Abstract
Processing bodies (P-bodies) are highly dynamic cytoplasmic granules conserved among eukaryotes. They are present under normal growth conditions and contain translationally repressed mRNAs together with proteins from the mRNA decay and microRNA (miRNA) machineries. We have previously shown that the core P-body components PatL1, LSm1, and DDX6 (Rck/p54) are required for hepatitis C virus (HCV) RNA replication; however, how HCV infection affects P-body granules and whether P-body granules per se influence the HCV life cycle remain unresolved issues. Here we show that HCV infection alters P-body composition by specifically changing the localization pattern of P-body components that are required for HCV replication. This effect was not related to an altered expression level of these components and could be reversed by inhibiting HCV replication with a polymerase inhibitor. Similar observations were obtained with a subgenomic replicon that supports only HCV translation and replication, indicating that these early steps of the HCV life cycle trigger the P-body alterations. Finally, P-body disruption by Rap55 depletion did not affect viral titers or HCV protein levels, demonstrating that the localization of PatL1, LSm1, and DDX6 in P-bodies is not required for their function on HCV. Thus, the HCV-induced changes on P-bodies are mechanistically linked to the function of specific P-body components in HCV RNA translation and replication; however, the formation of P-body granules is not required for HCV infection.
INTRODUCTION
Hepatitis C virus (HCV), a positive-strand RNA virus of the Flaviviridae family, is a major human pathogen. It has chronically infected around 170 million individuals worldwide and is a major cause of chronic liver disease and liver transplantation (24). Impressive advances in understanding the HCV life cycle have been achieved over the last years. These include the identification of cellular factors necessary for HCV entry into target cells, the elucidation of the viral multiplication requirements, and the description of cellular compartments necessary for particle assembly and progeny virus production (7). These advances not only increase our understanding of fundamental steps in HCV biology but also highlight the intimate relationship of virus components with cellular host factors.
Components of the so-called processing bodies (P-bodies) are among the host factors required for efficient HCV multiplication (5, 20, 26, 37, 38). P-bodies are discrete and highly dynamic granules present in the cytoplasm of eukaryotic cells under normal growth conditions (18, 23). They contain translationally repressed mRNAs together with multiple proteins from the mRNA decay pathway and the microRNA (miRNA) machinery. Once in P-bodies, mRNAs can be either degraded or stored for a later return into translation (9, 12, 15). Experiments involving fluorescence recovery after photobleaching (FRAP) revealed that P-body components rapidly cycle in and out of these granules, indicating that there is a constant exchange of molecules with the cytoplasm, where all P-body components are diffusely present (1, 3, 27). P-body formation requires translationally repressed, ribosome-free mRNAs and some core P-body proteins, such as decapping factors. Consequently, conditions that inhibit the interaction of mRNAs with ribosomes increase P-body formation, while P-bodies rapidly disappear when mRNAs are trapped in complexes with polysomes or when core P-body proteins are depleted (12, 15, 19, 22, 40).
There is growing evidence supporting a complex interplay between P-bodies and viruses (8). However, the precise significance of P-bodies in viral life cycles remains elusive. Since positive-strand RNA viruses have genomes that directly function as mRNA, their interface with P-bodies might be of particular importance. We and others have previously reported that the host P-body components PatL1, the LSm1-7 heptameric ring, and DDX6 (also designated Rck/p54) play essential roles in the HCV life cycle (26, 38). In noninfected cells, PatL1, LSm1-7, and DDX6 form a complex and promote mRNA decay by activating decapping in the 5′-3′ deadenylation-dependent mRNA decay pathway (21). The individual steps of this pathway include mRNA exit from translation, poly(A) tail shortening by deadenylases, decapping via the Dcp1/Dcp2 decapping enzyme, and 5′-3′ degradation via the exonuclease Xrn1. While the precise molecular functions of PatL1, LSm1-7, and DDX6 in this process are not fully understood, it has been suggested that these proteins promote the exit of cellular mRNAs from being actively translated to a translation-repressed state that allows the decapping complex to assemble (13, 14, 41). Additionally, PatL1 has been shown to directly stimulate the decapping enzyme (34). In contrast to this decay function in noninfected cells, PatL1, LSm1-7, and DDX6 are necessary for HCV RNA translation and replication in infected cells (38). Several observations support that this positive regulation is mediated by a direct and specific effect of these factors on HCV. First, depletion of PatL1, LSm1-7, and DDX6 acts on HCV without significantly affecting host physiology (38). Second, in vitro binding assays demonstrated direct interactions of LSm1-7 with essential translation/replication regulatory sequences in the 5′ and 3′ untranslated regions (UTRs) of the HCV genome (38). Third, in HCV-infected cells, DDX6 has been shown to interact with the viral genome and the HCV core protein by coimmunoprecipitation assays (26).
Since PatL1, LSm1-7, and DDX6 are core P-body components required for P-body formation and P-bodies are intimately linked to central steps of the cellular mRNA life cycle, two fundamental questions arise: Are P-bodies affected by an HCV infection? Are P-bodies or just their components required for HCV replication? Addressing these questions, we found that HCV alters P-body composition by specifically changing the localization pattern of P-body components that are required for HCV replication. However, disruption of P-bodies had no effect on HCV infection, indicating that P-body formation per se is not required for HCV infection.
MATERIALS AND METHODS
siRNAs.
Small interfering RNAs (siRNAs) were purchased from Dharmacon or Ambion. The sense sequences were as follows: siRap55, 5′-GGUCUUGGUUUCCGUGGUGuu-3′; siDDX6, 5′-GGAGGAGAGCAUUCCCAUUdtdt-3′; siIrr, 5′-UAGCGACUAAACACAUCAAdtdt-3′. Lowercase indicates noncomplementary nucleosides and nucleotides.
Antibodies.
The antibodies used in this work included the following: mouse anti-NS5A (a gift from Charles M. Rice, Rockefeller University, New York, NY); rabbit anti-DDX6 (MBL Co. Ltd.); rabbit anti-LSm1 and rabbit anti-PatL1 (gifts from Tilmann Achsel, Catholic University of Leuven, Belgium); chicken anti-LSm1 and mouse anti-β-actin (Sigma-Aldrich); rabbit anti-Dcp1, rabbit anti-Edc4, rabbit anti-Xrn1, and rabbit anti-Upf1 (gifts from Jens Lykke-Andersen, University of California San Diego, CA); mouse anti-Dcp1 and rabbit anti-Pcbp2 (Abnova); rabbit anti-Dcp2 (a gift from Megerditch Kiledjian, Rutgers University, New Brunswick, NJ); mouse anti-Rap55 (Novus Biologicals); human anti-Gw182 (a gift from Marvin J. Fritzler, University of Calgary, Canada); rabbit anti-Gemin5 (Bethyl Laboratories); mouse anti-HCV core (Thermo Scientific); rabbit anti-calnexin (Stressgen Bioreagents); Alexa 568 anti-rabbit, Alexa 488 anti-rabbit, Alexa 647 anti-mouse, and Alexa 568 anti-human (Invitrogen); peroxidase anti-human (Sigma-Aldrich); and IRDYE800 anti-rabbit, IRDYE800 anti-chicken, and IRDYE680 anti-mouse (Li-Cor).
Generation of HCV RNA and reporter RNA derivatives.
In vitro transcription was performed using the MEGAScript kit (Ambion) with T3 or T7 polymerase according to the manufacturer's instructions. The plasmids pFL-J6/JFH1/JC1 and pFL-J6/JFH1EMCVIRESaRlucNeo, kindly provided by Apath, LLC (St. Louis, MO) (29), were used to generate the full-length HCV-Jc1 RNA and the HCV subgenomic 2a replicon RNA, respectively. The pFL-J6/JFH1/JC1 plasmid contains the 5′ UTR and nonstructural regions NS3 through NS5B from the JFH1 (genotype 2a) strain (GenBank accession number AB047639) and the region from core through NS2 plus the 3′ UTR from the J6 (genotype 2a) strain (GenBank accession number AF177036). The pFL-J6/JFH1EMCVIRESaRlucNeo plasmid contains the HCV sequences derived from pFL-J6/JFH1/JC1 except that (i) the structural regions from the nucleotide 3281 in the capsid open reading frame (ORF) up to NS3 were replaced by the genes for Renilla luciferase (Renilla reniformis) and neomycin and (ii) the expression of HCV proteins is driven by an encephalomyocarditis virus (EMCV) internal ribosome entry site IRES).
Plasmid LUC cassette clone 2 (a kind gift from Fátima Gebauer) was used to generate Cap-Luc-poly(A) RNA (CAP-Luc-A). This RNA contains a 5′-capped nonviral 5′ UTR followed by the firefly luciferase gene (Photinus pyralis) and a poly (A) tail (38). Plasmid pFL-J6/JFH1/JC1p7Rluc2a (kindly provided by Apath, LLC, St. Louis, MO) was used to generate an HCVcc RNA derivative (Luc-HCV-GNN) that has the same composition as the one derived from pFL-J6/JFH1/JC1 except that (i) a Renilla luciferase gene is fused in frame to the N terminus of J6 NS2 and (ii) a GNN mutation is inserted in the active site of the NS5B gene.
Cell culture, HCV infection, and RNA transfections.
The hepatoma cell lines Huh7.5 and Huh7 (kindly provided by Apath, LLC, and F. Chisari, respectively) were described previously (10, 45). Cell infection was performed at a multiplicity of infection (MOI) of 0.5, using HCV stocks prepared by electroporation of Huh7.5 cells with HCV-Jc1 RNA. Titers ranged from 4 × 104 to 6 × 104 50% tissue culture infective doses (TCID50)/ml. Cells were seeded at 24 h prior to infection to a density of 1 × 105 to 1.6 × 105/ml. Subsequently, they were incubated with HCV stocks for 4 h, washed with phosphate-buffered saline (PBS), and cultured in Dulbecco modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (Sigma-Aldrich) and 1% nonessential amino acids (Invitrogen) until further analysis. RNA transfection was performed using Lipofectamine 2000 according to the manufacturer's recommendations (Invitrogen) or by electroporation as previously described (30).
Knockdown of host factors by RNA interference (RNAi) and drug treatment.
To knock down DDX6 and Rap55, cells were transfected with specific siRNAs at a concentration of 50 nM. Transfection efficiencies were determined by flow cytometry after transfecting cells with fluorescence-labeled siRNAs. Efficient silencing was achieved at 3 to 4 days after transfection (as tested by Western blotting). Cell viability was tested by following both cell growth and accumulation of intracellular ATP levels using CellTiterGlo (Promega). To decrease HCV propagation, cells were treated with the nucleoside analog 2′-C-methyladenosine (2′-C-Me-A), which was resuspended in dimethyl sulfoxide (DMSO) as described before (32).
Titration of infectious particles and luciferase assays.
Titration of infectious particles in the supernatant of HCV-infected cells was performed by limiting-dilution analysis as described previously (29). Replication of the HCV subgenomic 2a replicon was followed by measuring the luciferase activity at various time points after RNA transfection using the dual-luciferase reporter assay system (Promega) according to the manufacturer's recommendations. Values were normalized to the total amount of protein in the sample.
Indirect immunofluorescence, confocal imaging, and data analysis.
Immunofluorescence analysis was performed as previously described (25). Nuclei were labeled with 4′,6-diamidino-2-phenylindole (DAPI) or TOPRO-3 (Invitrogen). Cells were washed with PBS and mounted in Mowiol (Sigma-Aldrich). Images were acquired on a Leica SP2 confocal microscope (Leica). In the indicated experiments, Z-series from 30 randomly selected cells under each condition were collected. In every single cell, the P-body number was quantified with the ImageJ 1.45J software (National Institutes of Health [NIH]) by using the maximum-intensity projection image along the z axis. The quantification was accomplished by applying median and background filters and finally setting a threshold mask with “RenjyEntropy thresholding.” The particles counted were selected by pixel size of between 1 and 100 and circularity of 0.85. Under the setup conditions, every 8 pixels analyzed corresponded to 1 μm. In other experiments, at least 100 cells were analyzed under each condition, and P-body number and size were quantified following the same criteria mentioned above. In that case, the final values for P-body number per cell and their size were obtained from relating the number of P-bodies in each image to the number of nuclei present. To perform colocalization analyses, 30 randomly selected cells were single analyzed under each condition. The criterion to study colocalization was that the maximum distance between the centers of two P-bodies should be equal or to lower than 2 pixels.
RESULTS
HCV infection alters P-body composition.
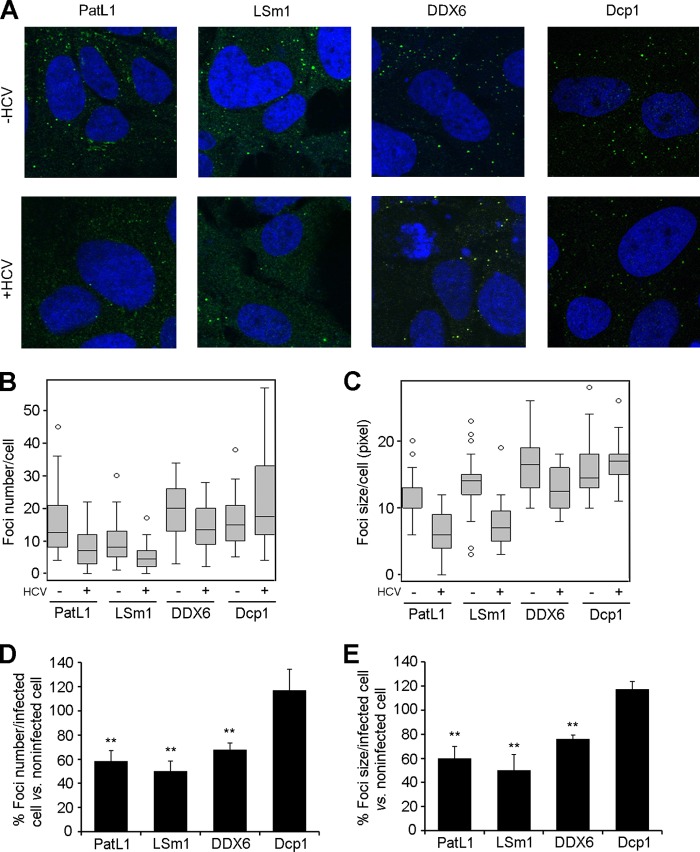
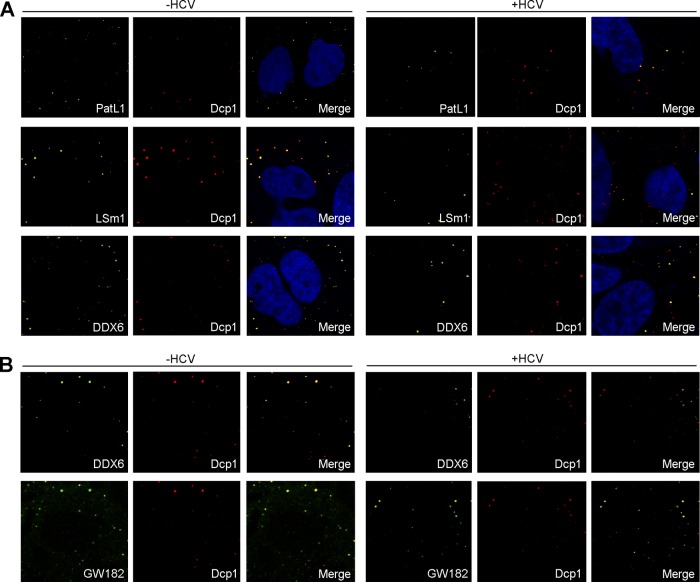
We have previously reported that the translation and replication of HCV RNA require the decapping activators PatL1, LSm1-7, and DDX6 but not the decapping complex Dcp1/Dcp2 or the 5′-3′ exonuclease Xrn1 (38). Since all of these host factors localize in P-bodies and since PatL1, LSm1-7, and DDX6 are core P-body components required for P-body formation in noninfected cells, we tested whether HCV infection affects P-body granules per se. For this, Huh7.5 cells were infected with HCV and then immunostained with antibodies against PatL1, LSm1, DDX6, or Dcp1 at 96 h postinfection, at which time point more than 90% of the cells were NS5A positive by immunostaining (data not shown). P-body size and number were analyzed with the Image J particle-counting software. To detect the LSm1-7 complex, we focused on the LSm1 subunit, since the other subunits when complexed with LSm8 are part of a nuclear complex involved in splicing. HCV infection reduced the abundance of P-bodies that contained PatL1, LSm1, or DDX6 by 42%, 50%, or 32% relative to that in noninfected cells, respectively. A similar pattern was obtained when P-body size was analyzed. Remarkably, no significant decrease in the number or size of Dcp1-containing P-bodies was observed (Fig. 1A to E). The last observation was unexpected, as depletion of PatL1, LSm1, and DDX6 was shown to disrupt P-bodies (18, 39). Thus, either some distinct P-bodies with Dcp1 that are not affected by HCV exist or HCV infection is able to induce alterations of P-body composition. We therefore analyzed the colocalization of PatL1, LSm1, and DDX6 with Dcp1 in noninfected and HCV-infected Huh7.5 cells (Fig. 2A). The percentage of Dcp1 colocalization with PatL1, LSm1, or DDX6 in HCV-infected cells was reduced by 55%, 63% or 53%, respectively, compared to that in noninfected cells. To verify that the remaining P-bodies were not just vestigial foci that contain only Dcp1 protein, a triple staining for Dcp1, DDX6, and GW182 was carried out in noninfected and HCV-infected cells. GW182 is another canonical P-body marker widely used to detect P-bodies. In contrast to the 2-fold reduction in the percentage of Dcp1-DDX6 colocalization, HCV infection had no significant effect on the percentage of Dcp1-GW182 colocalization (Fig. 2B). Together, these results strongly suggest that HCV infection alters P-body composition.
Fig 1.
HCV infection alters P-body composition. (A) Huh7.5 cells were infected for 96 h with HCV and immunostained with antibodies for PatL1, LSm1, DDX6, or Dcp1 (green). Nuclei were visualized using DAPI or TOPRO-3 (blue). Z-series from 30 randomly selected cells under each condition were collected. (B and C) P-body number (B) and size (C) were quantified using the maximum-intensity projection image along the z axis. Graphed are box plots showing medians, upper and lower quartiles, and outliers. (D and E) Results for the number (D) and size (E) of foci containing PatL1, LSm1, DDX6, or Dcp1 in HCV-infected cells were also plotted as percentages relative to those in noninfected cells. Error bars indicate the standard error of the mean (**, P < 0.005).
Fig 2.
Colocalization of different P-body markers after HCV infection. Huh7.5 cells were noninfected or infected with HCV. (A) After 96 h, cells were double immunostained with antibodies against Dcp1 (red) and PatL1, LSm1, or DDX6 (green) (B) or triple immunostained with antibodies against DDX6, GW182 (green), and Dcp1 (red) (B). Note that to clearly show colocalization patterns, the images were displayed as Dcp1-DDX6 and Dcp1-GW182 colocalization pairs. Images correspond to the same cells. Nuclei were visualized using DAPI (blue).
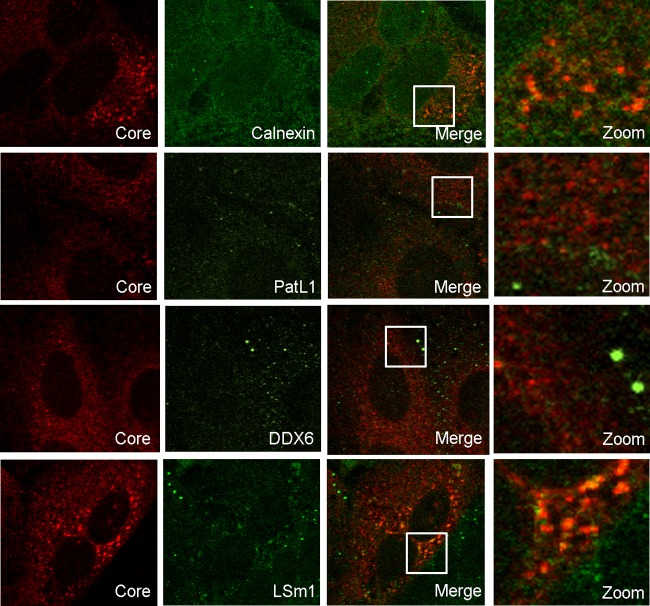
DDX6 has been reported to coimmunoprecipitate with the HCV core protein (26), a viral component that in the case of HCV-Jc1 virus localizes predominantly in the endoplasmic reticulum (ER) (11). In HCV-infected cells, PatL1, LSm1, and DDX6 diffusely relocated into the cytoplasm (Fig. 1A); however, when the intensity of the laser in the microscope was significantly increased, a colocalization of LSm1 with the core protein was visible in 24% of infected cells. In contrast, no colocalization with the core protein was detected for DDX6 or PatL1 (Fig. 3).
Fig 3.
LSm1 colocalizes with the HCV core protein in a subset of HCV-infected cells. HCV-infected Huh7.5 cells were coimmunostained for HCV core protein and calnexin, PatL1, DDX6, or LSm1. Indicated areas in the merged images are magnified in the right column.
P-body formation depends on the cytoplasmic concentration of the P-body components. Therefore, HCV infection could alter P-body composition either by specifically changing the localization pattern of a portion of them or by inhibiting their expression. Western blot analysis of 12 different P-body components, including PatL1, LSm1, DDX6, and Dcp1, showed that their expression levels were unchanged (Fig. 4). This indicates that HCV actively relocates those P-body components required for its replication.
Fig 4.
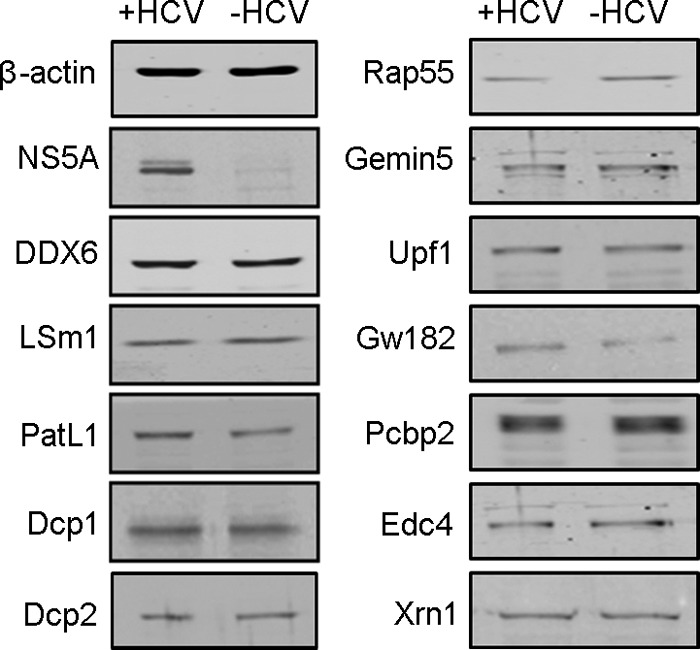
HCV infection does not affect the abundance of P-body components. Western blotting of core P-body components at 96 h after HCV infection is shown. NS5A and β-actin are shown as controls for infection and protein loading, respectively.
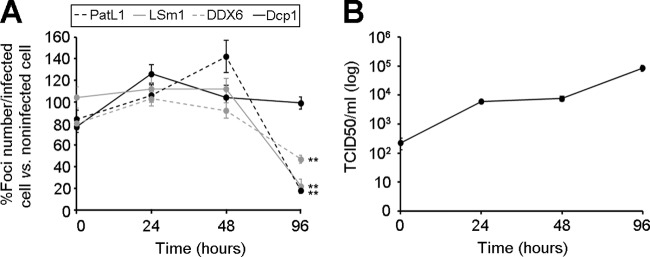
We next examined the effect of HCV at earlier time points postinfection. No significant differences in the amount of P-bodies containing PatL1, LSm1, DDX6, or Dcp1 were observed at 24 and 48 h, when HCV production was at least 10-fold lower than that at 96 h postinfection (Fig. 5). Thus, a certain level of HCV production is required to detect an effect on P-bodies in the HCV-infected cell population.
Fig 5.
Kinetics of P-body abundance during HCV infection. (A) Huh7.5 cells were infected with HCV and at different times postinfection were immunostained to quantify PatL1-, LSm1-, DDX6-, and Dcp1-containing P-bodies in at least 100 cells. Shown are the relative abundances of PatL1-, LSm1-, and DDX6- versus Dcp1-containing P-bodies in HCV-infected relative to noninfected cells during the first 96 h postinfection. The asterisks indicate the time point at which differences were statistically significant (**, P < 0.005). (B) Kinetics of HCVcc infectivity in the supernatant of HCV-infected cells in a representative experiment during the first 96 h of infection. Error bars indicate the standard error of the mean. Note that the error bars in panel B are too small to be visible for some time points.
Reduction of HCV RNA replication in HCV-infected cells restores P-body composition.
Current data support a direct interaction of some P-body components with HCV RNA or HCV proteins (26, 38). Thus, one would anticipate that a reduction of intracellular HCV levels will liberate the respective P-body components and restore P-body composition. To test this hypothesis, HCV-infected cells were treated with 2′-C-methyladenosine (2′-C-Me-A), a nucleoside analog that inhibits HCV replication by blocking the viral polymerase activity (32). A 49% reduction in NS5A expression levels was observed after 48 h of drug treatment, whereas no effect was observed in solvent-treated cells (Fig. 6A). To follow effects on P-body composition, the numbers of Dcp1- and DDX6-containing P-bodies in 2′-C-Me-A-treated versus solvent-treated cells were compared (Fig. 6B). No effect was found for Dcp1-containing P-bodies under any of the analyzed conditions. Importantly, treatment with 2′-C-Me-A specifically restored the amount of DDX6-containing P-bodies in infected cells to the level observed in noninfected cells. Thus, by partially reducing HCV replication, P-body composition could be restored. Again, this suggests that there is an HCV production threshold at which the P-body composition is altered.
Fig 6.
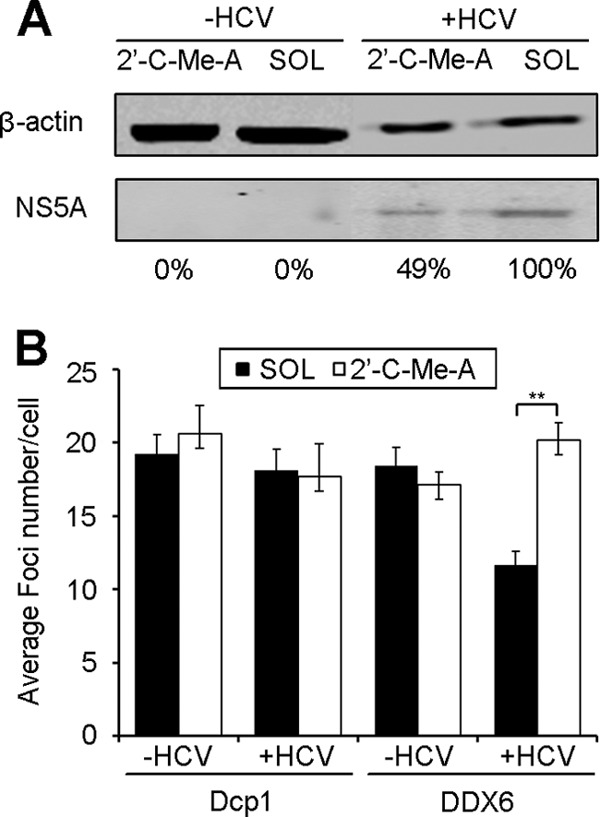
Treatment of HCV-infected cells with a polymerase inhibitor results in relocalization of DDX6 into P-bodies. Huh7.5 cells were infected with HCV for 84 h and treated at 48 h before sampling with 2′-C-methyladenosine (2′-C-Me-A), a nucleoside analog, or with the solvent (SOL) as a control. (A) NS5A protein levels were tested by Western blotting using β-actin as a control for protein loading. Numbers below the panel indicate the percentage of NS5A expression levels relative to those detected in solvent-treated cells. (B) Cells were immunostained as described for Fig. 1A, and P-body numbers in at least 100 cells were quantified. Error bars indicate the standard error of the mean (**, P < 0.005).
HCV RNA translation and replication cause the P-body alterations.
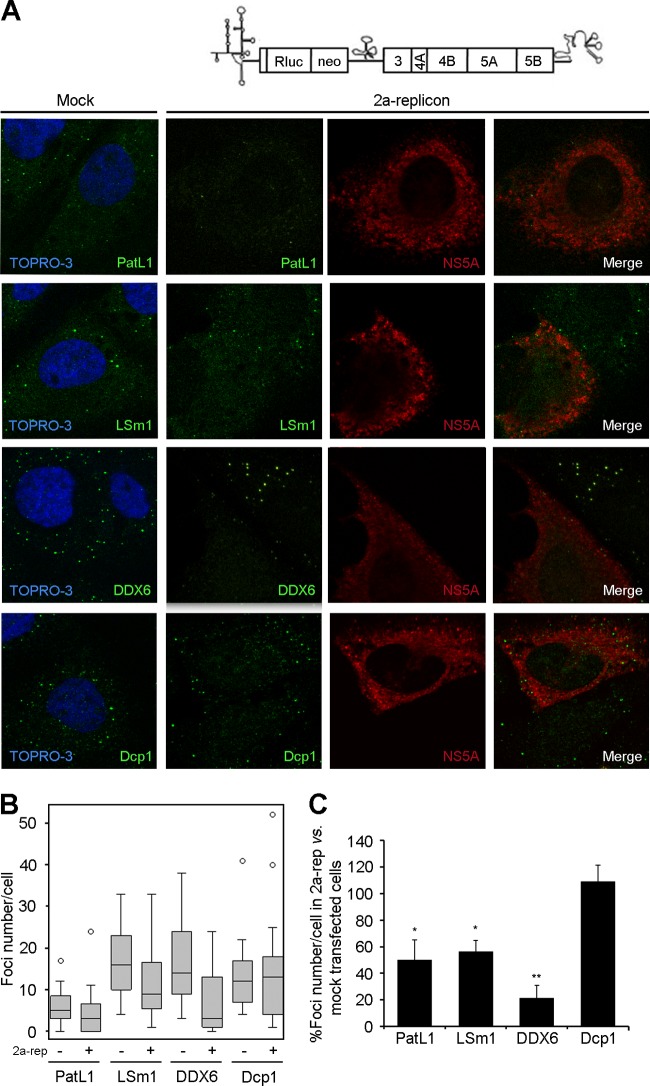
To investigate which steps of the HCV life cycle were responsible for the P-body alterations, we used a subgenomic 2a replicon. In this replicon, the viral structural proteins are replaced by a firefly luciferase reporter and a neomycin phosphotransferase selection marker; therefore, it supports only translation and replication of HCV RNA. Huh7.5 cells were electroporated with the HCV replicon, and P-bodies were analyzed 48 h later, when the maximum luciferase expression level was reached. Because only 20% of these cells harbored the 2a replicon, P-bodies were selectively quantified in replicon-containing cells by coimmunostaining for NS5A and PatL1, LSm1, DDX6, or Dcp1 (Fig. 7A to C). The results paralleled the ones obtained with the infectious virus. In NS5A-positive cells, a statistically significant decrease of the number of P-bodies that contained PatL1, LSm1, and DDX6, but not Dcp1, was observed compared to that in mock-transfected cells. This indicates that HCV RNA translation and/or replication is sufficient to selectively alter the localization pattern of P-body components.
Fig 7.
HCV RNA translation and replication alter P-body composition. (A) Huh7.5 cells were electroporated with a subgenomic 2a replicon RNA and coimmunostained after 48 h to detect NS5A and PatL1, LSm1, DDX6, or Dcp1. (B) Z-series from 30 randomly selected cells under each condition were collected. P-body number was quantified using the maximum-intensity projection image along the z axis. Graphed are box plots showing medians, upper and lower quartiles, and outliers. (C) Results for the number of foci containing PatL1, LSm1, DDX6, and Dcp1 in replicon-transfected NS5A-positive cells were also plotted as percentages relative to those in mock-transfected cells. Error bars indicate the standard error of the mean (*, P < 0.01; **, P < 0.005).
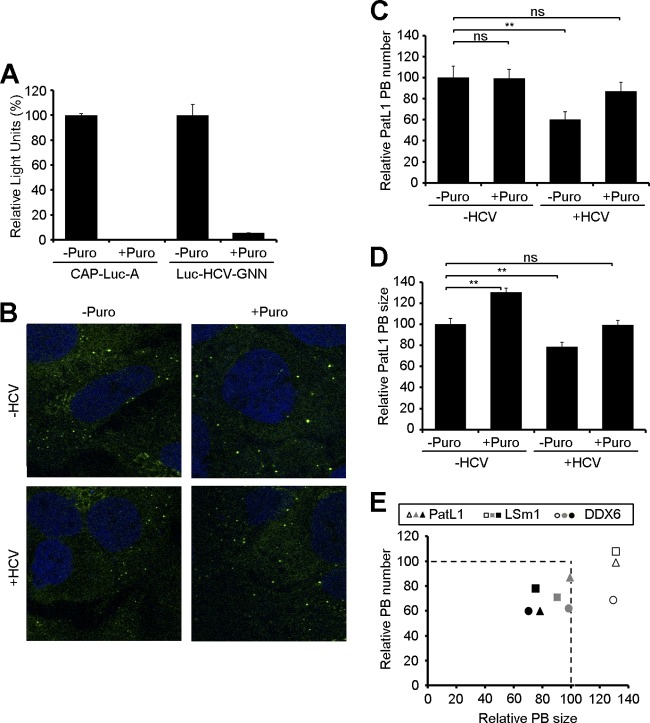
PatL1, LSm1, and DDX6 have been functionally associated with HCV RNA translation and replication. To further assess which of these steps is responsible for the observed changes in P-body composition, HCV-infected cells were incubated with puromycin. This drug inhibits translation by releasing the ribosomal subunits from the mRNAs, thereby increasing the levels of translationally repressed mRNAs, which in turn enhances the number and/or size of P-bodies (15, 19, 40). Consequently, if PatL1, LSm1, and DDX6 were solely associated with translating HCV RNAs, one would expect that puromycin would restore P-body composition. To evaluate the effect of puromycin on cellular and HCV mRNA translation, Huh7.5 cells were electroporated with a CAP-Luc-poly(A) derivative (CAP-Luc-A) or a nonreplicating HCVcc RNA that expresses luciferase protein (Luc-HCV-GNN) and treated for 1 h with puromycin. As expected, the puromycin treatment resulted in a complete translation inhibition of both mRNAs (Fig. 8A). Next, noninfected and HCV-infected Huh7.5 cells were treated with puromycin and the effects on P-body composition analyzed. In noninfected cells, puromycin increased the size of P-bodies compared to that in nontreated controls but did not affect their abundance. In HCV-infected cells, puromycin treatment restored the number of P-bodies to the levels found in noninfected puromycin-treated cells; however, P-body size was not completely recovered, as shown for the PatL1-containing P-bodies in Fig. 8B to D and for the complete P-body markers in Fig. 8E. This suggests that at least a subset of PatL1, LSm1, and DDX6 proteins might be associated with HCV RNA replication.
Fig 8.
Effect of puromycin treatment on P-bodies in HCV-infected cells. (A) Huh7.5 cells were electroporated with a CAP-Luc-poly(A) derivative (CAP-Luc-A) or a nonreplicating HCVcc RNA that expresses luciferase protein (Luc-HCV-GNN). After electroporation, cells were treated for 1 h with 100 μg/ml puromycin, and luciferase activity was measured. (B) Noninfected and HCV-infected cells were treated at 96 h postinfection with 100 μg/ml of puromycin for 1 h and immunostained with antibodies against PatL1. (C and D) The number (C) and size (D) of PatL1-containing P-bodies (PB) from at least 100 cells were quantified and compared to those detected in noninfected, nontreated cells. Error bars indicate the standard error of the mean (*, P < 0.01; **, P < 0.005). (E) Similar results were obtained for DDX6- and LSm1-containing P-bodies, as shown in a composite figure. The number and size of P-bodies containing PatL1 (triangles), LSm1 (squares), or DDX6 (circles) from at least 100 cells were quantified. Shown are the average values for HCV-infected, puromycin-treated cells (gray), HCV-infected, nontreated cells (black), and noninfected, puromycin-treated cells (white) relative to those for noninfected, nontreated cells.
HCV infection does not depend on P-body granules.
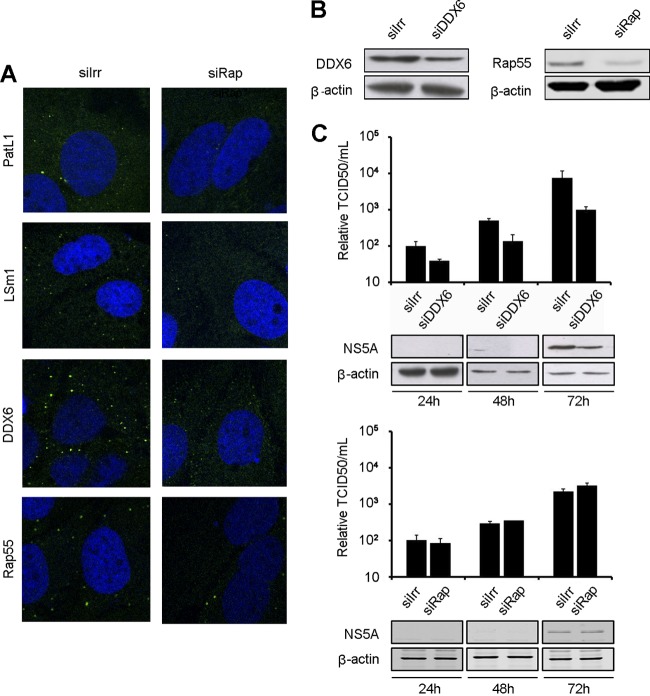
To determine whether the localization of PatL1, LSm1, and DDX6 in P-bodies is a prerequisite for their function in the HCV life cycle, P-body granules were disrupted by depleting a core P-body component, Rap55, and then HCV multiplication was measured. Rap55 is a translation repressor protein necessary for P-body formation but with an unknown function on HCV (42). Depleting Rap55 in Huh7 cells with a specific siRNA resulted in the disruption of P-bodies (Fig. 9A) but did not affect cell growth or intracellular ATP levels (data not shown). Subsequently, we tested whether Rap55 depletion affected HCV production. DDX6 depletion was included as a positive control, as it reduces HCV replication (26, 38). At the time of maximum silencing (Fig. 9B), Huh7.5 cells were transfected with HCV RNA, and NS5A expression and viral titers were analyzed at different time points (Fig. 9C). Rap55 depletion had no effect on either NS5A expression or HCV production. Taken together, these data indicate that disrupting P-bodies does not affect the HCV life cycle and that the localization of PatL1, LSm1, and DDX6 in P-body granules is not necessary for their function on HCV.
Fig 9.
P-body disruption does not affect HCV infection. Huh7.5 cells were transfected with siRNA targeting Rap55 (siRap) or DDX6 (siDDX6) or with nontargeting siRNA (siIrr). (A) Immunostaining with antibodies against DDX6, LSm1, PatL1, or Rap55 (green). Nuclei were visualized using DAPI or TOPRO-3 (blue). (B) Immunoblot analysis of DDX6, Rap55, and β-actin. (C) DDX6 (upper panel)- and Rap55 (lower panel)-silenced cells were transfected with HCV RNA at the time of maximum silencing, and infectivity in the supernatant was monitored for 72 h in a limiting-dilution assay. Intracellular accumulation of HCV NS5A and cellular β-actin was monitored by immunoblotting (panels below graphs). Error bars indicate the standard error of the mean.
DISCUSSION
HCV RNA translation and replication depend on the core P-body components PatL1, LSm1, and DDX6 (26, 38). In this report we demonstrate that HCV infection specifically alters the localization of these components in P-bodies and thereby changes P-body composition. The same observations were made with a subgenomic replicon that does not contain structural viral proteins. This indicates that the HCV-dependent PatL1, LSm1, and DDX6 relocation depends on HCV translation and/or replication and is independent of viral assembly and particle release from infected cells. By treating HCV-infected cells with the translation inhibitor puromycin, the equilibrium between actively translated mRNAs and nontranslated mRNAs located in P-bodies was pushed toward the latter. As this led to only a partial restoration of P-body composition, at least a fraction of PatL1, LSm1, and DDX6 proteins might be associated with HCV replication. These observations are consistent with studies on the plant brome mosaic virus, a model positive-strand RNA that replicates in yeast (2). Here, the yeast homologs of PatL1, LSm1, and DDX6 were shown to promote translation and also replication by selectively recruiting the viral genomes from the translation machinery to the replication complexes (31, 35), a process that requires translation repression.
P-body foci are in a dynamic equilibrium with their mRNA and protein components in the cytosol (1, 27). The simplest interpretation of the observed effect of HCV on P-body composition is that HCV breaks this equilibrium by sequestering PatL1, LSm1, and DDX6 in the cytosol, which thereby decreases the amount of available P-body proteins. This in turn would result in a depletion of PatL1, LSm1, and DDX6 from P-bodies. Two main observations support this view. First, disruption of P-body structures did not affect HCV infection, indicating that HCV can utilize cytosolic PatL1, LSm1, and DDX6 and does not require their prior localization into P-bodies. Second, detection of changes in P-body composition required a certain threshold of HCV components and was reversed when HCV infection levels were decreased, indicating that the equilibrium between P-bodies and their components can be shifted in both directions. In such a case, one may hypothesize that HCV infection could induce a complete P-body disruption under certain circumstances, such as very high replication rates or a sustained chronic infection level.
While our work was in progress, Ariumi et al. (6) reported that in HCV-infected cells, PatL1, LSm1, DDX6, and other P-body components—including Xrn1, a protein that does not affect HCV infection (38)—colocalize with the HCV core protein around lipid droplets (LDs). Since this was observed at advanced times postinfection and since LDs are proposed to act as platforms for HCV assembly (33), the authors suggested that a late step in HCV infection is responsible for relocating P-body components. Although our results do not exclude this possibility, we demonstrate that earlier infection steps, namely, HCV translation and replication, were sufficient to induce relocation of PatL1, LSm1, and DDX6 out of P-bodies. Moreover, our results do not support a colocalization with the HCV core protein in LDs. It is important to note that the core protein of JFH1, the HCV virus used by Ariumi et al., localizes predominantly in LDs, while the core protein of Jc1, the HCV virus used in this study, localizes mainly in ER membranes (11). We observed a colocalization of the core protein with LSm1 in the ER in a low percentage of HCV-infected cells, but we did not detect it for DDX6 or PatL1 (Fig. 3). In agreement with these results, Jangra et al. observed that a small fraction of the core protein coimmunoprecipitated with DDX6, but they did not detect colocalization by immunofluorescence (26). In any case, the interaction of PatL1, LSm1, and DDX6 with the HCV core protein is of uncertain meaning, since replication of HCV subgenomic replicons also depends on these proteins even though they do not contain the core protein (38). In line with this, DDX3, another P-body protein required for HCV replication, colocalizes with the JFH1 core protein in LDs. Interestingly, this requirement of DDX3 for HCV replication is unrelated to its interaction with the HCV core protein (4). All of these observations strongly suggest that the localization of P-body components in LDs is not related to their role in the HCV life cycle.
Modulation of P-body granules seems to be a common host cell response during viral infections, since several viruses have been shown to influence P-bodies. For example, the flaviviruses West Nile virus and dengue virus disrupt P-bodies by an unknown mechanism (17). A similar effect was observed in poliovirus infection, but in this case a virus-induced degradation of Pan3, a P-body component required for P-body formation, was detected (16). Interestingly, as for HCV, the cricket paralysis virus (CPV) and the prototype foamy virus (PFV) do not disrupt P-bodies; instead, they affect the localization of some P-body components. CPV infection affects the localization of Dcp1 and GW182; however, whether these proteins play a role in the CPV life cycle remains to be clarified (28). PFV infection affects the localization in P-bodies of DDX6 but not of other P-body components such as Dcp1 and GW182, among others. After infection, DDX6 relocalizes to the site of PFV assembly and is required for encapsidation of the viral genome (28, 43).
Numerous functional studies have investigated the importance of P-body components for viral life cycles. However, to our knowledge this is the first study to show that P-body components are used during HCV infection independently of their structural arrangement in P-body granules. These observations are in line with publications showing that translation repression and mRNA decay in noninfected cells also occur in the absence of P-bodies (18). Nonetheless, the conservation of P-body granules in all eukaryotes, from yeast to humans, indicates that they have an important physiological role. Some first hints on this came from a recent demonstration that P-bodies play a role in the long-term survival of yeast cells in the stationary phase, a model system for eukaryotic aging (36). Thus, while HCV replication is independent of P-body formation, virus-induced changes of P-body composition in infected individuals may well have consequences beyond virus multiplication. Interestingly, a study in mice has suggested that P-body disruption is correlated with an upregulation of global translation and inflammatory processes (44).
In conclusion, HCV utilizes the P-body components PatL1, LSm1, and DDX6 for viral translation and replication, thereby altering the P-body composition of infected cells. While the physiological consequences of such alterations in vivo are unknown, they may well be linked to inflammatory processes in the livers of infected individuals. This interesting possibility warrants further investigation.
ACKNOWLEDGMENTS
We thank Apath, LLC, C. Rice, J. Lykke-Andersen, M. Kiledjian, M. Fritzler, R. Luehrmann, T. Achsel, F. Chisari, P. Gastaminza, and F. Gebauer for reagents and A. Meyerhans and B. Lindenbach for critically reading the manuscript and for helpful discussions. We are also grateful to Xavier Sanjuan from the Advanced Light Microcopy Unit (Universitat Pompeu Fabra) for assistance with confocal microscopy and advice with analyses.
This work was supported by a grant from the Spanish Ministerio de Ciencia e Innovación (BFU2010-20803).
Footnotes
Published ahead of print 6 June 2012
REFERENCES
- 1. Aizer A, et al. 2008. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol. Biol. Cell 19:4154–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alves-Rodrigues I, Galao RP, Meyerhans A, Diez J. 2006. Saccharomyces cerevisiae: a useful model host to study fundamental biology of viral replication. Virus Res. 120:49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andrei MA, et al. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11:717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Angus AG, et al. 2010. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J. Gen. Virol. 91:122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ariumi Y, et al. 2007. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J. Virol. 81:13922–13926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ariumi Y, et al. 2011. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J. Virol. 85:6882–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartenschlager R, Cosset FL, Lohmann V. 2010. Hepatitis C virus replication cycle. J. Hepatol. 53:583–585 [DOI] [PubMed] [Google Scholar]
- 8. Beckham CJ, Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125:1111–1124 [DOI] [PubMed] [Google Scholar]
- 10. Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boson B, Granio O, Bartenschlager R, Cosset FL. 2011. A concerted action of hepatitis C virus p7 and nonstructural protein 2 regulates core localization at the endoplasmic reticulum and virus assembly. PLoS Pathog. 7:e1002144 doi:10.1371/journal.ppat.1002144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brengues M, Teixeira D, Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coller J, Parker R. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73:861–890 [DOI] [PubMed] [Google Scholar]
- 14. Coller J, Parker R. 2005. General translational repression by activators of mRNA decapping. Cell 122:875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cougot N, Babajko S, Seraphin B. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165:31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dougherty JD, White JP, Lloyd RE. 2011. Poliovirus-mediated disruption of cytoplasmic processing bodies. J. Virol. 85:64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Emara MM, Brinton MA. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U. S. A. 104:9041–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Erickson SL, Lykke-Andersen J. 2011. Cytoplasmic mRNP granules at a glance. J. Cell Sci. 124:293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. 2007. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27:3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fontanes V, Raychaudhuri S, Dasgupta A. 2009. A cell-permeable peptide inhibits hepatitis C virus replication by sequestering IRES transacting factors. Virology 394:82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Franks TM, Lykke-Andersen J. 2008. The control of mRNA decapping and P-body formation. Mol. Cell 32:605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Franks TM, Lykke-Andersen J. 2007. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 21:719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gimenez-Barcons M, Diez J. 2011. Yeast processing bodies and stress granules: self-assembly ribonucleoprotein particles. Microb. Cell Fact. 10:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gravitz L. 2011. A smouldering public-health crisis. Nature 474:S2–S4 [DOI] [PubMed] [Google Scholar]
- 25. Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8:1489–1501 [PMC free article] [PubMed] [Google Scholar]
- 26. Jangra RK, Yi M, Lemon SM. 2010. DDX6 (Rck/p54) is required for efficient hepatitis C virus replication but not for internal ribosome entry site-directed translation. J. Virol. 84:6810–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kedersha N, et al. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khong A, Jan E. 2011. Modulation of stress granules and P bodies during dicistrovirus infection. J. Virol. 85:1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lindenbach BD, et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 30. Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mas A, Alves-Rodrigues I, Noueiry A, Ahlquist P, Diez J. 2006. Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J. Virol. 80:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Migliaccio G, et al. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164–49170 [DOI] [PubMed] [Google Scholar]
- 33. Miyanari Y, et al. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097 [DOI] [PubMed] [Google Scholar]
- 34. Nissan T, Rajyaguru P, She M, Song H, Parker R. 2010. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell 39:773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Noueiry AO, Diez J, Falk SP, Chen J, Ahlquist P. 2003. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol. Cell. Biol. 23:4094–4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ramachandran V, Shah KH, Herman PK. 2011. The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol. Cell 43:973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Randall G, et al. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104:12884–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scheller N, et al. 2009. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc. Natl. Acad. Sci. U. S. A. 106:13517–13522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Scheller N, et al. 2007. Identification of PatL1, a human homolog to yeast P body component Pat1. Biochim. Biophys. Acta 1773:1786–1792 [DOI] [PubMed] [Google Scholar]
- 40. Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tharun S, Parker R. 2001. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell 8:1075–1083 [DOI] [PubMed] [Google Scholar]
- 42. Yang WH, Yu JH, Gulick T, Bloch KD, Bloch DB. 2006. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA 12:547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu SF, Lujan P, Jackson DL, Emerman M, Linial ML. 2011. The DEAD-box RNA helicase DDX6 is required for efficient encapsidation of a retroviral genome. PLoS Pathog. 7:e1002303 doi:10.1371/journal.ppat.1002303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhai Y, et al. 2008. Coordinated changes in mRNA turnover, translation, and RNA processing bodies in bronchial epithelial cells following inflammatory stimulation. Mol. Cell. Biol. 28:7414–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhong J, et al. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082–11093 [DOI] [PMC free article] [PubMed] [Google Scholar]