Abstract
Tomato spotted wilt virus (TSWV) is transmitted by Frankliniella occidentalis in a persistent propagative manner. Despite the extensive replication of TSWV in midgut and salivary glands, there is little to no pathogenic effect on F. occidentalis. We hypothesize that the first-instar larva (L1) of F. occidentalis mounts a response to TSWV that protects it from pathogenic effects caused by virus infection and replication in various insect tissues. A partial thrips transcriptome was generated using 454-Titanium sequencing of cDNA generated from F. occidentalis exposed to TSWV. Using these sequences, the L1 thrips proteome that resolved on a two-dimensional gel was characterized. Forty-seven percent of the resolved protein spots were identified using the thrips transcriptome. Real-time quantitative reverse transcriptase PCR (RT-PCR) analysis of virus titer in L1 thrips revealed a significant increase in the normalized abundance of TSWV nucleocapsid RNA from 2 to 21 h after a 3-h acquisition access period on virus-infected plant tissue, indicative of infection and accumulation of virus. We compared the proteomes of infected and noninfected L1s to identify proteins that display differential abundances in response to virus. Using four biological replicates, 26 spots containing 37 proteins were significantly altered in response to TSWV. Gene ontology assignments for 32 of these proteins revealed biological roles associated with the infection cycle of other plant- and animal-infecting viruses and antiviral defense responses. Our findings support the hypothesis that L1 thrips display a complex reaction to TSWV infection and provide new insights toward unraveling the molecular basis of this interaction.
INTRODUCTION
Insects and other arthropods, such as ticks and mites, are the primary vectors of several animal- and plant-infecting viruses. These vector-borne viruses rely on their invertebrate vector to be disseminated and to infect their primary animal or plant host. Both animal- and plant-infecting viruses have evolved different transmission strategies in that some attach to the cuticle lining of the mouthparts of the arthropod while others traverse different tissues of the vector's body, in which case virus replication within the invertebrate host may or may not occur. Usually, these arthropod-virus interactions are characterized by some degree of specificity. The cellular mechanisms that determine vector specificity and virus transmission are governed by the interactions between viral components and unknown molecules from the arthropod vector. In contrast to the extensive knowledge of the functions of viral genes and the biological aspects of these interactions, little is known about the molecular responses that arthropod vectors deploy during viral replication in different tissues within their body.
Viruses in the genus Tospovirus, the only plant-infecting members of the virus family Bunyaviridae, are transmitted exclusively by insects in the order Thysanoptera. Insects within this order (from the Greek thysanos and pteron, which mean fringe and wing, respectively) are extremely small, slender insects with fringed wings, commonly known as thrips. The order, which belongs to the hemipteroid assemblage, is composed of two suborders, the Tubulifera, with a single family, and the Terebrantia, with eight families (49). Thripidae is the largest family within the suborder Terebrantia, from which about 50 species are insect pests that cause damage to agricultural crops and to which all vectors of tospoviruses belong (52). Thysanopterans display a great diversity with respect to morphological structures, food preference, and behavioral characteristics. However, they share some common but unique characteristics, such as a postembryonic remetaboly development (34), a hapodiploid genetic system (reviewed in reference 18), and asymmetrical mouthparts that form a narrow stylet (35). Moreover, most vector species share several behavioral characteristics, such as high locomotory activity, high fecundity combined with a short generation time, a strongly female-biased sex ratio, a preference for concealed spaces, a wide range of host plants, and the habit of piercing and sucking from epidermal and mesophyll plant cells (50). Interestingly, of the 5,500 described species of Thysanoptera (53) only 14 are known vectors of tospoviruses, indicative of the specificity of these virus-vector interactions.
The western flower thrips, Frankliniella occidentalis (Pergande), is the most economically important insect pest among thysanopterans due to its extremely wide host range, broad geographical distribution, and competence to transmit 5 of the 17 recognized Tospovirus species. The polyphagous nature of F. occidentalis combined with its short reproductive cycle (∼12 days, egg to adult) and high fecundity (∼75 eggs per female) has contributed to the success of this insect pest as an invasive species. For decades, the use of insecticides has been the primary strategy for controlling this insect pest. However, several F. occidentalis populations from different geographical areas have developed insecticide resistance (9, 43, 59). Additionally, F. occidentalis displays thigmotactic behavior, whereby the insect prefers concealed spaces on or in plant organs, protecting it from harsh conditions. For all the aforementioned reasons, effective control of F. occidentalis is difficult to achieve using the current available strategies (51).
Tospoviruses are segmented, single-stranded, ambisense RNA viruses enclosed in a host-derived membrane. Tomato spotted wilt virus (TSWV) is the best-characterized tospovirus with respect to interactions with its most efficient vector, F. occidentalis. TSWV is transmitted in a persistent propagative fashion in which it journeys through the thrips' body and replicates in various organs of the insect vector. Acquisition of TSWV by thrips is restricted to the first and early second larval stages, and acquisition efficiency decreases as development proceeds. The virus persists through the prepupal and pupal life stages and adulthood. Once a virion is acquired from an infected plant by a larval thrips, the virus enters and replicates in the midgut epithelial cells and then moves to the muscle cells of the gut. Subsequently, the virus infects and replicates in the salivary glands. Both adult males and females will transmit the virus to a permissive plant host during nondestructive, brief probes.
During the early steps of virus infection of larval thrips, the virus particles most likely interact with proteins in the midgut lumen of the thrips vector to initiate the infection process. The virus replicates in the midgut epithelial cells, where it likely uses host cellular components to complete its replication cycle. Escape from the midgut involves traversing the basal lamina to spread to other organs of the insect. Although TSWV replicates in its thrips vector, pathological effects have not been observed in F. occidentalis (81), resulting in efficient transmission of TSWV by this thrips species. One possible explanation for the apparent lack of pathology of the insect vector is that the controlled expression of immune-related proteins by thrips successfully modulates the virus titer, as suggested or demonstrated for arthropod vectors of animal-infecting viruses (54, 63). The tight regulation of proteins involved in other biological processes, such as metabolism, development, growth and reproduction, membrane and protein trafficking, reduction/oxidation (redox), and homeostasis may also counterbalance some of the negative effects that virus infection might otherwise have in the vector.
Unlike mammals, insects solely possess an innate immune system to cope with infections caused by nematode, fungal, bacterial, viral, and protozoan pathogens. It has been shown for Drosophila melanogaster and other insects that the innate immunity is a multilayered system that involves production of antimicrobial peptides and reactive oxygen species (32, 47, 75), clotting and melanization (1, 65), encapsulation and phagocytosis (40, 79), apoptosis and autophagy (17, 67, 69), and RNA interference (RNAi) (27, 78). A better understanding of the molecular mechanisms that insects use to defend themselves against entomopathogenic and vector-borne viruses will enable the design of novel strategies to control these agriculturally and clinically important viruses and their vectors.
We hypothesize that the first-instar larva of F. occidentalis mounts a response during TSWV infection of various tissues and organs of the vector that protects the insect from pathogenic effects while allowing viral replication and spread to take place through the thrips' body for efficient virus transmission to occur. To begin to address the mechanisms underlying vector competency and lack of pathology in the TSWV-thrips interaction, we expanded the transcriptomic resources for F. occidentalis and utilized this large collection of coding sequences to identify proteins in healthy and TSWV-infected larval thrips. Two-dimensional (2-D) gel electrophoresis and matrix-assisted laser desorption ionization–tandem time of flight (MALDI-TOF/TOF) mass spectrometry coupled with computational analysis allowed us to identify 52% of the resolved proteins from healthy thrips, from which a higher proportion (47%) matched our F. occidentalis transcriptome collection. Fifteen proteins from naive thrips were functionally annotated as being part of the cell killing or immune system processes. The proteomic tools generated in this study were then used to study the F. occidentalis larva and its interaction with TSWV. We conducted an analysis of differentially expressed proteins between first-instar larvae of F. occidentalis that were exposed or not exposed to TSWV-infected leaves and harvested 24 h following a 3-h acquisition access period (AAP). Overall, we found that the newly generated F. occidentalis transcript sequence collection greatly improved the identification of thrips proteins over the existing arthropod sequences available and that TSWV infection resulted in differential expression of proteins involved in primary metabolic and cellular processes, as well as defense-related proteins of significance to a persistent propagative plant- and animal-infecting virus.
MATERIALS AND METHODS
F. occidentalis cultures.
Our virus-free colony of F. occidentalis originated from an isolate collected from the Kamilo Iki Valley on the island of Oahu, HI, in the early 1980s (3). Insects were reared and maintained on green bean pods (Phaseolus vulgaris) in 16-oz clear plastic deli cups with lids fitted with thrips-proof screens at 22°C (±2°C) under laboratory conditions as previously described (3, 76). To generate first-instar larvae for our experiments, beans were incubated with adult thrips for 3 days to allow females to oviposit. Adult thrips were then removed, and the beans were incubated at 23°C for 24 h to collect emergent larval thrips (0 to 24 h old).
Development of transcriptomic tools for F. occidentalis (Fo Seq).
To generate a large collection of cDNA sequences that represent expression of diverse genes during development and the virus infection cycle of the insect host, we prepared pools of TSWV-exposed first (L1)- and second (L2)-instar larvae and prepupal (P1), pupal (P2), and adult male and female thrips. Cohorts of 0- to 17-h-old L1s were given a 3-h AAP on TSWV-infected leaves of Emilia sonchifolia and then moved and reared to adulthood on green bean pods as described previously (60). The acquisition efficiency (number of L1s infected with TSWV) of our laboratory colony at the time ranged from 40 to 70%, and therefore each cohort represented both infected and noninfected insects. In order to collect adequate numbers of thrips from each developmental stage, eight cohorts of L1s were prepared and monitored over time. The L1 cohorts were collected 2 h after clearing on green bean pods, i.e., 5 h after release on TSWV-infected Emilia leaves and 24 h after release. P1 and P2 insects were collected when approximately 90% of the individuals within a cohort reached these stages. Adults were sampled 24, 72, and 144 h posteclosion, respectively. Each cohort of insects was collected in 1.5-ml nuclease-free microcentrifuge tubes, flash-frozen immediately, and stored at −80°C.
Total RNA was isolated from each of the eight cohorts of insects with TRIzol reagent (Invitrogen, Carlsbad, CA) to maximize yield. Frozen insect bodies were homogenized manually in 100 μl of TRIzol reagent for 30 s with a nuclease-free Kontes pestle (Fisher Scientific, Pittsburg, PA), followed by an additional 900 μl TRIzol reagent and brief homogenization. The remaining RNA isolation steps were performed using the standard manufacturer's protocol for TRIzol reagent. The RNA pellets were redissolved in 40 μl of diethyl pyrocarbonate (DEPC)-treated water and RNA was quantified by using a Nanodrop spectrophotometer (Agilent, Inc., Santa Clara, CA). RNA yields ranged from 274 to 1,729 ng μl−1 obtained for L1s 2 h after clearing from the AAP and adults collected 6 days posteclosion, respectively. Total RNA quality was determined by using an Agilent 2100 bioanalyzer using Nanochip technology (Agilent), revealing high-quality RNA (i.e., no evidence of degradation) for the eight cohort samples. Furthermore, real-time quantitative reverse transcriptase PCR (qRT-PCR) of TSWV nucleocapsid (N) RNA revealed the presence of TSWV in all of the eight cohorts, with normalized abundance ratios of TSWV N RNA to thrips actin RNA ranging from 7.8 × 10−3 to 1.5 × 10−1 for L1 2 h after clearing and for L2, respectively.
Two pooled samples of total RNA were prepared prior to mRNA isolation. In the first sample, 5 μg of RNA from each of the eight cohort samples was pooled and subsequently treated with DNase using the rigorous DNA removal protocol of the Turbo DNA-free kit (Applied Biosystems, Foster City, CA). The resulting amount of RNA was 33 μg. In the second sample, the remaining samples from seven of the eight cohorts (L1 24 h to adult) were treated separately with DNase, and then 8 μg of RNA from each cohort was pooled. mRNA was isolated from each of the two pooled samples using the MPG mRNA purification kit including 3-in-1 MPS (PureBiotech, LLC, Middlesex, NJ) with a few scale-down adjustments to increase yield and to enhance removal of rRNA contamination. Briefly, the manufacturer's protocol was performed as described, and mRNA was eluted from the beads in 40 μl release solution; then, a second round of isolation from the eluent was performed by scaling down the kit volumes by 7-fold and releasing mRNA in 30 μl solution. Quality and purity of each mRNA isolation were assessed using RNA 6000 Pico Chip technology on the Agilent 2100 bioanalyzer, and then the two isolations were pooled and subjected to one more round of mRNA isolation using the scaled-down-kit purification procedure to further clean up and concentrate mRNA. The quality and purity of the final mRNA sample were reconfirmed prior to cDNA synthesis.
A 200-ng sample of mRNA was sent to the KSU Integrated Genomics Facility (Kansas State University, Manhattan, KS) for preparation of a nonnormalized cDNA library with random hexamers and emulsion PCR amplification of cDNA fragments using the cDNA rapid library preparation method and GS FLX Titanium chemistry (454 Life Science/Roche, Branford, CT), respectively. One and one-half plates were prepared for pyrosequencing on the Genome Sequencer FLX system. The GS FLX Titanium Data Computing Cluster was used for processing the sequencing runs and generating base calls. The software program Newbler Metrics 2.3 (454 Life Sciences Corporation) was used to align reads and assemble contigs. Our sequencing effort yielded a total of 1,755,018 high-quality reads (average read length of 420 bases) composed of 625,551,775 bases, and 93% of the reads aligned and assembled into 26,324 contigs. Of these contigs, 70.4% (large contigs) ranged from 500 to 11,142 bases in length, with an average length of 1,368 bases; 96% of the contig lengths (i.e., total number of bases) received quality scores of at least 40.
Our transcript sequence (cDNA) database was constructed by combining the 454 contig sequences (26,324) and a previously described Sanger-sequenced expressed sequence tag (EST) collection (contigs and singletons) for F. occidentalis L1 insects generated by Sanger sequencing (61). The combined collection of 454 and Sanger sequences, herein referred to as Fo Seq, was clustered and assembled using the KSU ArthropodEST bioinformatics pipeline as described previously (61) to obtain one transcript sequence database. The sequences were compared to the NCBI nonredundant (nr) protein sequence database using BLASTx at an E value of ≤10−10. Gene ontology (GO) terms were mapped and sequences were annotated using the software program Blast2GO (http://www.blast2go.de).
Collection and protein extraction from healthy larval thrips for proteomic analysis.
Six hundred emergent larval thrips (0 to 24 h old) were manually collected using a fine paintbrush (000), pooled into a 1.7-ml nuclease- and protease-free microcentrifuge tube, flash-frozen in liquid nitrogen, and stored at −80°C until further use. Total proteins were extracted in 200 μl of rehydration/sample buffer containing 8 M urea, 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 50 mM 1,4-dithiothreitol (DTT), 0.2% Bio-Lyte 3/10 ampholyte, and 0.001% bromophenol blue (Bio-Rad, Hercules, CA) using a sterile Kontes pestle (Fisher Scientific). After centrifugation at 12,000 × g for 5 min, the protein supernatant was quantified for 2-D gel electrophoresis using the bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, Wilmington, DE), following the manufacturer's instructions.
Two-dimensional gel electrophoresis and MALDI-TOF/TOF mass spectrometry.
Following total protein extraction and quantification, 400 μg of protein supernatant was applied to an 11-cm IPG strip with a pH 3 to 10 gradient (Bio-Rad) for overnight passive rehydration at 25°C. For the first separation of proteins, the IPG strip was subjected to isoelectric focusing (IEF) using a Protean IEF cell (Bio-Rad). The IEF was carried out using the following voltage step-gradient program: 250 V for 2,000 Vh; 8,000 V for 18,000 Vh; and 8,000 V for 20,000 Vh at 20°C and a maximum current setting of 70 μA/strip. After IEF, the IPG strip was equilibrated for 15 min in equilibration buffer I (6 M urea, 2% SDS, 0.375 M Tris-HCl [pH 8.8], 20% glycerol, and 2% [wt/vol] DTT) (Bio-Rad), followed by another 15 min in equilibration buffer II (the same as equilibration buffer I but containing 2.5% iodoacetamide [IAA] instead of DTT) (Bio-Rad) to reduce and carbamidomethylate the focused proteins, respectively. For the second separation of proteins, the IPG strip was placed across a 10 to 20% Criterion Tris-HCl precast gel (Bio-Rad) and overlaid with agarose. Electrophoresis was run in Tris-glycine buffer (25 mM Tris, 192 mM glycine, and 0.1% SDS, pH 8.3) (Bio-Rad) using an electrophoresis Criterion unit (Bio-Rad) at a constant voltage of 150 V until the yellow dye front completely migrated out of the bottom of the gel. Using a clinical rotator at a low rotation speed, the 2-D gel was rinsed with double-distilled (dd) water for 15 min and then stained overnight with Bio-Safe Coomassie brilliant blue G-250 (Bio-Rad). After staining, the 2-D gel was washed with dd water until the blue background was no longer visible. The 2-D gel was kept at 4°C until further use.
The 2-D gel was scanned with a Bruker Daltonics Proteineer spII imaging system (Bruker Daltonics, Billerica, MA). All the clearly defined protein spots were manually excised from the 2-D gel using a cork borer and deposited individually in microcentrifuge tubes. One hundred microliters of 25 mM ammonium bicarbonate was added to each tube, which then was incubated for 30 min at room temperature to destain the protein spots. Proteins were destained with 50 μl of acetonitrile (ACN) (biotech-grade solvent > 99.93%) (Sigma-Aldrich, St. Louis, MO) for 5 min at room temperature. The gel pieces were dried by speed vacuum for 15 min at 4°C. The samples were subjected to tryptic digestion using 100 ng of proteomics-grade trypsin (Trypsin Gold; Promega, Madison, WI) in 20 μl of dd water at 37°C overnight (∼16 h). Trypsinized peptides were then extracted from the gel pieces with 5% trifluoroacetic acid (TFA) in 50% (vol/vol) ACN-water at 37°C for 30 min. The resulting peptide solutions were dried by speed vacuum for 4 h and then reconstituted in 10 μl of 20 mg/ml 2, 5-dihydroxylbenzonic acid in 33% ACN–0.1% TFA. Peptide solutions were then mixed (1:1) with matrix α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich). For each sample, 2 μl of peptide/matrix solution was dispensed onto a Bruker stainless steel matrix-assisted laser desorption ionization (MALDI) target plate (Bruker Daltonics). Mass spectra (MS and MS/MS) were obtained using a Bruker Daltonics Ultraflex II TOF/TOF mass spectrometer (Bruker Daltonics), and positively charged ions were acquired in the reflector mode over an m/z range of 500 to 1,500 using delayed extraction. The spectra were processed by the FlexAnalysis 3.0 and Biotools 3.0 software programs (Bruker Daltonics) without further smoothing or spectrum processing. Monoisotopic masses were obtained using the SNAP algorithm with at least 5 for the signal/noise ratio of peak intensity. Measurements were externally calibrated with 8 different peptides, ranging from 757.39 to 3,147.47 (Peptide Calibration Standard II; Bruker Daltonics). The generated peak lists were transferred to the ProteinScape 1.3 software program, which uses the MASCOT 2.2 (www.matrixscience.com) (Matrix Science, Boston, MA) search engine for protein identification.
Protein identification.
Peptide mass lists were compared to the Fo Seq and the Metazoan nr protein database of the National Center of Biotechnology Information (NCBI) for protein identification. Furthermore, we conducted a sequence search against the Prokaryotic nr protein database of NCBI to identify proteins from bacterial origin within the F. occidentalis resolved proteome. The NCBI database searches were conducted on 23 August 2010. For protein identification, we used an error tolerance search algorithm and the six possible translation open reading frames. Moreover, we used the following search parameters for protein identification: carbamidomethyl-cysteine as a fixed modification and methionine oxidation as a variable modification, with one missed tryptic cleavage allowed. The MS and MS/MS data collected were used to conduct a combined search. MASCOT 2.2 software was used to compare databases of peptide sequences and their theoretical fragmentation patterns with a given MS and MS/MS spectrum. Protein spots that had significant matches (P ≤ 0.05) were considered to be identified. Additionally, we considered to be identified protein spots that had marginally significant matches (P = 0.051 to 0.1) in order to provide a less stringent but conservative identification. Complete sequences from the Fo Seq that matched protein spots under the described criteria were subjected to a protein database search using BLASTp from NCBI to determine their homology to known proteins. Furthermore, we conducted a functional annotation of the sequences that matched the thrips proteins analyzed in our proteomic work using Blast2GO against the NCBI nr sequence database.
TSWV maintenance.
TSWV (isolate TSWV-MT2) was periodically maintained by thrips transmission on Emilia sonchifolia as previously described (77). Virus was mechanically inoculated onto young E. sonchifolia plants using thrips-inoculated Emilia leaf tissue. Systemically infected tissue from the mechanically inoculated plants was used as the source of virus acquisition by emergent larval thrips 12 days postinoculation (dpi).
Estimation of virus accumulation and acquisition efficiency of TSWV-exposed L1 thrips.
In order to select a time to sample cohorts of young L1s for our proteomic study of F. occidentalis response to TSWV infection, real-time qRT-PCR experiments were conducted to estimate virus activity, i.e., accumulation of viral RNA due to transcription and/or replication. The experiment consisted of two sampling time points, 2 and 21 h after clearing on healthy green bean pods following a 3-h AAP on TSWV-infected tissue, and three biological replications of the experiment. Young L1s (0 to 17 h old) were exposed to bouquets of TSWV-infected E. sonchifolia leaf tissue in the same type of cups used to maintain the thrips colony, and then the bouquets were replaced with healthy green bean pods in order for the larvae to feed and clear unbound virus particles from the gut lumen. At each time point, a group of 100 insects was collected using separate brushes for each treatment and then flash-frozen and stored at −80°C for analysis of virus titer. In addition, 10 individual insects were arbitrarily subsampled from the 21-h time point to assess the efficiency of acquisition by endpoint RT-PCR for virus detection.
Total RNA from the groups of thrips was isolated using TRIzol reagent as described above for the Fo Seq transcript collection. The RNA pellets were redissolved in 20 μl of DEPC-treated water, and RNA was quantified by using a Nanodrop spectrophotometer (Agilent, Inc., Santa Clara, CA). cDNA was synthesized from 1 μg of total RNA using the Verso cDNA synthesis kit (Thermo Scientific, Wilmington, DE) with a 3:1 ratio of random hexamers to anchored oligo(dT) primers and Verso RT Enhancer to remove contaminating DNA, following the manufacturer's protocol. After cDNA synthesis, the 20-μl reaction mixture was diluted up to 200 μl with DEPC-treated water.
Normalized real-time qRT-PCR was performed to estimate the abundance of TSWV N RNA relative to an internal standard at each time point using the same reaction components and conditions as previously described (60). TSWV N RNA represents both N gene mRNA and small (S) RNA of the virus. A sequence annotated as ribosomal protein 49 (RP49) in our Fo Seq transcript collection was tested and chosen as the internal reference gene because it was determined to be stably expressed between the sampling time points and between virus-infected and noninfected thrips (data not shown). Beacon Designer was used to design efficient RP49 primers (forward primer, 5′CAATAAGAACATCATCAAGAAGTC3′; reverse primer, 5′ATCGTCATGGTGCCTTTG3′) for use with iQ SYBER Green chemistry (Bio-Rad, Hercules, CA). Both the TSWV N and Fo Seq RP49 primer pairs were determined to produce PCR efficiencies (E) of 1.9 using the equation E = 10[−1/slope] (56). The normalized abundance of TSWV N RNA was calculated using the ratio ERP49CT(RP49)/ENCT(N), where E is the PCR efficiency of a primer pair, and CT is the fractional amplification cycle number at which florescence emitted during the reaction first exceeds background fluorescence and is inversely related to the initial template concentration in the PCR. Log10-transformed normalized abundance ratios were subjected to analysis of variance (ANOVA) to determine if there was a statistically significant difference between the abundance of TSWV N RNA in L1s at 2 and 21 h after clearing.
Acquisition efficiency, i.e., the proportion of TSWV-positive insects, was determined for the 21-h time point for each biological replicate. Ten individual insects were subsampled, frozen, and stored for endpoint RT-PCR amplification of TSWV N RNA. Total RNA was extracted from individual insects using a Chelex 100 (Bio-Rad) method developed by Boonham et al. (10). Ten microliters of the RNA sample served as the template for cDNA synthesis using the Verso cDNA synthesis kit. Negative and positive template controls were included in the cDNA and PCR steps. PCR was performed in 25-μl reaction mixtures using the PCR Go Taq system I (Promega, Madison, WI) at 95°C for 2 min followed by 40 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 5 min. The same primers used to amplify TSWV N cDNA for real-time qRT-PCR were used for endpoint RT-PCR (200 nM [each] in the final reaction). F. occidentalis actin endpoint PCR primers previously tested (10) were used for detection of insect cDNA to ensure that total RNA template was present in TSWV-negative insects. Amplified products were visualized in 1.5% agarose gels using ethidium bromide and a UV transilluminator (Bio-Rad).
Sample preparation for proteomic analysis of virus-exposed and nonexposed L1 thrips.
A virus acquisition experiment was conducted to determine the protein-level response of L1 thrips to TSWV infection. The experiment consisted of two treatments, virus-exposed and nonexposed L1s, and each experiment was conducted five independent times, i.e., biological replicates. Four replicates were used for differential protein analysis, and one replicate was used for Western blot analysis using the methods of Whitfield et al. (80).
Cohorts of emergent larval thrips (0 to 18 h old) were collected, starved for 30 min, and then exposed to either bouquets of virus-infected Emilia leaves as described above (exposed group) or bouquets of healthy, noninfected Emilia (nonexposed group) for a 3-h AAP. The bouquets were subsequently replaced by virus-free green bean pods, and the cohorts were allowed a 24-h clearing period. From each treatment, groups of 600 L1s (27 to 45 h old) were then collected manually and stored at −80°C as described above, and 10 insects were subsampled from each treatment to assess acquisition efficiency of the exposed group or virus contamination in the nonexposed group. We determined that acquisition efficiency of the L1 cohorts ranged from 80% to 90% (data not shown) and that nonexposed thrips were virus free. We chose cohorts with 90% infection as our four biological replicates for the proteome analysis and one replicate for the Western blot analysis.
Analysis of differentially expressed proteins.
To identify proteins that are differentially expressed in the first-instar larvae of F. occidentalis during TSWV infection, we subjected proteins extracted from virus-exposed and nonexposed L1 thrips by the trichloroacetic acid-acetone (TCA-A) extraction method used by Cilia et al. (15, 16) to 2-D electrophoresis. To standardize the relative abundance of proteins across treatments and biological replications, we used 375 μg of protein extract per sample. Images of the 2-D gels were generated using a Bruker Daltonics Proteineer spII imaging system (Bruker Daltonics). The images were uploaded into the software program Ludesi REDFIN 3 (http://www.ludesi.com/redfin/) for analysis of the differentially expressed proteins. The 2-D gels were manually warped and then automatically aligned to each other, and protein spot delineation was edited when necessary. Protein spots were selected as being differentially expressed between the two treatments if they showed an ANOVA P value of <0.1 (with a P value of <0.05 being significant and a P value of 0.051 to 0.1 being marginally significant) and a >1.1-fold change in spot density.
ESI-MS/MS and identification of differentially expressed proteins.
Protein spots that were identified as differentially expressed between TSWV exposed and nonexposed thrips were excised from one pair of 2-D gels and processed as previously described here. The gel-extracted peptides were reconstituted in a solution of 0.1% formic acid and 2% ACN and processed in a Bruker Daltonics HCT Ultra ion trap mass spectrometer (Bruker Daltonics) coupled with a microcolumn switching device (Switchos; LC Packings), an autosampler (Famos: LC Packings), and a nanogradient generator (UltiMate Nano HPLC; LC Packings) for electrospray ionization tandem (ESI-MS/MS) mass spectrometry to generate peptide sequences of extensive coverage. Samples were loaded on a C18 reversed-phase capillary column (75-μm inside diameter [i.d.] by 15 cm, PepMap; Dionex) in conjunction with an Acclaim C18 PepMap trapping column (300-μm i.d. by 10 mm, PepMap; Dionex). Peptides were separated by a nanoflow linear ACN gradient using buffer A (0.1% formic acid, 2% ACN) and buffer B (0.1% formic acid, 80% ACN), starting from 5% buffer B to 60% over 45 min at a flow rate of 200 nl/min. The column was washed with 95% buffer B for 5 min. The system control software, Hystar 3.2, was used to control the entire process. The eluted peptides were then injected into an HCT Ultra ion trap mass spectrometer (Bruker Daltonics). The mass spectrometer was set up in the data-dependent MS/MS mode to alternatively acquire full scans (m/z acquisition range from 300 to 1,500 Da). The four more intense peaks in any full scan were selected as precursor ions and fragmented by collision energy. MS/MS spectra were interpreted, and peak lists were generated by the DataAnalysis 3.4 and Biotools 3.0 software programs (Bruker Daltonics), respectively. Generated MS/MS spectra were compared to in silico-digested sequences from the Fo Seq and the Metazoan nr protein database of NCBI. Furthermore, we used the genome sequence of TSWV (accession numbers NC_002051 for the S RNA, NC_002050 for the M RNA, and NC_002052 for the L RNA) to identified protein spots that correspond to viral proteins in TSWV-exposed thrips. Other parameters used were as previously described here. Protein spots were considered identified when they had Mascot scores that represented a P value of <0.05. The sequences that matched proteins that were differentially expressed under the described parameters were also subjected to BLASTp from NCBI to obtain their homology to known proteins and to Blast2GO to assign them a provisional functional annotation.
RESULTS
Summary statistics of the transcript sequence database for Frankliniella occidentalis (Fo Seq).
A collection of assembled and annotated transcript sequences of F. occidentalis, Fo Seq, was generated and served as a subject database for identification of proteins in the larval thrips proteome. This database was constructed from previously described and available EST resources (Sanger sequence) for noninfected, healthy L1 thrips (61) and contigs generated from 454 pyrosequencing of cDNA synthesized from mRNA isolated from groups of young larvae to older adults exposed to TSWV in the present study. The “hybrid” assembly yielded 4,584 contigs (136 to 10,175 base lengths) composed of two or more ESTs and 454-derived contig sequences and 27,143 single EST or 454-derived contig sequences (100 to 8,888 base lengths). Sequence analysis revealed that of the 31,727 transcript sequences, 49% had significant matches (E < 10−10) to protein sequences in the NCBI nr sequence database. Analysis using Blast2GO revealed that 31% of the sequences were assigned annotations to known proteins. Our analysis revealed that the majority of the Fo Seq database sequences were nonannotated, high-quality sequences, revealing an overall high amino acid sequence variation (divergence) between F. occidentalis and members of insect orders that are well represented in sequence databases.
Characterization of the larval thrips proteome.
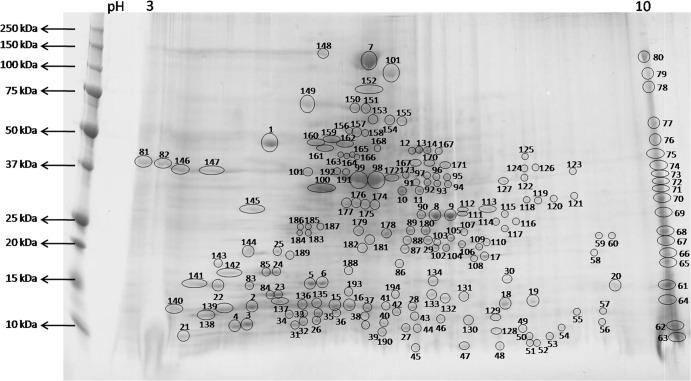
Using 400 μg of total protein extracted from 600 healthy L1s of F. occidentalis, we clearly resolved 194 protein spots in a 2-D gel with a pH range of 3 to 10 (Fig. 1). MALDI-TOF/TOF mass spectrometry coupled with computational analysis using the Fo Seq and the Metazoan nr protein database from NCBI allowed us to identify 52% of the 194 resolved protein spots (Table 1). The distribution of identified protein spots reflected that a higher proportion of peptides (47%) matched the Fo Seq, with a considerably lower proportion of peptides (23%) matching sequences from the Metazoan database (Fig. 2). Out of the 92 and 45 protein spots identified with the Fo Seq and the Metazoan database, respectively, 36 protein spots (19%) were identified with both databases. Fifty-six proteins (29%) were identified exclusively with the Fo Seq, while nine (5%) were identified with the Metazoan database only. It is worth mentioning that of the nine protein spots that were identified with the Metazoan database, two corresponded to cysteine proteases from F. occidentalis that were not represented in our subject database. Furthermore, there were 9 and 13 protein spots comprised of two different proteins as determined by comparison to the Fo Seq and the Metazoan database sequences, respectively (Table 1). Eighty protein spots (41%) had identities but no significant matches, and 13 spots (7%) were not identified with either of the databases searched (Fig. 2). The complete analysis of protein identification using the Fo Seq and Metazoan databases showing significant, not-significant, and not-identified proteins can be found in Table S1 in the supplemental material. Searches using the Prokaryote nr protein database from NCBI yielded 27 protein spots (14%) with identities (data not shown); however, these particular matches had less-significant Mascot scores than those obtained with the Fo Seq and Metazoan sequences. As such, we consider these proteins to be of insect origin and/or highly conserved across kingdoms.
Fig 1.
Two-dimensional (2-D) gel of the resolved proteome from healthy Frankliniella occidentalis first-instar larvae. Four hundred micrograms of total protein extracted from 600 pooled first-instar larvae of F. occidentalis was resolved in a 2-D gel using an 11-cm IPG strip with a pH 3 to 10 gradient, followed by standard SDS-polyacrylamide gel electrophoresis (10 to 20% polyacrylamide). The 2-D gel was stained by Coomassie brilliant blue G-250 for protein visualization. One hundred ninety-four protein spots were clearly resolved, excised, further processed, and subjected to MALDI-TOF/TOF mass spectrometry for protein identification. Molecular mass (in kilodaltons) is shown on the y axis, and pI (as pH range) is shown on the x axis.
Table 1.
Identified proteins from healthy Frankliniella occidentalis first-instar larvae
| Spot no. | Database used for identificationa | F. occidentalis transcript identifier | Protein IDd | Organism with sequence similarity | Accession no. | Mascot score |
No. of peptides matched |
||
|---|---|---|---|---|---|---|---|---|---|
| MS | MS/MS | MS | MS/MS | ||||||
| 1 | Fo | CL2461Contig1_S454 | Similar to calreticulin isoform 1 | Apis mellifera | XP_392689.2 | 100 | 524 | 21 | 6 |
| 1 | M | Calreticulin | Drosophila melanogaster | gi|6063416 | 51 | 1 | |||
| 3 | Fo | CL1605Contig1_S454 | Cuticular protein RR-1 family member 16 | Nasonia vitripennis | NP_001161311.1 | 362 | 3 | ||
| 3 | M | hCG1650121, isoform CRA_a | Homo sapiens | gi|119567926 | 43 | 1 | |||
| 4 | Fo | contig04259 | Cuticular protein RR-1 motif 43 | Bombyx mori | NP_001166711.1 | 148 | 1 | ||
| 5 | Fo | CL5080Contig1_S454 | Conserved hypothetical protein | Tribolium castaneum | XP_970222.2 | 90 | 370 | 9 | 4 |
| 6 | Fo | CL5080Contig1_S454 | Conserved hypothetical protein | Tribolium castaneum | XP_970222.2 | 80 | 250 | 6 | 4 |
| 6 | M | similar to ENSANGP00000015016 | Nasonia vitripennis | gi|156548106 | 53 | 1 | |||
| 7 | Fo | contig26596 | Myosin heavy chain, isoform N | Drosophila melanogaster | NP_001162990.1 | 166 | 87 | 23 | 1 |
| 7c | M | AGAP010147-PA | Anopheles gambiae str.e PEST | gi|158299190 | 94 | 26 | |||
| 7c | M | Myosin heavy chain, nonmuscle or smooth muscle | Aedes aegypti | gi|157110721 | 154 | 2 | |||
| 8 | Fo | CL4382Contig1_S454 | Cuticular protein RR-1 family member 39 | Nasonia vitripennis | NP_001161274.1 | 121 | 286 | 10 | 4 |
| 9 | Fo | CL4382Contig1_S454 | Cuticular protein RR-1 family member 39 | Nasonia vitripennis | NP_001161274.1 | 84 | 72 | 9 | 1 |
| 10 | Fo | contig17167 | Cuticular protein tweedle motif 1 | Bombyx mori | NP_001166628.1 | 54 | 1 | ||
| 10 | M | Integrin alpha 7A subunit | Homo sapiens | gi|2654173 | 44 | 1 | |||
| 11b | Fo | FOAA-aab46e12.g1 | 67 | 7 | |||||
| 11b | Fo | CL3474Contig1_S454 | Conserved hypothetical protein | Culex quinquefasciatus | XP_001867362.1 | 238 | 2 | ||
| 12 | Fo | contig14713 | Cuticular protein 111, RR-3 family | Acyrthosiphon pisum | XP_001950838.1 | 85 | 174 | 15 | 4 |
| 13 | Fo | contig14713 | Cuticular protein 111, RR-3 family | Acyrthosiphon pisum | XP_001950838.1 | 97 | 193 | 15 | 5 |
| 14 | Fo | contig14713 | Cuticular protein 111, RR-3 family | Acyrthosiphon pisum | XP_001950838.1 | 74 | 198 | 13 | 4 |
| 15 | Fo | CL3385Contig1_S454 | Cuticular protein 47Ef CG13214-PA | Tribolium castaneum | XP_968350.1 | 80 | 236 | 7 | 3 |
| 16 | Fo | CL3385Contig1_S454 | Cuticular protein 47Ef CG13214-PA | Tribolium castaneum | XP_968350.1 | 338 | 3 | ||
| 16 | M | CG14676 | Drosophila melanogaster | gi|24644452 | 50 | 1 | |||
| 18 | Fo | CL208Contig1_S454 | Monocarboxylate transporter | Pediculus humanus corporis | XP_002426810.1 | 108 | 1 | ||
| 19 | Fo | CL208Contig1_S454 | Monocarboxylate transporter | Pediculus humanus corporis | XP_002426810.1 | 65 | 1 | ||
| 20 | Fo | CL4854Contig1_S454 | Peptidyl-prolyl cis-trans isomerase 2 | Nasonia vitripennis | XP_001607048.1 | 47 | 1 | ||
| 22 | Fo | CL169Contig1_S454 | Calmodulin | Culex quinquefasciatus | XP_001849785.1 | 51 | 1 | ||
| 22c | M | KRT9 protein | Homo sapiens | gi|113197968 | 207 | 19 | |||
| 22c | M | Cytokeratin 9 | Homo sapiens | gi|435476 | 66 | 1 | |||
| 23 | Fo | CL54Contig1_S454 | Myosin 1 light chain | Apis mellifera | XP_393544.2 | 136 | 2 | ||
| 23 | M | Myosin 1 light chain-like protein | Maconellicoccus hirsutus | gi|121543987 | 110 | 1 | |||
| 24 | Fo | CL2263Contig1_S454 | Myosin regulatory light chain 2 | Bombyx mori | NP_001091813.1 | 71 | 1 | ||
| 25 | Fo | contig05467 | Cuticular protein 78, RR-1 family | Anopheles gambiae str. PEST | XP_318996.4 | 35 | 1 | ||
| 26 | Fo | CL4310Contig1_S454 | ATP synthase alpha subunit | Aedes aegypti | XP_001655906.1 | 82 | 214 | 13 | 3 |
| 26 | M | Putative mitochondrial ATP synthase alpha subunit precursor | Toxoptera citricida | gi|52630965 | 57 | 1 | |||
| 27 | M | Keratin 10 | Homo sapiens | gi|186629 | 99 | 14 | |||
| 28 | Fo | CL1680Contig1_S454 | Endocuticle structural glycoprotein SgAbd-2 | Pediculus humanus corporis | XP_002428599.1 | 464 | 4 | ||
| 29 | Fo | contig11827 | Cuticule protein | Pediculus humanus corporis | XP_002423073.1 | 225 | 3 | ||
| 30 | Fo | CL4382Contig1_S454 | Cuticular protein RR-1 family member 39 | Nasonia vitripennis | NP_001161274.1 | 83 | 328 | 9 | 5 |
| 30 | M | Heat shock 70-kDa protein 8 | Danio rerio | gi|94732277 | 71 | 8 | |||
| 31b | Fo | contig07826 | BRCA1-associated ring domain protein | Danaus plexippus | EHJ70219.1 | 63 | 7 | ||
| 31b | Fo | CL4704Contig1_S454 | cAMP-dependent protein kinase inhibitor beta | Pediculus humanus corporis | XP_002430533.1 | 40 | 1 | ||
| 31 | M | Unknown | Homo sapiens | gi|11692692 | 62 | 2 | |||
| 32 | Fo | CL4310Contig1_S454 | ATP synthase alpha subunit | Aedes aegypti | XP_001655906.1 | 55 | 2 | ||
| 33 | Fo | CL1081Contig1_S454 | Sortilin-related receptor L | Nasonia vitripennis | NP_001123523.1 | 75 | 2 | ||
| 34b | Fo | contig27831 | 67 | 9 | |||||
| 34b | Fo | contig00544 | Nicotinic acetylcholine receptor alpha 6 subunit 4 | Tribolium castaneum | NP_001153541.1 | 41 | 1 | ||
| 34 | M | Predicted protein | Nematostella vectensis | gi|156384837 | 77 | 10 | |||
| 35 | M | Ribosomal protein P2 isoform A | Lysiphlebus testaceipes | gi|62083357 | 65 | 1 | |||
| 38 | M | Chain B, complex between nucleosome core particle (H3, H4, H2a, H2b) and 146-bp-long DNA fragment | Xenopus laevis | gi|3745759 | 74 | 7 | |||
| 42 | Fo | CL3413Contig1_S454 | Cellular FABP-like protein 2 | Tribolium castaneum | NP_001164131.1 | 117 | 77 | 10 | 2 |
| 43 | Fo | CL798Contig1_S454 | Elongation factor 2 isoform 1 | Apis mellifera | XP_392691.2 | 95 | 1 | ||
| 43 | M | Elongation factor | Caenorhabditis elegans | gi|156279 | 95 | 1 | |||
| 44 | Fo | FOAA-aaa81c05.g1 | Vacuolar ATP synthase subunit G | Acyrthosiphon pisum | NP_001119628.1 | 67 | 1 | ||
| 44 | M | Vacuolar ATPase G subunit | Maconellicoccus hirsutus | gi|121543630 | 67 | 1 | |||
| 49 | Fo | contig26395 | 134 | 3 | |||||
| 50 | Fo | contig11827 | Cuticule protein | Pediculus humanus corporis | XP_002423073.1 | 147 | 2 | ||
| 51b | Fo | CL3568Contig1_S454 | Ubiquitin | Culicoides sonorensis | AAV84265.1 | 72 | 7 | ||
| 51b | Fo | FOAA-aab28b04.g1 | Hypothetical protein UM04588.1 | Ustilago maydis 521 | XP_760735.1 | 123 | 2 | ||
| 51c | M | Ubiquitin | Littorina littorea | gi|164510076 | 91 | 7 | |||
| 51c | M | Ubiquitin | Drosophila melanogaster | gi|158759 | 123 | 2 | |||
| 52b | Fo | CL3568Contig1_S454 | Ubiquitin | Culicoides sonorensis | AAV84265.1 | 68 | 6 | ||
| 52b | Fo | FOAA-aab28b04.g1 | Hypothetical protein UM04588.1 | Ustilago maydis 521 | XP_760735.1 | 131 | 2 | ||
| 52c | M | Ubiquitin | Littorina littorea | gi|164510076 | 84 | 6 | |||
| 52c | M | Ubiquitin | Drosophila melanogaster | gi|158759 | 131 | 2 | |||
| 53 | Fo | contig11827 | Cuticule protein | Pediculus humanus corporis | XP_002423073.1 | 54 | 1 | ||
| 56 | Fo | contig03553 | Similar to ENSANGP00000011747 | Nasonia vitripennis | XP_001599992.1 | 74 | 56 | 8 | 1 |
| 58 | Fo | contig25916 | Similar to CG2852-PA | Nasonia vitripennis | XP_001604234.1 | 64 | 10 | ||
| 59 | Fo | contig15005 | Hypothetical protein | Tribolium castaneum | XP_976233.2 | 77 | 1 | ||
| 61 | Fo | CL4854Contig1_S454 | Peptidyl-prolyl cis-trans isomerase 2 | Nasonia vitripennis | XP_001607048.1 | 63 | 95 | 14 | 2 |
| 62 | Fo | FOAA-aaa17c02.g1 | similar to H2A histone | Tribolium castaneum | XP_967411.1 | 36 | 1 | ||
| 63 | Fo | contig23633 | 51 | 1 | |||||
| 65b | Fo | CL66Contig1_S454 | Muscle protein 20-like protein | Anoplophora glabripennis | AAY68367.1 | 92 | 14 | ||
| 65b | Fo | contig17431 | Similar to sugar transporter | Nasonia vitripennis | XP_001604576.1 | 70 | 2 | ||
| 66 | Fo | CL198Contig1_S454 | Agrin | Schistosoma mansoni | XP_002577398.1 | 42 | 1 | ||
| 80 | Fo | contig00662 | Similar to CG13124-PA | Apis mellifera | XP_393579.2 | 34 | 1 | ||
| 81 | M | Actin | Drosophila melanogaster | gi|156763 | 253 | 3 | |||
| 90 | Fo | CL4382Contig1_S454 | Cuticular protein RR-1 family member 39 | Nasonia vitripennis | NP_001161274.1 | 75 | 48 | 9 | 1 |
| 92 | Fo | contig05881 | Hypothetical protein | Danio rerio | XP_001333755.3 | 69 | 7 | ||
| 93 | M | Melanoma-associated antigen E2 | Homo sapiens | gi|20162570 | 71 | 11 | |||
| 98 | M | Skeletal alpha-actin | Sparus aurata | gi|6653228 | 84 | 13 | |||
| 102 | Fo | CL1656Contig1_S454 | Glutathione S-transferase | Culex quinquefasciatus | XP_001847604.1 | 98 | 85 | 14 | 1 |
| 104 | Fo | contig26349 | Endocuticle structural glycoprotein SgAbd-2 | Pediculus humanus corporis | XP_002428599.1 | 63 | 126 | 6 | 3 |
| 105 | Fo | CL29Contig1_S454 | Similar to Cbp20 | Nasonia vitripennis | XP_001608109.1 | 104 | 1 | ||
| 105 | M | Similar to CG15006-PA | Apis mellifera | gi|110764439 | 47 | 1 | |||
| 109 | Fo | FOAA-aab00f06.g1 | GK13576 | Drosophila willistoni | XP_002072646.1 | 54 | 2 | ||
| 111 | Fo | CL4382Contig1_S454 | Cuticular protein RR-1 family member 39 | Nasonia vitripennis | NP_001161274.1 | 88 | 113 | 8 | 2 |
| 113 | Fo | CL3105Contig1_S454 | Glyceraldehyde-3-phosphate dehydrogenase | Bombyx mori | NP_001037386.1 | 84 | 7 | ||
| 114 | Fo | contig25068 | Hypothetical protein | Entamoeba dispar SAW760 | XP_001737751.1 | 66 | 6 | ||
| 116 | Fo | CL18Contig1_S454 | Heat shock protein cognate 4 | Apis mellifera | NP_001153522.1 | 118 | 35 | 10 | 1 |
| 116 | M | Heat shock 70-kDa protein 8 | Danio rerio | gi|94732277 | 103 | 7 | |||
| 132 | Fo | contig19365 | Superoxide dismutase | Culex quinquefasciatus | XP_001866335.1 | 70 | 6 | ||
| 134 | Fo | CL862Contig1_S454 | Phospholipid hydroperoxide glutathione peroxidase | Pediculus humanus corporis | XP_002429001.1 | 37 | 2 | ||
| 135 | Fo | CL3385Contig1_S454 | Cuticular protein 47Ef CG13214-PA | Tribolium castaneum | XP_968350.1 | 77 | 34 | 7 | 1 |
| 141 | Fo | contig16281 | 36 | 1 | |||||
| 141 | M | AGAP007997-PA | Anopheles gambiae str. PEST | gi|118789514 | 48 | 1 | |||
| 145 | Fo | CL1184Contig1_S454 | Nascent polypeptide-associated complex protein alpha subunit | Apis mellifera | XP_623555.1 | 101 | 1 | ||
| 145 | M | Nascent polypeptide-associated complex protein alpha subunit | Drosophila melanogaster | gi|1632784 | 69 | 1 | |||
| 146b | Fo | contig27610 | Actin | Hypochilus thorelli | ABZ91668.1 | 86 | 9 | ||
| 146b | Fo | contig01320 | Actin-related protein 1 | Nasonia vitripennis | NP_001157191.1 | 293 | 4 | ||
| 146c | M | Skeletal muscle alpha-actin | Cyprinus carpio | gi|37813312 | 145 | 14 | |||
| 146c | M | Unnamed protein product | Mus musculus | gi|74195718 | 309 | 5 | |||
| 147 | Fo | contig01320 | Actin-related protein 1 | Nasonia vitripennis | NP_001157191.1 | 98 | 84 | 11 | 2 |
| 147c | M | AGAP011516-PA | Anopheles gambiae str. PEST | gi|158293921 | 144 | 15 | |||
| 147c | M | Actin A3 | Bombyx mori | gi|5751 | 78 | 1 | |||
| 150 | Fo | contig00252 | Similar to ENSANGP00000012893 | Nasonia vitripennis | XP_001606463.1 | 63 | 7 | ||
| 151 | Fo | CL18Contig1_S454 | Heat shock protein cognate 4 | Apis mellifera | NP_001153522.1 | 66 | 13 | ||
| 153 | Fo | contig16955 | Similar to vATPase subunit A | Nasonia vitripennis | XP_001604685.1 | 120 | 171 | 22 | 4 |
| 153 | M | ATPase | Homo sapiens | gi|291866 | 94 | 130 | 15 | 3 | |
| 154 | Fo | contig17167 | Cuticular protein tweedle motif 1 | Bombyx mori | NP_001166628.1 | 64 | 6 | ||
| 157 | Fo | CL2000Contig1_S454 | Similar to ENSANGP00000014839 | Nasonia vitripennis | XP_001600045.1 | 125 | 102 | 14 | 2 |
| 157c | M | Similar to 60-kDa heat shock protein, mitochondrial precursor | Apis mellifera | gi|66547450 | 83 | 10 | |||
| 157c | M | 60-kDa heat shock protein | Drosophila melanogaster | gi|1653979 | 71 | 1 | |||
| 158 | Fo | CL2000Contig1_S454 | Similar to ENSANGP00000014839 | Nasonia vitripennis | XP_001600045.1 | 84 | 48 | 10 | 1 |
| 158 | M | Similar to 60-kDa heat shock protein, mitochondrial precursor | Drosophila melanogaster | gi|1653979 | 48 | 1 | |||
| 159 | Fo | contig04030 | Troponin T | Culex quinquefasciatus | XP_001851541.1 | 78 | 1 | ||
| 159 | M | RNA binding motif protein 25, isoform CRA_a | Homo sapiens | gi|119601492 | 47 | 1 | |||
| 160 | Fo | contig15776 | Similar to AGAP012407-PA | Tribolium castaneum | XP_975184.2 | 37 | 1 | ||
| 161b | Fo | CL618Contig1_S454 | Beta-tubulin 1 | Monochamus alternatus | ABY66392.1 | 103 | 17 | ||
| 161b | Fo | CL1556Contig1_S454 | Beta-tubulin | Bombyx mori | NP_001036964.1 | 70 | 4 | ||
| 161 | M | Beta-tubulin | Theromyzon tessulatum | gi|127906328 | 126 | 14 | |||
| 162 | Fo | CL788Contig1_S454 | Tubulin alpha-1 chain | Pediculus humanus corporis | XP_002429121.1 | 95 | 164 | 12 | 4 |
| 162c | M | Alpha-tubulin | Cryptocercus punctulatus | gi|119117127 | 103 | 12 | |||
| 162c | M | Alpha-tubulin at 84B | Drosophila melanogaster | gi|17136564 | 156 | 3 | |||
| 163 | Fo | CL321Contig1_S454 | ATP synthase beta subunit | Tribolium castaneum | NP_001164361.1 | 96 | 1 | ||
| 163c | M | Putative ATP synthase beta subunit | Maconellicoccus hirsutus | gi|124487966 | 109 | 11 | |||
| 163c | M | ATP synthase beta subunit | Drosophila melanogaster | gi|287945 | 270 | 3 | |||
| 164 | Fo | CL321Contig1_S454 | ATP synthase beta subunit | Tribolium castaneum | NP_001164361.1 | 83 | 1 | ||
| 164c | M | ATP synthase beta subunit | Aedes aegypti | gi|157132308 | 128 | 13 | |||
| 164c | M | beta-subunit | Bos taurus | gi|104 | 178 | 2 | |||
| 165 | Fo | CL321Contig1_S454 | ATP synthase beta subunit | Tribolium castaneum | NP_001164361.1 | 69 | 54 | 7 | 1 |
| 165c | M | Putative ATP synthase beta subunit | Maconellicoccus hirsutus | gi|124487966 | 149 | 14 | |||
| 165c | M | ATP synthase subunit family member (Atp-2) | Caenorhabditis elegans | gi|25144756 | 160 | 2 | |||
| 166 | Fo | CL321Contig1_S454 | ATP synthase beta subunit | Tribolium castaneum | NP_001164361.1 | 81 | 1 | ||
| 166c | M | ATP synthase beta subunit | Aedes aegypti | gi|157132308 | 103 | 11 | |||
| 166c | M | Beta-subunit | Bos taurus | gi|104 | 211 | 3 | |||
| 167 | Fo | CL1451Contig1_S454 | GI17663 | Drosophila mojavensis | XP_002002963.1 | 90 | 15 | ||
| 168b | Fo | contig02378 | Alpha-tubulin 1 | Monochamus alternatus | ABU24274.1 | 66 | 9 | ||
| 168b | Fo | CL1922Contig1_S454 | vATPase 55-kDa subunit B | Drosophila melanogaster | NP_476908.1 | 70 | 3 | ||
| 170 | Fo | CL4706Contig1_S454 | Similar to AGAP007827-PA | Tribolium castaneum | XP_967559.1 | 72 | 8 | ||
| 171 | Fo | CL4706Contig1_S454 | Similar to AGAP007827-PA | Tribolium castaneum | XP_967559.1 | 68 | 8 | ||
| 172 | Fo | contig01320 | Actin-related protein 1 | Nasonia vitripennis | NP_001157191.1 | 98 | 256 | 13 | 3 |
| 172c | M | Similar to actin-87E isoform 1 | Apis mellifera | gi|66509780 | 154 | 18 | |||
| 172c | M | RecName: full = actin, muscle-type; AltName: full = A2 | Molgula oculata | gi|3121741 | 303 | 4 | |||
| 173 | Fo | CL299Contig1_S454 | Similar to RE12057p isoform 2 | Acyrthosiphon pisum | XP_001943291.1 | 59 | 1 | ||
| 173 | M | Actin A3 | Bombyx mori | gi|5751 | 59 | 1 | |||
| 174 | M | Similar to tubulin, beta, 2 | Apis mellifera | gi|48095547 | 73 | 10 | |||
| 176 | Fo | contig01307 | Similar to pupal cuticle protein 78E | Tribolium castaneum | XP_969263.1 | 64 | 8 | ||
| 177 | Fo | contig01307 | Similar to pupal cuticle protein 78E | Tribolium castaneum | XP_969263.1 | 65 | 80 | 9 | 2 |
| 178 | Fo | contig17167 | Cuticular protein tweedle motif 1 | Bombyx mori | NP_001166628.1 | 65 | 186 | 6 | 3 |
| 179 | Fo | contig17167 | Cuticular protein tweedle motif 1 | Bombyx mori | NP_001166628.1 | 134 | 3 | ||
| 181 | Fo | CL357Contig1_S454 | Eukaryotic initiator factor 4a | Tribolium castaneum | NP_001177648.1 | 107 | 2 | ||
| 181 | M | Similar to eukaryotic initiation factor 4A (ATP-dependent RNA helicase eIF4A) (eIF-4A) isoform 2 | Apis mellifera | gi|66551115 | 100 | 2 | |||
| 184 | M | Cysteine protease CP7 precursor | Frankliniella occidentalis | gi|15593246 | 70 | 1 | |||
| 185 | M | Cysteine protease CP7 precursor | Frankliniella occidentalis | gi|15593246 | 104 | 2 | |||
| 187 | Fo | contig22086 | Similar to cuticular protein 47Ef CG13214-PA | Tribolium castaneum | XP_968350.1 | 53 | 1 | ||
| 192 | Fo | CL4870Contig1_S454 | Mitochondrial peptidase beta subunit | Culex quinquefasciatus | XP_001863592.1 | 90 | 14 | ||
| 193 | Fo | contig01320 | Actin-related protein 1 | Nasonia vitripennis | NP_001157191.1 | 71 | 1 | ||
| 193 | M | Beta-actin | Culex pipiens pipiens | gi|90811719 | 95 | 2 | |||
Protein identification was obtained using the Fo Seq (Fo) and the Metazoan nr protein database from NCBI (M).
Different F. occidentalis transcripts matched peptide sequences from a single protein spot within the 2-D gel of healthy larval thrips.
Different sequences from the Metazoan nr protein database matched the same peptide stretches from a single protein spot within the 2-D gel of healthy larval thrips.
ID, identification.
str., strain.
Fig 2.
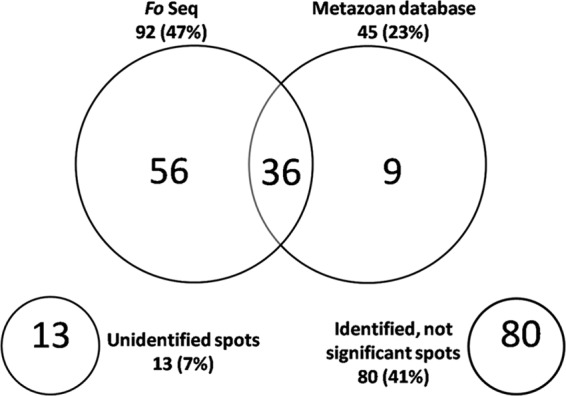
Venn diagram of the identified proteins from healthy Frankliniella occidentalis first-instar larvae. The large circles display the number and proportion (in parentheses) of proteins that were identified using the Fo Seq and the Metazoan nr protein database from NCBI. The overlap indicates the number of proteins that were identified with both databases, while the numbers inside each circle represent the proteins that were unique to either database. The small circles display the number and proportion (in parentheses) of proteins that were unidentified or identified without having significant matches.
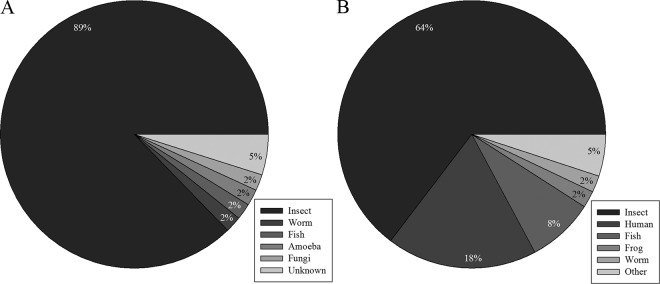
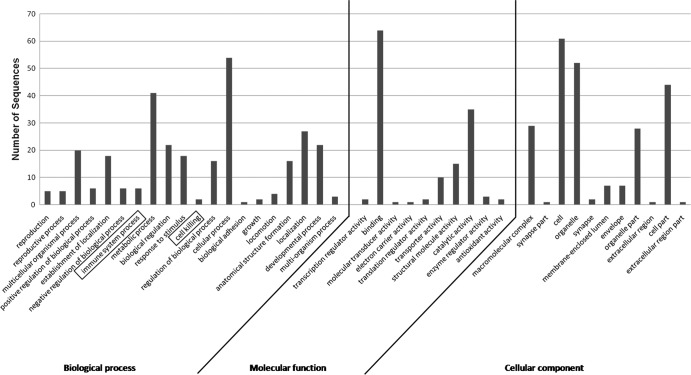
The subset of Fo Seq and Metazoan sequences that matched thrips larval proteins was subjected to BLASTp analysis to identify matches to protein sequences available at NCBI (Fig. 3). The majority of these Fo Seq and Metazoan sequences matched proteins of other insect species. Protein sequences of the red flour beetle, Tribolium castaneum, scored the highest and produced the greatest number of matches to the larval thrips proteins identified in this study. Seven and three proteins from F. occidentalis had sequence similarities to proteins of the body louse (Pediculus humanus corporis) and the pea aphid (Acyrthosiphon pisum), respectively, representing two other orders in the Hemipteroid assemblage. Moreover, 57% of our identified protein spots had provisional functional annotations obtained with Blast2GO analysis (Fig. 4). In relation to the host defense response, 15 protein spots were functionally annotated as proteins associated with cell killing or immune system processes. These spots comprised six different proteins, which are tubulin alpha-1 chain, beta tubulin, glutaredoxin 5, heat shock protein, cysteine protease, and lethal giant larvae homologue (Table 2).
Fig 3.
Distribution of classes of organisms with protein sequences similar to those of Frankliniella occidentalis proteins. These pie charts display the classes of organisms that had protein sequences similar to the sequences of the Fo Seq (A) or the Metazoan nr protein database from NCBI (B), used to identify proteins from healthy larval thrips.
Fig 4.
Distribution of Frankliniella occidentalis identified proteins by provisional biological processes, molecular functions, and cellular component categories. Each category represents a gene ontology (GO) term assigned by Blast2GO analysis, while the height of the bars indicates the number of proteins that were classified in each category.
Table 2.
Frankliniella occidentalis proteins from naive first-instar larvae with putative roles in insect innate immunity
| Protein ID | Putative role in defense | Spot no. | F. occidentalis transcript identifier(s) or NCBI accession no. |
|---|---|---|---|
| Tubulin alpha-1 chain | Phagocytosis | 28, 162 | CL1680Contig1_S454, CL788Contig1_S454/gi|17136564 |
| Beta tubulin | Regulation of apoptosis | 161, 174 | CL618Contig1_S454/CL1556Contig1_S454/gi|127906328, gi|48095547 |
| Glutaredoxin 5 | Cell redox homeostasis | 134 | CL862Contig1_S454 |
| Heat shock protein | Response to biotic/abiotic stress | 19, 25, 26, 30, 116, 157, 158 | CL208Contig1_S454, contig05467, CL4310Contig1_S454, gi|94732277, CL18Contig1_S454/gi|94732277, CL2000Contig1_S454/gi|66547450/gi|1653979, CL2000Contig1_S454//gi|1653979 |
| Cysteine protease | Ubiquitin cycle | 184, 185 | gi|15593246, gi|15593246 |
| Lethal giant larvae homologue | Induction of salivary gland cell autophagic cell death | 181 | CL357Contig1_S454 |
Significant accumulation of TSWV N RNA in F. occidentalis L1s.
Real-time qRT-PCR was performed to distinguish a reliable time frame to analyze the proteomic response to virus infection in young L1s. In three biological replicates, we calculated the normalized abundances of TSWV N RNA for two “virus clearing” time points (2 and 21 h after a 3-h TSWV exposure) to quantify increases in the virus titer. All three replications revealed a 70% acquisition efficiency, i.e., 7 of 10 insects chosen arbitrarily from the pool of L1s at 21 h of clearing had detectable levels of virus as determined by endpoint RT-PCR. Between the two time points, the normalized abundance of TSWV N RNA increased between 7- and 23-fold across replicates (Table 3), and on average, there was a significant 14-fold increase (P < 0.002). These data indicate that within 24 h after exposure to TSWV-infected plant tissue, L1s were infected and virus was transcribing and/or replicating during this time frame.
Table 3.
Real-time qRT-PCR estimation of TSWV N RNA accumulation in cohorts of F. occidentalis first-instar larvae (L1) given a 3-h acquisition access period on TSWV-infected leaf tissue and cleared on green beans
| Replicatea | L1 time point(h) |
CT value |
Normalized abundance of TSWV N RNAc |
||
|---|---|---|---|---|---|
| TSWV N | RP49b | Value for replicate | Avg for 3 replicates | ||
| 1 | 2 | 21.8 | 20.8 | 0.47 | |
| 2 | 2 | 22.7 | 21.0 | 0.30 | |
| 3 | 2 | 21.5 | 20.0 | 0.34 | 0.37a |
| 1 | 21 | 16.7 | 19.7 | 6.28 | |
| 2 | 21 | 17.5 | 20.7 | 6.89 | |
| 3 | 21 | 18.0 | 19.6 | 2.46 | 5.21b |
Biological replicates consisting of 100 larval thrips.
Ribosomal protein 49 RNA, identified in the Frankliniella occidentalis transcriptome (Fo Seq) and used as an internal reference gene for real-time qRT-PCR.
Normalized abundance of TSWV N RNA = (efficiency of RP49 primers)CT(RP49)/(efficiency of N primers)CT(N), where CT is the fractional cycle number when DNA amplification reaches a fixed threshold (56). Average values (n = 3) followed by different letters indicate a significant difference between normalized abundance of TSWV N RNA (i.e., titer) in L1 2-h and L1 21-h samples as determined by ANOVA (P = 0.002).
Identification of differentially expressed proteins in response to virus infection.
To identify F. occidentalis proteins that are differentially expressed in response to TSWV infection, a merged composite image of the 2-D gels for the virus-exposed and nonexposed treatments (eight in total) was generated by the software program Ludesi REDFIN 3. There were 488 protein spots present in the merged image (see Table S2 in the supplemental material). We identified 26 protein spots that varied in density between the virus-exposed and nonexposed treatments, of which 11 and 15 were found to be significantly (P < 0.05) and marginally (P = 0.051 to 0.1) different by at least 1.1-fold, respectively, between treatments as determined by ANOVA (Fig. 5 and Table 4). Among these 26 protein spots, 62% were downregulated in the TSWV-exposed treatment. ESI mass spectrometry coupled with Mascot search using the Fo Seq and the Metazoan nr protein database resulted in the identification of 37 proteins within the 26 spots. Moreover, one protein spot (spot 310) was not identified with either of the databases searched, and none of the differentially expressed spots contained peptides that matched TSWV proteins. The majority of the identified proteins had sequence similarities to proteins from other insects.
Fig 5.
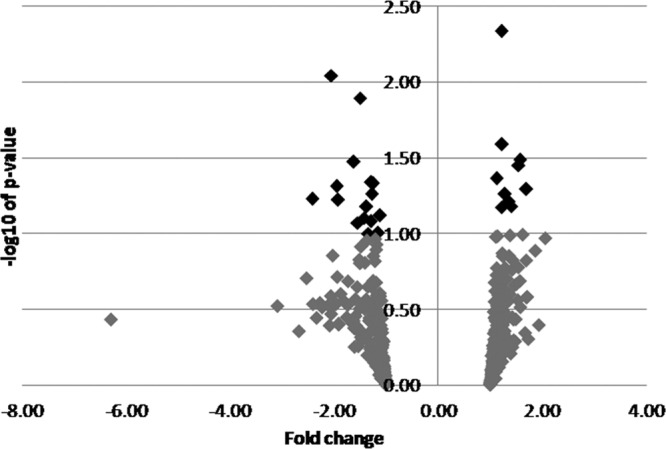
Volcano plot of protein spots from the analysis of differentially expressed proteins between TSWV-exposed and non-exposed Frankliniella occidentalis first-instar larvae. The relative abundances of the differentially expressed protein spots from the 4 biological replicates performed are presented in the volcano plot as dark diamonds. The direction of the change (up- or downregulation) of protein spots that were differentially expressed between treatments is shown as fold change on the x axis, while the magnitude of the change as −log10 P value is shown on the y axis.
Table 4.
Frankliniella occidentalis proteins differentially expressed between tomato spotted wilt virus-exposed and nonexposed first-instar larvae
| Finding and spot no. | P value from ANOVA | Fold change | Database searched (sequence matched) | Mascot score | Protein ID | Identified motifs | E value |
|---|---|---|---|---|---|---|---|
| Upregulated in response to TSWV infection | |||||||
| 201 | 0.0046 | 1.22 | Fo Seq (CL4589Contig1_S454) | 625 | Triosephosphate isomerase | TIM_phosphate binding | 5e−110 |
| NCBI (gi|259016078) | 151 | Triosephosphate isomerase | TIM_phosphate binding | 4e−41 | |||
| 47a | 0.0257 | 1.22 | Fo Seq (CL2263Contig1_S454) | 422 | Myosin 3 light chain | Ca+2 binding site, EFh | 2e−15 |
| NCBI (gi|312371061) | 334 | Hypothetical protein AND_22684 (similar to Skp1) | Skp1 | 4e−80 | |||
| 836a | 0.0326 | 1.57 | Fo Seq (CL63Contig1_S454) | 488 | Similar to voltage-dependent anion-selective channel 2 | Porin3, Porin3_Tom40 | 6e−118 |
| NCBI (RS3_AMBME) | 124 | 40S ribosomal protein S3 | 40S_S3_KH, KH-II, Ribosomal_S3_C | 1e−160 | |||
| 137 | 0.0355 | 1.54 | Fo Seq (CL4310Contig1_S454) | 264 | Mitochondrial ATP synthase α subunit | ATPase_alpha | 0.0 |
| NCBI (gi|52630965) | 261 | Mitochondrial ATP synthase α subunit | ATPase_alpha | 0.0 | |||
| 271a | 0.0433 | 1.12 | Fo Seq (CL4116Contig1_S454) | 499 | cxpwmw03 (similar to electron transfer flavoprotein subunit alpha) | ETF_alpha, AANH-like | 3e−153 |
| NCBI (gi|328786330) | 129 | Saccharopine dehydrogenase-like isoform 1 | GTP_cyclohydrol, TFold, NADB-Rossmann | 0.0 | |||
| 374a | 0.0508 | 1.69 | Fo Seq (contig16594) | 284 | Vitellogenin | lipoprotein N-terminal | 5e−113 |
| NCBI (gi|156763) | 155 | Actin | Actin | 0.0 | |||
| 277 | 0.0551 | 1.28 | Fo Seq (CL3352Contig1_S454) | 215 | Alpha-tocopherol transfer protein-like isoform 1 | SEC14 | 1e−164 |
| NCBI (no match) | |||||||
| 134 | 0.0616 | 1.36 | Fo Seq (CL2397Contig1_S454) | 557 | Vacuolar proton ATP synthase subunit E | V-ATP_synt-E | 4e−71 |
| NCBI (gi|46561760) | 300 | Vacuolar proton ATP synthase subunit E | V-ATP_synt-E | 7e−110 | |||
| 161 | 0.0662 | 1.40 | Fo Seq (CL4706Contig1_S454) | 782 | AGAP007827-PA isoform 1 (similar to enolase) | Enolase-like | 0.0 |
| NCBI (gi|157121051) | 434 | Enolase | Enolase-like | 0.0 | |||
| 72a | 0.0668 | 1.21 | Fo Seq (CL1656Contig1_S454) | 956 | Glutathione S-transferase | Thioredoxin-like, GST_Sigma | 4e−52 |
| NCBI (gi|66565444) | 149 | 60S ribosomal protein L9 | ribosomal_L6 | 2e−101 | |||
| Downregulated in response to TSWV infection | |||||||
| 244 | 0.0090 | 2.05 | Fo Seq (contig19201) | 389 | Cytochrome b-c1 complex subunit 2 | peptidase_M16, peptidase M16_C | 2e−103 |
| NCBI (no match) | |||||||
| 481a | 0.0128 | 1.50 | Fo Seq (contig15458) | 323 | Apolipoprotein D | lipocalin_2 | 4e−84 |
| NCBI (K1C10_HUMAN) | 534 | Keratin type I cytoskeletal 10 | Filament | 0.0 | |||
| 310 | 0.0338 | 1.62 | Fo Seq (no match) | ||||
| NCBI (no match) | |||||||
| 88 | 0.0466 | 1.26 | Fo Seq (CL423Contig1_S454) | 401 | Predicted hypothetical protein (similar to rad23) | RAD_23, UBA, XPC binding | 1e−77 |
| NCBI (gi|242023622) | 169 | UV excision repair protein rad23 | RAD_23, UBA, XPC binding | 3e−164 | |||
| 794 | 0.0460 | 1.29 | Fo Seq (contig30771) | 214 | |||
| NCBI (LYSC_CHICK) | 263 | Lysozyme C | LYZ1, Lys | 6e−110 | |||
| 378a | 0.0486 | 1.95 | Fo Seq (CL1213Contig1_S454) | 495 | AGAP009685-PA (similar to aspartate aminotransferase) | AAT_I | 0.0 |
| NCBI (gi|156763) | 215 | Actin | Actin | 0.0 | |||
| 319 | 0.0548 | 1.28 | Fo Seq (contig17796) | 232 | Peroxiredoxin 1-like | Thioredoxin-like | 5e−83 |
| NCBI (gi|58377838) | 88 | Thioredoxin-dependent peroxidase | Thioredoxin-like | 2e−111 | |||
| 819a | 0.0589 | 2.42 | Fo Seq (CL827Contig1_S454) | 360 | Zinc finger protein 1 | ZPR1_znf, zf-ZPR1, ZPR1 | 0.0 |
| NCBI (PDK3_HUMAN) | 264 | Pyruvate dehydrogenase kinase 3 | HATPase_C, BCDHK_Adom3, Bae5, Vick | 0.0 | |||
| 340a | 0.0597 | 1.92 | Fo Seq (CL2403Contig1_S454) | 523 | Hypothetical protein SINV_09553 (similar to stress-induced phosphoprotein 1) | Tad D, TPR | 0.0 |
| NCBI (gi|170027766) | 137 | Electron transfer flavoprotein; ubiquinone oxidoreductase | Pyr_Redox | 0.0 | |||
| 430a | 0.0667 | 1.38 | FoSeq (CL862Contig1_S454) | 425 | Phospholipid hydroperoxide glutathione peroxidase | Thioredoxin-like, GSH_peroxidase | 2e−67 |
| NCBI (gi|242008321) | 243 | Conserved hypothetical protein (similar to actin depolymerization factor 1) | ADF | 1e−81 | |||
| 196a | 0.0763 | 1.12 | Fo Seq (CL4854Contig1_S454) | 512 | Cyclophilin | cyclophilin_ABH, CLD | 6e−83 |
| NCBI (gi|307643755) | 228 | Glyceraldehyde-3-phosphate dehydrogenase | Gp_dh_C | 3e−109 | |||
| 803 | 0.0787 | 1.43 | Fo Seq (CL18Contig1_S454) | 362 | Heat shock protein 70 cognate 4 | HSP70, Dnak, HscA | 0.0 |
| NCBI (HSP7C_BOVIN) | 218 | Heat shock cognate 71 protein | HSP70, Dnak, HscA | 0.0 | |||
| 283a | 0.0833 | 1.29 | Fo Seq (contig16705) | 625 | Protein yellow-like | MRJP | 2e−138 |
| NCBI (gi|157134067) | 248 | 26S protease regulatory subunit 7 | P-loop NTPase | 3e−162 | |||
| 296 | 0.0851 | 1.55 | FoSeq (contig14504) | 240 | Arginine kinase | phosphagen_ kinase | 2e−129 |
| NCBI (gi|161088212) | 227 | Arginine kinase | phosphagen_ kinase | 4e−130 | |||
| 206 | 0.0997 | 1.15 | FoSeq (contig14504) | 640 | Arginine kinase | phosphatase_kinase | 1e−103 |
| NCBI (gi|161088180) | 496 | Arginine kinase | phosphatase_kinase | 1e−104 | |||
| 182 | 0.0998 | 1.35 | Fo Seq (CL3105Contig1_S454) | 422 | Glyceraldehyde-3-phosphate dehydrogenase | Gp_dh_C/N | 3e−101 |
| NCBI (gi|156547538) | 435 | Glyceraldehyde-3-phosphate dehydrogenase | Gp_dh_C/N | 2e−173 |
Different proteins were identified within a single spot using the Fo Seq and the Metazoan nr protein database from NCBI.
Eighty-six percent of the identified proteins were provisionally annotated using Blast2GO (data not shown). Special attention was given to GO terms with particular relevance to biological processes and molecular functions associated with the life cycle of the insect vector and to a persistent propagative vector-borne virus. Among the 32 provisionally annotated proteins, 2 are proteins involved in reproduction and growth, 8 in development, 21 in metabolic process, 9 in localization and transport, 14 in response to stimuli, 8 in reduction/oxidation and homeostasis, 22 in binding, and 21 in catalytic activity. The provisional annotations of these differentially expressed thrips proteins indicate the multifunctionality of these proteins in several aspects of the insect life cycle and most likely during virus infection. Among the 14 proteins in the category of response to stimuli, 9 proteins were clearly associated with innate immune defenses (Skp1, 40S ribosomal protein S3, mitochondrial ATP synthase α subunit, actin, lysozyme C, thioredoxin-dependent peroxidase, pyruvate dehydrogenase kinase 3, stress-induced phosphoprotein 1, and heat shock cognate 71 protein).
DISCUSSION
Despite the importance of F. occidentalis as a worldwide insect pest and vector of tospoviruses, the thrips response to virus infection at the molecular level is just beginning to be studied. In 2010, the first EST sequencing project for F. occidentalis L1 was reported (61). The current study builds on this initial transcriptome characterization with extended coverage including the five insect life stages following TSWV acquisition and infection, and here we demonstrate its utility for exploring the thrips proteome. Additionally, we have conducted a proteomic analysis of the young L1 stage of F. occidentalis and determined the numbers and identities of differentially expressed proteins between TSWV-exposed and nonexposed L1s using our transcriptome collection as the primary database. The Fo Seq therefore provides a unique sequence resource not only for studying F. occidentalis and other thrips species but also for better understanding the evolutionary biology of the overlooked insect order Thysanoptera with regard to other insects and arthropods in general.
The proteomic analysis of the L1 stage allowed the identification of more than half of the resolved protein spots, with a high proportion of peptides matching the Fo Seq compared to those peptides identified with the Metazoan nr protein database from NCBI. Moreover, 48% of the resolved proteins were not identified with any of the databases searched because peptide identification relies on in silico digestion of protein sequences present in public or private databases (84). The inability to identify proteins spots from 2-D gels is a common phenomenon for organisms without a sequenced genome or those having a partial transcriptome only. Interestingly, 29% of the proteins with significant matches were identified exclusively with the Fo Seq, indicating that these might be proteins unique to the insect order Thysanoptera or species-specific proteins.
Several proteins resolved and annotated in this study may have potential relevance to virus entry and the host response to virus infection. An integrin, a known receptor for some viruses in the family Bunyaviridae, was identified from naive L1s (spot 10). Integrins are a large family of heterodimeric transmembrane glycoproteins that mediate cell adhesion of and binding to the extracellular matrix (37). It has been shown that several diverse families of viruses use integrins as receptors to bind to and enter into their host cells. Several hantaviruses (family Bunyaviridae) have been found to use integrins as their cellular receptors to enter and infect their host cells (28, 29). Further characterization of thrips integrins and possible interactions with TSWV glycoproteins is warranted.
With regard to the insect host defense response to infectious agents, six types of putative immune-related proteins were identified for naive L1s. Both alpha- and beta- tubulin were identified in the L1 proteome, and various tubulin proteins have been shown to play a role in phagocytosis (31, 41) and in regulation of apoptosis (42). A protein spot that matched lethal giant larvae homologue was found, and one possible function for this protein is the induction of salivary gland cell autophagic cell death. Autophagy is required for degradation of the cytoplasmic contents of salivary gland cells during development, and it is an essential component of antiviral defenses against vesicular stomatitis virus (VSV) in D. melanogaster (6, 69). Two protein spots were identified as cysteine proteases, which also play a role in apoptosis and the ubiquitin cycle (2, 73). Glutaredoxin 5, which is involved in cell redox homeostasis to maintain cell integrity and functionality (4), was also classified as an immune-related protein in healthy F. occidentalis larvae. We also identified several protein spots as heat shock proteins, which are expressed in response to biotic and abiotic stresses (19). It is well known that many of these proteins play roles other than those involved in defense mechanisms against foreign organisms. Thus, it is plausible that their expression in naive larval thrips reflects their multifaceted roles in different aspects of the thrips life cycle or that there is a basal level of expression before a pathogen invasion takes place.
The high vector competency of F. occidentalis for TSWV transmission combined with the lack of measurable pathological effects on the insect vector despite virus replication in different insect tissues (81), led us to study the response of the F. occidentalis L1 during the early stages of TSWV infection. Analysis of virus titer in L1s at 2 and 21 h post-virus access indicated that the relative abundance of virus significantly increased during this time frame (Table 3). The average 14-fold increase in relative abundance of TSWV N RNA suggests that the viral polymerase is active within 24 h of thrips exposure to virus and transcription and/or replication may be occurring at the time point sampled for proteomic analysis. Findings from our analysis of titer by real-time qRT-PCR are similar to increases in virus titer observed using serological techniques (74). We also detected TSWV N protein in thrips 24 h after virus exposure (see Fig. S1 in the supplemental material). In all, we found that 26 spots containing 37 proteins were up- or downregulated in response to TSWV infection; however, in the majority of the cases, TSWV had a negative effect on thrips gene expression at the protein level (Table 4). As observed in other proteome studies using 2-D gels (85), we found several single spots that contained two different proteins. At this time, it is not known if one or both of these proteins were differentially regulated by TSWV infection. Using a finer-scale pH gradient to separate these comigrating proteins in future studies may aid in a better resolution of these proteins.
Our statistical analysis of protein expression over four biological replications of the experiment revealed that only 5.3% of the protein spots (26/488) resolved from the pools of 600 insects were differentially regulated by virus infection. This relatively low but significant number of responsive proteins may be explained by the following: (i) inclusion of noninfected thrips in the cohorts of thrips (10% have no detectable virus), (ii) inclusion of noninfected tissues, as is the case with whole insect bodies (only the midgut epithelial and muscle cells are infected during the early stages of the TSWV replication cycle in larval thrips [82]), and/or (iii) inclusion of viruliferous insects harboring low titers of virus that may result in little to no effect on protein expression. Using cutoff significance P values of 0.05 to 0.1 in addition to a P value of <0.05 as our primary criteria for differential expression allowed the detection of protein changes that otherwise might be missed due to the aforementioned reasons. Moreover, our use of four biological replications (i.e., independent experiments with inherent natural and technical variation) to identify differentially expressed proteins provides confidence in the effects due to virus infection.
Among the 21 proteins potentially involved in metabolism, spot 201 was identified as a triosephosphate isomerase, an enzyme which plays an important role in glycolysis and is essential for efficient energy production (39). Interestingly, triosephosphate isomerase deficiency, a unique glycolytic enzymopathy known to affect humans and other mammals, is characterized by susceptibility to pathogen infections (66), which contrasts with the upregulation of this protein in TSWV-infected thrips. The upregulated spot 137 matched a mitochondrial ATP synthase alpha subunit that is a membrane-bound enzyme involved in ATP synthesis and/or hydrolysis and transport of protons across membranes (20). A mitochondrial ATP synthase alpha subunit of the brown planthopper, Laodelphax striatellus, was found to be upregulated at the transcript level during infection with Rice stripe virus (RSV) (Tenuivirus) (86). Furthermore, an enolase, which is a metalloenzyme, was identified in spot 161 as being upregulated by TSWV infection. Consistent with our findings, a putative enolase transcript was also identified as upregulated in RSV-infected L. striatellus compared to results for the naive counterparts (86). Matrix metalloproteases have recently been documented to play a role in baculovirus escape from gut tissue through activation of effector caspases (45). The resulting remodeling of the basal lamina lining of tracheal cells associated with the intestine allows baculoviruses to escape from the midgut epithelial cells and establish systemic infections in their lepidopteran hosts (45). Larval acquisition of TSWV is required for transmission of virus; however, when adult thrips feed on virions, they enter the midgut and replicate but do not escape the gut tissues (76). Thus, a midgut escape barrier is hypothesized to govern vector specificity and competency. Remodeling of the midgut basal lamina in larval thrips during TSWV infection, similar to the described mechanism of escape for baculoviruses, could be one explanation for the differences in virus dissemination that have been observed in larval versus adult thrips.
Virus infection of insects by entomopathogenic and vector-borne viruses can cause both positive and negative effects on the development of the insect host (13, 58). We found vitellogenin and actin (both within spot 374) to be upregulated in the TSWV-exposed thrips. Vitellogenin is the precursor of the lipoproteins and phosphoproteins that make up most of the protein content of yolk. Interestingly, spot 378, which also contains actin, was downregulated during TSWV infection (in contrast with actin being upregulated in spot 374). These contradictory results reflect the complex response that F. occidentalis displays during TSWV infection. It is well known that members of the actin gene family are widely dispersed in the genome of metazoans and plants and that they can be differentially regulated under various conditions (26, 44). A 60S ribosomal protein, L9 (spot 72), which is involved in translation elongation and termination, was found to be upregulated in the TSWV-exposed cohorts. Overall, these findings suggest that TSWV might have a direct positive effect at least to some extent on the development and metabolism of its most efficient insect vector, F. occidentalis. For example, it was reported that larval thrips exposed to TSWV reached larger body sizes than their nonexposed counterparts, which ultimately resulted in a shorter period of vulnerability to two species of predatory mites (5). Our findings of five proteins involved in thrips development being upregulated during virus infection combined with results obtained by Belliure and colleagues (5) suggest that TSWV infection of its thrips vector is responsible at least in part for the positive effects observed in this virus-vector interaction, such as a faster larval growth and no detrimental effect during virus infection.
We identified nine proteins putatively involved in localization and membrane and protein transport as being differentially expressed. These proteins are potentially important for the viral infection cycle steps of entry and exit from insect vector cells. For example, vacuolar proton ATP synthase (v-ATPase) subunit E, identified within spot 134, was upregulated by virus infection. v-ATPases are membrane-bound enzymes that catalyze ATP hydrolysis to transport solutes across membranes and lower the pH in organelles (55). Upregulation of v-ATPase might have implications in TSWV entry to the midgut epithelial cells of its thrips vector, since this protein is involved in clathrin-coated vesicle trafficking (25), the known mechanism of entry for other bunyaviruses (38, 70). An apolipoprotein, a protein that binds to and transports lipids through the hemolymph of insects and the circulatory system of mammals, was identified in spot 481 as being downregulated by TSWV infection. A glyceraldehyde-3-phosphate dehydrogenase (G3PDH) was identified in spot 182 and within spot 196 as being downregulated by TSWV infection in both spots. In contrast, this protein was consistently found upregulated in three different vector-competent genotypes of the greenbug (16), and G3PDH has been found to be potentially involved in the transcytosis of beet western yellows virus in the green peach aphid (Myzus persicae) (68). G3PDH has been found to be important in the infection process of other plant- and animal-infecting viruses. For example, G3PDH negatively regulates the replication of bamboo mosaic virus and its associated satellite RNA in vitro (57), and G3PDH mutants directly inhibit the accumulation and plus-strand synthesis of tomato bushy stunt virus in yeast and plants (36). It is also known that G3PDH binds to membranes and regulates endocytosis (71) and exocytosis (30). Additional research of TSWV entry and exit in thrips will aid in understanding the relevance of these proteins in the virus infection process.
Insects rely solely on the innate immune responses to defend themselves against invading microbes. We found 14 proteins that were ascribed a functional annotation associated with response to stimuli, which includes responses to biotic and abiotic stresses. For example, spot 88 was identified as a UV excision repair protein, designated Rad23, which is involved in a multiprotein and proteosome complex that targets nucleotide excision repair on the genome and ubiquitinated proteins for degradation, respectively, during abiotic stresses (7, 8, 88). Among the 14 proteins within the category of response to stimuli, nine are potentially involved in the defense response of F. occidentalis to TSWV infection. Interestingly, 5 of these 9 proteins were downregulated in the TSWV-exposed cohorts. A thioredoxin-dependent peroxidase (also peroxiredoxin) that among several functions acts as an inhibitor of apoptosis (87) was identified in spot 319, which was downregulated by TSWV infection. A protein showing similarity to a stress-induced phosphoprotein 1, which plays a role in signaling in the Jak-STAT pathway (reviewed in reference 33), was identified in spot 340. The Jak-STAT pathway is a major component of the innate immune response of mammals and insects against viruses (21, 23, 72). Lysozyme C, which was identified in spot 794 as being downregulated by TSWV infection, has been well documented to play a role in killing bacteria (24). Moreover, the downregulated spot 803 was identified as heat shock cognate 71 protein, which has been shown to be modulated during influenza virus infection (48). In contrast, spots 836 and 137 were both upregulated during virus infection. Spot 836 was identified as 40S ribosomal protein S3, which plays a role in apoptosis, and it is involved in the negative regulation of NF-κB transcription factor activity, which includes the well-studied Toll and Imd pathways. These two signaling pathways have been shown to play an important antiviral role in insects (reviewed in reference 62). The virus-upregulated spot 137 was identified as a mitochondrial ATP synthase alpha subunit, which GO term suggests a possible role in engulfment during phagocytosis to clear pathogen infections. Characterizing the components of the innate immune system that recognize and respond to TSWV may be important for understanding vector competency and designing novel control strategies.
We found that 5 of the 37 identified proteins did not have a Blast2GO annotation, but their identity (based on protein sequence similarities found within the NCBI nr protein database) suggests possible roles for these proteins. Spot 196, which was downregulated in the virus-exposed group, was identified as cyclophilin. Cyclophilins have been shown to play a role in virus infection and have been implicated as a determinant of insect vector competency. For example, incorporation of cyclophilin A into the newly synthesized human immunodeficiency virus 1 (HIV-1) particles is required for virion attachment to host cells (64), while its recruitment by the GAG polyprotein during HIV-1 infection (11) promotes HIV-1 replication (12). Cyclophilins were identified in greenbugs (Schizaphis graminum) that are vectors of Cereal yellow dwarf virus (family Luteoviridae) but not in nonvectors of the same aphid species (85). Two different proteins coresolved within spot 283, the protein yellow-like and 26S protease regulatory subunit 7, which have been associated with melanization and behavior of D. melanogaster (22, 83) and the ATP-dependent degradation of ubiquitinated proteins (14), respectively. The identification of several proteins with roles in ubiquitin-mediated protein degradation suggests that this pathway may be differentially activated in F. occidentalis during TSWV infection.
Despite detection of TSWV in 90% of the thrips subsampled from 4 biological replicates, we did not detect viral proteins in our analysis of differentially expressed proteins between TSWV-exposed and nonexposed larval thrips. A possible explanation for the inability to identify virus proteins in our current analysis may be the early time point used to collect larval thrips, the detection limit of a 2-D gel stained with Coomassie brilliant blue G-250, or the comigration of viral proteins with other proteins. However, we detected TSWV N in Western blots of 2-D gels from virus-exposed larvae (600 individuals with a 90% infection rate) and purified virus (positive control) but not in the nonexposed cohort (see Fig. S1 in the supplemental material). This indicates that viral proteins are expressed in our virus-exposed cohorts and that changes in their protein profile are indeed attributed to TSWV infection. Moreover, we did not identify most of the immune-related proteins that were identified by Medeiros et al. (46) during thrips infection by TSWV, with the exception of lectin C. In this previous study, the authors used a virus isolate and thrips population different from those used in this study. Furthermore, they conducted a macroarray analysis of subtractive cDNA libraries of TSWV-infected second-instar larvae (96 h post-AAP), which may have been the cause of the different findings obtained in the current study using proteomic tools to identify not only immune-related proteins but all proteins responsive to viral infection.
Our proteomic analysis of the response of F. occidentalis to TSWV infection has identified a suite of candidate proteins that respond to virus infection. Further analysis of the transcriptome responses will enable a meta-analysis and comparison of changes at the mRNA and protein levels that occur in virus-infected thrips. At this time, functional genomic assays using RNA interference (RNAi) have not been developed for thrips. The development of RNAi technology for thrips will enable studies to describe the functional roles of the differentially expressed proteins in TSWV-F. occidentalis interactions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michelle Cilia for insightful discussions about proteomic techniques.
This work was supported by the National Research Initiative of the USDA National Institute of Food and Agriculture, grant 2007-35319-18326. I.E.B.-V. was partially supported by an Ecological Genomics Graduate Fellowship. We also acknowledge support provided by the K-State Targeted Excellence Functional Genomics Consortium and Arthropod Genomics Center.
This is contribution no. 12-467-J from the Kansas Agricultural Experiment Station.
Footnotes
Published ahead of print 13 June 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Agaisse H, Petersen U, Boutros M, Mathey-Prevot B, Perrimon N. 2003. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell 5:441–450 [DOI] [PubMed] [Google Scholar]
- 2. Balakirev M, Tcherniuk S, Jaquinod M, Chroboczek J. 2003. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 4:517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bautista RC, Mau RFL, Cho JJ, Custer DM. 1995. Potential of Tomato spotted wilt tospovirus plant nests in Hawaii as virus reservoirs for transmission by Frankliniella occidentalis (Thysanoptera, Thripidae). Phytopathology 85:953–958 [Google Scholar]
- 4. Belli G, Molina M, Garcia-Martinez J, Perez-Ortin J, Herrero E. 2004. Saccharomyces cerevisiae glutaredoxin 5-deficient cells subjected to continuous oxidizing conditions are affected in the expression of specific sets of genes. J. Biol. Chem. 279:12386–12395 [DOI] [PubMed] [Google Scholar]
- 5. Belliure B, Janssen A, Sabelis MW. 2008. Herbivore benefits from vectoring plant virus through reduction of period of vulnerability to predation. Oecologia 156:797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berry DL, Baehrecke EH. 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131:1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bertolaet B, et al. 2001. UBA domains mediate protein-protein interactions between two DNA damage-inducible proteins. J. Mol. Biol. 313:955–963 [DOI] [PubMed] [Google Scholar]
- 8. Bertolaet B, et al. 2001. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 8:417–422 [DOI] [PubMed] [Google Scholar]
- 9. Bielza P. 2008. Insecticide resistance management strategies against the western flower thrips, Frankliniella occidentalis. Pest Manag. Sci. 64:1131–1138 [DOI] [PubMed] [Google Scholar]
- 10. Boonham N, et al. 2002. Detection of Tomato spotted wilt virus (TSWV) in individual thrips using real time fluorescent RT-PCR (Taqman). J. Virol. Methods 101:37–48 [DOI] [PubMed] [Google Scholar]
- 11. Braaten D, Ansari H, Luban J. 1997. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 71:2107–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Braaten D, Luban J. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burand JP, Park EJ. 1992. Effect of nuclear polyhedrosis virus infection on the development and pupation of gypsy moth larvae. J. Invertebr. Pathol. 60:171–175 [Google Scholar]
- 14. Chen Y, Sharp ZD, Lee W. 1997. HEC binds to the seventh regulatory subunit of the 26 S proteasome and modulates the proteolysis of mitotic cyclins. J. Biol. Chem. 272:24081–24087 [DOI] [PubMed] [Google Scholar]
- 15. Cilia M, et al. 2009. A comparison of protein extraction methods suitable for gel-based proteomic studies of aphid proteins. J. Biomol. Tech. 20:201–215 [PMC free article] [PubMed] [Google Scholar]
- 16. Cilia M, et al. 2011. Genetics coupled to quantitative intact proteomics links heritable aphid and endosymbiont protein expression to circulative polerovirus transmission. J. Virol. 85:2148–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clem R, Fechheimer M, Miller L. 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254:1388–1390 [DOI] [PubMed] [Google Scholar]
- 18. Crespi B. 1992. Eusociality in Australian gall thrips. Nature 359:724–726 [Google Scholar]
- 19. De Maio A. 1999. Heat shock proteins: facts, thoughts, and dreams. Shock 11:1–12 [DOI] [PubMed] [Google Scholar]
- 20. Devenish R, Prescott M, Boyle G, Nagley P. 2000. The oligomycin axis of mitochondrial ATP synthase: OSCP and the proton channel. J. Bioenerg. Biomembr. 32:507–515 [DOI] [PubMed] [Google Scholar]
- 21. Dostert C, et al. 2005. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat. Immunol. 6:946–953 [DOI] [PubMed] [Google Scholar]
- 22. Drapeau M, Radovic A, Wittkopp P, Long A. 2003. A gene necessary for normal male courtship, yellow, acts downstream of fruitless in the Drosophila melanogaster larval brain. J. Neurobiol. 55:53–72 [DOI] [PubMed] [Google Scholar]
- 23. Dupuis S, et al. 2003. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33:388–391 [DOI] [PubMed] [Google Scholar]
- 24. Fleming A. 1922. On a remarkable bacteriolytic element found in tissues and secretions. Proc. R. Soc. Lond. B 93:306–317 [Google Scholar]
- 25. Forgac M. 1999. Structure and properties of the clathrin-coated vesicle and yeast vacuolar V-ATPases. J. Bioenerg. Biomembr. 31:57–65 [DOI] [PubMed] [Google Scholar]
- 26. Fyrberg E, Bond B, Hershey N, Mixter K, Davidson N. 1981. The actin genes of Drosophila—protein coding regions are highly conserved but intron positions are not. Cell 24:107–116 [DOI] [PubMed] [Google Scholar]
- 27. Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. 2006. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat. Immunol. 7:590–597 [DOI] [PubMed] [Google Scholar]
- 28. Gavrilovskaya IN, Brown EJ, Ginsberg MH, Mackow ER. 1999. Cellular entry of hantaviruses which cause hemorrhagic fever with renal syndrome is mediated by beta 3 integrins. J. Virol. 73:3951–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gavrilovskaya I, Shepley M, Shaw R, Ginsberg M, Mackow E. 1998. Beta(3) integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc. Natl. Acad. Sci. U. S. A. 95:7074–7079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Glaser P, Gross R. 1995. Rapid plasmenylethanolamine-selective fusion of membrane bilayers catalyzed by an isoform of glyceraldehyde-3-phosphate dehydrogenase—discrimination between glycolytic and fusogenic roles of individual isoforms. Biochemistry 34:12193–12203 [DOI] [PubMed] [Google Scholar]
- 31. Gotthardt D, et al. 2006. Proteomics fingerprinting of phagosome maturation and evidence for the role of a G alpha during uptake. Mol. Cell. Proteomics 5:2228–2243 [DOI] [PubMed] [Google Scholar]
- 32. Ha E, et al. 2009. Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in Drosophila gut. Nat. Immunol. 10:949–957 [DOI] [PubMed] [Google Scholar]
- 33. Haura E, Turkson J, Jove R. 2005. Mechanisms of disease: insights into the emerging role of signal transducers and activators of transcription in cancer. Nat. Clin. Pract. Oncol. 2:315–324 [DOI] [PubMed] [Google Scholar]
- 34. Heming BS. 1973. Metamorphosis of pretarsus in Frankliniella fusca (Hinds) (Thripidae) and Haplothrips verbasci (Osborn) (Phlaeothripidae) (Thysanoptera). Can. J. Zool. 51:1211–1234 [Google Scholar]
- 35. Heming BS. 1980. Development of the mouthparts in embryos of Haplothrips verbasci (Osborn) (Insecta, Thysanoptera, Phlaeothripidae). J. Morphol. 164:235–263 [DOI] [PubMed] [Google Scholar]
- 36. Huang T, Nagy PD. 2011. Direct inhibition of tombusvirus plus-strand RNA synthesis by a dominant negative mutant of a host metabolic enzyme, glyceraldehyde-3-phosphate dehydrogenase, in yeast and plants. J. Virol. 85:9090–9102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hynes R. 1992. Integrins—versatility, modulation, and signaling in cell-adhesion. Cell 69:11–25 [DOI] [PubMed] [Google Scholar]
- 38. Jin M, et al. 2002. Hantaan virus enters cells by clathrin-dependent receptor-mediated endocytosis. Virology 294:60–69 [DOI] [PubMed] [Google Scholar]
- 39. Jung J, Yoon T, Choi E, Lee K. 2002. Interaction of cofilin with triose-phosphate isomerase contributes glycolytic fuel for Na, K-ATPase via Rho-mediated signaling pathway. J. Biol. Chem. 277:48931–48937 [DOI] [PubMed] [Google Scholar]
- 40. Kocks C, et al. 2005. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 123:335–346 [DOI] [PubMed] [Google Scholar]
- 41. Lim J, et al. 2007. An essential role for talin during alpha(M)beta(2)-mediated phagocytosis. Mol. Biol. Cell 18:976–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin Y, Tsai W, Liu H, Liang P. 2009. Intracellular beta-tubulin/chaperonin containing TCP1-beta complex serves as a novel chemotherapeutic target against drug-resistant tumors. Cancer Res. 69:6879–6888 [DOI] [PubMed] [Google Scholar]
- 43. López-Soler N, Cervera A, Moores GD, Martínez-Pardo R, GarcerÁ MD. 2008. Esterase isoenzymes and insecticide resistance in Frankliniella occidentalis populations from the south-east region of Spain. Pest Manag. Sci. 64:1258–1266 [DOI] [PubMed] [Google Scholar]
- 44. Meagher R. 1991. Divergence and differential expression of actin gene families in higher plants. Int. Rev. Cytol. 125:139–163 [DOI] [PubMed] [Google Scholar]
- 45. Means JC, Passarelli AL. 2010. Viral fibroblast growth factor, matrix metalloproteases, and caspases are associated with enhancing systemic infection by baculoviruses. Proc. Natl. Acad. Sci. U. S. A. 107:9825–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Medeiros RB, Resende RDO, de Avila AC. 2004. The plant virus Tomato spotted wilt tospovirus activates the immune system of its main insect vector, Frankliniella occidentalis. J. Virol. 78:4976–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meister M, Lemaitre B, Hoffmann J. 1997. Antimicrobial peptide defense in Drosophila. Bioessays 19:1019–1026 [DOI] [PubMed] [Google Scholar]
- 48. Melville M, et al. 1999. The cellular inhibitor of the PKR protein kinase, P58(IPK), is an influenza virus-activated co-chaperone that modulates heat shock protein 70 activity. J. Biol. Chem. 274:3797–3803 [DOI] [PubMed] [Google Scholar]
- 49. Moritz G, Morris DCJ, Mound LA. 2001. ThripsID: pest thrips of the world. An interactive identification and information system. ACIAR and CSIRO Publishing, Melbourne, Australia [Google Scholar]
- 50. Moritz G, Kumm S, Mound L. 2004. Tospovirus transmission depends on thrips ontogeny. Virus Res. 100:143–149 [DOI] [PubMed] [Google Scholar]
- 51. Morse JG, Hoddle MS. 2006. Invasion biology of thrips. Annu. Rev. Entomol. 51:67–89 [DOI] [PubMed] [Google Scholar]
- 52. Mound L. 1996. The thysanopteran vector species of tospoviruses, p 298–309 In Kuo G. (ed), Tospoviruses and thrips of floral and vegetable crops: an international symposium, Taichung, Taiwan International Society for Horticultural Science, Asian Vegetable Research and Development Center, Shanhua, Taiwan [Google Scholar]
- 53. Mound LA. 2002. So many thrips—so few tospoviruses? p 15–18 In Marullo R, Mound LA. (ed), Thrips tospovirus: Proceedings of the 7th International Symposium on Thysanoptera Australian National Insect Collection, Canberra, Australia [Google Scholar]
- 54. Myles KM, Wiley MR, Morazzani EM, Adelman ZN. 2008. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc. Natl. Acad. Sci. U. S. A. 105:19938–19943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nelson N, et al. 2000. The cellular biology of proton-motive force generation by V-ATPases. J. Exp. Biol. 203:89–95 [DOI] [PubMed] [Google Scholar]
- 56. Pfaffl M. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 doi:10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Prasanth KR, et al. 2011. Glyceraldehyde 3-phosphate dehydrogenase negatively regulates the replication of Bamboo mosaic virus and its associated satellite RNA. J. Virol. 85:8829–8840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Reese SM, Beaty MK, Gabitzsch ES, Blair CD, Beaty BJ. 2009. Aedes triseriatus females transovarially infected with La Crosse virus mate more efficiently than uninfected mosquitoes. J. Med. Entomol. 46:1152–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Robb KL, Newmann J, Virzi JK, Parrella MP. 1995. Insecticide resistance in western flower thrips, p 341–346 In Parker BL, Skinner M, Parrella MP. (ed), Thrips biology and management, vol 276 NATO ASI series, series A. Life sciences Plenum Press, New York, NY [Google Scholar]
- 60. Rotenberg D, et al. 2009. Variation in Tomato spotted wilt virus titer in Frankliniella occidentalis and its association with frequency of transmission. Phytopathology 99:404–410 [DOI] [PubMed] [Google Scholar]
- 61. Rotenberg D, Whitfield AE. 2010. Analysis of expressed sequence tags from Frankliniella occidentalis, the western flower thrips. Insect Mol. Biol. 19:537–551 [DOI] [PubMed] [Google Scholar]
- 62. Sabin LR, Hanna SL, Cherry S. 2010. Innate antiviral immunity in Drosophila. Curr. Opin. Immunol. 22:4–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sanchez-Vargas I, et al. 2009. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito's RNA interference pathway. PLoS Pathog. 5:e1000299 doi:10.1371/journal.ppat.1000299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saphire A, Bobardt M, Gallay P. 1999. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 18:6771–6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Scherfer C, et al. 2006. The Toll immune-regulated Drosophila protein Fondue is involved in hemolymph clotting and puparium formation. Dev. Biol. 295:156–163 [DOI] [PubMed] [Google Scholar]
- 66. Schneider A. 2000. Triosephosphate isomerase deficiency: historical perspectives and molecular aspects. Best Pract. Res. Clin. Haematol. 13:119–140 [DOI] [PubMed] [Google Scholar]
- 67. Seay M, Dinesh-Kumar S, Levine B. 2005. Digesting oneself and digesting microbes: autophagy as a host response to viral infection, p 245–279 In Palese P. (ed), Modulation of host gene expression and innate immunity by viruses. Springer, New York, NY [Google Scholar]
- 68. Seddas P, et al. 2004. Rack-1, GAPDH3, and actin: proteins of Myzus persicae potentially involved in the transcytosis of Beet western yellows virus particles in the aphid. Virology 325:399–412 [DOI] [PubMed] [Google Scholar]
- 69. Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. 2009. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 30:588–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Simon M, Johansson C, Mirazimi A. 2009. Crimean-Congo hemorrhagic fever virus entry and replication is clathrin-, pH- and cholesterol-dependent. J. Gen. Virol. 90:210–215 [DOI] [PubMed] [Google Scholar]
- 71. Sirover M. 1997. Role of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in normal cell function and in cell pathology. J. Cell. Biochem. 66:133–140 [PubMed] [Google Scholar]
- 72. Souza-Neto J, Sim S, Dimopoulos G. 2009. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. U. S. A. 106:17841–17846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sun L, Chen Z. 2004. The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 16:119–126 [DOI] [PubMed] [Google Scholar]
- 74. Tsuda S, Fujisawa I, Ohnishi J, Hosokawa D, Tomaru K. 1996. Localization of Tomato spotted wilt tospovirus in larvae and pupae of the insect vector Thrips setosus. Phytopathology 86:1199–1203 [Google Scholar]
- 75. Tzou P, et al. 2000. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13:737–748 [DOI] [PubMed] [Google Scholar]
- 76. Ullman DE, Cho JJ, Mau RFL, Wescot DM, Custer DM. 1992. A midgut barrier to Tomato spotted wilt virus acquisition by adult western flower thrips. Phytopathology 82:1333–1342 [Google Scholar]
- 77. Ullman DE, German TL, Sherwood JL, Wescot DM, Cantone FA. 1993. Tospovirus replication in insect vector cells: immunocytochemical evidence that the nonstructural protein encoded by the S RNA of Tomato spotted wilt tospovirus is present in thrips vector cells. Phytopathology 83:456–463 [Google Scholar]
- 78. Wang XH, et al. 2006. RNA interference directs innate immunity against viruses in adult Drosophila. Science 312:452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Washburn J, Kirkpatrick B, Volkman L. 1996. Insect protection against viruses. Nature 383:767 [Google Scholar]
- 80. Whitfield AE, Ullman DE, German TL. 2004. Expression and characterization of a soluble form of Tomato spotted wilt virus glycoprotein GN. J. Virol. 78:13197–13206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wijkamp I, Goldbach R, Dand Peters 1996. Propagation of Tomato spotted wilt virus in Frankliniella occidentalis does neither result in pathological effects nor in transovarial passage of the virus. Entomol. Exp. Appl. 81:285–292 [Google Scholar]
- 82. Wijkamp I, van Lent J, Kormelink R, Goldbach R, Peters D. 1993. Multiplication of Tomato spotted wilt virus in its insect vector, Frankliniella occidentalis. J. Gen. Virol. 74:341–349 [DOI] [PubMed] [Google Scholar]
- 83. Wittkopp P, True J, Carroll S. 2002. Reciprocal functions of the Drosophila Yellow and Ebony proteins in the development and evolution of pigment patterns. Development 129:1849–1858 [DOI] [PubMed] [Google Scholar]
- 84. Wolschin F, Amdam GV. 2007. Plasticity and robustness of protein patterns during reversible development in the honey bee (Apis mellifera). Anal. Bioanal. Chem. 389:1095–1100 [DOI] [PubMed] [Google Scholar]
- 85. Yang X, et al. 2008. Coupling genetics and proteomics to identify aphid proteins associated with vector-specific transmission of polerovirus (Luteoviridae). J. Virol. 82:291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhang F, et al. 2010. Massively parallel pyrosequencing-based transcriptome analyses of small brown planthopper (Laodelphax striatellus), a vector insect transmitting Rice stripe virus (RSV). BMC Genomics 11:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang P, et al. 1997. Thioredoxin peroxidase is a novel inhibitor of apoptosis with a mechanism distinct from that of Bcl-2. J. Biol. Chem. 272:30615–30618 [DOI] [PubMed] [Google Scholar]
- 88. Zhao G, et al. 2006. Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways. J. Biol. Chem. 281:13751–13761 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.