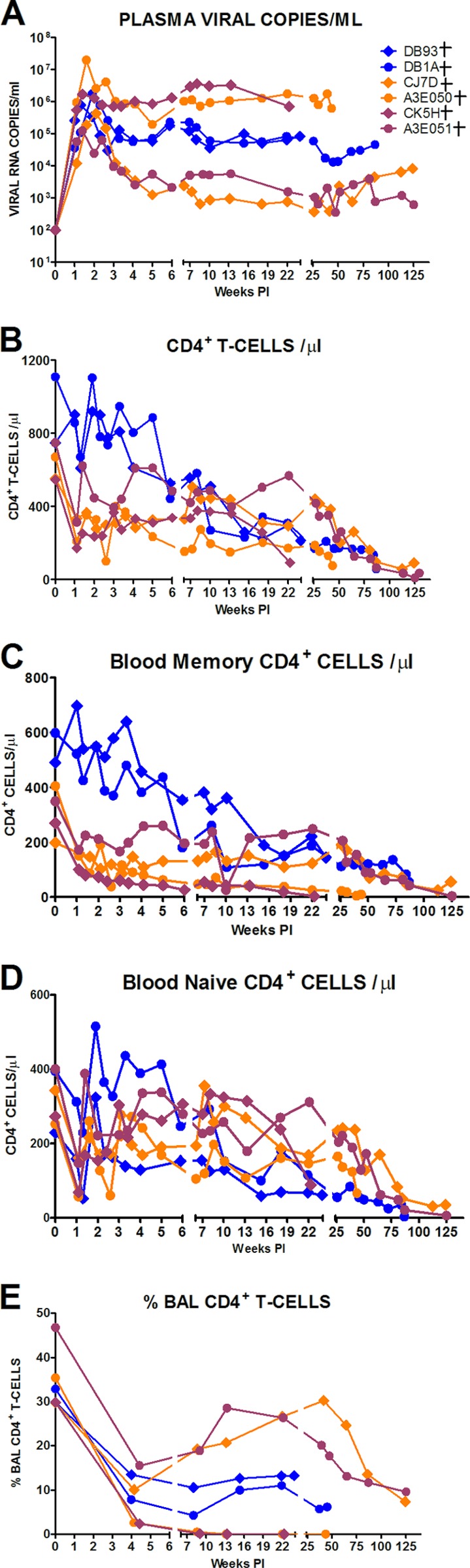
SHIVAD8 induces sustained plasma viremia and loss of CD4+ T cells in intravenously inoculated rhesus macaques. The levels of plasma viremia (A); absolute numbers of peripheral CD4+ T cells (B), memory CD4+ T cells (C), and naïve CD4+ T cells (D); and percentages of BAL fluid CD4+ T cells (E) are shown. Note that purple symbols denote animals infected with SHIVAD8-CE8J, maroon symbols denote animals infected with SHIVAD8-CL98, and orange symbols denote animals infected with SHIVAD8-CK15. Macaques euthanized with AIDS are indicated (†).
The specific targeting of memory CD4+ T cells at an effector site by the new SHIVAD8 stocks is best illustrated by the marked depletion of CD4+ T lymphocytes in BAL specimens during acute infection (Fig. 2E). In four of these monkeys (DB93, DB1A, CJ7D, and A3E051), the levels of BAL fluid CD4+ T lymphocytes stabilized or partially recovered, whereas in the remaining two (A3E050 and CK5H), this memory CD4+ T cell subset was markedly depleted (Fig. 2E).
In summary, 4 of 6 animals inoculated intravenously with three new swarm SHIVAD8 virus stocks exhibited a normal-progressor (NP) clinical course characterized by durably maintained set point virus loads and the gradual depletion of circulating CD4+ T lymphocytes. The remaining 2 SHIVAD8-infected macaques (CK5H and DB93) experienced a more rapid clinical course associated with very high levels of plasma viremia and very low levels of circulating and effector site memory CD4+ T cells and required euthanasia at weeks 23 and 24 p.i., respectively.
Mucosal transmission of new SHIVAD8 swarm stocks.
A major aim of this study was to determine the minimal SHIVAD8 inoculum resulting in the establishment of infection in rhesus macaques challenged by the i.r. route. As shown in Table 1, one (CL5E) of two animals inoculated with 100 TCID50 of the SHIVAD8-CE8J stock became infected following a single i.r. inoculation. A second macaque (DA24), inoculated two times with 100 TCID50 of SHIVAD8-CE8J, failed to become infected. Infection was established in this monkey when the challenge dose was increased to 1,000 TCID50. Assuming that 1,000 TCID50 might represent the threshold dose for successfully infecting monkeys with SHIVAD8-CE8J by the i.r. route, four additional macaques (DBJE, DBZL, CZH, and DBX9) were inoculated with 1,000 TCID50, and all four became infected following a single virus challenge (Table 1). The peak virus loads in five of the six SHIVAD8-CE8J-infected animals ranged from 1.0 × 106 to 1.1 × 107 viral-RNA copies/ml of plasma; the peak viral-RNA level in the sixth macaque (DBZL) was delayed and notably lower (6 × 103 copies/ml of plasma) (Fig. 3A).
Fig 3.
Intrarectal inoculation of SHIVAD8 viral stocks in rhesus macaques. The colors in the graphs represent the challenge virus as described in the legend for Fig. 2. Macaques euthanized with AIDS are indicated (†).
A similar approach was used to determine the minimal i.r. inoculum size for the new SHIVAD8-CL98 and SHIVAD8-CK15 stocks. As shown in Table 1, infection was not established in two monkeys (DA55 and DB18) inoculated with 100 TCID50 of SHIVAD8-CL98. Successful virus acquisition in these 2 animals required increasing the inoculum size to 1,000 TCID50 of SHIVAD8-CL98. Furthermore, two additional animals (CL5B and DB0F) resisted infection even following two i.r. exposures to 1,000 TCID50, but each became infected after receiving a single inoculation of 10,000 TCID50 of SHIVAD8-CL98. Once infected, all four of the SHIVAD8-CL98-inoculated macaques generated peak viral-RNA loads ranging from 1.4 × 106 to 1.2 × 107 copies/ml of plasma and levels of set point viremia ranging from 1.3 × 103 to 8.0 × 105 viral-RNA copies/ml of plasma (Fig. 3C).
Based on these results, 6 animals were inoculated by the i.r. route with 1,000 TCID50 of the third virus stock (SHIVAD8-CK15). As indicated in Table 1, four of six monkeys became infected following a single i.r. inoculation and developed persistent viremia. The two remaining animals (BIH and ENF) resisted infection with 1,000 TCID50 of SHIVAD8-CK15 but did become infected when the inoculum size was increased to 10,000 TCID50 (Table 1). Plasma viral loads in the six SHIVAD8-CK15-infected monkeys are shown in Fig. 3E.
No correlation was observed between inoculum size and peak viral loads/time to peak viremia following i.r. inoculation with any of the three new SHIVAD8 preparations.
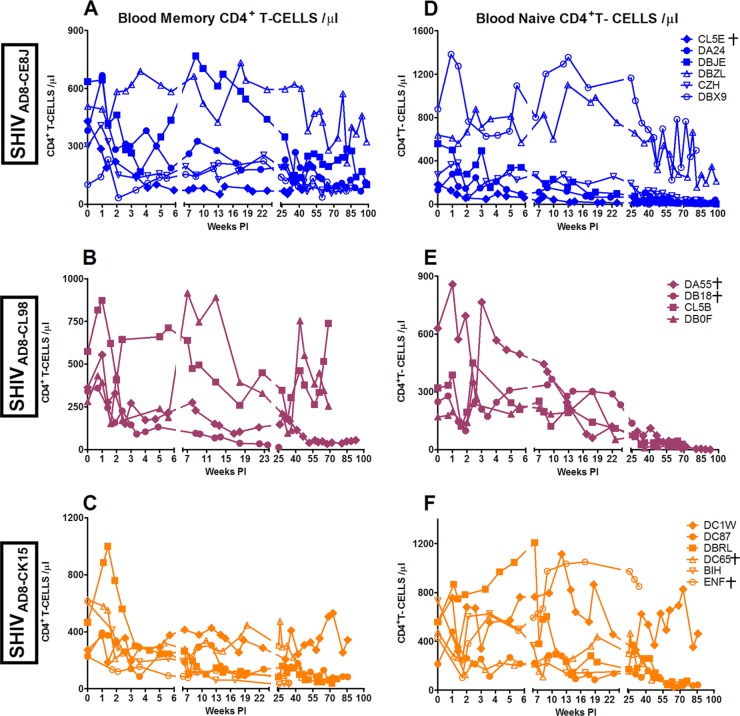
CD4+ T cell depletion in macaques inoculated by the i.r. route with SHIVAD8.
The absolute numbers of CD3+ CD4+ T cells in peripheral blood markedly declined in a majority of animals challenged by the i.r. route during the 80- to 90-week observation period (Fig. 3), and 12 of 16 macaques sustained depletions of circulating memory CD4+ T cells to the 200-cell/μl level by week 60 p.i. (Fig. 4). This was accompanied by the loss of naïve CD4+ T cells to below the 100-cell/μl level in most of the monkeys in the same time frame (Fig. 4). Targeting and loss of memory CD4+ T lymphocytes at an effector site (BAL samples) during acute infection was observed once again in a majority of the macaques inoculated by the mucosal route (Fig. 5). In summary, the decline of CD4+ T lymphocyte subsets in the circulation or at an effector site following inoculation by the mucosal route (Fig. 3 and 5) paralleled the pattern previously observed for SHIVAD8 administered intravenously (Fig. 2).
Fig 4.
Loss of CD4+ T cell lymphocyte subsets following i.r. inoculation of SHIVAD8 virus stocks. Absolute numbers of memory CD4+ T cells (A, B, and C), and naïve CD4+ T cells (D, E, and F) are shown. The colors in the graphs represent the challenge virus as described in the legend for Fig. 2. Macaques euthanized with AIDS are indicated (†).
Fig 5.
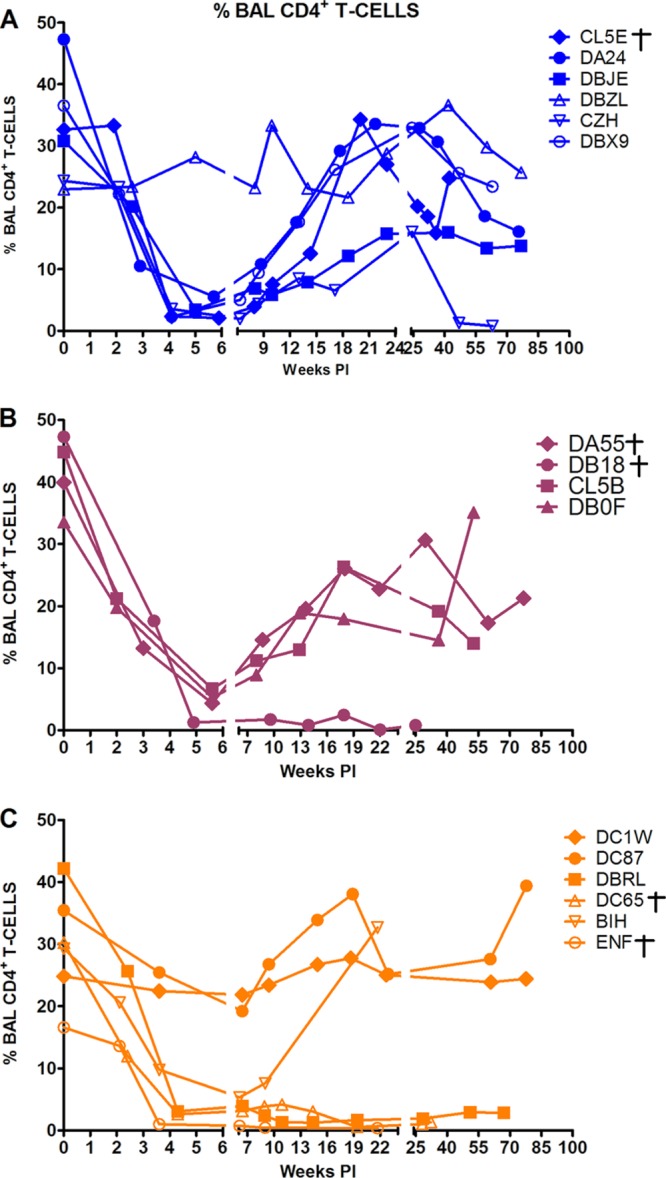
Loss of CD4+ T cell lymphocytes at an effector site (BAL fluid). Macaques euthanized with AIDS are indicated (†).
Immune responses to SHIVAD8 infection.
Anti-SHIVAD8 Gag-specific CD4+ and CD8+ T lymphocyte responses were determined by flow cytometry by measuring intracellular staining of cells expressing tumor necrosis factor alpha (TNF-α) and/or interferon gamma (IFN-γ) following stimulation with a 15-mer peptide pool spanning SIVmac239 Gag. The levels of virus-specific memory CD8+ T cells in rhesus macaques inoculated by the i.r. route ranged from 0.38 to 1.78% (see Table S1 in the supplemental material). A similar analysis of Gag-specific responses in memory CD4+ T cells in the same animals indicated that 0.55 to 4.02% expressed TNF-α and/or IFN-γ (see Table S1 in the supplemental material).
Anti-HIV-1 gp120 antibodies in macaques inoculated by the i.r. route with the three swarm SHIVAD8 stocks became detectable by ELISA between weeks 3 and 12 p.i. (see Fig. S2 in the supplemental material). The levels of these antiviral binding antibodies remained high in most of these animals throughout the course of infection. Two macaques (DB18 and ENF) that developed very high levels of set point viremia and experienced a rapid and irreversible loss of memory CD4+ T cells at an effector site (in BAL specimens) failed to generate detectable anti-gp120 antibodies and required euthanasia at weeks 25 and 35, respectively, because of their deteriorating clinical condition. This constellation of findings fulfilled the definition of “rapid progressors” assigned to SIV-infected macaques exhibiting a similar clinical phenotype (3).
A system for categorizing the neutralization sensitivities of HIV-1 isolates to plasma pools collected from individuals infected with geographically and genetically diverse virus strains has described three HIV-1 subgroups: tier 1 (very sensitive), tier 2 (moderate sensitivity), and tier 3 (neutralization resistant). In this classification system, tissue culture-adapted tier 1 viruses represent HIV-1 strains that can be neutralized by sera with minimal NAb activity, whereas tier 2 viruses exhibit sensitivities commonly observed with circulating strains of HIV-1 (44). The neutralization phenotypes of the three new SHIVAD8 swarm stocks are shown in Table 2. In contrast to its very resistant parental HIV-1AD8 (tier 3) and the highly sensitive HIV-1MN (tier 1), all three SHIVAD8 stocks exhibited a tier 2 phenotype. The sensitivities of these three stocks to a panel of anti-HIV-1 monoclonal antibodies are shown in Table 3. All three SHIVAD8 stocks are resistant to the recently described PG9 and PG16 MAbs, as well as the high-mannose glycan-targeting 2G12 MAb. SHIVAD8-CE8J alone is resistant to the PGT121 MAb.
Table 2.
Neutralization phenotype of SHIVAD8 viral stocks
| Sample ID | ID50 in TZM-bl cellsa |
HIV IG (μg/ml) | Tier phenotype | ||||||
|---|---|---|---|---|---|---|---|---|---|
| S321 | C500 | B520 | G435 | T520b | M263 | M600c | |||
| SHIVAD8-CK15 | 63 | 112 | 145 | 36 | 145 | 85 | 64 | >2,500 | 2 |
| SHIVAD8-CE8J | 91 | 94 | 112 | 43 | 147 | 137 | 65 | >2,500 | 2 |
| SHIVAD8-CL98 | 77 | 54 | 47 | 63 | 198 | 92 | 89 | 1,870 | 2 |
| HIVAD8 | 189 | <20 | <20 | <20 | <20 | <20 | 25 | 2,032 | 3 |
| MN.3 | 13,944 | 9,152 | 822 | 8,432 | 3,968 | 43,722 | 1,709 | 1.81 | 1 |
| CAAN5342.A2 | 84 | <20 | 27 | <20 | <20 | 77 | 185 | 638 | 2 |
ID50, serum dilution at which relative luminescence units were reduced 50% compared to virus control wells (no test sample).
Table 3.
Neutralization assessment of SHIVAD8 virus stocks by known MAbs targeting independent epitopes on the Env glycoprotein
| Virus | Parameter | Value (μg/ml)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV-IG | b12 | VRC01 | VRC03 | VRC-PG04 | CD4-Ig | PG9 | PG16 | 2G12 | PGT121 | PGT123 | PGT128 | ||
| SHIVAD8-CE8J | IC50 | 2,412 | 8.04 | 0.917 | 0.088 | 0.317 | 1.89 | >50 | 0.098 | >50 | >50 | 34.4 | 0.148 |
| IC80 | >5000 | 26.5 | 2.41 | 0.315 | 1.48 | 5.95 | >50 | >50 | >50 | >50 | >50 | 2.76 | |
| SHIVAD8-CK15 | IC50 | 1,331 | 4.37 | 2.34 | 0.184 | 1.73 | 1.72 | >50 | >50 | >50 | 0.086 | 0.116 | 0.081 |
| IC80 | >5000 | 11.9 | 6.61 | 0.542 | 7.03 | 3.98 | >50 | >50 | >50 | 0.232 | 0.324 | 0.201 | |
| SHIVAD8-CL98 | IC50 | 1,805 | 6.16 | 2.84 | 0.468 | 2.23 | 1.69 | 30.6 | 22.9 | >50 | 0.097 | 0.123 | 0.048 |
| IC80 | >5000 | 18.4 | 8.06 | 1.59 | 7.33 | 4.01 | >50 | >50 | >50 | 0.254 | 0.306 | 0.123 | |
| SHIVSF162 (P3) | IC50 | 2,884 | 2.19 | 2.01 | 0.388 | >50 | 3.06 | >50 | >50 | >50 | ND | ND | ND |
| IC80 | >5000 | 4.94 | 5.87 | 0.893 | >50 | 8.25 | >50 | >50 | >50 | ND | ND | ND | |
ND, not determined.
In a recently published study describing the extraordinarily broad and high-titer NAbs generated by one animal inoculated intravenously with an intermediate SHIVAD8 derivative, we reported that, irrespective of the set point, only 20 to 25% of SHIVAD8-infected monkeys develop autologous NAbs during the course of their infections (50). When detectable, autologous neutralization activity directed against SHIVAD8 emerged after 30 to 35 weeks p.i. and was usually low titer (<1:100) and transient. In the present study, only 3 (DA24, DBJE, and DA55) of 19 monkeys inoculated with the three new SHIVAD8 stocks developed low levels of autologous NAbs (see Table S2 in the supplemental material). In two of these macaques (DA24 and DBJE), the neutralization activity measured at weeks 41 and 42 became undetectable at weeks 57 and 58 p.i. Furthermore, in this group of 19 animals, there was no obvious relationship between the levels of plasma viremia or preservation of CD4+ T cell subsets, which distinguishes the three monkeys able to generate autologous NAbs from those that were not.
SGA of transmitted SHIVAD8.
The SGA-plus-sequencing approach has been previously used to determine the number of transmitted/founder viruses present in macaques following i.r. inoculation of the uncloned SIVmac251 and SIVsmE660 swarm stocks (28, 46). At low inoculum doses, one or only a few founder viruses present in the swarm virus preparation were detected in plasma collected during the early phase of acute infection of inoculated monkeys. A similar SGA analysis was performed using plasma samples collected on day 10 p.i. from two animals (DBX9 and DBJE) that became infected following a single i.r. inoculation with 1,000 TCID50 of SHIVAD8-CE8J (Table 1 and Fig. 3). A total of 58 SGA-derived 3.5-kb sequences (including the entire env gene) were obtained from the plasma sample of each macaque or from the SHIVAD8-CE8J stock used for inoculation. The viral-RNA loads in macaques DBX9 and DBJE on day 14 p.i. were 8.03 × 106 and 1.64 × 106 RNA copies/ml, respectively. As shown in Fig. 6, a single SHIV env variant was detected in each animal. In both instances, the variant detected corresponded to an individual and unique virus component present in the SHIVAD8-CE8J stock, which overall contained a maximum diversity of 2.0%.
Fig 6.
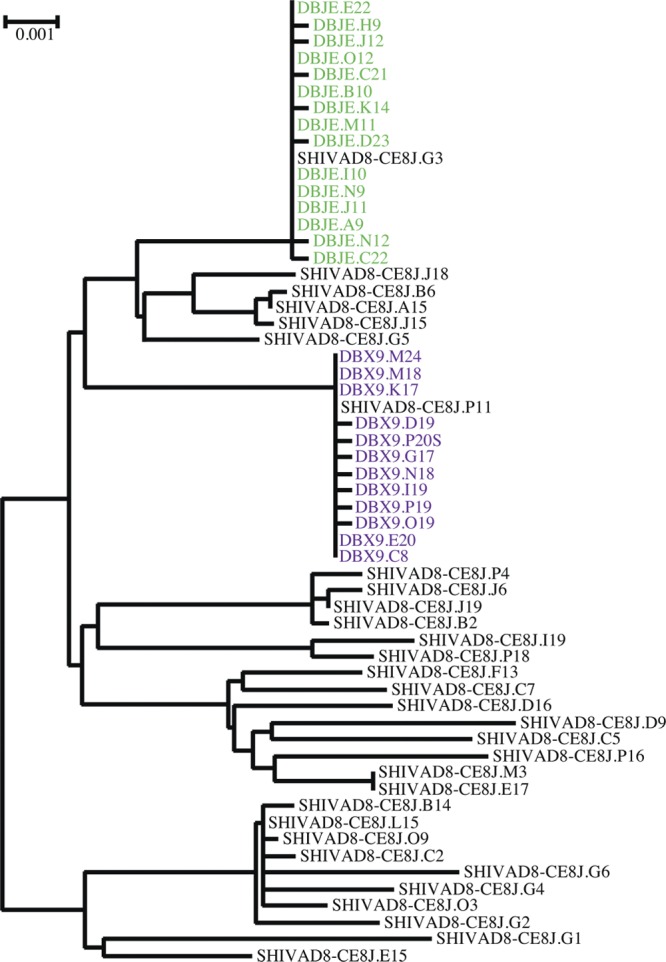
Composite phylogenic tree of env gene sequences present in the SHIVAD8-CE8J stock and in plasma from macaques (DBJE and DBX9) on day 14 after i.r. inoculation. Animal-derived sequences from plasma at peak viremia are color coded, and inoculum-derived sequences are shown in black.
Some SHIVAD8-infected macaques progressed to AIDS.
The clinical status and disease outcomes of all 22 animals inoculated by the i.v. or i.r. route with new SHIVAD8 stocks are presented in Table 4. Eleven animals were euthanized with symptoms of AIDS, including uncontrolled diarrhea, enteritis, typhocolitis, renal and mesenteric arteriolopathy, pulmonary thrombosis, and varying degrees of thrombocytopenia. Histopathological studies performed on specimens collected at the time of necropsy revealed the presence of Pneumocystis, Candida, and Mycobacterium species in individual macaques.
Table 4.
Clinical and pathological findings in rhesus macaques infected with SHIVAD8 virus stocks
| Animal | Inoculum | Clinical data/pathological findings | No. of plateletsb (103/μl) |
|---|---|---|---|
| DB93a | SHIVAD8-CE8J | Euthanized (wk 24); rapid progressor | 29 |
| DB1Aa | SHIVAD8-CE8J | Euthanized (wk 87); arteriolopathy in kidneys and mesentery | 37 |
| CL5Ea | SHIVAD8-CE8J | Euthanized (wk 51); pulmonary thrombosis | 66 |
| DA24 | SHIVAD8-CE8J | Total CD4+ T cells, 79/mm3 (wk 94) | 223 |
| DBJE | SHIVAD8-CE8J | Total CD4+ T cells, 183/mm3 (wk 95) | 196 |
| DBZL | SHIVAD8-CE8J | Total CD4+ T cells, 796/mm3 (wk 95) | 341 |
| CZH | SHIVAD8-CE8J | Total CD4+ T cells, 106/mm3 (wk 81) | 183 |
| DBX9 | SHIVAD8-CE8J | Total CD4+ T cells, 583/mm3 (wk 81) | 423 |
| CJ7Da | SHIVAD8-CK15 | Euthanized (wk 125); enteritis | 482 |
| A3E050a | SHIVAD8-CK15 | Euthanized (wk 44); uncontrolled diarrhea | 201 |
| DC1W | SHIVAD8-CK15 | Total CD4+ T cells, 1419/mm3 (wk 94) | 464 |
| DC87 | SHIVAD8-CK15 | Total CD4+ T cells, 173/mm3 (wk 94) | 301 |
| DBRL | SHIVAD8-CK15 | Total CD4+ T cells, 211/mm3 (wk 84) | 66 |
| DC65a | SHIVAD8-CK15 | Euthanized (wk 43); Mycobacterium intracellulare | 48 |
| BIH | SHIVAD8-CK15 | Total CD4+ T cells, 312/mm3 (wk 38) | 110 |
| ENFa | SHIVAD8-CK15 | Euthanized (wk 35), Mycobacterium avium | 125 |
| CK5Ha | SHIVAD8-CL98 | Euthanized (wk 23); rapid progressor; uncontrolled diarrhea, viral enterotyphocolitis | 65 |
| A3E051a | SHIVAD8-CL98 | Euthanized (wk 125); enterotyphocolitis | 358 |
| DA55a | SHIVAD8-CL98 | Euthanized (wk 95); lymphoma | 36 |
| DB18a | SHIVAD8-CL98 | Euthanized (wk 25); Candida and Pneumocystis | 126 |
| CL5B | SHIVAD8-CL98 | Total CD4+ T cells, 696/mm3 (wk 69) | 372 |
| DB0F | SHIVAD8-CL98 | Total CD4+ T cells, 268/mm3 (wk 69) | 261 |
Euthanized with simian AIDS.
Normal range is 260 × 103 to 361 × 103/μl.
DISCUSSION
In this report, we have described the biological and transmission properties of three SHIVAD8 swarm stocks. Although each possesses distinct characteristics, their in vivo properties are similar to those reported (34) for “intermediate” SHIVAD8 derivatives, which include (i) sustained but variable levels (102.5 to 106 RNA copies/ml) of set point viremia; (ii) induced depletion of circulating CD4+ T cells, even in animals with modest (<103 RNA copies/ml) plasma virus loads; (iii) targeting of memory CD4+ T lymphocytes for depletion at an effector site during acute infection; (iv) losses of memory and naïve CD4+ T cells in the blood; (v) development of the “rapid-progressor” clinical phenotype in a few inoculated macaques; and (vi) induction of symptomatic AIDS in 9/20 normally progressing monkeys by week 125 p.i. As a consequence of their immunodeficiency, some animals acquired opportunistic infections (Mycobacteria, Pneumocystis, and Candida species), and several developed thrombocytopenia, with platelet counts of <70,000/μl (Table 4).
The genetic signatures of the three SHIVAD8 swarm stocks permitted an SGA analysis of founder viruses transmitted following i.r. inoculation. In two animals administered 1,000 TCID50, a single and different SHIV variant, present in the original uncloned virus stock, was transmitted to each monkey. Although an exhaustive in vivo titration of the three new SHIVAD8 stocks was not performed, the virus acquisition results presented in Table 1 suggest that the SHIVAD8-CE8J and SHIVAD8-CK15 preparations have higher infectivity titers for i.r. inoculation of rhesus macaques than the SHIVAD8-CL98 stock. When the method of Reed and Muench (40) is used to determine the in vivo infectivity of the SHIVAD8-CE8J and SHIVAD8-CK15 stocks from the acquisition data, we calculate that 1,000 TCID50, measured in vitro, is equivalent to approximately 3 50% animal infectious doses (AID50). Thus, a single inoculation of 1,000 TCID50 of SHIVAD8-CE8J and SHIVAD8-CK15 might be close to the minimal challenge dose required for successful virus acquisition. Interestingly, the 1,000-TCID50 (∼107 RNA copies) threshold for establishing SHIVAD8-CE8J and SHIVAD8-CK15 infections is also in the same range as that needed for virus acquisition with a single inoculation of SIVmac251 and SIVsmE660 (11, 22, 28).
We previously determined the coreceptor properties of the viruses used to prepare the three swarm SHIVs (SHIVAD8-CE8J, SHIVAD8-CK15, and SHIVAD8-CL98) described in this study following their isolation from animals CE8J, CK15, and CL98 at the time of their euthanasia with clinical symptoms of immunodeficiency (see Fig. S2 in reference 34). All three SHIV stocks exclusively utilized CCR5. Because CCR5 is not expressed on naïve CD4+ T lymphocytes, one could ask why this T cell subset is lost during SHIVAD8 infections of macaques. It is now recognized that depletion of this T lymphocyte subset is a prominent feature of chronic HIV-1 infections in the absence of coreceptor switching (23, 42) and has been reported to occur in SIVsm543-infected rhesus macaques (25, 32), but not in SIVmac239-infected monkeys (39). Loss of naive CD4+ T cells is also characteristic of R5 SHIVAD8 infections in normally progressing rhesus macaques (34). In contrast, coreceptor switching has been observed only during the rapid and irreversible loss of naïve CD4+ T cells in some R5-tropic SHIV rapid progressors (14, 15, 34, 47), but never in SIV rapid progressors.
The presence of the HIV-1 envelope glycoprotein on all SHIVs makes them particularly attractive challenge viruses to evaluate the protective effects of NAbs in the macaque model. In this regard, we recently reported that a single rhesus monkey (CE8J), inoculated with an intermediate SHIVAD8 derivative, generated extremely high-titer and unusually broad autologous NAbs directed against viruses from different HIV-1 clades (50). Mapping studies revealed that the HIV-1 gp120 epitopes targeted for neutralization were dependent on the glycan present at amino acid 332. In the present study, our characterization of the neutralization properties of the three new swarm SHIVAD8 challenge stocks indicated that each exhibited a tier 2 phenotype (Table 2) and would therefore be an ideal surrogate for circulating HIV-1 strains in vaccine experiments. Furthermore, the three SHIVAD8 preparations exhibited similar sensitivities to several broadly reacting anti-HIV-1 neutralizing MAbs (Table 3). However, SHIVAD8-CE8J, isolated at the time of euthanasia from macaque CE8J, the animal producing potent cross-clade anti-HIV-1 NAbs, was uniquely resistant to the recently described PGT121 and PGT123 MAbs (49). Perhaps the previously reported emergence of neutralization escape SHIV variants in monkey CE8J (50) also conferred the resistance of SHIV-AD8-CE8J to the PGT121 and PGT123 MAbs.
At present, we have no explanation for the relatively small fraction of SHIVAD8-infected animals that develop sustained levels of autologous NAbs (see Table S2 in the supplemental material). We previously noted that only 9 macaques in our cohort of 40 monkeys inoculated with SHIVAD8 (intermediate derivatives, swarm stocks, or a pathogenic molecular clone) generated detectable autologous antibodies. One of these 9 infected animals was macaque CE8J, described above, which developed high-titer and cross-HIV-1 clade autologous neutralizing activity. The low frequency of SHIVAD8-infected monkeys generating autologous NAbs contrasts with results obtained for X4-tropic SHIVs (8, 26) and the R5-tropic SHIVSF162P4 (24), all of which develop high levels of autologous NAbs. This property of the R5-tropic SHIVAD8 could reflect the tier 3 neutralization phenotype of its HIV-1AD8 parent (Table 2) and is reminiscent of the failure of SIVmac239-infected monkeys to produce significant levels of autologous NAbs (21). It is worth noting, however, that all SHIVAD8-infected macaques are able to generate cross-reactive NAbs against some tier 1 and 2 HIV-1 strains (M. Shingai and M. A. Martin, unpublished data).
Our data indicate that these new SHIVAD8 viruses (SHIVAD8-CE8J, SHIVAD8-CK15, and SHIVAD8-CL98) reproducibly establish infections in rhesus macaques following the mucosal transmission of one founder virus, consistently cause the loss of memory CD4+ T lymphocytes, possess a tier 2 phenotype, and are sensitive to several potent cross-reacting anti-HIV-1 neutralizing MAbs. Although other R5-tropic SHIVs have been constructed, the levels of sustained viremia, depletion of CD4+ T cells, and development of immunodeficiency in inoculated animals are frequently variable (35–37). These inconsistencies could affect the outcomes and interpretations of passive transfer or vaccine experiments when such R5 SHIVs are used as challenge viruses. We are also in the process of deriving a pathogenic molecular clone from SHIVAD8-infected animals. This would provide a reagent that can readily be used to identify neutralization escape virus variants that emerge following a SHIVAD8 challenge or during the course of vaccine experiments.
Supplementary Material
ACKNOWLEDGMENTS
We thank Keiko Tomioka, Robin Kruthers, and Ranjini Iyengar for determining viral-RNA loads. We appreciate the contribution of Boris Skopets, Rahel Petros, and Erin O'Sheil in diligently assisting in the maintenance of animals and assisting with procedures.
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Disease, National Institutes of Health. The work was also supported in part with federal funds from the National Cancer Institute under contract HHSN266200400088C. Plasma samples for determining the neutralization phenotype of virus stocks were provided by Áine McKnight through the Bill and Melinda Gates Foundation's Collaboration for AIDS Vaccine Discovery/Comprehensive Antibody Vaccine Immune Monitoring Consortium (grant number 38619).
Footnotes
Published ahead of print 30 May 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Amara RR, et al. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69–74 [DOI] [PubMed] [Google Scholar]
- 2. Barouch DH, et al. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486–492 [DOI] [PubMed] [Google Scholar]
- 3. Brown CR, et al. 2007. Unique pathology in simian immunodeficiency virus-infected rapid progressor macaques is consistent with a pathogenesis distinct from that of classical AIDS. J. Virol. 81:5594–5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burton DR, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 5. Chen Z, et al. 2002. CD4+ lymphocytopenia in acute infection of Asian macaques by a vaginally transmissible subtype-C, CCR5-tropic simian/human immunodeficiency virus (SHIV). J. Acquir. Immune Defic. Syndr. 30:133–145 [DOI] [PubMed] [Google Scholar]
- 6. Dewar RL, et al. 1994. Application of branched DNA signal amplification to monitor human immunodeficiency virus type 1 burden in human plasma. J. Infect. Dis. 170:1172–1179 [DOI] [PubMed] [Google Scholar]
- 7. Dreja H, et al. 2010. Neutralization activity in a geographically diverse East London cohort of human immunodeficiency virus type 1-infected patients: clade C infection results in a stronger and broader humoral immune response than clade B infection. J. Gen. Virol. 91:2794–2803 [DOI] [PubMed] [Google Scholar]
- 8. Endo Y, et al. 2000. Short- and long-term clinical outcomes in rhesus monkeys inoculated with a highly pathogenic chimeric simian/human immunodeficiency virus. J. Virol. 74:6935–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fauci AS, et al. 2008. HIV vaccine research: the way forward. Science 321:530–532 [DOI] [PubMed] [Google Scholar]
- 10. Feinberg MB, Moore JP. 2002. AIDS vaccine models: challenging challenge viruses. Nat. Med. 8:207–210 [DOI] [PubMed] [Google Scholar]
- 11. Fenizia C, et al. 2011. TRIM5alpha does not affect simian immunodeficiency virus SIV(mac251) replication in vaccinated or unvaccinated Indian rhesus macaques following intrarectal challenge exposure. J. Virol. 85:12399–12409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gautam R, et al. 2009. Simian immunodeficiency virus SIVrcm, a unique CCR2-tropic virus, selectively depletes memory CD4+ T cells in pigtailed macaques through expanded coreceptor usage in vivo. J. Virol. 83:7894–7908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harouse JM, Gettie A, Tan RC, Blanchard J, Cheng-Mayer C. 1999. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science 284:816–819 [DOI] [PubMed] [Google Scholar]
- 14. Ho SH, et al. 2007. Coreceptor switch in R5-tropic simian/human immunodeficiency virus-infected macaques. J. Virol. 81:8621–8633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ho SH, Trunova N, Gettie A, Blanchard J, Cheng-Mayer C. 2008. Different mutational pathways to CXCR4 coreceptor switch of CCR5-using simian-human immunodeficiency virus. J. Virol. 82:5653–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hsu M, et al. 2003. Increased mucosal transmission but not enhanced pathogenicity of the CCR5-tropic, simian AIDS-inducing simian/human immunodeficiency virus SHIV(SF162P3) maps to envelope gp120. J. Virol. 77:989–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Humbert M, et al. 2008. SHIV-1157i and passaged progeny viruses encoding R5 HIV-1 clade C env cause AIDS in rhesus monkeys. Retrovirology 5:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Igarashi T, et al. 2003. Macrophage-tropic simian/human immunodeficiency virus chimeras use CXCR4, not CCR5, for infections of rhesus macaque peripheral blood mononuclear cells and alveolar macrophages. J. Virol. 77:13042–13052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Igarashi T, et al. 1999. Emergence of a highly pathogenic simian/human immunodeficiency virus in a rhesus macaque treated with anti-CD8 mAb during a primary infection with a nonpathogenic virus. Proc. Natl. Acad. Sci. U. S. A. 96:14049–14054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joag SV, et al. 1996. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J. Virol. 70:3189–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson WE, et al. 2003. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 77:9993–10003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keele BF, et al. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Khoury G, Rajasuriar R, Cameron PU, Lewin SR. 2011. The role of naive T-cells in HIV-1 pathogenesis: an emerging key player. Clin. Immunol. 141:253–267 [DOI] [PubMed] [Google Scholar]
- 24. Kraft Z, et al. 2007. Macaques infected with a CCR5-tropic simian/human immunodeficiency virus (SHIV) develop broadly reactive anti-HIV neutralizing antibodies. J. Virol. 81:6402–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuwata T, et al. 2009. Association of progressive CD4(+) T cell decline in SIV infection with the induction of autoreactive antibodies. PLoS Pathog. 5:e1000372 doi:10.1371/journal.ppat.1000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laird ME, Igarashi T, Martin MA, Desrosiers RC. 2008. Importance of the V1/V2 loop region of simian-human immunodeficiency virus envelope glycoprotein gp120 in determining the strain specificity of the neutralizing antibody response. J. Virol. 82:11054–11065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li M, et al. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu J, et al. 2010. Low-dose mucosal simian immunodeficiency virus infection restricts early replication kinetics and transmitted virus variants in rhesus monkeys. J. Virol. 84:10406–10412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luciw PA, Pratt-Lowe E, Shaw KE, Levy JA, Cheng-Mayer C. 1995. Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc. Natl. Acad. Sci. U. S. A. 92:7490–7494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. National Institutes of Health 1985. Guide for the care and use of laboratory animals. Department of Health and Human Services publication no. NIH 85-23 National Institutes of Health, Bethesda, MD [Google Scholar]
- 31. Nishimura Y, et al. 2005. Resting naive CD4+ T cells are massively infected and eliminated by X4-tropic simian-human immunodeficiency viruses in macaques. Proc. Natl. Acad. Sci. U. S. A. 102:8000–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nishimura Y, et al. 2007. Loss of naive cells accompanies memory CD4+ T-cell depletion during long-term progression to AIDS in simian immunodeficiency virus-infected macaques. J. Virol. 81:893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nishimura Y, et al. 2011. Recombination-mediated changes in coreceptor usage confer an augmented pathogenic phenotype in a nonhuman primate model of HIV-1-induced AIDS. J. Virol. 85:10617–10626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nishimura Y, et al. 2010. Generation of the pathogenic R5-tropic simian/human immunodeficiency virus SHIVAD8 by serial passaging in rhesus macaques. J. Virol. 84:4769–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pahar B, Wang X, Dufour J, Lackner AA, Veazey RS. 2007. Virus-specific T cell responses in macaques acutely infected with SHIV(sf162p3). Virology 363:36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pal R, et al. 2003. Characterization of a simian human immunodeficiency virus encoding the envelope gene from the CCR5-tropic HIV-1 Ba-L. J. Acquir. Immune Defic. Syndr. 33:300–307 [DOI] [PubMed] [Google Scholar]
- 37. Parren PW, et al. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pejchal R, et al. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Picker LJ, et al. 2004. Insufficient production and tissue delivery of CD4+ memory T cells in rapidly progressive simian immunodeficiency virus infection. J. Exp. Med. 200:1299–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. J. Am. Hyg. 27:493–497 [Google Scholar]
- 41. Reimann KA, et al. 1996. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J. Virol. 70:6922–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rickabaugh TM, et al. 2011. The dual impact of HIV-1 infection and aging on naive CD4 T-cells: additive and distinct patterns of impairment. PLoS One 6:e16459 doi:10.1371/journal.pone.0016459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rose NF, et al. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539–549 [DOI] [PubMed] [Google Scholar]
- 44. Seaman MS, et al. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shu Y, et al. 2007. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine 25:1398–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stone M, et al. 2010. A limited number of simian immunodeficiency virus (SIV) env variants are transmitted to rhesus macaques vaginally inoculated with SIVmac251. J. Virol. 84:7083–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tasca S, Ho SH, Cheng-Mayer C. 2008. R5X4 viruses are evolutionary, functional, and antigenic intermediates in the pathway of a simian-human immunodeficiency virus coreceptor switch. J. Virol. 82:7089–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trkola A, et al. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Walker LM, et al. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Walker LM, et al. 2011. Rapid development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies in an R5 SIV/HIV chimeric virus infected macaque. Proc. Natl. Acad. Sci. U. S. A. 108:20125–20129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Willey RL, et al. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 62:139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu X, et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu X, et al. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.