Abstract
The HIV-1 capsid (CA) protein plays essential roles in both early and late stages of virl replication and has emerged as a novel drug target. We report hybrid structure-based virtual screening to identify small molecules with the potential to interact with the N-terminal domain (NTD) of HIV-1 CA and disrupt early, preintegration steps of the HIV-1 replication cycle. The small molecule 4,4′-[dibenzo[b,d]furan-2,8-diylbis(5-phenyl-1H-imidazole-4,2-diyl)]dibenzoic acid (CK026), which had anti-HIV-1 activity in single- and multiple-round infections but failed to inhibit viral replication in peripheral blood mononuclear cells (PBMCs), was identified. Three analogues of CK026 with reduced size and better drug-like properties were synthesized and assessed. Compound I-XW-053 (4-(4,5-diphenyl-1H-imidazol-2-yl)benzoic acid) retained all of the antiviral activity of the parental compound and inhibited the replication of a diverse panel of primary HIV-1 isolates in PBMCs, while displaying no appreciable cytotoxicity. This antiviral activity was specific to HIV-1, as I-XW-053 displayed no effect on the replication of SIV or against a panel of nonretroviruses. Direct interaction of I-XW-053 was quantified with wild-type and mutant CA protein using surface plasmon resonance and isothermal titration calorimetry. Mutation of Ile37 and Arg173, which are required for interaction with compound I-XW-053, crippled the virus at an early, preintegration step. Using quantitative PCR, we demonstrated that treatment with I-XW-053 inhibited HIV-1 reverse transcription in multiple cell types, indirectly pointing to dysfunction in the uncoating process. In summary, we have identified a CA-specific compound that targets and inhibits a novel region in the NTD-NTD interface, affects uncoating, and possesses broad-spectrum anti-HIV-1 activity.
INTRODUCTION
The HIV-1 capsid (CA) protein has gained attention as a promising therapeutic target owing to its pivotal structural and regulatory roles in HIV-1 replication. In addition, the seminal finding that retroviral species-specific host cell restriction factors target the incoming capsid core highlights the potential of the capsid protein as a target for antiviral agents (9, 30, 39, 40). The feasibility of disrupting the capsid using small molecules as a potential anti-HIV-1 strategy was first demonstrated by Tang and coworkers (42) with the discovery of the assembly inhibitor CAP-1. Since that initial work, several groups have used either rational modification of CAP-1 or high-throughput screening to identify compounds with anti-HIV-1 activities that function by targeting this pocket (1, 6, 14, 20, 36, 44). These small-molecule CA inhibitors fall into two main functional categories, those that target assembly and those that target uncoating.
Although several structures for the HIV-1 CA protein exist, only recently have the structures of the CA hexameric and pentameric building blocks of the viral capsid been resolved (31). These structures reveal that six N-terminal domains (NTDs) form the rigid core of hexameric CA, and six C-terminal domains (CTDs) form the hexamer's much more flexible outer ring (32). Dimeric interactions between CTDs of neighboring hexamers hold the capsid together, while NTD-NTD interactions are responsible for the formation of the HIV-1 CA hexameric configuration. The NTD-NTD interactions are mediated through helices 1, 2, and 3, which associate as an 18-helix bundle in the center of the hexamer. The interface is primarily stabilized by hydrophilic contacts but also contains a small core of hydrophobic residues (32). Since this initial finding, the atomic-level details of the CA lattice have been refined and complemented by structural insights obtained with other techniques (11, 27, 33, 37, 46).
Extensive mutagenesis of the CA NTD has been performed. In addition to the mutations that perturb normal particle assembly, specific mutations in the NTD that either stabilize or destabilize the structure of the CA hexamer have adverse affects on viral replication at an early, postentry stage (8, 10, 45). The inhibitory effects of the mutations that modulate the stability of the capsid further highlight the need for a very delicate balance of favorable and unfavorable interactions within HIV-1 CA to allow assembly but also facilitate the uncoating process following infection.
The availability of atomic-level details of the NTD-NTD interface, coupled with mutagenesis studies demonstrating that disruption of this interface is deleterious to the virus, underscores the potential of this site as a target for the design of new HIV-1 inhibitors. In the studies reported here, we used the hybrid structure-based (HSB) screening method (18), which employs both structural and biochemical information, to design inhibitors of the HIV-1 CA NTD-NTD interface. An initial screen identified 4,4′-[dibenzo[b,d]furan-2,8-diylbis(5-phenyl-1H-imidazole-4,2-diyl)]dibenzoic acid (CK026) as a promising starting point for design of CA inhibitors that bind to the NTD-NTD interface and interfere with HIV-1 replication at an early, preintegration stage. Further optimization of CK026 led to the discovery and characterization of the novel small-molecule inhibitor I-XW-053, which binds to a site in the NTD of the CA protein and affects the uncoating events required in viral replication. This study demonstrates that both the NTD of CA and early CA-mediated events, such as uncoating, can be effectively targeted by small molecules, thereby providing new chemical scaffolds for the development of novel small-molecule HIV-1 inhibitors.
MATERIALS AND METHODS
Virtual screening using the hybrid structure-based method.
An iterative in silico-in vitro HSB method was employed for screening small molecules that inhibit the formation of the CA NTD-NTD interface (18). The protocol has been described in detail elsewhere (4, 27, 28). Electronic libraries of 3 million small molecules acquired from commercial vendors are screened using a hybrid pharmacophore. The hybrid pharmacophore is generated using the interactions of residues that form the NTD-NTD interface derived from Protein Databank entry 3H4E (32) in a dynamic mode. A four-point three-dimensional pharmacophore consisting of three hydrophobic and one hydrogen bond donor-acceptor feature was designed using interactions involving residues Ala42, Met39, Arg173, and Leu20 from the neighboring CA protomer. The pharmacophore-based screening resulted in 900 hit molecules that were then subjected to a modified Lipinski's “rule of five” to identify “drug-like” molecules (21). Lipinski's rules were relaxed to include molecules with molecular mass of up to 700 Da and a log(P) of ≤7 in order to identify a wide range of chemical cores. Chemical core analysis using clustering and principal-component analysis resulted in 300 molecules that were then docked to the binding site of CA-NTD using the GOLD program (Genetic Optimisation for Ligand Docking, version 4.1) (15). The docked receptor-ligand complexes were then scored using a customizable knowledge-based scoring function that is based on the nature of the interaction of every atom within the NTD-NTD docking pharmacophore (18). A consensus scoring scheme that involves GoldScore, ChemScore, contact score, and a shape-weighted scoring scheme (17) was then used to rank the compounds. The best-ranking complexes were visually inspected to include compounds that not only interacted with the specified residues but also had extended volume to maximize the inhibition of the NTD-NTD interface.
Chemicals.
Compounds CK026, DMJ-I-073, I-XW-091, I-XW-053, and NBD-556 were synthesized as described in the supplemental material. All other chemicals were purchased from commercial suppliers.
Generation of recombinant HIV-1-expressing luciferase.
Using the Effectene transfection reagent (Qiagen, Germantown, MD), 293T human embryonic kidney cells were cotransfected with plasmids expressing the pCMVΔP1ΔenvpA HIV-1 Gag-Pol packaging construct, the wild-type or mutant HIV-1YU-2 envelope glycoproteins or the envelope glycoproteins of the control amphotropic murine leukemia virus (AMLV), and the firefly luciferase-expressing vector at a DNA ratio of 1:1:3 μg. For the production of viruses pseudotyped with the AMLV glycoprotein, a rev-expressing plasmid was added to ensure the expression of the HIV-1 proteins from the packaging plasmids. The single-round, replication-defective viruses were harvested in the supernatants 24 to 30 h after transfection, filtered (0.45 μm), aliquoted, and frozen at −80°C until further use. The reverse transcriptase (RT) activities of all viruses were measured as described previously (35).
Assay of virus infectivity and drug sensitivity.
Cf2Th/CD4-CCR5 target cells were seeded at a density of 6 × 103 cells/well in 96-well luminometer-compatible tissue culture plates (Perkin Elmer, Waltham, MA) 24 h before infection. On the day of infection, compounds of interest (1 to 100 μM) were added to recombinant viruses (10,000 RT units) in a final volume of 50 μl and incubated at 37°C for 30 min. The medium was removed from the target cells, which were then incubated with the virus-drug mixture for 2 to 4 h at 37°C. At the end of this period, complete medium was added to a final volume of 150 μl and incubated for 48 h at 37°C. The medium was removed from each well, and the cells were lysed with 30 μl of passive lysis buffer (Promega, Madison, WI) by three freeze-thaw cycles. An EG&G Berthold Microplate LB 96V luminometer (Berthold Detection Systems, Pforzheim, Germany) was used to measure luciferase activity in each well after the addition of 100 μl of luciferin buffer (15 mM MgSO4, 15 mM KPO4 [pH 7.8], 1 mM ATP, 1 mM dithiothreitol) and 50 μl of 1 mM d-luciferin potassium salt (BD Pharmingen, San Diego, CA).
Anti-HIV efficacy evaluation in MAGI cell lines.
P4-R5 MAGI cells (NIH AIDS Research & Reference Reagent Program, catalog number 3580) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, sodium bicarbonate (0.05%), and antibiotics (penicillin, streptomycin, and kanamycin at 40 μg/ml each and puromycin at 1 μg/ml). HIV-1 infection of P4-R5 MAGI reporter cells, which express CD4, CXCR4, and CCR5, results in long-terminal-repeat-directed β-galactosidase expression, which can be readily and accurately quantified. Approximately 18 h prior to the experiment, P4-R5 MAGI cells were plated at a concentration of 1.2 × 104 cells/well in a flat-bottom 96-well plate. On the day of the experiment, cells were infected in quadruplicate with HIV-1 strain IIIB (Advanced Biotechnologies, Columbia, MD) in the presence or absence of putative CA inhibitors at the indicated concentrations. After 48 h of incubation at 37°C, cells were assayed for infection using the Galacto-Star One-Step β-galactosidase reporter gene assay system (Applied Biosystems, Bedford, MA). Each IC50 (concentration at which exposure to the compound resulted in a 50% decrease in infection relative to mock-treated, HIV-1-infected cells) was calculated using the Forecast function of Microsoft Excel (Redmond, WA).
Anti-HIV-1 efficacy evaluation in human peripheral blood mononuclear cells.
Assays involving HIV-1 infection of human peripheral blood mononuclear cells (PBMCs) were performed as described previously (19, 34). Fresh PBMCs, seronegative for HIV-1 and hepatitis B virus, were isolated from blood samples of the screened donors (Biological Specialty Corp., Colmar, PA) by using lymphocyte separation medium (density, 1.078 ± 0.002 g/ml; Cellgro; Mediatech, Manassas, VA) and following the manufacturer's instructions. Cells were stimulated by incubation in 4 μg/ml phytohemagglutinin (PHA; Sigma) for 48 to 72 h. Mitogenic stimulation was maintained with the addition of 20 μM/ml recombinant human interleukin-2 (R&D Systems, Minneapolis, MN) to the culture medium. PHA-stimulated PBMCs from at least two donors were pooled, diluted in fresh medium, and added to 96-well plates at 5 × 104 cells/well. Cells were infected (final multiplicity of infection [MOI] of ∼0.1) in the presence of nine different concentrations of test compounds (triplicate wells for each concentration) and incubated for 7 days. To determine the level of virus inhibition, cell-free supernatant samples were collected for analysis of RT activity (4). Following removal of supernatant samples, compound cytotoxicity was measured by the addition of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (CellTiter 96 reagent; Promega) according to the manufacturer's instructions.
Virus isolates.
All virus isolates were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (see the supplemental material for details).
Overproduction and purification of His-tagged wild-type and mutant HIV-1 CA.
The expression vector for C-terminally His-tagged HIV-1NL4-3 CA was a generous gift from Eric Barklis (Department of Molecular Microbiology and Immunology, Oregon Health and Science University, Portland, OR). CA-H6 protein was overproduced in Escherichia coli strain BL21(DE3) Codon+-RIL (Stratagene, La Jolla, CA) by autoinduction overnight in ZYP-5052 medium (41). The CA-H6 protein was subsequently purified using immobilized metal affinity chromatography on a Talon cobalt resin affinity column (Clontech Laboratories, Mountain View, CA), dialyzed against 20 mM Tris-HCl (pH 8.0), concentrated to 120 μM, flash frozen in liquid nitrogen, and stored at −80°C until further use. Individual alanine mutations were introduced in to the wild-type C-terminally His-tagged HIV-1NL4-3 CA expression vector by site-directed mutagenesis. Mutant CA proteins were purified as described above for the wild-type CA protein.
Surface plasmon resonance (SPR) binding assays.
Interaction analyses were performed on a Biacore 3000 optical biosensor (Biacore, Piscataway, NJ) with simultaneous monitoring of two flow cells. Immobilization of the CA protein to CM7 sensor chips was performed following the standard amine coupling procedure according to the manufacturer's specifications. A reference surface on which the nonspecific anti-gp120 antibody 17b (43) was immobilized was used as a background to correct nonspecific binding and for instrument and buffer artifacts.
Direct binding of compounds to HIV-1 CA.
Stock solutions of I-XW-053 and NBD-556 were prepared by dissolving them in 100% dimethyl sulfoxide (DMSO) to a final concentration of 10 mM. To prepare the sample for analysis, 30 μl of the compound stock solution was added to sample preparation buffer (25 mM Tris-HCl, 150 mM NaCl [pH 7.5]) to a final volume of 1 ml and mixed thoroughly. Preparation of analyte in this manner ensured that the concentration of DMSO matched that of running buffer with 3% DMSO. Lower concentrations of each compound were then prepared by 2-fold serial dilutions in running buffer (25 mM Tris-HCl, 150 mM NaCl, 3% DMSO [pH 7.5]). These compound dilutions were then injected over the control and CA surfaces at a flow rate of 50 μl min−1, for a 2-min association phase, followed by a 5-min dissociation phase. Specific regeneration of the surfaces between injections was not needed owing to the nature of the interaction.
Binding site analysis via SPR.
Wild-type and mutant HIV-1 CA proteins were attached to the surface of a CM7 chip by standard amine chemistry as described above. Compound I-XW-053 was injected over these surfaces at a concentration of 27.5 μM at a flow rate of 50 μl min−1, for a 2-min association phase, followed by a 5-min dissociation phase, and the response at equilibrium recorded. For comparison, and to take into account minor differences in the ligand density of the mutant surfaces, responses were normalized to the theoretical Rmax (maximum analyte binding capacity of the surface in RU), assuming a 2:1 interaction.
SPR data analysis.
Data analysis for the direct binding studies was performed using BIAEvaluation 4.0 software (Biacore Life Sciences). The responses of a buffer injection and responses from the reference flow cell were subtracted to account for nonspecific binding. In order to obtain the equilibrium dissociation constants (KD), experimental data were fitted globally to the heterogeneous ligand model. The average parameters generated from a minimum of four data sets were used to define the equilibrium dissociation constants (KD1 and KD2). In studies of the mutant CA protein, the maximum response was recorded from a minimum of six data sets and was used to define the average maximum response for compound I-XW-053 binding to wild-type and mutant HIV-1 CA proteins.
Isothermal titration calorimetry.
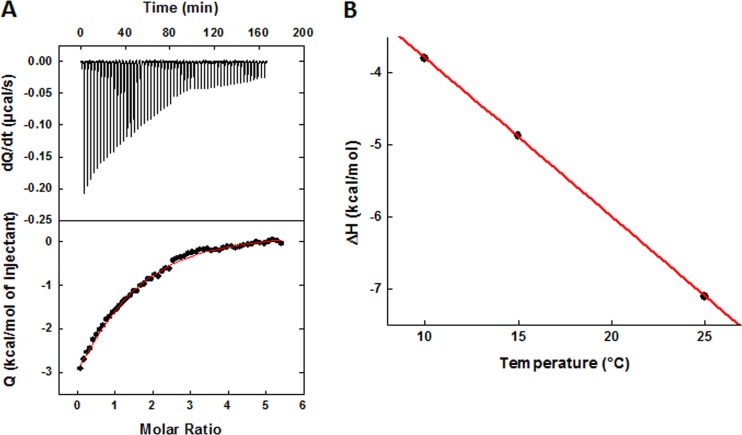
Isothermal titration calorimetric experiments were performed at 10°C, 15°C, and 25°C using a high-precision ITC200 (isothermal titration calorimetry) system from MicroCal LLC/GE Healthcare (Northampton, MA). All titrations were performed by adding I-XW-053 in steps of 1.4 μl. All solutions contained within the calorimetric cell and injector syringe were prepared in the same buffer (25 mM Tris-HCl [pH 7.5], with 150 mM NaCl and 3% DMSO). The concentrations of CA and I-XW-053 were 35 and 600 μM, respectively, at 10°C and 15°C and 65 and 800 μM, respectively, at 25°C. In order to saturate CA with the inhibitor, the syringe had to be refilled after 28 injections. The heat evolved upon injection of I-XW-053 was obtained from the integral of the calorimetric signal. The heat associated with the binding reaction was obtained by subtracting the heat of dilution from the heat of reaction. The individual heat values were plotted against the molar ratio, and the values for the number of binding sites (n), the enthalpy change (ΔH), and dissociation constant (KD = 1/KA) were obtained by nonlinear regression of the data.
Quantitative real-time PCR of viral reverse transcripts.
Purified monocytes from PBMCs of normal healthy donors were purchased from the University of Pennsylvania Immunology core facility. The monocytes were differentiated by using medium containing 50 ng/ml macrophage colony-stimulating factor for 5 to 7 days to obtain adherent monocyte-derived macrophages (MDMs). MDMs, P4-R5 cells, and U87.CD4.CXCR4 cells were treated with 100 μM I-XW-053 or 250 nM azidothymidine (AZT) (NIH AIDS Research and Reference Reagent Program) or DMSO and infected with HIV-1 particles pseudotyped with the BaL envelope for 24 h. Following infection, full genomic DNA was isolated from infected MDMs or cell lines using a blood and tissue kit from Qiagen (Valencia, CA). To detect U5Ψ late products (amplification of a product corresponding to the completion of reverse transcription prior to nuclear import), PCR was carried out using primers MH531 (5′-TGTGTGCCCGTCTGTTGTGT-3′) and MH532 (5′-GAGTCCTGCGTCGAGAGATC-3′) along with probe LRT-P (5′-FAM-CAGTGGCGCCCGAACAGGGA-TAMRA-3′).
With each experiment, a standard curve of serially diluted HIV backbone plasmid pNL4-3 plus a no-template control were run for absolute quantitation of copy number in the experimental samples using the method of Liszewski et al. (22).
All reactions were performed using 200 ng of DNA with Applied Biosystems (Carlsbad, CA) TaqMan Universal Master Mix and run using an Applied Biosystems 7300 real-time PCR system as described elsewhere (5).
RESULTS
Identification of CK026 as an early-stage inhibitor of HIV-1 replication.
Small-molecule inhibitors targeted to the NTD-NTD hexameric interface of HIV-1 CA were designed using the HSB method. Screening with the hybrid pharmacophore resulted in 900 hits that were filtered for drug-like properties. The molecules were also screened using principal-component analysis to identify those with unique chemical cores, which resulted in 300 hits. These molecules were then docked into the NTD-NTD interface region of a CA monomer and scored using GoldScore, ChemScore, and a customized scoring scheme. From the 300 docked complexes, the 25 best-ranking molecules were purchased for analysis of antiviral activity using single-round infection assays. Details of the single-round infection assay have been published in detail elsewhere, and the method has been routinely used for phenotypic characterization of HIV-1 envelope glycoproteins and studies of inhibitors of HIV-1 replication (23, 24, 38). Effects on early-stage events by the compounds were determined by producing virus in the absence of compound and then exposing target cells to virus in the absence or presence of various concentrations of compounds.
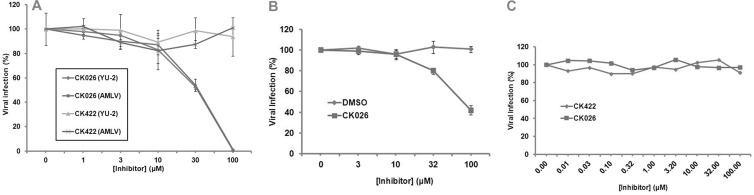
From this initial screen, one compound, 4,4′-[dibenzo[b,d]furan-2,8-diylbis(5-phenyl-1H-imidazole-4,2-diyl)]dibenzoic acid (CK026), was identified as having anti-HIV-1 activity with an IC50 of 33.3 ± 0.31 μM on the infection of recombinant luciferase-containing HIV-1 viruses (HIV-1NL4-3 backbone) pseudotyped with the envelope protein from HIV-1YU-2. CK026 also inhibited HIV-1 pseudotyped with the envelope glycoprotein from AMLV, indicating that its action was postentry (Fig. 1A). Although production of pseudovirions by transfection and the ability to analyze inhibition in a single-round infection are advantageous for addressing the inhibitory effect of a given compound, this type of assay cannot address the effects of multiple rounds of infection and cell-to-cell spread on the efficacy of the test compounds. We therefore sought to address whether CK026 could inhibit the replication of fully infectious virus (Fig. 1B). The compound was assessed against fully infectious HIV-1IIIB replicating in the P4-R5 MAGI cell line. This analysis demonstrated that CK026 did inhibit the replication of this isolate with an IC50 of 89 ± 3.2 μM. The P4-R5 MAGI cell line is an HeLa derivative and is therefore not a natural target cell type. Moreover, HIV-1IIIB is a laboratory-adapted virus that has been multiply passaged in culture and lacks some of the accessory proteins. We therefore wished to test whether CK026 could inhibit a primary isolate, HIV-192BR030, replicating in primary PBMCs. However, the compound displayed no activity in the PBMC assay (Fig. 1C), despite being stable in the medium over the course of the experiment.
Fig 1.
Effect of compound CK026 on the replication of single- and multiple-round infectious HIV-1. (A) Disruption of infection by CK026 at early and postentry stages as shown by single-round infection assays. Shown are the effects of compound CK026 on the infection of Cf2Th-CCR5 cells by recombinant luciferase-expressing HIV-1 bearing the envelope glycoprotein of the HIV-1YU-2 strain or amphotropic murine leukemia virus (AMLV). Virus infection is expressed as the percentage of infection (measured by luciferase activity in the target cells) observed in the presence of compound relative to the level of infection observed in the absence of the compound. Data are means from three replicates. IC50 for compound CK026 against HIV-1 was demonstrated to be 33.3 ± 0.31 μM. Compound CK422 was included as a compound control, as it has previously been determined not to have any effect on HIV-1 infection. (B) Effect of CK026 on replication of HIV-1IIIB in the primary peripheral HeLa P4-R5 MAGI cell line. (C) CK026 does not affect replication of HIV-192BR030 in primary peripheral blood mononuclear cells. CK422 is a compound that was tested in the screen and did not have early-stage activity. Error bars show 1 standard deviation.
Size reduction and optimization of CK026 (identification of I-XW-053).
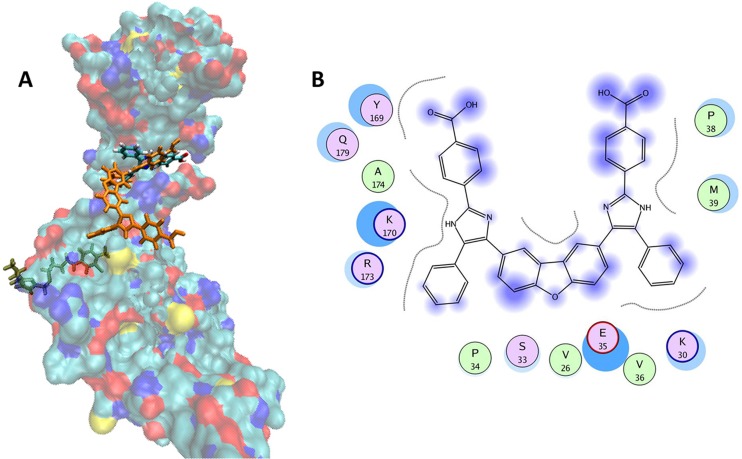
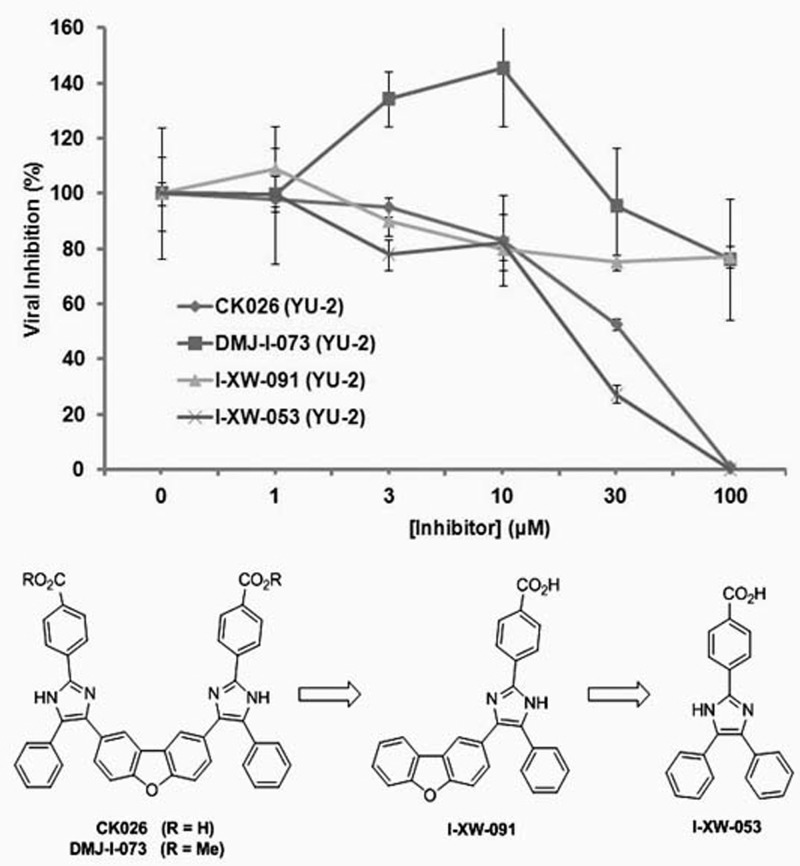
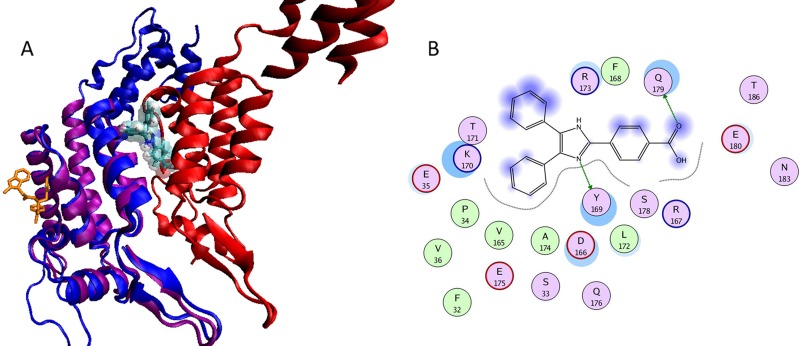
Because compound CK026 displayed activity in single- and multiple-round infection assays using cell lines, we reasoned that the compound may be poorly permeative across the PBMC membrane. This is probably a function of the poor drug-like properties (high octanol-water partition coefficient [log(P)] of 9.31, as determined using the weighted log(P) function in Jchem; ChemAxon, Budapest, Hungary) and the high molecular mass (692 Da). Therefore, we sought to reduce the size of the compound, optimize the physicochemical properties, and improve the permeability while retaining the antiviral activity. CK026 has C2 symmetry along the central dibenzofuran ring. Docking results suggested that the proposed binding area of CK026 spans the entire NTD-NTD interface region, including the junction between the N- and C-terminal lobes (Fig. 2). Based on the docking model, the upper arm of CK026 is proposed to interact with residues Arg173, Asp166, Lys170, Tyr169, Glu180, Gln179, Ser33, and Pro34 and the lower arm with Pro38, Met39, Glu35, Lys30, and Val36. Owing to the symmetry, docking solutions indicated that either arm could occupy either of the two sites; there was no preference for one arm over the other. In order to test these docking observations, two analogues of CK026 (Fig. 3; also, see Scheme 1 in the supplemental material) were designed. The first, I-XW-091, is composed of the furan ring linker region attached to one arm of the parental molecule (Fig. 3; also, see Scheme 2 in the supplemental material), whereas the other, I-XW-053, corresponds to only the arm structure (Fig. 3; also, see Scheme 3 in the supplemental material). In addition, the benzoic acid moiety on CK026 was predicted from the docking pose to form hydrogen bond interactions with Gln179 and Glu180 of CA. We therefore tested this prediction—specifically, how critical these potential interactions are to the antiviral activity of CK026—with the synthesis of DMJ-I-073, a dimethyl ester variant of CK026, which removed the hydrogen-bonding capability at this region (Fig. 3; also, see Scheme 4 in the supplemental material). These compounds were then subjected to antiviral analysis using the single-round infection assay. DMJ-I-073 lost all activity in the single-round infection assay (Fig. 3), indicating that potential hydrogen bonds formed by the benzoic acid moiety in CK026 are critical for activity. I-XW-091 also lost antiviral activity, perhaps owing to poor solubility in aqueous media. Compound I-XW-053, however, retained all of the activity of the parental molecule, reducing the replication of both YU-2 and AMLV (data not shown) pseudotyped viral particles. I-XW-053, which retained the antiviral activity of CK026, represents a significant reduction in molecular mass (692 versus 340 Da) and an improvement in physicochemical properties [log(P) of 9.31 versus 5.05]. We therefore decided to test I-XW-053 in the PBMC assay. I-XW-053 had IC50s for inhibition of a broad range of HIV-1 isolates replicating in primary PBMCs comparable to that exhibited by the parental CK026 against HIV-1IIIB replicating in P4-R5 MAGI cells (see Table S1 in the supplemental material). Moreover, this inhibition was specific to HIV-1, as I-XW-053 displayed no appreciable efficacy against the related retrovirus SIVmac239 (simian immunodeficiency virus), or against a panel of viruses from different classes (see Table S2 in the supplemental material).
Fig 2.
Proposed binding mode of CK026 to the HIV-1NL4-3 capsid protein. (A) Surface representation of the monomeric unit of CA protein colored by atom type (C in cyan, O in red, N in blue, and S in yellow). CK026, I-XW-053, and a known CA inhibitor, CAP-1, are docked in their predicted binding sites and are shown as licorice models that are orange, colored by atom type, and green, respectively. (B) Proposed binding mode of CK026 in CA. The binding site residues are colored by their nature, with hydrophobic residues in green, polar residues in purple, and charged residues highlighted with bold contours. Blue spheres and contours indicate matching regions between ligand and receptors. The figure was generated using the MOE ligX module.
Fig 3.
Comparison of the effects of compounds CK026, DMJ-I-073, I-XW-091, and I-XW-053 on the infection of Cf2Th-CCR5 cells by recombinant luciferase-expressing HIV-1 bearing the envelope glycoprotein of the HIV-1YU-2 strain. Virus infection is expressed as the percentage of infection (measured by luciferase activity in the target cells) observed in the presence of compound relative to the level of infection observed in the absence of the compound. Data are means from three replicates. Error bars show 1 standard deviation. The IC50 for compound I-XW-053 against HIV-1 was demonstrated to be 22.5 ± 1.1 μM. The chemical structures of CK026, DMJ-I-073, I-XW-091, and I-XW-053 are shown.
I-XW-053 directly binds to HIV-1 CA: mutation of residues required for interaction affects early, preintegration events in the viral life cycle.
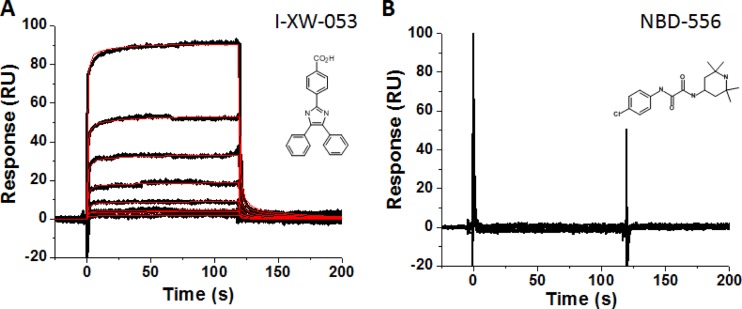
Compound I-XW-053 is predicted to interact with the NTD of HIV-1 CA, based on computer docking solutions. However, it is possible that the compound could exert the inhibitory activity via another mechanism not involving CA. We therefore sought to confirm the interaction of I-XW-053 with HIV-1 CA using SPR. Wild-type HIV-1 CA protein was purified as outlined in Materials and Methods and immobilized on the surface of a high-capacity CM7 sensor chip. A surface to which the monoclonal antibody 17b (a generous gift from James E. Robinson, Department of Pediatrics, Tulane University Medical Center, New Orleans, LA) was immobilized was used to correct for background binding and instrument and buffer artifacts. I-XW-053 directly interacted with sensor chip-immobilized HIV-1 CA (Fig. 4A). In contrast, the small-molecule CD4 mimetic compound NBD-556 displayed no such interaction with HIV-1 CA, establishing the specificity of I-XW-053 for HIV-1 CA (Fig. 4B). Interestingly, fitting of the SPR data indicated that I-XW-053 interacts with HIV-1 CA with a 2:1 stoichiometry.
Fig 4.
Sensorgrams depicting the interaction of (A) I-XW-053 and (B) NBD-556 with sensor chip-immobilized HIV-1NL4-3 CA. I-XW-053 at concentrations in the range from 0.86 to 110 μM are shown. Black lines indicate experimental data, whereas red lines indicate fitting to the heterogeneous ligand model. The individual rate constants were out of the dynamic range of the instrument. The equilibrium dissociation constants are as follows: KD1 = 66.3 ± 4.8 μM; KD2 = 66.3 ± 5.2 μM. The chemical structures of each compound are shown as insets.
The binding of I-XW-053 to HIV-1 CA was further characterized thermodynamically by ITC. Figure 5A shows the calorimetric titration of HIV-1NL4-3 CA with I-XW-053 at 25°C in Tris-HCl, 150 mM NaCl with 3% DMSO (the exact buffer used for the SPR experiment). The experimental data fitted to a binding model wherein two molecules of I-XW-053 bind to one CA molecule with equal affinity, supporting the 2:1 stoichiometry obtained from SPR analysis. The affinity of the I-XW-053–CA interaction at 25°C was determined to be 85 μM, corresponding to a change in Gibbs energy of −5.6 kcal/mol of I-XW-053. The changes in enthalpy (ΔH) and entropy (ΔS) were −7.1 kcal/mol and −5.0 cal/(K × mol), respectively, and the change in heat capacity, calculated from temperature dependence of the enthalpy, was −220 cal/(K × mol) (Fig. 5B). The measured thermodynamic parameters are typical for a small molecule that binds without inducing major conformational changes in the target protein (26, 29).
Fig 5.
(A) Calorimetric titration of HIV-1NL4-3 CA with I-XW-053 at 25°C in Tris-HCl, 150 mM NaCl with 3% DMSO. The concentration of CA was 35 μM, and the syringe contained I-XW-053 at a concentration of 600 μM. The experimental data fit with a binding model whereby two molecules of I-XW-053 bind to one CA, each with a binding affinity of 85 μM, which corresponds to a change in Gibbs energy of −5.6 kcal/mol. The changes in enthalpy (ΔH) and entropy (ΔS) are −7.1 kcal/mol and −5.0 cal/(K × mol), respectively. (B) Temperature dependence of the enthalpy of binding of I-XW-053 to CA. The slope corresponds to a heat capacity change of −220 cal/(K × mol).
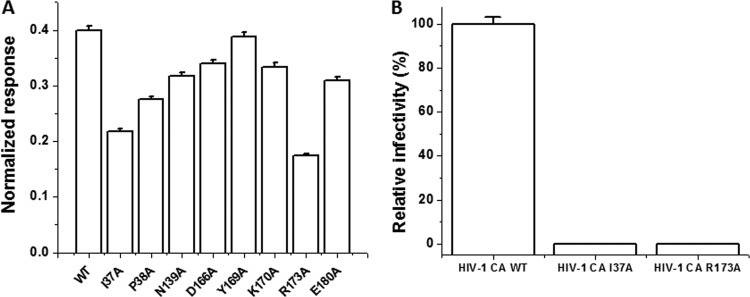
The combined results from SPR and ITC studies indicated that compound I-XW-053 binds to HIV-1 CA with a 2:1 stoichiometry. This finding is consistent with results obtained from docking studies using I-XW-053 that imply that I-XW-053 could potentially interact with both upper and lower regions of the NTD. Therefore, to further investigate the potential binding site(s) of I-XW-053 on CA, we created mutations in the HIV-1 CA protein based on the docking models. Residues Ile37, Pro38, Asn139, Asp166, Tyr169, Lys170, Arg173, and Glu180 were individually mutated to alanine, and the effect of the mutagenesis on the binding of I-XW-053 at a single concentration (27.5 μM) relative to wild-type CA was assessed using SPR. From this analysis, mutation of residues Ile37, Pro38, Asn139, or Arg173 reduced the binding of I-XW-053 compared with wild-type CA to various degrees, with residues Ile37 and Arg173 having the most pronounced effect (a 2-fold reduction or greater) (Fig. 6A). To test whether mutation of these residues translated to a reduction in antiviral efficacy of the compound, we introduced two CA mutations (Ile37Ala and Arg173Ala; these mutations resulted in the most pronounced decrease in I-XW-053 binding) into the proviral backbone and performed single-round infection assays. Both mutations caused only a moderate reduction in the amount of virus produced, as judged by RT content, but showed dramatic reduction in infectivity (Fig. 6B). The pronounced reduction in infectivity precluded testing the efficacy of the compound against these mutated viruses. A similar situation with NTD mutations was recently observed (28). Our results therefore demonstrate that the binding of I-XW-053 to HIV-1 CA is dependent on interactions with residues within the NTD and that mutation of residues required for interaction with the compound negatively affects early, preintegration events. Moreover, these results point to a site(s) of interaction not seen among previously discovered CA inhibitors (Fig. 7).
Fig 6.
(A) Effect of mutation of capsid residues in and around the proposed I-XW-053 binding site on compound binding. The interaction of I-XW-053 at a concentration of 27.5 μM with wild-type and mutant versions of the CA protein was assessed using SPR. To allow comparison, responses at equilibrium were normalized to the theoretical Rmax, assuming a 2:1 interaction. (B) Infection efficiency of mutations made within the HIV-1NL4-3 CA provirus. 293T cells were transfected with either wild-type pNL4-3 CA or pNL4-3 backbone containing a single amino acid change within CA (Ile37Ala or Arg173Ala). Pseudoviral supernatants were normalized to reverse transcriptase content before infection of U87.CD4.CXCR4 target cells. Results were measured by luciferase activity and were normalized to the infectivity of wild-type pNL4-3 CA and plotted, with standard deviations shown by error bars. Note that the standard deviation for the I37A mutant was 0.002 and that for the R173A mutant was 0.001 (not visible on the histograms).
Fig 7.
(A) Comparison of the proposed binding site of I-XW-053 with the binding site of compound PF74. Structural superpositioning of cocrystallized PF74 with NTD of CA protein on CA dimers. The protein is represented in a cartoon model, and the protomers are colored purple, green, and red. PF74 is represented in an orange licorice model, and I-XW-053 is docked at the proposed binding site and is represented in a licorice model and colored by atom type (C in cyan, N in blue, O in red, and H in white). The two binding sites are distinct and opposite to each other. The van der Waals surface model of I-XW-053 clearly shows that I-XW-053 sterically clashes with one of the CA protomers (in red) and hence blocks the assembly of the CA protein. (B) I-XW-053 in the binding site of CA monomer. The binding site residues are colored by their nature, with hydrophobic residues in green, polar residues in purple, and charged residues highlighted with bold contours. Blue spheres and contours indicate matching regions between ligand and receptors. Hydrogen-bonded interactions are shown by green arrows. The figure was generated using the MOE ligX module.
Viruses in the presence of I-XW-053 cannot effectively complete reverse transcription.
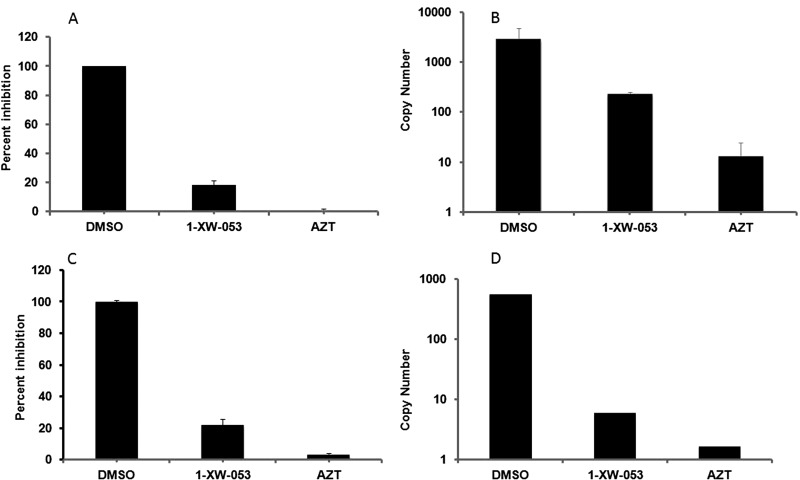
Having demonstrated that I-XW-053 blocks HIV replication at an early, preintegration stage, using the single-round infection assay, and that the compound interacts with HIV-1 CA, we sought to link the mechanism of action to early, CA-mediated processes. Uncoating and reverse transcription have been shown to be intimately related, with the hallmark of the disruption of uncoating being the inability of the virus to complete reverse transcription (8, 13). Unintegrated viral DNA synthesized during HIV-1 infection includes linear and circular forms, and each of these distinct viral cDNAs can be used as a surrogate marker for events surrounding the completion of reverse transcription and for nuclear import of viral DNA during replication (25). We therefore performed real-time PCR to monitor the amounts of HIV-1 late RT products, to assess the effect of compound I-XW-053 upon completion of reverse transcription in different cell types. As can be seen in Fig. 8 and in Fig. S2 in the supplemental material, infection of P4-R5, U87.CD4.CXCR4, or MDMs in the presence of 100 μM I-XW-053 resulted in a drastic reduction in the amount of late RT products that could be detected. Although there are cell type-specific variations in the levels of U5Ψ products, treatment with I-XW-053 reduces the amount of late RT products by an order of magnitude or more.
Fig 8.
I-XW-053 treatment inhibits HIV-1 reverse transcription in target cells. P4-R5 and U87.CD4.CXCR4 cells were inoculated with HIV-1 particles in the presence of I-XW-053 (100 μM) or 250 nM AZT, and infectivity was measured using a luciferase assay. Inhibition of P4-R5 cells (A) and U87.CD4.CXCR4 cells (C) is shown. Following infection, DNA was harvested from these cells and assayed by quantitative PCR for late RT products as described in Materials and Methods. The level of real-time U5Ψ late RT products formed against copy numbers is shown for P4-R5 cells (B) and for U87.CD4.CXCR4 cells (D). Values are means of duplicate determinations, with error bars depicting one standard deviation.
DISCUSSION
The HIV-1 CA protein plays essential roles in both the early and late stages of viral replication and has recently emerged as an attractive target for drug discovery and development. In the present study, we employed the HSB method to identify small molecules that bind to the capsid NTD-NTD hexamerization interface and that are capable of disrupting HIV-1 replication at an early, preintegration event. Using the HSB method, we obtained 900 hits by pharmacophore-based screening and filtering of a database of more than 3 million compounds. Of these 900 hits, 300 molecules belonging to different chemical cores (identified using principal-component analysis on molecular descriptors derived from MOE [molecular operating environment]) were subjected to further docking and scoring. Finally, the best-ranked complexes were visually inspected for their potential not only to interact with CA but also to effectively disrupt the interaction of CA monomers with each other. Antiviral testing of the top-ranked 25 compounds using a single-round infection assay identified 4,4′-[dibenzo[b,d]furan-2,8-diylbis(5-phenyl-1H-imidazole-4,2-diyl)]dibenzoic acid (CK026) as a CA-targeted compound with the ability to disrupt HIV-1 replication at a postentry, preintegration event. This compound retained the ability to inhibit fully infectious HIV-1IIIB replicating in P4-R5 MAGI cells but was unable to inhibit the primary isolate HIV-192BR030 replicating in PBMCs. We therefore reasoned that this lack of efficacy in the PBMC assay was due to poor permeability in that system resulting from the unfavorable physicochemical properties and the large size of the compound. We therefore attempted to reduce the size of the compound and improve the physicochemical properties, while retaining its antiviral activity. Based upon the docking pose of CK026 to HIV-1 CA, three analogues were designed and synthesized: DMJ-I-073, I-XW-091, and I-XW-053. We subsequently assessed these analogues in the single-round infection assay and found that I-XW-053, the smallest of the analogues, retained all of the inhibitory capability of the parental compound. Having identified I-XW-053, we performed secondary screening using primary isolates replicating in PBMCs. Unlike the parent, CK026, I-XW-053 prevented a range of primary HIV-1 isolates from replicating in PBMCs. The combined use of SPR and ITC demonstrated a specific interaction of I-XW-053 with HIV-1 capsid and implicated residues in the NTD as being critical for this interaction. Moreover, compound I-XW-053 demonstrated little or no cytotoxicity over the concentration range tested (up to 100 μM). Thus, I-XW-053 is HIV-1 specific, showing no inhibition of SIV nor of a panel of nonretroviral DNA and RNA viruses when tested at a highest concentration of 100 μM (see Table S1 in the supplemental material) and possesses broad-spectrum anti-HIV-1 activity (see Table S2 in the supplemental material).
Mutational analysis of potential I-XW-053-interacting residues on CA indicates that residues in the NTD and, more specifically, in the NTD-NTD interfacing region are required for interaction of this compound with the target. This site includes residue Arg173, which is completely conserved across all HIV-1 strains. Importantly, this binding region is in stark contrast to the binding sites of the other previously discovered CA inhibitors. CAP-1, a small-molecule compound initially discovered by the Summers group, was demonstrated by structural analyses to bind in a hidden pocket adjacent to the NTD-CTD interface and to prevent assembly by altering the local geometry required to make NTD-CTD interactions within the hexamer (16, 32). The CAP-1 binding site has subsequently been targeted by other improved CAP-1 derivatives (14, 48) and has also been found to be the binding site for new, more efficacious molecules (20).
Blair et al. (1), using high-throughput screening, identified the novel compound PF-3450074 (PF74). This compound targets CA and inhibits HIV-1 at an early stage. This compound, unlike previous CA inhibitors, apparently increased the rate of assembly of CA in solution (1). Subsequent mechanism-of-action studies, however, demonstrated that PF74 destabilized the preformed capsid cores in vitro and exerted antiviral effects by triggering a premature uncoating of HIV-1 (36). Intriguingly, the binding site of PF74, determined by X-ray crystallography, is a preformed pocket in HIV-1 CA bounded by helices 3, 4, 5, and 7 and situated in the NTD of the CA protein. Despite the similarities in the mechanism of action of PF74 and I-XW-053 (interaction with the NTD and destabilization of the capsid), our data suggest that the I-XW-053 binding site is distinct from that of PF74. Despite the fact that I-XW-053 was found to be able to disrupt CA assembly in vitro (see Fig. S1 in the supplemental material), the action of this compound only on the early stages of infection using the single-round infection assay, in conjunction with the concomitant reduction in late RT products in a number of cell lines, implies a mechanism in which I-XW-053 preferentially disrupts the incoming capsid by interacting with the hexamerization interface. Although these results are seemingly at odds with each other, NYAD1, which was found to prevent the in vitro assembly of the CA protein, was also found to preferentially target HIV-1 replication at an early stage (47). Further studies by our group are under way to determine the mechanism of action of I-XW-053, to define the precise binding site of the compound on HIV-1 CA, and to improve the pharmacokinetic and affinity properties.
In summary, we have identified a new CA inhibitor that interacts with and functions through a novel binding site on HIV-1 CA: the NTD-NTD hexamerization interface. Although this compound is active against HIV-1 in the high micromolar range, it exhibits broad-spectrum anti-HIV-1 activity, inhibiting all of the genetically diverse isolates tested in this study. The compound is also highly specific to HIV-1 in that it has no activity against SIV or other unrelated viruses. These properties, coupled with the compound's promising thermodynamic signature and novel mechanism of action, suggest that I-XW-053 will serve as a good starting point for the development of high-efficacy analogues through optimization by medicinal chemistry approaches. The broad-spectrum activity of this compound is particularly promising, further highlighting the HIV-1 CA protein as a viable viral target with significant therapeutic potential.
Supplementary Material
ACKNOWLEDGMENTS
This project was funded by a grant obtained from the Pennsylvania Department of Health (Tobacco Settlement Formula Funds, SK and SC).
We thank Eric Barklis (Department of Molecular Microbiology and Immunology, Oregon Health and Science University, Portland, OR) for generously providing the CA-H6 expression vector and James E. Robinson (Department of Pediatrics, Tulane University Medical Center, New Orleans, LA) for providing monoclonal antibody 17b. We also thank Robert Siliciano for providing the HIV plasmid for generating integration standards, Una O'Doherty and Erin Graf for their advice and assistance with the real-time PCR experiments, and Diana Winters (Academic Publishing Services, Drexel University College of Medicine) for proofreading the manuscript.
Footnotes
Published ahead of print 30 May 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Blair WS, et al. 2010. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog. 6:e1001220 doi:10.1371/journal.ppat.1001220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reference deleted.
- 3. Reference deleted.
- 4. Buckheit RW, Jr., Swanstrom R. 1991. Characterization of an HIV-1 isolate displaying an apparent absence of virion-associated reverse transcriptase activity. AIDS Res. Hum. Retroviruses 7:295–302 [DOI] [PubMed] [Google Scholar]
- 5. Butler SL, Hansen MS, Bushman FD. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631–634 [DOI] [PubMed] [Google Scholar]
- 6. Curreli F, et al. 2011. Virtual screening based identification of novel small-molecule inhibitors targeted to the HIV-1 capsid. Bioorg. Med. Chem. 19:77–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reference deleted.
- 8. Forshey BM, von Schwedler U, Sundquist WI, Aiken C. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76:5667–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ganser-Pornillos BK, et al. 2011. Hexagonal assembly of a restricting TRIM5alpha protein. Proc. Natl. Acad. Sci. U. S. A. 108:534–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. 2004. Assembly properties of the human immunodeficiency virus type 1 CA protein. J. Virol. 78:2545–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Han Y, et al. 2010. Solid-state NMR studies of HIV-1 capsid protein assemblies. J. Am. Chem. Soc. 132:1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reference deleted.
- 13. Hulme AE, Perez O, Hope TJ. 2011. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc. Natl. Acad. Sci. U. S. A. 108:9975–9980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jin Y, et al. 2010. SAR and molecular mechanism study of novel acylhydrazone compounds targeting HIV-1 CA. Bioorg. Med. Chem. 18:2135–2140 [DOI] [PubMed] [Google Scholar]
- 15. Jones G, Willett P, Glen RC, Leach AR, Taylor R. 1997. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267:727–748 [DOI] [PubMed] [Google Scholar]
- 16. Kelly BN, et al. 2007. Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein. J. Mol. Biol. 373:355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kortagere S, Chekmarev D, Welsh WJ, Ekins S. 2009. Hybrid scoring and classification approaches to predict human pregnane x receptor activators. Pharm. Res. 26:1001–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kortagere S, Welsh WJ. 2006. Development and application of hybrid structure based method for efficient screening of ligands binding to G-protein coupled receptors. J. Comput. Aided Mol. Des. 20:789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lanier ER, et al. 2010. Development of hexadecyloxypropyl tenofovir (CMX157) for treatment of infection caused by wild-type and nucleoside/nucleotide-resistant HIV. Antimicrob. Agents Chemother. 54:2901–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lemke CT, et al. 2012. Distinct effects of two HIV-1 capsid assembly inhibitor families that bind the same site within the N-terminal domain of the viral CA protein. J. Virol. 86:6643–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. 2001. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46:3–26 [DOI] [PubMed] [Google Scholar]
- 22. Liszewski MK, Yu JJ, O'Doherty U. 2009. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 47:254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Madani N, Hubicki AM, Perdigoto AL, Springer M, Sodroski J. 2007. Inhibition of human immunodeficiency virus envelope glycoprotein-mediated single cell lysis by low-molecular-weight antagonists of viral entry. J. Virol. 81:532–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Madani N, et al. 2008. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure 16:1689–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mbisa JL, Delviks-Frankenberry KA, Thomas JA, Gorelick RJ, Pathak VK. 2009. Real-time PCR analysis of HIV-1 replication post-entry events. Methods Mol. Biol. 485:55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mobley DL, Dill KA. 2009. Binding of small-molecule ligands to proteins: “what you see” is not always “what you get.” Structure 17:489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Monroe EB, Kang S, Kyere SK, Li R, Prevelige PE., Jr 2010. Hydrogen/deuterium exchange analysis of HIV-1 capsid assembly and maturation. Structure 18:1483–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noviello CM, et al. 2011. Second-site compensatory mutations of HIV-1 capsid mutations. J. Virol. 85:4730–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ohtaka H, Freire E. 2005. Adaptive inhibitors of the HIV-1 protease. Prog. Biophys. Mol. Biol. 88:193–208 [DOI] [PubMed] [Google Scholar]
- 30. Pertel T, et al. 2011. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472:361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pornillos O, Ganser-Pornillos BK, Banumathi S, Hua Y, Yeager M. 2010. Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus. J. Mol. Biol. 401:985–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pornillos O, et al. 2009. X-ray structures of the hexameric building block of the HIV capsid. Cell 137:1282–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pornillos O, Ganser-Pornillos BK, Yeager M. 2011. Atomic-level modelling of the HIV capsid. Nature 469:424–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ptak RG, et al. 2008. Inhibition of human immunodeficiency virus type 1 replication in human cells by Debio-025, a novel cyclophilin binding agent. Antimicrob. Agents Chemother. 52:1302–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rho HM, Poiesz B, Ruscetti FW, Gallo RC. 1981. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology 112:355–360 [DOI] [PubMed] [Google Scholar]
- 36. Shi J, Zhou J, Shah VB, Aiken C, Whitby K. 2010. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J. Virol. 85:542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shin R, Tzou YM, Wong HC, Krishna NR. 2011. (1)H, (15)N, and (13)C resonance assignments for a monomeric mutant of the HIV-1 capsid protein. Biomol. NMR Assign. [Epub ahead of print.] doi:10.1007/s12104-011-9340-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Si Z, et al. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 101:5036–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stremlau M, et al. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853 [DOI] [PubMed] [Google Scholar]
- 40. Stremlau M, et al. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. U. S. A. 103:5514–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207–234 [DOI] [PubMed] [Google Scholar]
- 42. Tang C, et al. 2003. Antiviral inhibition of the HIV-1 capsid protein. J. Mol. Biol. 327:1013–1020 [DOI] [PubMed] [Google Scholar]
- 43. Thali M, et al. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tian B, et al. 2009. Synthesis and antiviral activities of novel acylhydrazone derivatives targeting HIV-1 capsid protein. Bioorg. Med. Chem. Lett. 19:2162–2167 [DOI] [PubMed] [Google Scholar]
- 45. von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77:5439–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yeager M. 2011. Design of in vitro symmetric complexes and analysis by hybrid methods reveal mechanisms of HIV capsid assembly. J. Mol. Biol. 410:534–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang H, et al. 2008. A cell-penetrating helical peptide as a potential HIV-1 inhibitor. J. Mol. Biol. 378:565–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y, Qian H, Love Z, Barklis E. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.