Abstract
Despite a central role in immunity, antibody neutralization of virus infection is poorly understood. Here we show how the neutralization and persistence of adenovirus type 5, a prevalent nonenveloped human virus, are dependent upon the intracellular antibody receptor TRIM21. Cells with insufficient amounts of TRIM21 are readily infected, even at saturating concentrations of neutralizing antibody. Conversely, high TRIM21 expression levels decrease the persistent fraction of the infecting virus and allows neutralization by as few as 1.6 antibody molecules per virus. The direct interaction between TRIM21 and neutralizing antibody is essential, as single-point mutations within the TRIM21-binding site in the Fc region of a potently neutralizing antibody impair neutralization. However, infection at high multiplicity can saturate TRIM21 and overcome neutralization. These results provide insight into the mechanism and importance of a newly discovered, effector-driven process of antibody neutralization of nonenveloped viruses.
INTRODUCTION
Antibody-mediated immunity forms a crucial part of the antiviral immune response, and its induction is a principal objective of vaccination. Reduced antibody (Ab) production, as occurs in X-linked agammaglobulinemia, hypogammaglobulinemia, and dysgammaglobulinemia, leads to persistent bacterial and viral infection (30, 31). In vitro, the binding of Abs to virus causes a reduction in infectious titer, termed neutralization, which is independent of effector mechanisms such as complement fixation or Fc-mediated phagocytosis (5). Neutralizing antibodies (NAbs) are thought to play an important role in antiviral immunity, since the passive transfer of strongly neutralizing Abs is associated with both antiviral protection (10, 12) and the abrogation of disease (7, 34). However, modeling and prediction of neutralization are not straightforward (29). For instance, it is unclear how the binding of one or a few Ab molecules per virus is sufficient for neutralization (4). An average of 1.4 NAb molecules is capable of neutralizing human adenovirus (AdV) type 2 (39), an apparently paradoxical finding given that IgG molecules are considerably smaller than adenovirus particles and occupy only a fraction of the viral surface when bound. The binding of a single NAb was also reported to neutralize poliovirus (13, 38). A second neutralization phenomenon that is poorly understood is the persistent fraction (PF), i.e., the level of infection that remains at high NAb concentrations. The cause of the PF was previously attributed to aggregated virus, low-affinity Abs, viral heterogeneity, and polyclonal interference (2, 19).
Recently, we showed that Abs can mediate neutralization intracellularly by recruiting the cytosolic Ig receptor TRIM21 (23). The engagement of NAb-virus complexes by TRIM21 promotes the degradation of both Ab and virus by the proteasome, a process termed antibody-dependent intracellular neutralization (ADIN) (25). In this study, we describe the mechanistic requirements for ADIN. We quantitatively examine both NAb-virus stoichiometry and TRIM21 levels in order to probe their influence on the efficiency of neutralization and the PF of viral infection. We find that in cells depleted of TRIM21, neutralization is inefficient, requiring a higher NAb-virus stoichiometry, and is accompanied by a higher level of remaining infectivity. Conversely, interferon (IFN)-stimulated cells have robust TRIM21 expression, efficient neutralization, and low levels of persistent infection. The importance of ADIN in virus neutralization is confirmed by the genetic knockout (KO) of TRIM21 in mouse cells and antibody site-directed mutagenesis to ablate TRIM21 binding, either of which is sufficient to substantially diminish the potency of a strongly neutralizing monoclonal Ab. However, while ADIN provides efficient protection at low levels of viral infection, coinfection experiments reveal that it can be overcome at high levels of virus exposure.
MATERIALS AND METHODS
Cell lines and viruses.
Mouse embryonic fibroblast (MEF) cells were obtained from the ATCC (ATCC SCRC 1008) or prepared from wild-type (WT) C57BL/6 embryos or embryos with TRIM21 replaced by an enhanced green fluorescent protein (GFP) construct (42). The level of expression of GFP in cells from knockout mice was sufficiently low to permit the detection of adenovirus-delivered GFP. MEF and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a 5% CO2 atmosphere. For stable TRIM21 knockdown, HeLa cells were transduced with retroviral particles encoding small hairpin RNA (shRNA) directed to the human TRIM21 sequence GCAGCACGCTTGACAATGA, and transduced cells were selected with puromycin at 2 μg/ml as previously described (23). For transient knockdown by small interfering RNA (siRNA), cells were transfected with equal amounts of siRNA oligonucleotides T21siRNA1 (UCAUUGUCAAGCGUGCUGC; Dharmacon) and T21siRNA2 (UGGCAUGGAGGCACCUGAAGGUGG; Invitrogen) by using Oligofectamine (Invitrogen) as directed by manufacturer and as previously described (23). Replication-deficient, E1-deleted GFP- and red fluorescent protein (RFP)-expressing human adenovirus 5 stocks (3) were prepared by two rounds of CsCl centrifugation as previously described (23).
Quantification of adenovirus.
In order to determine virus stock concentrations, samples were diluted 1:20 in 0.1% SDS and heated to 98°C for 2 min, and the absorbance at 260 and 280 nm was measured. The number of particles per ml was calculated by multiplying the absorbance at 260 nm by the dilution factor and dividing by the extinction coefficient for AdV (9.09 × 10−13 ml cm virion−1) (22, 27). The ratio of the absorbance at 260 nm to the absorbance at 280 nm was 1.3, as expected for pure, intact AdV preparations. In order to calculate the concentration of hexon trimers, the virus concentration was multiplied by 240. To determine virus titers by quantitative PCR (qPCR), DNA was extracted by using a DNeasy blood and tissue kit (Qiagen) and assayed by using primers and a probe directed to the GFP transgene of AdV-GFP (see “Quantitative PCR”). A dilution series of virus stocks of known concentrations was used as a calibration curve in order to calculate the virus concentration.
Antibodies.
Anti-AdV hexon antibody 9C12 (32, 37) was purified from the hybridoma supernatant on a protein G column (GE Healthcare). 9C12 was labeled with Alexa Fluor 488 (9C12-A488) by using a protein-labeling kit as directed by the manufacturer (Invitrogen). Absorbance measurements were taken at 495 and 280 nm, permitting the calculation of the concentration in mg/ml by using the formula [A280 − (0.11 × A495)]/1.4. For the cloning of mouse monoclonal IgG 9C12, cDNA was prepared from 9C12 hybridoma cells (Developmental Studies Hybridoma Bank) and was subjected to PCR amplification by using a strategy based on that described previously by Li et al. (21). Mixtures of 5′ degenerate primers that target the signal peptide-coding regions were used with reverse primers that target the 3′ end of the constant domains. The light-chain amplicon was digested with SalI and XbaI for cloning into the cytomegalovirus (CMV) promoter of pBUD CE4.1 (Invitrogen), while the heavy chain was digested with NotI and KpnI for cloning into the EF1α promoter of the same vector. For all primers, R is A or G; Y is C or T; M is A or C; K is G or T; S is C or G; W is A or T; V is A, C, or G; and N is A, C, G, or T. For the light chain, the 5′ primers used were VL1S (acgtGTCGACCCACCATGGAGACAGACACACTCCTGCTAT), VL2S (acgtGTCGACCCACCATGGATTTTCAAGTGCAGATTTTCAG), VL3S (acgtGTCGACCCACCATGGAGWCACAKWCTCAGGTCTTTRTA), VL4S (acgtGTCGACCCACCATGKCCCCWRCTCAGYTYCTKGT), and VL5S (acgtGTCGACCCACCATGAAGTTGCCTGTTAGGCTGTTG) (the SalI site is underlined, and the Kozak sequence is in boldface type). For the light chain, the 3′ primer used was CLX (catgtctagaCTAACACTCATTCCTGTTGAAGC) (the XbaI site is underlined, and the stop codon is in boldface type). For the heavy chain, the 5′ primers used were VH1N (aataGCGGCCGCCACCATGGRATGSAGCTGKGTMATSCTCTT), VH2N (aataGCGGCCGCCACCATGRACTTCGGGYTGAGCTKGGTTTT), VH3N (aataGCGGCCGCCACCATGGCTGTCTTGGGGCTGCTCTTCT), and VH4N (aataGCGGCCGCCACCATGATRGTGTTRAGTCTTYTGTRCCTG) (the NotI site is underlined, and the Kozak sequence is in boldface type). Lowercase type denotes stuffer sequences that are not complementary to the target DNA. The reverse primer used was CHKpn (catgGGTACCTCATTTACCAGGAGAGTGGGAG) (the KpnI site is underlined, and the stop codon is in boldface type). Point mutations were introduced by using QuikChange site-directed mutagenesis (Stratagene) according to the manufacturer's instructions. Protein was produced by the transfection of plasmids into 293F cells (Invitrogen), using FreeStyle Max transfection reagent. The supernatant was harvested after 6 days and purified on a protein G-Sepharose column (GE Healthcare). Purified goat anti-Ad5 hexon polyclonal IgG preparations were obtained from AbD Serotec (pAb1) and Millipore (pAb2). Immunoblotting for TRIM21 was performed, as previously described (23), with mouse monoclonal antibody D12 (Santa Cruz Biotechnology). The β-actin level was detected with rabbit polyclonal serum (Cell Signaling). Immunoblot bands were quantified by using ImageQuant TL software (GE Healthcare).
Quantitative PCR.
For quantitative real-time PCR (qPCR), cDNA was prepared from target cells concurrently with infection by using a TaqMan Gene Expression Cells-to-Ct kit (Applied Biosystems) according to the manufacturer's instructions, including a DNase treatment step to remove genomic DNA. For human cells, the absolute quantification of TRIM21 was performed by using a TRIM21 TaqMan gene expression assay (Hs00172616_m1; Applied Biosystems) with primers that span the boundary of exons 4 to 5 against a serial dilution of a plasmid encoding human TRIM21 cDNA. Mouse TRIM21 mRNA was detected with primers designed to span the boundary of exons 2 to 3, mT21F (TGGGTGTGTGCCCAGTCT) and mT21R (CATCGTGAGATCCATTTCCA), and probe mT21P (6-carboxyfluorescein [FAM]–ACCAGGAGAAGATCCACGTG–6-carboxytetramethylrhodamine [TAMRA]). The TRIM21 locus is disrupted by a GFP transgene in TRIM21 KO mice (42). mRNA from this transgene was detected by using primers directed to GFP, GFPF (CAACAGCCACAACGTCTATATCAT) and GFPR (ATGTTGTGGCGGATCTTGAAG) and probe GFPP (FAM-CCGACAAGCAGAAGAACGGCATCAA-TAMRA). The control β-actin gene was quantified by using an ACTB gene expression assay (catalog number 4387430; Applied Biosystems). Thermal cycling and fluorescent measurements were performed by using a 7900HT Realtime PCR system (Applied Biosystems), and cycling conditions were chosen according to gene expression assay instructions.
Virus neutralization assays.
Cells were seeded into 6-well plates at 1 × 105 cells/well and allowed to adhere overnight. Where appropriate, human or mouse IFN-α (Sigma-Aldrich) was added at the time of plating at 1,000 U/well unless otherwise indicated. Virus was diluted to 3 × 104 IU in 10 μl in phosphate-buffered saline (PBS), mixed with an equal volume of antibody at the stated concentration, and incubated for 1 h to allow binding to reach equilibrium. NAb-virus mixes were diluted 100-fold into cell culture medium and added to cells. Siliconized reaction tubes and pipette tips were used throughout to minimize nonspecific interactions of protein and plasticware. GFP-positive cells were analyzed by flow cytometry after 24 h of incubation at 37°C. Levels of infection in the absence of NAb were in the range of 30 to 50%. Infectivity was calculated by converting the percent GFP-positive cells to a multiplicity of infection (MOI) by using the equation MOI = −ln{[100 − (% GFP-positive cells)]/100}. The remaining infectivity (I/I0) was calculated by dividing the infectivity by that of a PBS-treated control infection. x-axis values are expressed as μg/ml of the input Ab (the concentration in the reaction mixture is half the stated concentration).
Quantitative electron microscopy (EM) and neutralization assays.
A total of 9 × 108 particles of AdV-GFP and antibody 9C12 at the appropriate concentration were mixed 1:1 in a 20-μl reaction mixture for 1 h. Ten microliters of the reaction mixture was removed for the infection of HeLa or MEF cells after the appropriate dilution in PBS. The remaining 10 μl of the virus-Ab reaction mixture was incubated with 1 μl undiluted 10-nm-gold-labeled anti-mouse secondary antibody (Sigma-Aldrich) for 1 h before the loading of 5 μl of ternary complexes onto non-glow-discharged carbon-coated grids. Samples were stained with 1% uranyl acetate and examined with a Philips model EM208S microscope. Seven images, with an average of 12.6 virions per image, were taken at each primary Ab concentration at a ×28,000 magnification. Virions were scored for the number of gold particles per virion. To calculate the number of primary Ab sites per virion, the background level of labeling in the absence of 9C12 was subtracted (0.503 gold particles per virus) and divided by the average number of gold particles per labeled site (1.403 gold particles per site). To calculate the number of NAb molecules required to neutralize a virion (λ), the number of NAb molecules at the lowest concentration of 9C12 (0.5 μg/ml) was chosen for downstream calculations, as any effects of the saturation of the secondary antibody are minimized. The number of NAb molecules per virion was assumed to increase linearly with the 9C12 concentration. This is an approximation, since each virus will eventually become saturated, and the curve is therefore not linear. However, within the range of 9C12 concentrations used, the linear approximation lies within 3% of those predicted by equilibrium dynamics. The number of NAb molecules per virus associated with 1 natural logarithm of neutralization on IFN-stimulated cells was then calculated by linear regression.
Quantitative fluorescent pelleting assay.
9C12-A488 was mixed 1:1 with 2 × 109 virus particles in a 200-μl reaction mixture and permitted to reach equilibrium by incubation at room temperature for 2 h. Extended incubations (overnight) were also performed but showed no greater binding or significant impact on the Kd (dissociation constant) (data not shown). As a control, 9C12-A488 was incubated with PBS only. Virus–9C12-A488 or control mixtures were carefully overlaid onto a 600-μl 30% sucrose cushion and centrifuged at 21,000 × g for 4 h. The uppermost 300 μl was removed, and the top of the sucrose cushion was washed twice with 300 μl PBS to reduce the amount of potentially contaminating unbound 9C12-A488. The remaining liquid was carefully removed, and the pellet was resuspended by periodic vortexing in 100 μl PBS over 2 h. The fluorescence at 520 nm was measured after excitation at 485 nm on a BMG Pherastar FS plate reader. The concentration of 9C12-A488 in the resuspended pellets was quantified by comparison to a calibration curve of 9C12-A488 at known concentrations. The amount of 9C12-A488 that was specifically pelleted by the interaction with AdV was calculated by subtracting the nonspecific 9C12-A488–PBS control values. The molar concentration of 9C12-A488 was calculated by using a molecular mass of IgG of 150,000 g mol−1, and x-axis values in Fig. 2F represent nanomolar concentrations of 9C12 in the NAb-virus-binding reaction. The concentration of virus in the pellet was calculated by qPCR for GFP, calibrated against a dilution series of virus at known titers in terms of particles per milliliter (see above). These data permitted the calculation of the stoichiometric ratio of 9C12-A488 and adenovirus.
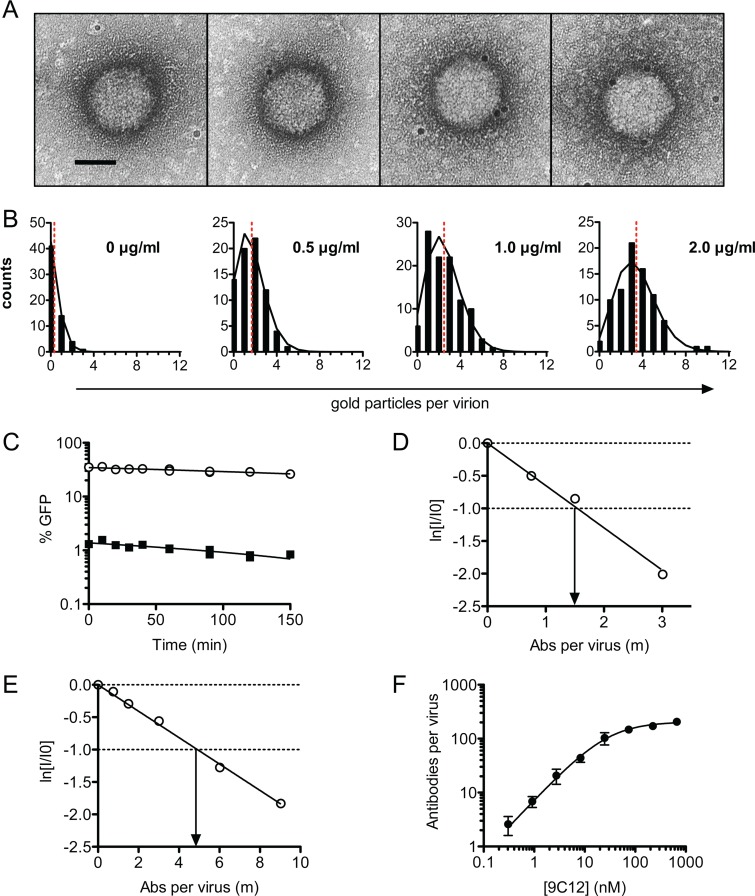
Fig 2.
ADIN mediates neutralization by few antibody molecules. (A) 9C12-adenovirus complexes were immunogold labeled and examined by electron microscopy. Gold particles are visible as black dots around the AdV capsid and show 0, 1, 2, and 3 regions labeled, respectively (from left to right). Scale bar, 50 nm. (B) Virions were scored for the number of gold particles per virus at increasing concentrations of 9C12. The distribution of gold particles per virus (black bars) was found to approximate a Poisson distribution (black lines), given the mean number of gold particles per virus (red dashed line). These data were used to calculate the average number of NAb molecules per virion. (C) PBS (circles) or 9C12 at 1.5 μg/ml (squares) was added to AdV for 1 h before 100-fold dilution into complete DMEM and incubation for various amounts of time before addition to cells. No increase in titer was observed for NAb-labeled virus over time, confirming that complexes do not dissociate upon dilution. (D and E) The infectivity of diluted NAb-virus complexes was assayed with MEF (D) and HeLa (E) cells treated with IFN-α, permitting a direct quantification of the average number of NAb molecules per virion at 1/e neutralization. Data represent the natural logarithm of remaining infectivity, ln(I/I0). (F) Stoichiometry of 9C12-A488–adenovirus after pelleting of complexes. Virus and antibody concentrations were quantified by qPCR and fluorescence spectroscopy, respectively. Data are fitted to a Michaelis-Menten curve (R2 = 0.94), giving a Kd of 28 nM and a maximum binding of 205 antibody molecules per virus (standard error of the mean, ±10). Data are represented as means ± standard errors of the means. A parallel infection assay with IFN-treated HeLa cells estimated the number of antibody molecules required for 1 natural logarithm of neutralization (λ) to be 5.2.
Prediction of NAb-virus stoichiometry by equilibrium dynamics.
We used the law of mass action to predict the binding of antibody to virus at equilibrium. The corresponding equations follow a previously reported methodology (18).
Viral antigen (V) and antibody (A) are proposed to bind, to give a complex (VA):
| (1) |
The rate of the forward reaction is governed by a constant, kon, while the rate of the reverse reaction is governed by koff. If the concentration of the viral antigen is [V] and the concentration of the NAb is [A], then the concentration of complex [VA] will be governed by the dissociation constant, Kd:
| (2) |
At equilibrium, the concentrations of viral antigen and antibody will depend on the initial concentrations, denoted [V0] and [A0], respectively. A conservation condition is applied, such that
| (3) |
and
| (4) |
Substituting the conservation equations into equation 1, we can state the rate of change of virus-NAb complex formation as
| (5) |
At equilibrium, the relationship between free and complexed virus and antibody takes the form of the quadratic equation
| (6) |
Solving the quadratic equation permits the calculation of the concentration of the antibody-virus complex at equilibrium:
| (7) |
The proportion of viral antigen bound (θ) is equal to
| (8) |
We can then calculate the average number of antibodies per virus (m) as
| (9) |
where S is the number of bound antigens per virus at maximum capacity. For these calculations, we let [V] equal the concentration of hexon trimers, as measured by the optical density. A value of 205 was used for S, and a value of 28 nM was used for Kd, parameters which were obtained from the quantitative fluorescence pelleting assay.
Saturation and postadsorption neutralization assays.
For saturation experiments, AdV-GFP at 1.6 × 109 IU/ml was incubated 1:1 with 9C12 at 1 μg/ml for 1 h before the addition of increasing volumes of Ab-virus complexes to HeLa cells in 6-well plates, as described above. The MOI for AdV-GFP is expressed as the equivalent of unneutralized infectious units per cell. Concurrently, AdV-RFP was incubated 1:1 with 9C12 at 1 μg/ml for 1 h and added to wells (MOI of ∼0.1) immediately after the addition of AdV-GFP. For postadsorption neutralization (PAN) assays, cells were cooled to 4°C before the addition of virus to the medium. Cells were incubated at 4°C for 1 h, before 3 cold-PBS washes. One milliliter of cold complete DMEM with 9C12 was then added to cells for a further 1 h, before washing as described above and the addition of warm complete DMEM. Cells were incubated at 37°C for a further 24 h before analysis of infection by flow cytometry.
All statistical analyses were performed with Prism 5 (GraphPad Software).
RESULTS
ADIN is an important component of antibody neutralization.
To investigate the importance of ADIN in virus neutralization, mouse embryonic fibroblast (MEF) cells were prepared from wild-type (WT) (TRIM21+/+) and TRIM21 knockout (K21) (TRIM21−/−) mice (42). Cells were infected with replication-deficient human adenovirus type 5-GFP that had been preincubated with a titration of a mouse monoclonal antihexon IgG1, 9C12 (32, 37). 9C12 neutralized the infection of WT cells but had only a limited effect on the infection of K21 cells (Fig. 1A). We have previously shown that interferon (IFN) upregulates TRIM21 expression, resulting in an enhanced ADIN phenotype (23). Repeating the neutralization in the presence of IFN resulted in a highly potent block to infection in WT cells, while K21 cells remained readily infected. The quantification of TRIM21 mRNA transcripts reveals a 30-fold increase in the transcript abundance after IFN stimulation of WT cells (Fig. 1B). We confirmed that K21 cells remained responsive to IFN by measuring the abundance of transcripts from the transgene that disrupts the TRIM21 locus (Fig. 1C). Thus, in the absence of TRIM21, cells are poorly protected from infection by 9C12 and, moreover, do not display an enhanced ADIN phenotype after IFN stimulation.
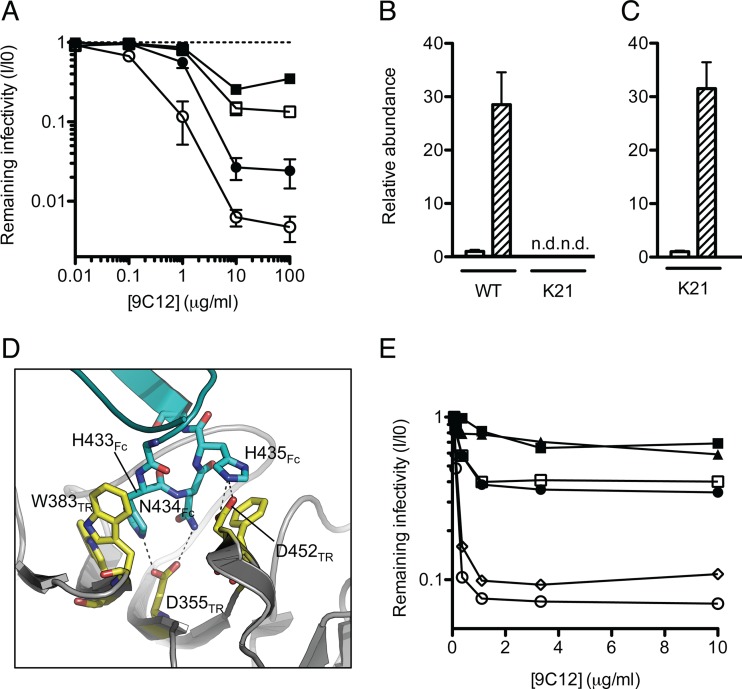
Fig 1.
ADIN is essential for efficient neutralization of adenovirus. (A) Antihexon monoclonal antibody 9C12 was titrated against adenovirus on clonal fibroblast (MEF) lines derived from wild-type (WT) (n = 8) (circles) or TRIM21 knockout (K21) (n = 8) (squares) mice. Cells were either untreated (closed symbols) or treated with IFN-α (open symbols). Data represent mean remaining infectivities (I/I0) from the addition of antibody versus a PBS-treated control infection ± standard errors of the means. (B) TRIM21 mRNA levels were quantified by qPCR in unstimulated (white bars) or IFN-α-stimulated (hatched bars) MEF cells. n.d., not detected. (C) Levels of the transgene that disrupts the TRIM21 locus in K21 cells were measured by qPCR. Shading is as described above for panel B. (D) The previously solved structure of the TRIM21-IgG Fc complex (14) showing interactions between TRIM21 (yellow) (residues labeled TR) and the IgG Fc HNH motif (blue) (residues labeled Fc). (E) Antihexon monoclonal antibody 9C12 was mutated at three positions or expressed as the wild type. Antibodies were titrated against adenovirus on HeLa cells. Open circles, WT 9C12; closed circles, H433A mutant; open squares, N434A mutant; closed squares, N434D mutant; triangles, H435A mutant; diamonds, K219R mutant.
To demonstrate that neutralization by 9C12 depends on a specific interaction between TRIM21 and NAb, we performed structure-guided mutagenesis of the binding interface. The previously solved crystal structures of human and mouse TRIM21-IgG complexes and the accompanying isothermal titration calorimetry data show that TRIM21 residues W381, W383, D355, and D452 bind to the “HNH” (positions 433 to 435) motif within the Fc (Fig. 1D), and the mutation of any one of these TRIM21 residues ablates binding (14). Here we made single-point mutants of the antibody HNH motif, replacing each residue with alanine as well as replacing N434 with aspartic acid. As can be seen in Fig. 1E, each single-point mutant reversed neutralization, whereas a mutation outside the TRIM21-binding domain (K219R) did not impair neutralization. Indeed, the mutation of H435 (a residue that makes hydrogen bond interactions with TRIM21) to alanine increased the remaining infectivity from 8% to 79% at 1 μg/ml 9C12. Meanwhile, the mutation of N434 to aspartic acid, which introduces a repulsive charge opposite TRIM21 residue D355, impaired neutralization beyond that observed for the mutation of the same residue to alanine. This finding confirms that a specific interaction between NAb and TRIM21 is required for neutralization and demonstrates that neutralization by this monoclonal NAb is severely diminished when its TRIM21-binding motif is mutated.
ADIN mediates neutralization by few antibody molecules per virus.
The existence of an effector mechanism for neutralization, such as ADIN, may explain why neutralization can sometimes be observed with very few antibody molecules. To determine how many NAb molecules are required for ADIN-mediated neutralization, we quantified NAb-virus stoichiometry by two independent quantitative techniques: an immunogold electron microscopy (EM) assay and a dye-conjugated NAb pelleting assay. The former technique provides information concerning the quantity and distribution of NAb molecules on the virus and can detect phenomena such as virus aggregation, while the latter technique provides a robust measure of NAb-virus stoichiometry over a wide range of NAb concentrations. With both approaches, parallel infection experiments permit the quantification of the number of NAb molecules (λ) required to neutralize a virus. We define λ as the number of NAb molecules bound at 1 natural logarithm of neutralization. Assuming a Poisson distribution of antibody binding, this means that the maximum potential neutralization is a λ value of 1, with the remaining infectivity equal to the fraction of unbound virus (19). We chose 9C12 as the neutralizing Ab, since it does not block entry (37) and therefore permits the specific study of ADIN-mediated neutralization.
For the EM approach, virus was incubated with 9C12, followed by incubation with an excess of gold-labeled secondary Ab, and the number of Ab molecules per virion was calculated (Fig. 2A). The binding of NAb to virus was found to closely approximate a Poisson distribution at all concentrations of NAb assayed (Fig. 2B). We confirmed that the dilution of preformed NAb-virus complexes does not result in the dissociation of complexes within the time frame of the experiment (Fig. 2C). Finally, and in agreement with previous findings (37), 9C12 was not found to aggregate virus, since virions remained largely monodispersed when 9C12 was present. Parallel infection experiments were performed on IFN-stimulated HeLa and wild-type MEF cells, allowing neutralization to be plotted as a function of the number of NAb molecules per virus. The data showed that in MEF and HeLa cells, 1.6 and 4.8 molecules of 9C12, respectively, are sufficient to neutralize adenoviral infection by 1 natural logarithm (Fig. 2D and E). Thus, when using a mouse Ab on mouse cells and in the presence of abundant TRIM21, neutralization approaches its theoretical maximum.
To confirm that low Ab-virus stoichiometry is sufficient for ADIN, we also measured the number of Abs per virus by complexing virus with Alexa Fluor 488-labeled 9C12 (9C12-A488) at a range of concentrations and pelleting through a sucrose cushion. Pelleted complexes were resuspended, the amount of bound dye was quantified by measurements of the fluorescence intensity, and the number of viruses was measured by qPCR, permitting the calculation of the stoichiometric ratio of Ab to virus. The resulting data were fit to a Michaelis-Menten curve (Fig. 2F), to give a dissociation constant (Kd) for the 9C12 interaction with adenovirus of 28 nM. We also observed that a maximum of 205 NAb molecules (standard error, ±10) were able to bind per virion. Given that there are 240 hexon trimers per virion, this finding suggests that the majority of hexon trimers can be occupied by 9C12 at saturation. Parallel neutralization experiments on IFN-treated HeLa cells revealed that 5.2 NAb molecules per virion were sufficient for neutralization, comparable to the value of 4.8 NAb molecules per virion obtained by EM. Together, the data show that high TRIM21 levels resulting from IFN stimulation permit neutralization at a low Ab-virus stoichiometry.
TRIM21 levels influence neutralization efficiency and the level of the persistent fraction.
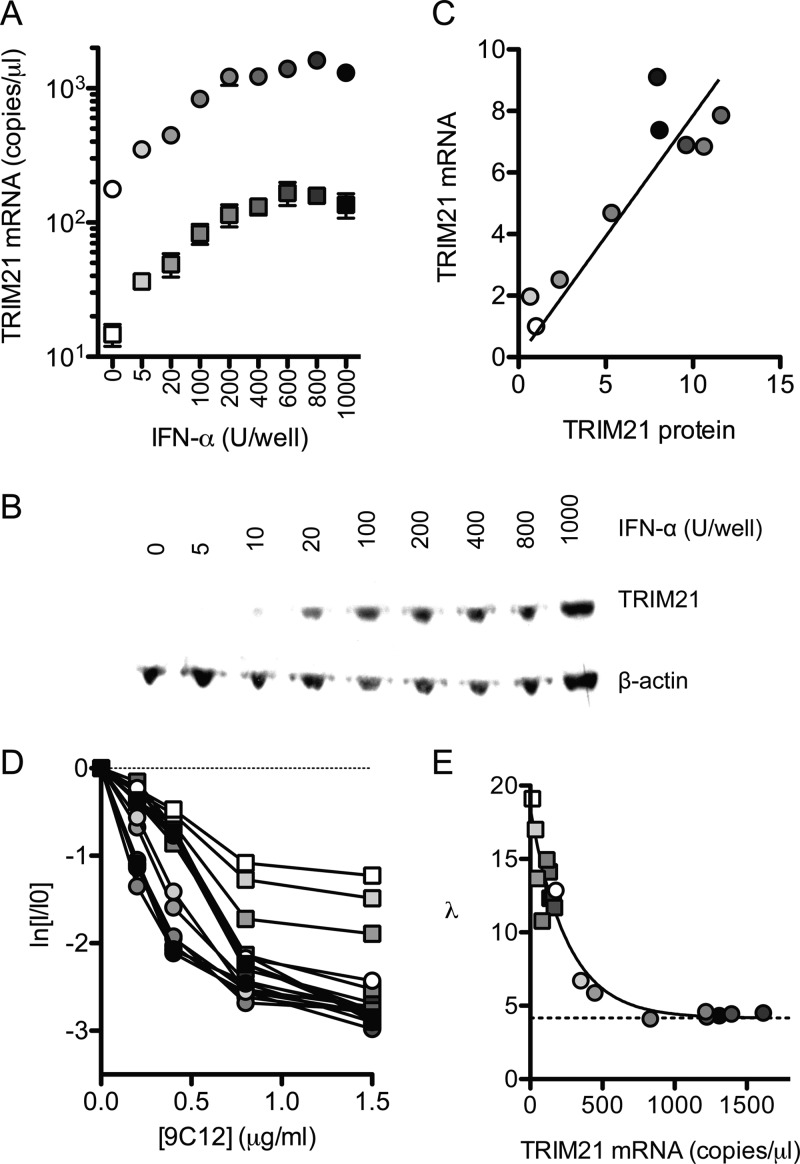
Next, we sought to analyze the impact of TRIM21 levels on the efficiency of ADIN. TRIM21 expression was manipulated by IFN-α titration and the retroviral transduction of HeLa cells with a TRIM21-directed shRNA. Using this approach, we were able to obtain cells with TRIM21 mRNA levels that varied over 2 orders of magnitude and with a corresponding distribution of protein levels (Fig. 3A to C). Neutralization experiments were carried out with these cells by using 9C12 at concentrations that yielded neutralization but not a saturation of binding (less than 20% maximum occupancy). Cells with the least amounts of TRIM21 gave a shallow gradient, reflecting a requirement for higher levels of NAb binding in order to elicit an equivalent level of neutralization (Fig. 3D). In contrast, high levels of TRIM21 permitted more efficient neutralization, reflecting a lower NAb-virus stoichiometry being sufficient for neutralization. We used equilibrium dynamics to calculate the number of antibodies per virus, using the parameters obtained from the fluorescence pelleting method, and plotted the number of Ab molecules required for 1 natural logarithm of neutralization against the TRIM21 transcript levels (Fig. 3E). At the lowest concentration of TRIM21, 18 Ab molecules were required for 1 natural logarithm of neutralization. As TRIM21 levels were increased, the number of Ab molecules required decreased rapidly until reaching a plateau corresponding to ∼4.2 NAb molecules per virus. Importantly, the data suggest that TRIM21 levels, rather than other effects of IFN, dictate the efficiency of neutralization, since IFN stimulation of TRIM21 knockdown cells restores the neutralization efficiency only to the level of unstimulated HeLa cells. Thus, where NAbs operate via ADIN, the number of Ab molecules required for neutralization can be described as a function of the TRIM21 concentration, λ(TR), which varies according to an exponential decay (Fig. 3E):
| (10) |
where TR is the TRIM21 concentration, B is the value of λ at the asymptote, A is the value of λ at a TR of 0 minus the asymptote value B, and c is the decay parameter. These findings imply that at low levels of cellular TRIM21, large gains in the efficiency of ADIN can be made by small increases in TRIM21 levels. However, as TRIM21 concentrations increase further, gains in neutralization efficiency will be reduced.
Fig 3.
TRIM21 levels determine neutralization efficiency. (A) IFN-α was added to HeLa cells (circles) or HeLa cells transduced with TRIM21 shRNA (squares), and TRIM21 mRNA levels were quantified by RT-qPCR. (B) TRIM21 protein levels were quantified by immunoblotting and normalized to β-actin levels. (C) Relative TRIM21 mRNA and protein levels are closely correlated (P < 0.001 by Pearson's product-moment correlation; R2 = 0.83). (D) Virus neutralization assays were performed on these cells, which display an initial gradient of neutralization that varies with TRIM21 levels. (E) The number of antibody molecules per virus at each concentration of 9C12 was calculated by using binding parameters obtained as described above (Fig. 2F). Linear regression was used in order to calculate the number of molecules of 9C12 bound at 1/e neutralization (λ). λ was found to decrease exponentially with the TRIM21 concentration, reaching a plateau at 4.2 antibody molecules per virus (dashed line). For panels D and E, circles indicate HeLa cells, and squares indicate HeLa cells expressing TRIM21-directed shRNA. Points for panels A and C to E are shaded white to black according to IFN levels.
The above-described data show that at a low NAb-virus stoichiometry, robust TRIM21 expression is essential for efficient neutralization. To determine the importance of TRIM21 at a high NAb-virus stoichiometry, we performed neutralization experiments over a wide range of 9C12 concentrations with cells with various TRIM21 expression levels. In HeLa cells, independent of TRIM21 levels, we observed that the addition of 9C12 beyond a concentration of 2 μg/ml (equivalent to 6.7 nM 9C12 in the binding reaction) yielded no further increase in neutralization (Fig. 4A). The PF remained constant even after a further 50-fold increase in the Ab concentration to 100 μg/ml (Fig. 4A). A similar phenomenon was observed for MEF cells, where the addition of 9C12 above 10 μg/ml gave little further neutralization (Fig. 1A). These extremes of 9C12 concentrations (100 μg/ml) are equivalent to a 12-fold excess over the Kd and are predicted by equilibrium dynamics to provide a 93% saturation of hexon binding sites. In both HeLa and MEF cells, the level of remaining infectivity at which the PF occurs is dependent on the cellular TRIM21 concentration. In HeLa cells depleted of TRIM21, the PF occurs when the remaining infectivity is 32%, and increasing the level of TRIM21 results in a proportional decrease in the PF (Fig. 4B). Interestingly, there appears to be a threshold after which increasing TRIM21 levels further fails to reduce the PF further. We hypothesize that this represents the point at which other ADIN components become limiting. The onset of the PF occurs before the saturation of virus with Ab (Fig. 4C), coinciding with fewer than 50 NAb molecules per virus, indicating that even though virus particles can accommodate more NAb molecules, an increasing NAb concentration does not result in higher levels of neutralization. Taken together, the data suggest that TRIM21 levels remain important at a high NAb-virus stoichiometry to lower the persistent fraction. Furthermore, increasing the Ab concentration further yields diminishing returns and cannot compensate for low levels of TRIM21.
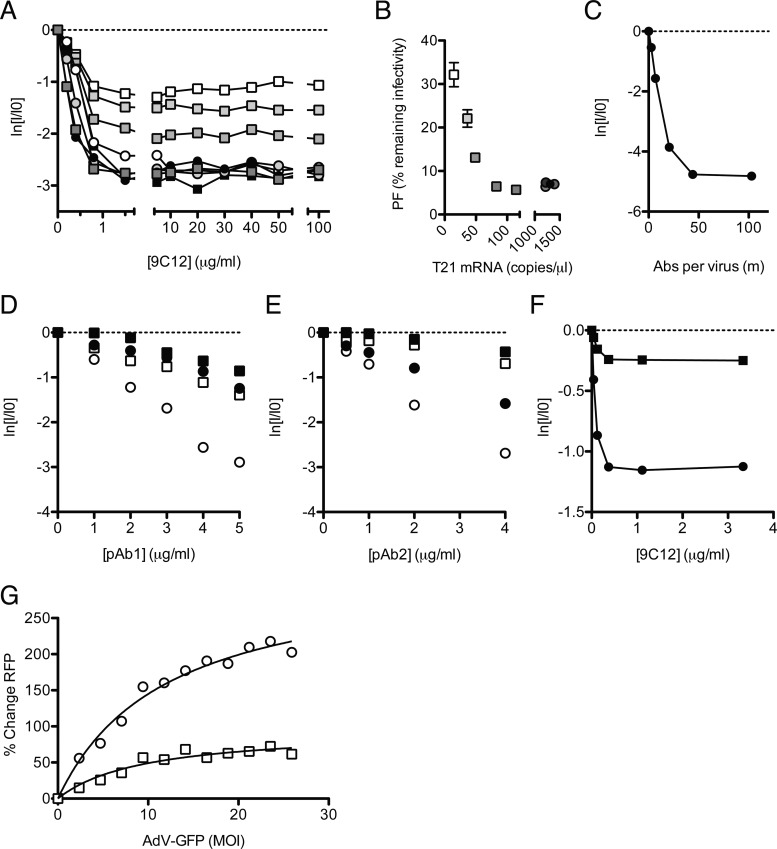
Fig 4.
Infection and persistence are influenced by the neutralization mechanism and viral MOI. (A) Neutralization data from Fig. 3D are shown with the x axis extended to show the persistent fraction range. The PF remains approximately constant within the range of 1.5 to 100 μg/ml 9C12, but its level is determined by the cellular TRIM21 concentration. (B) The average PF, calculated from all data at ≥5 μg/ml 9C12 (n = 7) (±standard errors of the means), decreases with TRIM21 levels before remaining constant at ∼6% remaining infectivity. Symbols and shading as for Fig. 3D (C) Remaining infectivity of adenovirus, assayed alongside a fluorescence pelleting assay, permitting quantification of the number of antibody molecules per virus in the persistent fraction. (D and E) Two polyclonal sera, pAb1 (D) and pAb2 (E), were titrated against AdV on HeLa cells (circles) or HeLa cells treated with siRNA directed toward TRIM21 (squares), which were either IFN stimulated (open symbols) or untreated (closed symbols). (F) Remaining infectivity after AdV was permitted to adsorb to the surface of HeLa cells before the addition of neutralizing monoclonal antibody 9C12. Symbols are as described above for panels D and E. (G) AdV-GFP, preincubated with 9C12, was added to HeLa (circles) or TRIM21 shRNA knockdown (squares) cells at a range of MOIs. A high MOI of AdV-GFP relieved the neutralization of 9C12-labeled AdV-RFP in HeLa cells but to a lesser extent in TRIM21 knockdown cells. Michaelis-Menten curve fitting (black lines) reveals that the maximal saturation values are significantly different between the two conditions (P = 0.0008 by F test).
A feature of 9C12 neutralization in the above-described experiments is that it is linear and commences without a discernible lag (Fig. 2D and E). However, in a previous experiment with polyclonal sera, we observed evidence of a lag when TRIM21 was depleted (23). We repeated these experiments using two different polyclonal sera against adenovirus and over an extended range of antibody concentrations. As before, potent neutralization was observed at high TRIM21 expression levels and was markedly diminished when TRIM21 was depleted (Fig. 4D and E). In addition to a drop in the neutralization potency, the depletion of TRIM21 results in a clear change in the shape of the neutralization curve: in IFN-stimulated cells, infectivity decreases linearly with the NAb concentration, whereas a lag or shoulder is observed for TRIM21-depleted cells. Classically, the presence of a lag has been interpreted as evidence that neutralization depends on multiple NAb-binding events, with a threshold number of NAb molecules being required to bind the virus before infection can be prevented (19). Threshold-based neutralization mechanisms are likely to include entry blocking, which typically requires that available receptor interaction sites are masked. The data therefore suggest that while neutralization by polyclonal sera is mediated principally by ADIN, other mechanisms of neutralization are also present. The data also imply that the number of Abs required for a given level of neutralization may depend upon the mechanism of neutralization. For example, the addition of 2 μg/ml pAb2 gives nearly 2 natural logs of neutralization on IFN-stimulated cells but almost no observable neutralization in cells depleted of TRIM21, presumably because in the latter case, the critical number of NAb molecules required to block entry has not been reached (Fig. 4E).
Examination of neutralization profiles is not the only way of differentiating between neutralization mechanisms. Abs have been reported to neutralize without preventing viral attachment to cells, a phenomenon referred to as postadsorption neutralization (PAN). In a previous study by Emini et al., it was found that six out of seven monoclonal Abs against poliovirus failed to prevent viral entry but were nevertheless potently neutralizing (8). This kind of phenomenon may be indicative of neutralization by an intracellular process such as ADIN. To determine whether ADIN provides a mechanism for PAN, we added AdV-GFP to target cells at 4°C, a temperature that permits virus attachment but not uptake. Antibody 9C12 was then introduced, and the temperature was increased to 37°C to allow infection. We observed efficient PAN, in that the Ab neutralized infection despite being unable to block receptor binding (Fig. 4F). Furthermore, PAN occurred without a lag and was dependent, in terms of both the neutralization efficiency and the level of persistence, on TRIM21.
High levels of infection can overcome ADIN.
In the above-described experiments, we have shown how Ab and TRIM21 levels operate synergistically to influence the efficiency of neutralization and the persistent fraction. The third important component that determines cellular infection is the viral dose or multiplicity of infection (MOI). We hypothesized that at a high MOI, TRIM21 and the ADIN machinery may become saturated. To test this, we simultaneously infected cells with two NAb-bound adenoviruses encoding GFP or RFP reporter genes (Fig. 4G). NAb-labeled AdV-GFP was added to cells in increasing amounts at a high MOI, while NAb-labeled AdV-RFP was added at a constant, low MOI (0.1). We observed a dose-dependent rescue of AdV-RFP infection with increasing AdV-GFP levels. To confirm that increased infection is due specifically to the saturation of TRIM21, we also performed the experiment with cells depleted of TRIM21 by shRNA. The ability of AdV-GFP to rescue AdV-RFP was significantly reduced in cells with reduced TRIM21 levels. The data therefore support a model in which TRIM21 can be titrated by virus-NAb complexes at high MOIs and provide a parallel to other antiviral restriction factors, TRIM5α and Lv1, which are also saturated by high levels of infection (11).
DISCUSSION
This study suggests that ADIN provides an important component of viral neutralization, to the extent that neutralization can be substantially impaired in cells that lack the ADIN receptor TRIM21 or when an antibody is mutated to prevent TRIM21 binding. We have determined the mechanistic requirements of ADIN in terms of how antibody, TRIM21, and virus contribute to the efficiency of neutralization and the level of the persistent fraction. These mechanistic determinants are useful in distinguishing ADIN from other forms of neutralization. In particular, ADIN generates characteristic linear neutralization profiles and is detectable in postadsorption neutralization assays.
Our data indicate that ADIN is remarkably efficient, capable of mediating neutralization in the presence of as few as 1.6 Ab molecules per virus in a cognate murine TRIM21-IgG system. Such low-stoichiometry neutralization may be particularly important in vivo when NAb concentrations are low, such as during the initial stages of an adaptive immune response or in the intercellular spaces of infected tissue. The difference observed for human cells, where a minimum of 5 Ab molecules is required, may reflect the ∼10-fold-lower affinity of human TRIM21 for mouse IgG (15). The conservation of ADIN demonstrated here with human and mouse cells, as well as the conservation of the TRIM21-IgG molecular interaction between distantly related mammalian species (15), suggests that ADIN is broadly conserved as an antiviral mechanism. Further studies are required to determine the prevalence of ADIN among other mammalian species as well as the in vivo contribution of ADIN to antiviral immunity.
Neutralizing antibodies can fail to completely prevent infection, even at a stoichiometric excess. We find that this persistent fraction of infection can be dependent on cellular levels of TRIM21, since reductions in TRIM21 levels increase the level of the PF. However, neutralization also becomes unresponsive to increases in TRIM21 expression levels beyond a threshold level. This “double-plateau” region, where neutralization is unresponsive to changes in both antibody and TRIM21 levels, is suggestive of a maximum rate of virus disposal within the cell, possibly because other factors become limiting. This interpretation is favored over a model in which the plateau is a result of the saturation of the NAb binding of the virus surface, since our data suggest that the PF commences at Ab concentrations substantially lower than those required for saturation. This in turn implies that the incoming virions could accommodate more TRIM21 molecules under higher levels of NAb labeling, given that TRIM21 dimers bind IgG at a 1:1 ratio (14). We therefore predict that the PF will be sensitive to the pharmacological or biological modulation of ADIN components acting downstream of TRIM21. Moreover, limiting concentrations of TRIM21 and its cofactors provide a potential mechanism to explain why the PF has been observed to vary between cell lines (16). ADIN may also help to explain other neutralization phenomena such as postadsorption and postentry neutralization. For instance, the ability of an Ab to neutralize infection after virus adsorption to the cell surface or after entry has been documented for many nonenveloped viruses, including adenovirus, papillomavirus, and poliovirus (1, 6, 8, 9, 24, 32, 36, 37, 40), but has remained largely unexplained. Furthermore, we speculate that antibodies are not the only extracellular antiviral molecules to exert their effects within cells. Indeed, the postentry neutralization of adenovirus has been demonstrated by the antimicrobial peptides of the defensin family, which prevent virus uncoating (28, 33).
Despite the efficiency of the ADIN system, our data suggest that high levels of infection are capable of saturating TRIM21 and overcoming neutralization. Thus, a high multiplicity of infection may promote escape from neutralization in vivo. Given that the burst yield of an AdV-infected cell is typically more than 1,000 IU (an infected cell may contain in excess of 10,000 particles [41]) and that virions are released relatively synchronously upon cell lysis, localized virus titers may result in effective multiplicities of infection that exceed those required for saturation in this study. Future in vivo studies are needed to confirm whether the saturation of TRIM21 represents a mechanism of evasion of neutralization.
We observed the most efficient neutralization when cells were activated by IFN. This emphasizes the importance of immune stimulation in the process of neutralization and is an example of synergy between innate and adaptive immunity. Synergy between IFN and neutralization was previously noted for other nonenveloped viruses (enterovirus 70, coxsackievirus A24, and adenovirus type 3) (20). Of note, such synergy was not observed for an enveloped virus, herpes simplex virus. Viruses will differ in their susceptibilities to ADIN according to their specific entry mechanisms, capsid compositions, and life cycles. Most significantly, viruses with envelopes are likely to avoid ADIN altogether by shedding both their lipid coats and attached Ab molecules during entry. It is therefore important to note that unusual neutralization phenomena have been observed for enveloped viruses, including variations of the PF between cell lines (17) and low-stoichiometry neutralization (26, 35). While TRIM21 is unlikely to be directly involved, our data suggest that considering neutralization, at least in part, as an effector-driven process may be important. Thus, where evidence for such phenomena exists, we do not rule out that other receptors may mediate or facilitate the neutralization of these pathogens.
Footnotes
Published ahead of print 30 May 2012
REFERENCES
- 1. Booy FP, Roden RB, Greenstone HL, Schiller JT, Trus BL. 1998. Two antibodies that neutralize papillomavirus by different mechanisms show distinct binding patterns at 13 A resolution. J. Mol. Biol. 281:95–106 [DOI] [PubMed] [Google Scholar]
- 2. Burton DR, Saphire EO, Parren PW. 2001. A model for neutralization of viruses based on antibody coating of the virion surface. Curr. Top. Microbiol. Immunol. 260:109–143 [DOI] [PubMed] [Google Scholar]
- 3. de Martin R, Raidl M, Hofer E, Binder BR. 1997. Adenovirus-mediated expression of green fluorescent protein. Gene Ther. 4:493–495 [DOI] [PubMed] [Google Scholar]
- 4. Dimmock NJ. 1984. Mechanisms of neutralization of animal viruses. J. Gen. Virol. 65(Pt 6):1015–1022 [DOI] [PubMed] [Google Scholar]
- 5. Dimmock NJ. 1995. Update on the neutralization of animal viruses. Rev. Med. Virol. 5:165–179 [Google Scholar]
- 6. Dorner AJ, Dorner LF, Larsen GR, Wimmer E, Anderson CW. 1982. Identification of the initiation site of poliovirus polyprotein synthesis. J. Virol. 42:1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edghill-Smith Y, et al. 2005. Smallpox vaccine-induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat. Med. 11:740–747 [DOI] [PubMed] [Google Scholar]
- 8. Emini EA, Kao SY, Lewis AJ, Crainic R, Wimmer E. 1983. Functional basis of poliovirus neutralization determined with monospecific neutralizing antibodies. J. Virol. 46:466–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emini EA, Ostapchuk P, Wimmer E. 1983. Bivalent attachment of antibody onto poliovirus leads to conformational alteration and neutralization. J. Virol. 48:547–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Emini EA, et al. 1992. Prevention of HIV-1 infection in chimpanzees by gp120 V3 domain-specific monoclonal antibody. Nature 355:728–730 [DOI] [PubMed] [Google Scholar]
- 11. Hatziioannou T, Cowan S, Goff SP, Bieniasz PD, Towers GJ. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hessell AJ, et al. 2007. Fc receptor but not complement binding is important in antibody protection against HIV. Nature 449:101–104 [DOI] [PubMed] [Google Scholar]
- 13. Icenogle J, et al. 1983. Neutralization of poliovirus by a monoclonal antibody: kinetics and stoichiometry. Virology 127:412–425 [DOI] [PubMed] [Google Scholar]
- 14. James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. 2007. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc. Natl. Acad. Sci. U. S. A. 104:6200–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keeble AH, Khan Z, Forster A, James LC. 2008. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc. Natl. Acad. Sci. U. S. A. 105:6045–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kjellen L. 1985. A hypothesis accounting for the effect of the host cell on neutralization-resistant virus. J. Gen. Virol. 66(Pt 10):2279–2283 [DOI] [PubMed] [Google Scholar]
- 17. Kjellen LE, Schlesinger RW. 1959. Influence of host cell on residual infectivity of neutralized vesicular stomatitis virus. Virology 7:236–239 [DOI] [PubMed] [Google Scholar]
- 18. Klasse PJ, Moore JP. 1996. Quantitative model of antibody- and soluble CD4-mediated neutralization of primary isolates and T-cell line-adapted strains of human immunodeficiency virus type 1. J. Virol. 70:3668–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klasse PJ, Sattentau QJ. 2002. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 83:2091–2108 [DOI] [PubMed] [Google Scholar]
- 20. Langford MP, Villarreal AL, Stanton GJ. 1983. Antibody and interferon act synergistically to inhibit enterovirus, adenovirus, and herpes simplex virus infection. Infect. Immun. 41:214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li J, Wang Y, Wang Z, Dong Z. 2000. Influences of amino acid sequences in FR1 region on binding activity of the scFv and Fab of an antibody to human gastric cancer cells. Immunol. Lett. 71:157–165 [DOI] [PubMed] [Google Scholar]
- 22. Maizel JV, Jr, White DO, Scharff MD. 1968. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology 36:115–125 [DOI] [PubMed] [Google Scholar]
- 23. Mallery DL, et al. 2010. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21). Proc. Natl. Acad. Sci. U. S. A. 107:19985–19990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mandel B. 1967. The interaction of neutralized poliovirus with HeLa cells. I. Adsorption. Virology 31:238–247 [DOI] [PubMed] [Google Scholar]
- 25. McEwan WA, Mallery DL, Rhodes DA, Trowsdale J, James LC. 2011. Intracellular antibody-mediated immunity and the role of TRIM21. Bioessays 33:803–809 [DOI] [PubMed] [Google Scholar]
- 26. McLain L, Dimmock NJ. 1994. Single- and multi-hit kinetics of immunoglobulin G neutralization of human immunodeficiency virus type 1 by monoclonal antibodies. J. Gen. Virol. 75(Pt 6):1457–1460 [DOI] [PubMed] [Google Scholar]
- 27. Mittereder N, March KL, Trapnell BC. 1996. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J. Virol. 70:7498–7509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nguyen EK, Nemerow GR, Smith JG. 2010. Direct evidence from single-cell analysis that human alpha-defensins block adenovirus uncoating to neutralize infection. J. Virol. 84:4041–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reading SA, Dimmock NJ. 2007. Neutralization of animal virus infectivity by antibody. Arch. Virol. 152:1047–1059 [DOI] [PubMed] [Google Scholar]
- 30. Rezaei N, Hedayat M, Aghamohammadi A, Nichols KE. 2011. Primary immunodeficiency diseases associated with increased susceptibility to viral infections and malignancies. J. Allergy Clin. Immunol. 127:1329–1341.e2; quiz 1342-1343. doi:10.1016/j.jaci.2011.02.047 [DOI] [PubMed] [Google Scholar]
- 31. Sanna PP, Burton DR. 2000. Role of antibodies in controlling viral disease: lessons from experiments of nature and gene knockouts. J. Virol. 74:9813–9817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith JG, Cassany A, Gerace L, Ralston R, Nemerow GR. 2008. Neutralizing antibody blocks adenovirus infection by arresting microtubule-dependent cytoplasmic transport. J. Virol. 82:6492–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith JG, Nemerow GR. 2008. Mechanism of adenovirus neutralization by human alpha-defensins. Cell Host Microbe 3:11–19 [DOI] [PubMed] [Google Scholar]
- 34. Takada A, Ebihara H, Jones S, Feldmann H, Kawaoka Y. 2007. Protective efficacy of neutralizing antibodies against Ebola virus infection. Vaccine 25:993–999 [DOI] [PubMed] [Google Scholar]
- 35. Taylor HP, Armstrong SJ, Dimmock NJ. 1987. Quantitative relationships between an influenza virus and neutralizing antibody. Virology 159:288–298 [DOI] [PubMed] [Google Scholar]
- 36. Toogood CI, Crompton J, Hay RT. 1992. Antipeptide antisera define neutralizing epitopes on the adenovirus hexon. J. Gen. Virol. 73(Pt 6):1429–1435 [DOI] [PubMed] [Google Scholar]
- 37. Varghese R, Mikyas Y, Stewart PL, Ralston R. 2004. Postentry neutralization of adenovirus type 5 by an antihexon antibody. J. Virol. 78:12320–12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wetz K, Willingmann P, Zeichhardt H, Habermehl KO. 1986. Neutralization of poliovirus by polyclonal antibodies requires binding of a single IgG molecule per virion. Arch. Virol. 91:207–220 [DOI] [PubMed] [Google Scholar]
- 39. Wohlfart C. 1988. Neutralization of adenoviruses: kinetics, stoichiometry, and mechanisms. J. Virol. 62:2321–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wohlfart CE, Svensson UK, Everitt E. 1985. Interaction between HeLa cells and adenovirus type 2 virions neutralized by different antisera. J. Virol. 56:896–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wold WSM, Horwitz MS. 2007. Adenoviruses, p 2395–2436 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 42. Yoshimi R, et al. 2009. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. J. Immunol. 182:7527–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]