Abstract
Severe fever with thrombocytopenia syndrome (SFTS) is an emerging infectious disease characterized by high fever, thrombocytopenia, multiorgan dysfunction, and a high fatality rate between 12 and 30%. It is caused by SFTS virus (SFTSV), a novel Phlebovirus in family Bunyaviridae. Although the viral pathogenesis remains largely unknown, hemopoietic cells appear to be targeted by the virus. In this study we report that human monocytes were susceptible to SFTSV, which replicated efficiently, as shown by an immunofluorescence assay and real-time reverse transcription-PCR. We examined host responses in the infected cells and found that antiviral interferon (IFN) and IFN-inducible proteins were induced upon infection. However, our data also indicated that downregulation of key molecules such as mitochondrial antiviral signaling protein (MAVS) or weakened activation of interferon regulatory factor (IRF) and NF-κB responses may contribute to a restricted innate immunity against the infection. NSs, the nonstructural protein encoded by the S segment, suppressed the beta interferon (IFN-β) and NF-κB promoter activities, although NF-κB activation appears to facilitate SFTSV replication in human monocytes. NSs was found to be associated with TBK1 and may inhibit the activation of downstream IRF and NF-κB signaling through this interaction. Interestingly, we demonstrated that the nucleoprotein (N), also encoded by the S segment, exhibited a suppressive effect on the activation of IFN-β and NF-κB signaling as well. Infected monocytes, mainly intact and free of apoptosis, may likely be implicated in persistent viral infection, spreading the virus to the circulation and causing primary viremia. Our findings provide the first evidence in dissecting the host responses in monocytes and understanding viral pathogenesis in humans infected with a novel deadly Bunyavirus.
INTRODUCTION
Severe fever with thrombocytopenia syndrome (SFTS) is an emerging infectious disease with a high fatality rate of 12 to 30% and was initially identified in several provinces of central China in 2007 (59). The disease was characterized by sustained high fever with thrombocytopenia, leukocytopenia, and gastrointestinal symptoms among villagers in rural areas (57). The disease was originally suspected to be human granulocytic anaplasmosis (59), but its causative agent, Anaplasma phagocytophilum, were not isolated from clinical samples. Specific nucleotides against A. phagocytophilum and other known pathogens were not detected in the SFTS patients. It was not until recently that a novel Bunyavirus genome was identified in most patient sera and the virus subsequently isolated. The causative agent was named severe fever with thrombocytopenia syndrome virus (SFTSV) (28, 57). One recent study showed an antibody prevalence rate of 3.6% among humans and 46% among animals, indicating that the virus has spread widely in areas of endemicity (19).
In underdeveloped areas of the world viral febrile diseases are often caused by viruses in the families Bunyaviridae, Arenaviridae, Filoviridae, Flaviviridae, and Togaviridae (12). Phylogenetic analysis showed that SFTSV is closely related to viruses of Bunyaviridae. Among the genera Orthobunyavirus, Phlebovirus, Hantavirus, Nairovirus, and Tospovirus of Bunyaviridae, all SFTSV isolates are phylogenetically closest to the genus Phlebovirus (28, 57). Within the phleboviruses, SFTSV is unique for being equidistant from the Sandfly fever group (Rift Valley fever virus [RVFV] and Punta Toro virus) and the Uukuniemi group (52), suggesting that SFTSV is a novel virus falling in a new, third group of this genus. Like RVFV, SFTSV can cause human infections (although ticks, rather than mosquitoes, may be the vector), while Uukuniemi virus is rarely pathogenic to humans (11).
Obvious loss of leukocytes is a critical clinical symptom of many hemorrhagic virus infections (12). However, the target cells of SFTSV in peripheral blood have not been determined. It is certain that the virus targets hemopoietic cells, but there have been no studies on viral pathogenesis in SFTS patients, and virus-host interaction is largely unknown.
SFTSV is a negative sense, single-stranded RNA virus, composed of three segmented genomes. The segments of L, M, and S encode viral RNA polymerase, glycoproteins (Gn and Gc), nucleoprotein (N), and nonstructural (NSs) proteins, respectively. N and NSs are expressed by separate open reading frames in opposite orientations on the S segment, which has 1744 nucleotides of ambisense RNA. NSs proteins have been found with variable sizes and coding strategies in the genera Bunyavirus, Phlebovirus, and Tospovirus of the family Bunyaviridae (5, 6). In the genus Phlebovirus, NSs of RVFV is located in fibrillar structures in the nuclei of infected cells (53), but NSs of Uukuniemi virus is distributed throughout the cytoplasm (39). Studies have shown that NSs of RVFV can target cellular TFIIH transcription factor (25), block interferon (IFN) production through various mechanisms (4, 41, 46), and contribute to viral pathogenesis (7). However, NSs could control the activity of viral polymerase and suppress viral replication as well (47). Although no function is known for NSs in SFTSV, we postulate that it may also play an important role in viral replication and the modulation of host responses.
In this report we show that SFTSV can infect human monocytic THP-1 cells, in which the virus replicated efficiently but no cell death occurred. We examined host responses in the infected cells and found that IFN and NF-κB signaling were activated, but our data also demonstrate that viral NSs and N proteins inhibited IFN signaling. While our study indicates that viral NSs and N proteins suppress the NF-κB pathway, we found that NF-κB signaling facilitates SFTSV replication in human monocytic cells.
MATERIALS AND METHODS
Cells, viruses, and reagents.
THP-1 cells were maintained in RPMI 1640 medium (Invitrogen/Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO), 1% antibiotic-antimycotic solution (Gibco), 1 mM sodium pyruvate (Amresco, Solon, OH), 0.1 mM nonessential amino acids (Gibco), and 55 μM 2-mercaptoethanol (Amresco). Human embryonic kidney 293T (HEK293T) cells and African green monkey kidney Vero cells were grown in Dulbecco modified Eagle medium (DMEM; Gibco) with 8% serum and antibiotics. Cell cultures were incubated at 37°C with 5% CO2. SFTSV strain JS-2010-014 (19), isolated from peripheral blood samples of a patient in Jiangsu, China, was propagated in Vero cells in a biosafety level 3 laboratory, Jiangsu Provincial Centers for Disease Control, and used in the present study. The sequences of the viral L, M, and S segments have been deposited at GenBank, National Center for Biotechnology Information; the accession numbers are JQ317169, JQ317170, and JQ317171, respectively. An H9N2 avian influenza virus, A/ph/CA/2373/98 (H9N2) (51), was propagated in 10-day-old chicken embryos and later titrated in MDCK cells as described previously (50). All viral aliquots were stored at −80°C. Antibodies for caspase-3, caspase-6, caspase-7, NF-κB, TBK1, MAVS, MyD88, phosphor-p65, phospho-NF-κB2, and anti-IκB-α antibodies were purchased from Cell Signaling Technology (Danvers, MA). Anti-pro-caspase-3, MDA5, and anti-β-actin antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Flag M2 antibody was obtained from Sigma-Aldrich. NF-κB and IκB kinase (IKK) inhibitors, 6-amino-4-(4-phenoxyphenylethylamino)quinazoline (catalog no. 481406) and wedelolactone (catalog no. 401474), respectively, were supplied by EMD Chemicals (Gibbstown, NJ).
Quantitative real-time PCR.
Portions (1 μg) of total RNA extracted from SFTSV- or mock-infected cells with an RNeasy kit (Qiagen, Hilden, Germany) were used for reverse transcription using a PrimeScript reagent kit (TaKaRa, Shiga, Japan). Quantitative real-time PCR was performed with SYBR Premix Ex Taq II (TaKaRa) according to the manufacturer's instructions. Relative gene expressions were standardized by a GAPDH (glyceraldehyde-3-phosphate dehydrogenase) control. The fold change was calculated according to the formula: 2ΔCT(gene) − ΔCT(GAPDH) (50). For SFTSV S gene, the standard curve was performed with a plasmid containing full-length nucleotides of S gene according to a protocol described previously (29).
NSs and N cDNA cloning and expression.
The full length of the S segment cDNA of the SFTSV virus was synthesized by PCR from the reverse transcription product as described above with specific primers. NSs and N were amplified by PCR procedures and then subcloned into a plasmid pRK5 with either a flag or hemagglutinin tag for mammalian expression under a cytomegalovirus promoter. HEK293T cells were transfected with pRK5-NSs or pRK5-N with Lipofectamine 2000 reagents (Invitrogen) for the expression of NSs and N proteins.
Immunoblot analysis and immunoprecipitation.
Mock- or SFTSV-infected THP-1 cells were treated with precooled 1% NP-40 lysis buffer containing 2 mM phenylmethylsulfonyl fluoride (PMSF), 2 mM NaF, 1 mM Na3VO4, 1 μg of leupeptin/ml, and 1 μg of aprotinin/ml (Sigma-Aldrich) for 20 min on ice. After low-speed centrifugation (2,500 rpm, 5 min at 4°C), cell lysates were measured with a BCA assay (Pierce, Rockford, IL) before being subjected to SDS-PAGE and transferred to an immunoblot polyvinylidene difluoride membrane (Millipore, Billerica, MA), followed by incubation with primary antibodies overnight. After four washes, membranes were incubated with alkaline phosphatase- or horseradish peroxidase-conjugated secondary antibodies for another 90-min incubation. After additional washes, BCIP (5-bromo-4-chloro-3-indolylphosphate)/nitroblue tetrazolium or enhanced chemiluminescence reagents (Invitrogen) were used for signal development. The β-actin levels were detected as an input control.
Cell lysates, prepared as described above, were pretreated with isotype mouse IgG and then with protein A/G-agarose beads. The pretreated supernatants were incubated with the anti-Flag antibody at 4°C overnight and incubated with protein A/G-agarose beads at 4°C for 90 min. After four washes with NP-40 lysis buffer, the immunoprecipitates were denatured and subjected to SDS-PAGE and Western blot analyses.
Dual-luciferase reporter assay.
HEK293T cells were seeded in 24-well plates at a density of 2.5 × 105 cells per well. The next day, cells were transfected with pRK5 control plasmid or the plasmids expressing NSs or N, along with pGL3-IFNβ-Luc or pGL3-Igκ-Luc, and pRL8-SV40 using Lipofectamine 2000. The total amount of DNA was kept constant by adding empty control plasmid. At 24 h after transfection, the cells were stimulated by 50 μg of poly(I·C)/ml or infected with the avian influenza virus H9N2 for 6 h, and cell lysates were used to determine the firefly and Renilla luciferase activities (Promega, Madison, WI) according to the manufacturer's instructions.
Microarray and pathway analysis.
Total RNAs prepared from samples at each time point after infection were pooled in equivalent amounts and subjected to microarray analyses. For all microarray experiments, cyanine 3-CTP-labeled cRNA probes were generated from 1 μg of total RNA following Agilent one-color microarray-based gene expression analysis (Quick Amp Labeling; Agilent). Human 4x44K slides (Agilent) were used for hybridization, followed by scanning with an Agilent scanner (G2565BA). Each microarray experiment was performed with technical duplicates for infected or uninfected control samples. Changes in the levels of mRNA of any gene were marked significant only when the following two criteria were met: (i) the alteration in expression was statistically significant (P value for paired Student t test of ≤ 0.05), and (ii) the change was at least 50% (equivalent to a 1.5-fold change where the value for no change is 0) above or below the baseline expression level. The baseline was calculated as the expression level of the 0 h (uninfected control) for a particular gene. Gene transcription data, which have been sent to GenBank for deposition, were further analyzed with GeneData Expressionist (www.genedata.com) for differential expression and heat map construction. Differential gene expression data were uploaded into Ingenuity systems (Ingenuity, Redwood City, CA) for pathway and functional analyses.
Indirect immunofluorescence assay and virus titration.
THP-1 cells, mock infected or infected with SFTSV (JS-2010-014), were fixed at various time points with 4% paraformaldehyde for 30 min. The cells were permeabilized with 0.1% Triton X-100 on ice for 10 min, followed by three washes with phosphate-buffered saline (PBS). The cells were then incubated with an anti-SFTSV serum collected from a patient (19), who was confirmed clinically and serologically, at a 1:100 dilution at 4°C overnight or 37°C for 30 min. After several washes with PBS, the cells were incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-human IgG(H+L) (Beyotime, Hangzhou, China) at a 1:200 dilution at 37°C for 1 h and then stained with 1 μg of DAPI/ml at room temperature for 5 min. The cells were washed and resuspended in PBS, smeared on a glass slide, and observed under an Olympus confocal microscope.
Cell media were collected at various times points from THP-1 cells infected with the virus for infectious virus titration [50% tissue culture infective dose(s) (TCID50)]. Tenfold serial dilutions were performed with DMEM to dilute the cultural media, which were used to inoculate Vero cells in 12-well plates. The cells were transferred to glass cover slides at 8 or 18 h postinfection (hpi), air dried, fixed, and permeabilized with 4% paraformaldehyde and 0.1% Triton X-100, followed by staining with anti-SFTSV serum and FITC-conjugated secondary antibody as described above. Infectious virus titers (TCID50/ml) were calculated according to the Reed and Muench method.
Statistical analysis.
A two-tailed Student t test was used to evaluate the data by SPSS software (IBM SPSS, Armonk, NY). An χ2 analysis was used to calculate significant differences of the data, with a probability of 0.05 considered a significant deviation.
RESULTS
SFTSV replicates in human monocytes without inducing apoptosis.
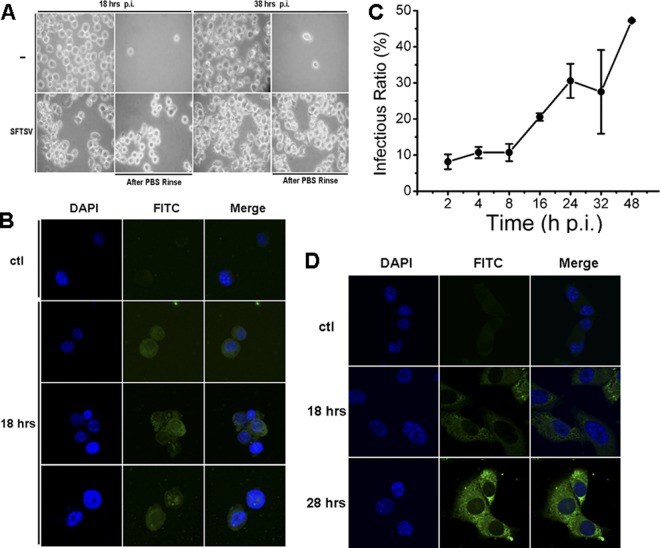
SFTSV appears to target human hemopoietic cells, but its exact target cells are unclear. In order to determine whether SFTSV infects and replicates in human monocytes, SFTSV isolated from a patient's peripheral blood of febrile phase and propagated in Vero cells was used to infect human THP-1 cells. No obvious cytopathic effect was observed at 18 and 38 hpi. However, monocytes appeared to respond to the infection, as shown in Fig. 1A, where THP-1 cells adhered to the culture dish and extended their pseudopodia after 18 h, in contrast to the cells in the mock control.
Fig 1.
Human THP-1 monocytes were susceptible to SFTSV infection. (A) THP-1 cells were mock infected or infected with SFTSV. More adhered cells were observed at 18 and 38 hpi in infected cells (magnification, ×100). Uninfected and infected cells were washed with PBS three times at different time points. (B) IFA for SFTSV antigens in infected THP-1 cells. The cells were mock infected or infected with the virus and fixed at 18 or 28 hpi. After permeabilization with Triton X-100, the cells were incubated with a human anti-SFTSV serum at a dilution of 1:100, followed by a staining with FITC anti-human IgG. (C) Percentage of THP-1 cells during the course of infection was positive with viral antigens at various time points postinfection. (D) IFA for SFTSV antigens in infected Vero cells.
To examine viral antigen expression in infected cells, we performed immunofluorescence analysis (IFA) and stained the infected THP-1 cells, which were fixed at different times postinfection, with a diluted human anti-SFTSV serum. Infected monocytes were stained positively at 18 or 28 hpi as shown in Fig. 1B. Through the course of infection, we quantified the percentage of the THP-1 cells that were positive in the IFA and found that within the initial 8 hpi only 10% of the cells were positive. The numbers of positive cells rose to 30 and 50% at 24 and 48 h, respectively, when the cells were infected with 0.1 multiplicity of infection (MOI) (Fig. 1C). The culture media of the infected THP-1 cells were also used to inoculate Vero cells, which were examined later by IFA. Characteristic viral antigen staining was exhibited in infected Vero cells (Fig. 1D), and the infection of Vero cells was used thereafter for infectious virus titration (TCID50).
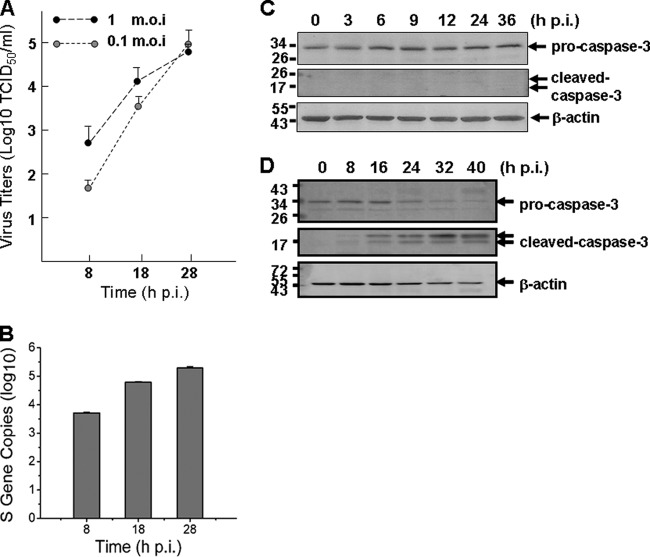
To measure viral replication in THP-1 cells, culture media of the cells infected with SFTSV at MOIs of 1 or 0.1, respectively, were taken at 8, 18, and 28 hpi for infectious virus titration in Vero cells and then examined by IFA. The results indicated that virus titers in the media of infected THP-1 cells reached up to 105 TCID50/ml at 28 hpi when infected with an MOI of either 1 or 0.1 (Fig. 2A). To further confirm a successful infection and viral replication, we extracted total RNA from the cells at various time points postinfection and measured the viral S gene copies by real-time reverse transcription-PCR (RT-PCR). As early as 8 hpi, we detected S gene copies of 5.1 × 103 in 1 μl of cDNA, and the copy numbers increased to 6.2 × 104 and 1.9 × 105 at 18 and 28 hpi, respectively (Fig. 2B), indicating that the virus replicated in monocytes. The oligonucleotide primers specific for viral S gene, as well as cellular genes used in the present study, are listed in Table 1.
Fig 2.
Replication of SFTSV in human monocytes without causing apoptosis. (A) Replicative curve of SFTSV in the culture media of infected THP-1 cells. Culture media of infected THP-1 cells were taken at 8, 18, and 28 hpi, serially diluted, and inoculated in Vero cells. Infectious virus titers were determined after the cells were examined with IFA and calculated based on the Reed and Muench method. (B) Efficient replication of SFTSV in monocytes. Total RNA were extracted from either control or infected THP-1 cells at 8, 18, and 28 hpi and reverse transcribed before cDNAs were subjected to real-time PCR and the S gene copies were quantified. (C) No apoptosis was induced during the infection. Western blot analysis was performed to determine that no cleavage/activation of caspase-3 occurred in SFTSV-infected cells. (D) Apoptotic control. Caspase-3 was cleaved and activated in etoposide (10 μM)-treated THP-1 cells.
Table 1.
Sequences of the primers used for detecting genes by real-time PCR
| Gene | Primer sequence |
|
|---|---|---|
| 5′ | 3′ | |
| S (Jiangsu-014) | ACATTTTCCCTGATGCCTTG | GCTGAAGGAGACAGGTGGAG |
| IFN-α | GGAGTTTGATGGCAACCAGT | TCTCCTCCTGCATCACACAG |
| IFN-β | TGCTCTGGCACAACAGGTAG | CAGGAGAGCAATTTGGAGGA |
| IFN-γ | ATCCCATGGGTTGTGTGTTT | CAAACCGGCAGTAACTGGAT |
| MX1 | ACCACAGAGGCTCTCAGCAT | CTCAGCTGGTCCTGGATCTC |
| OAS1 | CCAAGCTCAAGAGCCTCATC | TGGGCTGTGTTGAAATGTGT |
| RIG-1 | AGATTTTCCGCCTTGGCTAT | ACTCACTTGGAGGAGCCAGA |
| MDA-5 | CAGGGAGTGGAAAAACCAGA | TTTCCAGGCTCAGATGCTTT |
| FasL | GGCCTGTGTCTCCTTGTGAT | TGCCAGCTCCTTCTGTAGGT |
| GAPDH | ACAGTCAGCCGCATCTTCTT | ACGACCAAATCCGTTGACTC |
Intact morphology of infected cells during the course of infection suggests that no cell death or apoptosis was induced. To confirm that no apoptosis was initiated, we analyzed lysates of SFTSV-infected cells at 3, 6, 9, 12, 24, and 36 hpi for pro-caspase-3 and cleaved caspase-3 protein levels. The results showed that the level of pro-caspase-3 was not decreased and the cleaved form of caspase-3 was not detected, indicating that no apoptosis was induced by SFTSV infection in monocytes (Fig. 2C), unlike the control cells treated with etoposide (10 μM), in which cleaved caspase-3 was detected at various time points posttreatment (Fig. 2D).
SFTSV induces attenuated innate immune responses.
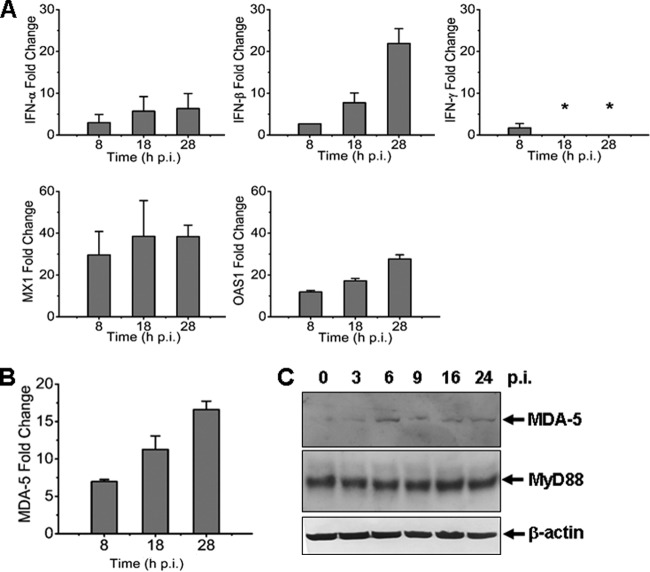
To examine how host antiviral responses are induced during infection, we measured mRNA transcripts of IFNs, IFN-inducible myxovirus resistance 1 (MX1), and 2′,5′-oligoadenylate synthetase 1 (OAS1) in THP-1 cells infected with the virus at an MOI of ∼1. Mild induction of IFN-β mRNA transcripts was detected up to 8- and 22-fold at 8 and 28 hpi (Fig. 3A). We observed a moderate increase of antiviral MX1 and OAS1, which rose up to 40- and 30-fold, respectively. These data demonstrate that antiviral host responses were mounted, but the levels were moderate in SFTSV-infected monocytes, suggesting that the responses may be somehow restrained. We also measured the transcripts of MDA-5, a member of RIG-I-like receptors (RLR) involved in IFN responses and innate immunity, and found that its level increased starting 8 hpi, whereas MyD88 remained unchanged in monocytes (Fig. 3B and C).
Fig 3.
Antiviral responses induced in SFTSV-infected human monocytes. Total RNA were prepared from noninfected and infected cells at various time points postinfection, and real-time RT-PCR was performed to measure the transcript levels of IFN-α, IFN-β, IFN-γ, MX1, and OAS1 (A) and MDA-5 (B) at 8, 18, and 28 hpi, respectively. (C) Induction of MDA-5 expression. Cell lysates were prepared and subjected to SDS-PAGE and Western blot analysis with antibodies against MDA-5, MyD88, and β-actin at the time points as indicated.
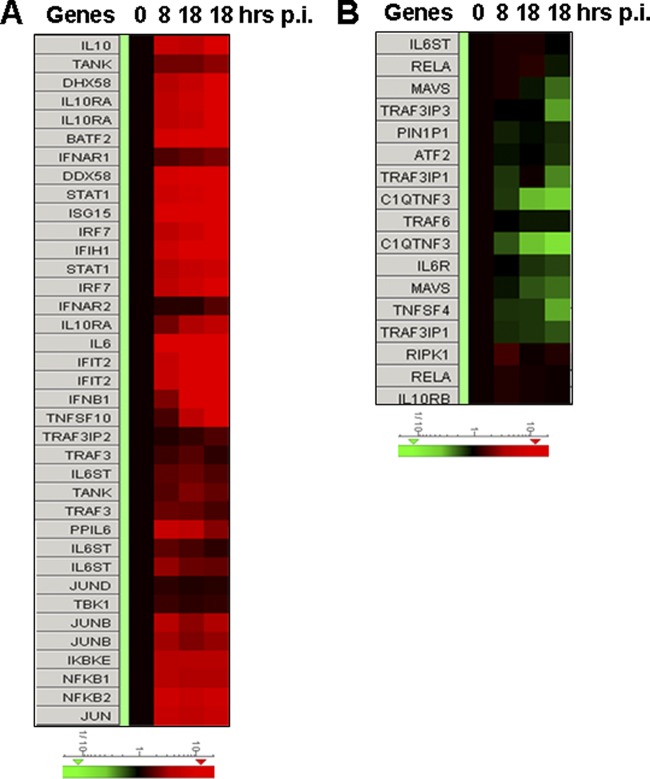
To broaden our understanding of how monocytes react in antiviral responses to SFTSV, we analyzed the regulation of IFN-inducible proteins with Agilent expression microarrays. As shown in Fig. 4 and Table 2, the induction of antiviral MX1 and OAS1 in addition to MDA-1 (IFIH1), and another RLR member, RIG-I (DDX58), was confirmed. More IFN-inducible proteins were detected upregulated, including IFI35, IFI44, IFI44L, IFI6, IFIT1, IFIT3, IFIT5, IFITM1, IRF1, IRF9, ISG15, MX2, and OAS2. Some of these have recently been confirmed to be antiviral (38). Some inducible proteins have important roles in positive feedback of IFN responses, such as STAT1 and STAT2, whereas others, such as SOCS1, may provide a negative-feedback signal (31, 44). Pim1, a serine/threonine kinase, may be another IFN-inducible protein upregulated during SFTSV infection. In addition, a few IFN-inducible proteins seemed to be involved in initiating or regulating adaptive immunity, including CD274 (10), TAP1 (2), and proteasome subunit beta type-8 (PSMB8). Adding to the complexity of the role of IFN-inducible proteins, adenosine deaminase acting on RNA (ADAR) was moderately upregulated, and it has recently been shown to be an enhancer for viral replication (38). Nevertheless, induction of SERPING1 (8, 24), a complement C1 esterase inhibitor, may have an effect on viral pathogenesis since its main function is the inhibition of the complement system. The levels of IFN-β (IFNB) transcripts were also induced, albeit only marginally (2.28-fold) at 8 hpi (Table 3), and rose to 12.36- and 31.96-fold at later stages of infection, which is in accordance with our real-time RT-PCR results (Fig. 3A).
Fig 4.
Heat maps showing the expression profiles of genes in IFN and NF-κB signaling pathways in SFTSV-infected monocytes. Total RNA from noninfected and infected cells at 8, 18, and 28 hpi were prepared, and cRNA was labeled with Cy3-CTP before they were subjected to hybridization on A4x44 microarray slides (Agilent) and scanning. The GeneData Expressionist platform was used to calculate the relative fold changes of genes that are involved in IFN and NF-κB responses (A and B). Log ratios are depicted in red (upregulated) or green (downregulated).
Table 2.
Fold changes of IFN-Inducible genes upon infection
| Genea | Accession no.b | Fold changec at: |
||
|---|---|---|---|---|
| 8 hpi | 18 hpi | 28 hpi | ||
| ADAR* | NM_001111.4 | 2.33 | 2.63 | 2.86 |
| CD274† | NM_014143.3 | 5.52 | 10.56 | 29.50 |
| DDX58 | NM_014314.3 | 7.71 | 13.41 | 23.71 |
| IFIH1 | NM_022168.2 | 6.80 | 10.03 | 14.77 |
| IFI35 | NM_005533.4 | 2.26 | 4.56 | 8.05 |
| IFI44 | NM_006417.4 | 19.71 | 39.64 | 52.97 |
| IFI44L | NM_006820.2 | 24.63 | 61.67 | 111.40 |
| IFI6 | NM_022872.2 | 8.48 | 22.81 | 45.84 |
| IFIT1 | NM_001548.3 | 23.45 | 44.02 | 81.82 |
| IFIT3 | NM_001549.4 | 14.86 | 36.20 | 73.39 |
| IFIT5 | NM_012420.2 | 11.17 | 17.64 | 24.40 |
| IFITM1 | NM_003641.3 | 5.23 | 11.81 | 19.28 |
| IFNAR1 | NM_000629.2 | 1.63 | 1.88 | 2.13 |
| IFNGR1 | NM_000416.2 | 2.32 | 2.58 | 2.75 |
| IFNGR2 | NM_005534.3 | 3.77 | 3.59 | 4.29 |
| IRF1 | NM_002198.2 | 1.87 | 2.21 | 3.72 |
| IRF9 | NM_006084.4 | 2.26 | 2.15 | 2.52 |
| ISG15 | NM_005101.3 | 10.14 | 14.12 | 27.40 |
| JUN | NM_002228.3 | 5.57 | 4.82 | 5.16 |
| MX1 | NM_002462.3 | 13.91 | 21.48 | 27.96 |
| MX2 | NM_002463.1 | 5.88 | 7.77 | 12.53 |
| OAS1 | NM_002534.2 | 9.17 | 14.42 | 22.58 |
| OAS2 | NM_002535.2 | 10.39 | 14.44 | 25.30 |
| PIM1‡ | NM_002648.3 | 4.33 | 4.46 | 8.80 |
| PSMB8† | NM_004159.4 | 1.12 | 1.71 | 2.12 |
| PTPN2 | NM_002828.3 | 1.57 | 2.27 | 2.55 |
| SERPING1 | NM_000062.2 | 1.47 | 11.64 | 37.47 |
| SOCS1‡ | NM_003745.1 | 1.88 | 3.01 | 7.36 |
| STAT1 | NM_007315.3 | 6.32 | 8.31 | 13.74 |
| STAT2 | NM_005419.3 | 2.11 | 2.47 | 3.77 |
| TAP1† | NM_000593.5 | 4.19 | 5.49 | 7.89 |
Role: *, enhancer or viral replications; †, adaptive immunity; ‡, suppressor of IFN signaling.
Accession number according to the NCBI database.
That is, the fold changes of IFN-inducible genes induced at 8, 18, and 28 hpi in SFTSV-infected human monocytes. All fold changes were ≥1.5 except for the underscored values. hpi, hours post infection.
Table 3.
Fold changes of genes involved in the regulation of the IFN and NF-κB signaling
| Gene | Accession no.a | Fold changeb at: |
||
|---|---|---|---|---|
| 8 hpi | 18 hpi | 28 hpi | ||
| BATF2 | NM_138456.3 | 7.73 | 15.19 | 25.49 |
| DDX58 | NM_014314.3 | 7.71 | 13.41 | 23.71 |
| IFIH1 | NM_022168.2 | 6.80 | 10.03 | 11.77 |
| IFIT2 | NM_001547.4 | 5.64 | 17.50 | 47.01 |
| IFNAR1 | NM_000629.2 | 1.63 | 1.88 | 2.13 |
| IFNAR2 | NM_207585.1 | 1.21 | 1.21 | 1.63 |
| IFNB | NM_002176.2 | 2.28 | 12.36 | 31.96 |
| IKBKB | NM_001556.2 | 1.00 | –1.45 | –1.47 |
| IKBKE | NM_014002.3 | 3.89 | 3.57 | 4.22 |
| IL10 | NM_000572.2 | 5.79 | 4.96 | 11.53 |
| IL10RA | NM_001558.3 | 4.88 | 6.92 | 12.78 |
| IRF3 | NM_001571.5 | 1.00 | –1.14 | –1.11 |
| IRF7 | NM_004031.2 | 5.10 | 6.50 | 9.20 |
| ISG15 | NM_005101.3 | 10.14 | 14.12 | 27.40 |
| JUN | NM_002228.3 | 5.57 | 4.82 | 5.16 |
| JUNB | NM_002229.2 | 3.51 | 2.54 | 3.43 |
| JUND | NM_005354.4 | 1.28 | 1.16 | 1.21 |
| MAVS | NM_020746.4 | –1.19 | –1.90 | –2.17 |
| NFKB1 | NM_003998.3 | 3.84 | 3.24 | 3.44 |
| NFKB2 | NM_002502.3 | 6.52 | 5.68 | 6.46 |
| NFKBIA | NM_020529.2 | 7.11 | 7.10 | 7.68 |
| NFKBIB | NM_002503.4 | 2.72 | 2.71 | 1.86 |
| NFKBIE | NM_004556.2 | 2.35 | 2.18 | 2.25 |
| NFKBIZ | NM_031419.3 | 7.42 | 8.64 | 8.09 |
| RELA | NM_021975.3 | 1.08 | 1.21 | –1.23 |
| STAT1 | NM_007315.3 | 6.32 | 8.31 | 13.74 |
| STAT2 | NM_005419.3 | 2.11 | 2.47 | 1.82 |
| SOCS1 | NM_003745.1 | 1.88 | 3.01 | 7.36 |
| TANK | NM_004180.2 | 2.02 | 2.05 | 2.50 |
| TBK1 | NM_013254.3 | 1.39 | 1.27 | 1.29 |
| TNF | NM_000594.2 | 20.86 | 17.71 | 21.60 |
| TNFSF10 | NM_003810.3 | 1.55 | 4.96 | 14.06 |
| TNFAIP3 | NM_006290.2 | 20.74 | 17.02 | 23.18 |
| TRAF3 | NM_145725.2 | 1.35 | 1.68 | 1.27 |
| TRAF6 | NM_145803.2 | –1.05 | –1.22 | –1.23 |
That is, the accession number according to the NCBI database.
That is, the fold changes of genes that are involved in the regulation of IFN and NF-κB signaling. All fold changes were ≥1.5 except for the underscored values. hpi, hours post infection.
Regulation of IRF and NF-κB signaling in SFTSV-infected monocytes.
To explore the mechanism by which IFN signaling pathways were upregulated, we further analyzed the data obtained from the Agilent microarray analysis. Induction of IFN is regulated by IRF3/7 and NF-κB signaling pathways. As shown in Fig. 4 and Table 3, among those with significant fold changes during the infection are transcription factors involved in IFN induction and NF-κB signaling, including NF-κB1 (3.24 to 3.84), NF-κB2 (5.68 to 6.52), IRF7 (5.10 to 9.20), Jun (4.82 to 5.57), and JunB (2.54 to 3.51), but IRF3 (0.82 to 1.00) and RELA (RelA, 0.81 to 1.08) were unchanged. Activating kinase IKBKB (IKKβ, 0.68 to 1.00) remained unchanged, while IKBKE (IKKε, 3.57 to 4.22) was upregulated up to 4-fold, binding to TANK-binding kinase 1 (TBK1, 1.27 to 1.39), to form a complex (34) important in the activation of both IFN (IRF3/7) and noncanonical NF-κB signaling. On the other hand, the inhibitors of NF-κB signaling, IκBα (NFKBIA, 7.10 to 7.18) and IκBβ (NFKBIB, 1.86 to 2.72), both IκB inhibitor proteins, were upregulated, as were the levels of NFKBIE (2.18 to 2.35) and NFKBIZ (7.42 to 8.64), which inhibit NF-κB signaling (54). Although RIG-I (DDX58, 7.71 to 23.71) and MDA-5 (IFIH1, 6.80 to 11.77) were both upregulated, MAVS (IPS-1, 0.84 to 0.46), a key player on mitochondria membrane transmitting RIG-I and MDA-5 signaling, was downregulated. While multiple players were either up- or downregulated, IFN signaling was activated and IFN-β was induced, leading to the induction of many IFN-inducible proteins. Likewise, NF-κB signaling was evidently activated in increased transcription of the target genes such as TNFSF10 (TRAIL), interleukin-6 (IL-6), and IFN, as well as RIG-1 and MDA-5, albeit the increased transcription of a number of inhibitory factors to NF-κB signaling was also demonstrated.
In addition, we observed that the levels of MAVS, RIPK1, TRAF3, TRAF6, and Rel A, which play important roles in RIG-like receptor and Toll-like receptor pathways, were unchanged or even downregulated in SFTSV-infected monocytes (Fig. 4B and Table 3), suggesting that increased IFN and NF-κB signaling could be negatively affected in SFTSV infection, especially when the inhibitors from the host cells, such as NFKBIA, NFKBIAE, and NFKBIZ, or the virus were upregulated or produced.
Pathway analysis in SFTSV-infected human monocytes.
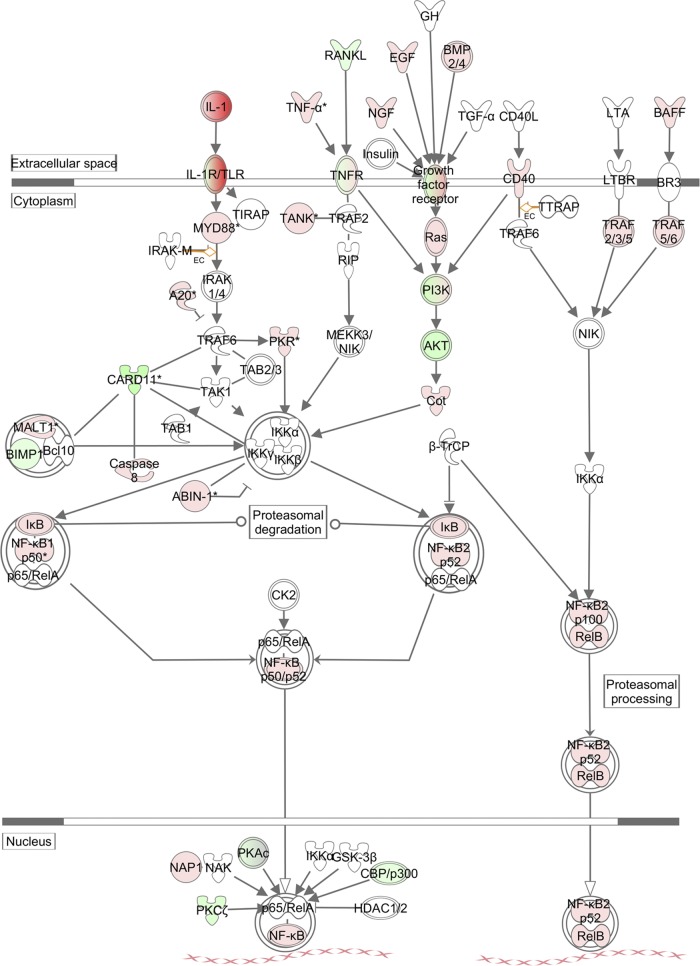
We next used the Ingenuity pathway analysis (IPA) platform to analyze the networking and biological functions of those genes that were associated with the NF-κB signaling pathway. As shown in Fig. 5, NF-κB was activated in both canonical and noncanonical ways and upregulated TLR, tumor necrosis factor alpha (TNF-α), growth factors, and BAFF activated these pathways through similarly upregulated MyD88, Ras, CD40, and TRAF2/3/5/6, respectively. Also upregulated were NF-κB1 and NF-κB2, which are processed to become p50 or p52 and translocated into the nucleus after binding to RelA (p65) and RelB. However, a number of proteins were downregulated, including those in the phosphatidylinositol 3-kinase (PI3K) pathway. These include PI3K and Akt, along with upregulated A20, a strong suppressor of TRAF6, and IκB, the suppressor of NF-κB. These upregulated inhibitors suggest that NF-κB signaling was in fact attenuated or suppressed to a certain degree as a negative-feedback mechanism during the infection of monocytes.
Fig 5.
NF-κB signaling analysis in THP-1 cells infected with SFTSV. The IPA platform was used to analyze components involved in the NF-κB response and their relationships and interactions. Fold changes of transcript expression levels of the genes in NF-κB signaling at 18 hpi were imported from the Agilent microarray analysis described above. Fold changes are shown in red (upregulation) and green (downregulation).
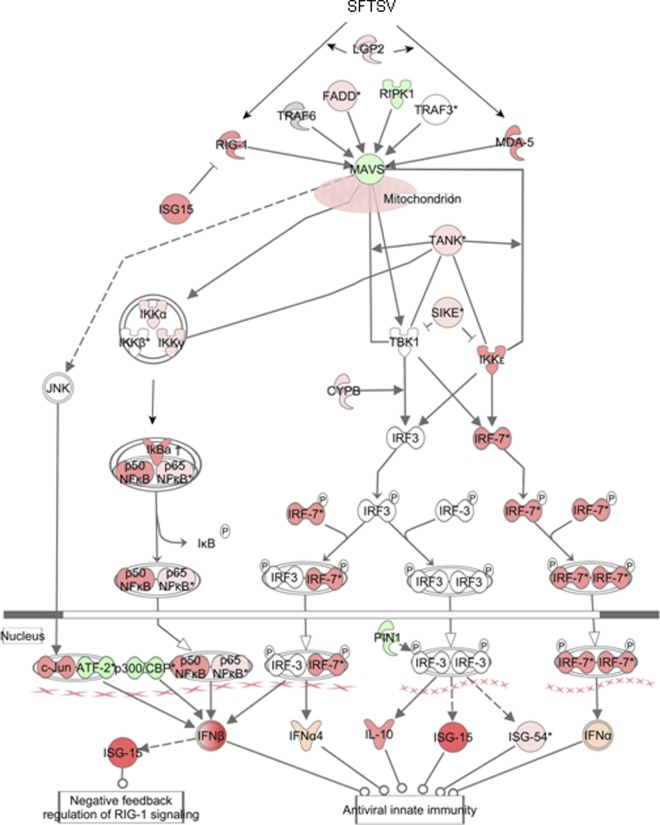
As for IFN signaling (Fig. 6), at the earlier stage of infection, MAVS (IPS-1) was activated by RIG-I and MDA-5, which were further induced. Another member of the RLR family, LGP2, was also upregulated, along with FADD, both of which activated MAVS. Induced downstream kinases and regulators, including IKKγ, CYPB, TANK, and IRF7, were critical to IRF3/7 pathway activation, leading to the induction of IFN and inflammatory cytokines (Fig. 6). However, downregulation of MAVS from 18 hpi, which was also negatively regulated by upregulated IFN-inducible ISG15 as a negative feedback, may restrain further induction of IFN at later stages of infection.
Fig 6.
Pathway analysis of the antiviral IFN response in infected THP-1 cells. The IPA platform was used to analyze components of the IFN response involved in both IRF and NF-κB signaling. Fold changes of transcript expression levels of the genes in IRF and NF-κB signaling at 18 hpi were imported from the Agilent microarray analysis. Fold changes are shown in red (upregulation) and green (downregulation).
NSs and N protein suppress IFN-β promoter activation.
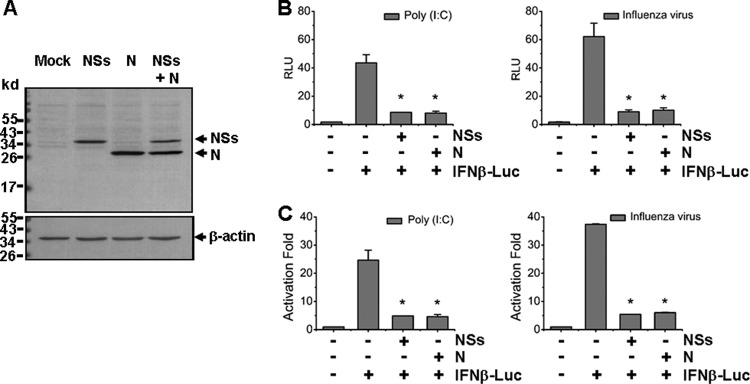
To assess whether viral proteins are involved in the modulation of host responses, we cloned cDNAs of viral nonstructural protein (NSs) and nucleoprotein (N), both encoded by the S segment. The cDNAs were subcloned into an expression vector, pRK5, respectively, and NSs (38-kDa) and N (31-kDa) proteins were expressed in HEK293 cells (Fig. 7A). We examined whether these viral proteins have any effect on host responses. First, we investigated whether NSs affects the IFN-β promoter activation. IFN-β promoter activity was determined by a dual-luciferase reporter assay. HEK293T cells were cotransfected with the reporter plasmids, along with pRK5-Flag-NSs or a control plasmid, respectively. At 24 h posttransfection, the cells were stimulated with 50 μg of synthetic poly(I·C) dsRNA/ml or with the avian influenza virus H9N2 at an MOI of 0.5. Cell lysates were prepared at 6 h after stimulation and analyzed for luciferase activities. Our results demonstrated that while either poly(I·C) or influenza virus infection enhanced the relative luciferase units (RLU) and activation fold of IFN-β promoter activity up to 40- to 60-fold, NSs protein significantly reduced IFN-β promoter activity down to 20% of the original level. We also tested viral nucleoprotein (N). To our surprise, when the cells were transfected with pRK5-Flag-N, N protein suppressed IFN-β promoter activities, induced by either poly(I·C) or the influenza virus infection, down to 20% level as well (Fig. 7B and C).
Fig 7.
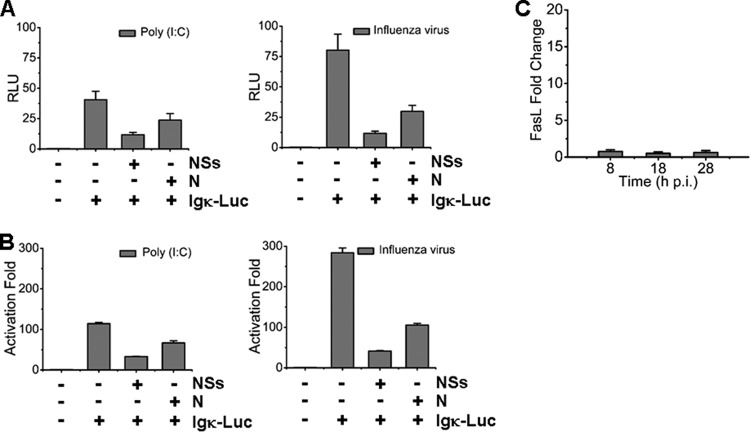
NSs and N proteins inhibited IFN-β promoter activity. (A) HEK293T cells were transfected with the plasmids expressing NSs, N, or both, and Western blot analysis was performed to examine the expression levels of NSs and N proteins. (B and C) IFN-β promoter activity was suppressed by NSs and N proteins in a luciferase assay. Cells were cotransfected with pRK5-NSs or pRK5-N plasmid, along with pGL3-IFNβ-Luc and pRL8 for 24 h, followed by stimulation with 50 μg of poly(I:C)/ml or infection with 0.5 MOI of an avian influenza virus for another 6 h. Cell lysates were subjected to lysis and the lysates were measured for luciferase activities. The results are shown as RLU (B) and activation fold (C).
NF-κB signaling is suppressed by NSs and N proteins.
We next examined whether NSs and N proteins could also suppress NF-κB activation. A reporter assay was performed to measure Igκ light-chain promoter activity with a plasmid, pLuc-Igκ, which is composed of the responsive elements to activated NF-κB signaling. NSs protein suppressed Igκ promoter activity down to 25% of the basal level in response to poly(I·C) stimulation and down to 16% of the basal level in response to the influenza virus infection in transfected HEK293T cells. Similarly, transfected N protein reduced NF-κB activity down to 60% of the basal level in response to poly(I·C) and down to 35% of the basal level in response to the influenza virus infection (Fig. 8A and B). These data indicate that viral NSs and N proteins may suppress NF-κB activation and its target gene expressions in SFTSV-infected monocytes. We also measured expression of a few NF-κB target genes, including FasL, and found no induction, which may be attributed to suppressed NF-κB signaling through viral proteins during the infection (Fig. 8C).
Fig 8.
NSs and N proteins suppressed NF-κB promoter activity. HEK293T cells were cotransfected with plasmids expressing NSs or N, along with pGL3-Igκ-Luc and pRL8, followed by stimulation with 50 μg of poly(I:C)/ml or 0.5 MOI of the avian influenza virus for additional 6 h. The results of luciferase activities were analyzed as RLU (A) and activation fold (B). (C) Transcript expression levels of FasL in infected cells were determined by real-time RT-PCR.
Attenuated NF-κB responses in SFTSV-infected human monocytes.
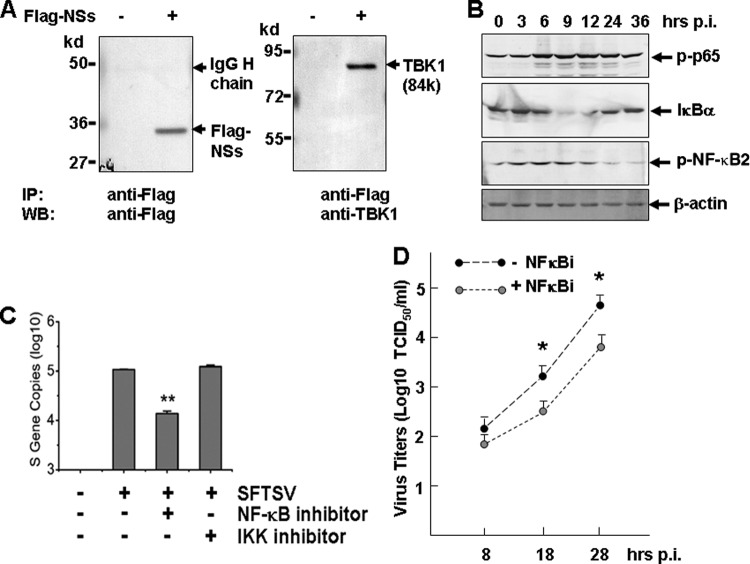
To further understand the mechanism by which NSs may suppress IRF and NF-κB responses, we began by searching cellular proteins critical in innate immunity, IRF, and NF-κB signaling pathways, which may interact with NSs. We transfected HEK293T cells with pRK5-Flag-NSs and then used the cell lysates for coimmunoprecipitation with anti-Flag antibodies and Western blot analyses with antibodies specific for a number of cellular proteins. Among the proteins that were positive in coimmunoprecipitation, TANK-binding kinase 1 (TBK1), which is important in activating IRF and NF-κB signaling pathways, was found to be associated with NSs (Fig. 9A).
Fig 9.
NSs association with TBK1 and transient NF-κB activation. (A) Coimmunoprecipitation of NSs and TBK1. Pretreated cell lysates in pRK5-Flag-NSs-transfected HEK293 were immunoprecipitated with the anti-Flag antibody. The immunoprecipitates were denatured and subjected to SDS-PAGE and Western blot analyses with anti-Flag and anti-TBK1 antibodies, respectively. (B) Transient phosphorylation and activation of NF-κB p65 (RelA) and NF-κB2, and degradation of IκB-α in infected THP-1 cells. Cell lysates at various time points postinfection were prepared from noninfected or infected cells, and subsequently subjected to SDS-PAGE and Western blot analyses. (C) Inhibition of NF-κB signaling suppressed SFTSV replication. THP-1 cells were pretreated with inhibitors of NF-κB (100 nM) or IKK (10 μM) for 1 h, and subsequently infected with SFTSV. At 24 hpi, total RNA was prepared for reverse transcription and cDNA were subjected to real-time PCR for quantifying S gene copies in the infected cells. (D) Infectious virus titers in NF-κB inhibitor-treated THP-1 cells were lower than those in nontreated cells. Culture media from infected THP-1 cells, treated or not treated with the inhibitor, were diluted and subsequently used to infect Vero cells in 12-well plates for infectious virus titration (TCID50, Student t test; *, P < 0.05).
As discussed earlier, SFTSV infection markedly increased the expression levels of both NF-κB1 and NF-κB2 (Table 3), albeit suppressive effects by NSs and N were also found in the present study. NF-κB activation was evident, as demonstrated in cell lysates prepared from various time points postinfection with Western blot analyses. Increased phosphorylation of NF-κB p65 (RelA) and NF-κB2 were both observed (Fig. 9B), even though the expression level of p65 (RelA) remained unchanged (Table 3). However, these activations appeared to be transient since the levels of both phosphorylated NF-κB p65 and NF-κB2 decreased at a later stage of infection. The transient activation of NF-κB was mirrored by the degradation of IκBα, which was also transiently reduced in the middle of the infection, a sign of elevated NF-κB activation. However, the levels of IκB bounced back at a later stage of infection (Fig. 9B). This indicates that the activation of NF-κB signaling may indeed be transient and could be somewhat attenuated later in the infection. Increased expression of the inhibitors such as IκBα, including NFKBIA, NFKBIB, NFKBIE, and NFKBIZ, and reduced levels of MAVS may contribute to decreased activation of NF-κB. A significant increase of TNF-α-induced protein 3 (TNFAIP3) (33), i.e., 20.74- to 23.18-fold at 8 and 28 hpi, respectively, as shown in Table 2, could be a contributing factor as well, since this is an important protein capable of terminating TNF-induced NF-κB responses by binding to TRAF2/6 (16, 40) and IKKγ (60). NF-κB responses will be further decreased by viral NSs and N protein when the virus replicates efficiently in the cell.
NF-κB signaling facilitates SFTSV replication.
THP-1 cells, pretreated with inhibitors of NF-κB (100 nM) and IKKαβ (10 μM) for 1 h prior to infection, were infected with SFTSV. Although no obvious toxicity was observed in the cells treated with either NF-κB or IKK inhibitor, the S gene copies were significantly reduced in the cells treated with specific inhibitor of NF-κB (Fig. 9C). Infectious virus titration also confirmed that virus titers in NF-κB inhibitor-treated THP-1 cultures were significantly lower than those in nontreated cells (Fig. 9D), suggesting that NF-κB is critical to virus replication.
DISCUSSION
As a newly emerging infectious disease, SFTS poses a serious public health concern because it causes high mortality and its transmission routes are not entirely clear. Most phleboviruses and arboviruses are transmitted by mosquitoes, ticks, and phlebotomine sandflies (37). SFTSV has been isolated in ticks, and vector-borne transmission appears to be responsible for most cases. However, recent studies also indicate that SFTS may be transmitted from human to human, and body fluids from patients contain high titers of infectious viruses (3). The most common clinical symptoms in patients are fever (100%), thrombocytopenia (95%), and leukocytopenia (86%) (57). However, details about the viral pathogenesis are largely unknown. Our data demonstrate that human monocytes are susceptible to SFTSV, which replicates efficiently during the first few days of infection. Since the infected monocytes remain intact, these cells may also support persistent viral infection. SFTSV is capable of replicating in many cell types, but usually no cell death is observed (58), which is similar to many other bunyaviruses such as Hantaan virus. Apoptosis may be sufficiently suppressed in the infected cells, but the mechanisms are unknown. It is important to note that SFTSV replicates in monocytes, which may be implicated in viral pathogenesis, especially when the cells remain intact during the initial days of infection. Resident or trafficking monocytes can transport the virus through lymphatic drainage to regional lymph nodes and then spread into the circulation and cause primary viremia. We are exploring whether other types of hemopoietic cells are also susceptible to SFSTV.
Production of type I IFNs is one of the defense mechanisms of the host innate immune system against virus infection (21, 42). IFN signaling is initiated by Toll-like (TLR) and RIG-like (RLR) receptors, and IFN mRNA induction is controlled by transcription factors, including NF-κB, IRF3, IRF7, and c-Jun (AP-1) (21, 43, 56). Clinically, there were almost no detectable levels of IFN observed in patients' blood serum (28), indicating that IFN production may be suppressed, especially at a later stage of the infection, although IFN induction could be transient, and its level may be detectable earlier. In the present study we explored how this suppression may occur in infected monocytes and found that some mechanism may explain dysregulation of IFN induction in patients. For example, during the infection in monocytes, IFN and relevant transcription factors were moderately upregulated at the transcription level due to the activation of IRF and NF-κB responses. However, several upstream molecules, including MAVS, TRAF6, and TRAF3, were unchanged or downregulated. These molecules are critical in TLR/RLR signaling and/or important in the activation of IRF3/7 or NF-κB pathways, essential for IFN induction. Although NF-κB1 and NF-κB2 were both upregulated, the inhibitors of NFKB, such as NFKBIA (IκBα), NFKB IB, and NFKBIZ, were also upregulated, which suppressed the activation of NF-κB directly. Our data also demonstrated that the activation of p65 (RelA) and NF-κB2 appeared to be transient, which matched the increased level of IκBα during the late stage of infection (Fig. 9B). Activation of NF-κB could also be attenuated upstream by TNFAIP3 (Table 3) and ISG15 (Table 2 and Fig. 6), which were all upregulated and acted in a negative-feedback manner.
Our data further demonstrated that the nonstructural protein, NSs of SFTSV, might be implicated in the suppression of the IFN induction, as shown in a reporter assay. IFN-β promoter activity induced by either an avian influenza virus infection or poly(I·C) was efficiently suppressed by ectopically expressed NSs in vitro. Many viruses evolve to encode specific components that counteract IFN induction to evade host antiviral responses. Well-known IFN antagonists are the NS1 protein of influenza A virus, the NS3-4A protein of hepatitis C virus, the V protein of paramyxoviruses, the P protein of rabies virus, the M protein of vesicular stomatitis virus, the G1 protein of hantaan virus, the VP35 protein of Ebola virus, and the proteases of picornaviruses (48). Two NSs protein of bunyaviruses, including Bunyamwera of the orthobunyaviruses and RVFV of the phleboviruses, have also been shown to be effective suppressors of IFN induction (4, 20, 45). On the other hand, due to significant sequence differences, the NSs of different bunyaviruses appear to function with different mechanisms. Our finding showed that NSs of SFTSV interacted with TBK1, a homolog of IKKε, critical in the activation of IRF and NF-κB pathways. Studies are under way to characterize how this interaction, unique among NSs proteins of bunyaviruses, may affect the function of TBK1 and its subsequent activation of the downstream pathways.
Suppression of host protein synthesis has been considered the main mechanism for NSs to inhibit IFN induction (27). However, since no inhibitory effect on the activation of PKR (41), IRF2, NF-κB, and AP1 (4) was found, NSs was considered to inhibit transcriptional mechanism at the terminal end, which would show no specificity. However, NSs of RVFV can bind to SAP30 in the nucleus to form an inhibitory complex, which is bound to, and specifically inhibits, the promoter region of IFN-β (26). It has recently been shown that NSs of RVFV can suppress IFN production by targeting the upstream component of the signaling pathway, in this case, PKR, which is specific (14, 17, 18). NSs of another Phlebovirus, sandfly fever Sicilian virus, and Orthobunyavirus La Cross virus, do not have this function.
Surprisingly, we found that N protein of SFTSV also exhibited a suppressive capacity on IFN-β promoter activity. Like other helical RNA viruses, nucleoprotein (N) encapsidates the segmented or nonsegmented RNA genomes (36). Unexpected antagonism of N proteins to host IFN induction in viral infections is an intriguing phenomenon and a recent focus of intensive study; it has been found that N proteins of SARS-CoV and mouse hepatitis coronavirus antagonized IFN responses (30, 55). N protein of Lassa virus, a member of the Arenaviridae, also exhibits strong suppression of host cytokine responses, including IFNs (35). Mechanisms of the IFN suppression are variable and in many cases unknown. However, it is interesting that the suppression of wide host cytokine responses by N protein of Lassa virus is dependent on its intrinsic RNase activity, which cleaves viral RNA and blunts the activation of RIG-I signaling (35). Here, we expand our view and propose that the N protein of a Bunyavirus has also potential to inhibit IFN responses, although the mechanism is currently unclear.
An obvious dilemma presented by our research is that an apparent IFN induction occurred in THP-1 cells after infection and IFN-inducible genes were produced, even though NSs and nucleoproteins were shown to be suppressive. A reasonable explanation would be that the suppression of IFN and NF-κB responses by NSs and nucleoprotein is complete in the infected human monocytes. However, the presence of a large portion of the THP-1 cells that remained negative in viral antigens detected by IFA (Fig. 1C) indicates that these cells were uninfected, or infected at a very early stage with the expression of NSs or N protein at very low levels. Thus, this portion of the cells was in fact stimulated in response to signaling from the infected cells and contributed to upregulated IFN and NF-κB responses. Since we did not have specific antibodies to viral N and NSs proteins at this stage and were unable to detect expression levels of these viral proteins during infection, it was difficult for us to measure how efficiently NSs or nucleoprotein suppressed IFN or NF-κB responses in infected cells. We believe that this issue could be appropriately addressed when mutant viruses are generated and used to study this phenomenon.
NF-κB signaling is critical to host proinflammatory, apoptotic, and antiviral responses in viral infection. We observed that both NF-κB1 and NF-κB2 were upregulated, in addition to IKKε (Table 3), an NF-κB-activating kinase in a noncanonical pathway. Increased phosphorylation of p65 (RelA) and NF-κB2 at the early stage of infection indicates the activation of NF-κB signaling (Fig. 9B). Despite the transcriptional upregulation of IKK, including IKKγ and IKKε, we failed to detect the activation of IKKα/β (data not shown), suggesting that NF-κB signaling may be activated through other mechanisms, such as activating TBK1/TRIF through TLR signaling. Our data also indicate that the activation of NF-κB may be transient, as discussed above. Obviously, regulation of NF-κB signaling and its target genes is complicated in SFTSV-infected monocytes. At a molecular level, we also observed that NSs of SFTSV was suppressive to the NF-κB target activation, as shown in a luciferase assay, in which the NF-κB promoter activity, induced by either the avian influenza virus infection or poly(I·C) stimulation, was inhibited (35). Interestingly, we also found that the N protein of SFTSV possesses the inhibitory effect on NF-κB promoter activity as well. NF-κB signaling appears to be an attractive target for many viruses (9, 15). Suppression of the NF-κB pathway in SFTSV infection may contribute to its restrictive host antiviral responses, including IFN induction and unchanged proapoptotic FasL expression, which may extend host cell survival and benefit the virus in replicating.
It is also interesting that NF-κB signaling promotes viral replication, as shown in SFTSV-infected monocytes in our study. Although we have no data yet to explain the mechanism, NF-κB is required for efficient replication of other viruses (such as influenza A viruses), probably through its involvement in vRNA synthesis, but not the corresponding cRNA or mRNA synthesis (23). Restrained NF-κB signaling in infected monocytes may reduce SFTSV viral load, likely contributing to persistent viral infection at lower levels both in vitro and in infected tissues. We speculate that infected cells will eventually be targeted by specific CD8+ T cells and that free viruses will be cleared by neutralizing antibodies if the patient survives. More study is needed to ascertain the molecular events involved in viral pathogenesis, which will provide in-depth understanding of the disease and ideas for therapeutic and preventive strategies for this fatal emerging disease.
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (NSFC 30971450) and the State Key Laboratory of Pharmaceutical Biotechnology of Nanjing University (KFGW-200902) to Z.X. and the Jiangsu Province Key Medical Talent Foundation (RC2011084) and the “333” Projects of Jiangsu Province to X.Q. Z.X. was supported by a grant-in-aid from the University of Minnesota at Twin Cities.
We thank Sandy Shanks for her expertise in editing the manuscript.
Footnotes
Published ahead of print 23 May 2012
REFERENCES
- 1. Bachmann M, Moroy T. 2005. The serine/threonine kinase Pim-1. Int. J. Biochem. Cell Biol. 37:726–730 [DOI] [PubMed] [Google Scholar]
- 2. Bahram S, Arnold D, Bresnahan M, Strominger JL, Spies T. 1991. Two putative subunits of a peptide pump encoded in the human major histocompatibility complex class II region. Proc. Natl. Acad. Sci. U. S. A. 88:10094–10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bao CJ, et al. 2007. A family cluster of infections by a newly recognized bunyavirus in eastern China: further evidence of person-to-person transmission. Clin. Infect. Dis. 53:1208–1214 [DOI] [PubMed] [Google Scholar]
- 4. Billecocq A, et al. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78:9798–9806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bishop DH. 1986. Ambisense RNA genomes of arenaviruses and phleboviruses. Adv. Virus Res. 31:1–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bishop DH. 1986. Ambisense RNA viruses: positive and negative polarities combined in RNA virus genomes. Microbiol. Sci. 3:183–187 [PubMed] [Google Scholar]
- 7. Bridgen A, Weber F, Fazakerley JK, Elliott RM. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 98:664–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cicardi M, Zingale L, Zanichelli A, Pappalardo E, Cicardi B. 2005. C1 inhibitor: molecular and clinical aspects. Springer Semin. Immunopathol. 27:286–298 [DOI] [PubMed] [Google Scholar]
- 9. Cotter CR, et al. 2011. The virion host shut-off (Vhs) protein of HSV-1 blocks the replication-independent activation of NF-κB in dendritic cells in the absence of type I interferon signaling. J. Virol. 85:12662–12672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dong H, Zhu G, Tamada K, Chen L. 1999. B7–H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 5:1365–1369 [DOI] [PubMed] [Google Scholar]
- 11. Flick R, Elgh F, Pettersson RF. 2002. Mutational analysis of the Uukuniemi virus (Bunyaviridae family) promoter reveals two elements of functional importance. J. Virol. 76:10849–10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Geisbert TW, Jahrling PB. 2004. Exotic emerging viral diseases: progress and challenges. Nat. Med. 10:S110–S121 [DOI] [PubMed] [Google Scholar]
- 13. Glynne R, et al. 1991. A proteasome-related gene between the two ABC transporter loci in the class II region of the human MHC. Nature 353:357–360 [DOI] [PubMed] [Google Scholar]
- 14. Habjan M, et al. 2009. NSs protein of rift valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 83:4365–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hang do TT, Song JY, Kim MY, Park JW, Shin YK. 2011. Involvement of NF-κB in changes of IFN-γ-induced CIITA/MHC-II and iNOS expression by influenza virus in macrophages. Mol. Immunol. 48:1253–1262 [DOI] [PubMed] [Google Scholar]
- 16. Heyninck K, Beyaert R. 1999. The cytokine-inducible zinc finger protein A20 inhibits IL-1-induced NF-κB activation at the level of TRAF6. FEBS Lett. 442:147–150 [DOI] [PubMed] [Google Scholar]
- 17. Ikegami T, et al. 2009. Dual functions of Rift Valley fever virus NSs protein: inhibition of host mRNA transcription and posttranscriptional downregulation of protein kinase PKR. Ann. N. Y. Acad. Sci. 1171(Suppl 1):E75–E85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ikegami T, et al. 2009. Rift Valley fever virus NSs protein promotes posttranscriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog. 5:e1000287 doi:10.1371/journal.ppat.1000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiao Y, et al. 2011. Preparation and evaluation of recombinant severe fever with thrombocytopenia syndrome virus (SFTSV) nucleocapsid protein for detection of total antibodies in human and animal sera by double antigen sandwich ELISA. J. Clin. Microbiol. 50:372–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kalveram B, Lihoradova O, Ikegami T. 2011. NSs protein of rift valley fever virus promotes posttranslational downregulation of the TFIIH subunit p62. J. Virol. 85:6234–6243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kawai T, Akira S. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131–137 [DOI] [PubMed] [Google Scholar]
- 22. Kelly A, et al. 1991. Second proteasome-related gene in the human MHC class II region. Nature 353:667–668 [DOI] [PubMed] [Google Scholar]
- 23. Kumar N, Xin ZT, Liang Y, Ly H. 2008. NF-κB signaling differentially regulates influenza virus RNA synthesis. J. Virol. 82:9880–9889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lappin D, Whaley K. 1989. Regulation of C1-inhibitor synthesis by interferons and other agents. Behring Inst. Mitt. 1989:180–192 [PubMed] [Google Scholar]
- 25. Le May N, et al. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541–550 [DOI] [PubMed] [Google Scholar]
- 26. Le May N, et al. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus-infected cells. PLoS Pathog. 4:e13 doi:10.1371/journal.ppat.0040013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leonard VH, Kohl A, Hart TJ, Elliott RM. 2006. Interaction of Bunyamwera orthobunyavirus NSs protein with mediator protein MED8: a mechanism for inhibiting the interferon response. J. Virol. 80:9667–9675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li DX. 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 25:81–84 (In Chinese.) [PubMed] [Google Scholar]
- 29. Li J, Cardona CJ, Xing Z, Woolcock PR. 2008. Genetic and phenotypic characterization of a low-pathogenicity avian influenza H11N9 virus. Arch. Virol. 153:1899–1908 [DOI] [PubMed] [Google Scholar]
- 30. Lu X, Pan J, Tao J, Guo D. 2011. SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes 42:37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Minamoto S, et al. 1997. Cloning and functional analysis of new members of STAT induced STAT inhibitor (SSI) family: SSI-2 and SSI-3. Biochem. Biophys. Res. Commun. 237:79–83 [DOI] [PubMed] [Google Scholar]
- 32. Nakitare GW, Elliott RM. 1993. Expression of the Bunyamwera virus M genome segment and intracellular localization of NSm. Virology 195:511–520 [DOI] [PubMed] [Google Scholar]
- 33. Opipari AW, Jr, Boguski MS, Dixit VM. 1990. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J. Biol. Chem. 265:14705–14708 [PubMed] [Google Scholar]
- 34. Pomerantz JL, Baltimore D. 1999. NF-κB activation by a signaling complex containing TRAF2, TANK, and TBK1, a novel IKK-related kinase. EMBO J. 18:6694–6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qi X, et al. 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468:779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Raymond DD, Piper ME, Gerrard SR, Smith JL. 2010. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc. Natl. Acad. Sci. U. S. A. 107:11769–11774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmaljohn CH, et al. 2001. Bunyaviridae: the viruses and their replication, 4th ed Lippincott Williams & Wilkin, Philadelphia, PA [Google Scholar]
- 38. Schoggins JW, et al. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Simons JF, Persson R, Pettersson RF. 1992. Association of the nonstructural protein NSs of Uukuniemi virus with the 40S ribosomal subunit. J. Virol. 66:4233–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song HY, Rothe M, Goeddel DV. 1996. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-κB activation. Proc. Natl. Acad. Sci. U. S. A. 93:6721–6725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Streitenfeld H, et al. 2003. Activation of PKR by Bunyamwera virus is independent of the viral interferon antagonist NSs. J. Virol. 77:5507–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takeuchi O, Akira S. 2009. Innate immunity to virus infection. Immunol. Rev. 227:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thompson AJ, Locarnini SA. 2007. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol. Cell Biol. 85:435–445 [DOI] [PubMed] [Google Scholar]
- 44. Ungureanu D, Saharinen P, Junttila I, Hilton DJ, Silvennoinen O. 2002. Regulation of Jak2 through the ubiquitin-proteasome pathway involves phosphorylation of Jak2 on Y1007 and interaction with SOCS-1. Mol. Cell. Biol. 22:3316–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Knippenberg I, Carlton-Smith C, Elliott RM. 2010. The N terminus of Bunyamwera orthobunyavirus NSs protein is essential for interferon antagonism. J. Gen. Virol. 91:2002–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weber F, et al. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 76:7949–7955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weber F, Dunn EF, Bridgen A, Elliott RM. 2001. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology 281:67–74 [DOI] [PubMed] [Google Scholar]
- 48. Weber F, Haller O. 2007. Viral suppression of the interferon system. Biochimie 89:836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Won S, Ikegami T, Peters CJ, Makino S. 2007. NSm protein of Rift Valley fever virus suppresses virus-induced apoptosis. J. Virol. 81:13335–13345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xing Z, et al. 2009. Differential regulation of antiviral and proinflammatory cytokines and suppression of Fas-mediated apoptosis by NS1 of H9N2 avian influenza virus in chicken macrophages. J. Gen. Virol. 90:1109–1118 [DOI] [PubMed] [Google Scholar]
- 51. Xing Z, et al. 2008. Modulation of the immune responses in chickens by low-pathogenicity avian influenza virus H9N2. J. Gen. Virol. 89:1288–1299 [DOI] [PubMed] [Google Scholar]
- 52. Xu F, et al. 2007. Antigenic and genetic relationships among Rift Valley fever virus and other selected members of the genus Phlebovirus (Bunyaviridae). Am. J. Trop. Med. Hyg. 76:1194–1200 [PubMed] [Google Scholar]
- 53. Yadani FZ, Kohl A, Prehaud C, Billecocq A, Bouloy M. 1999. The carboxy-terminal acidic domain of Rift Valley Fever virus NSs protein is essential for the formation of filamentous structures but not for the nuclear localization of the protein. J. Virol. 73:5018–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamazaki S, Muta T, Takeshige K. 2001. A novel IκB protein, IκB-zeta, induced by proinflammatory stimuli, negatively regulates nuclear factor-κB in the nuclei. J. Biol. Chem. 276:27657–27662 [DOI] [PubMed] [Google Scholar]
- 55. Ye Y, Hauns K, Langland JO, Jacobs BL, Hogue BG. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 81:2554–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yoneyama M, Fujita T. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 227:54–65 [DOI] [PubMed] [Google Scholar]
- 57. Yu XJ, et al. 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 364:1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang L, et al. 2008. Nosocomial transmission of human granulocytic anaplasmosis in China. JAMA 300:2263–2270 [DOI] [PubMed] [Google Scholar]
- 59. Zhang SQ, Kovalenko A, Cantarella G, Wallach D. 2000. Recruitment of the IKK signalosome to the p55 TNF receptor: RIP and A20 bind to NEMO (IKKγ) upon receptor stimulation. Immunity 12:301–311 [DOI] [PubMed] [Google Scholar]