Abstract
Responses to alpha interferon (IFN-α)-based treatment are dependent on both host and viral factors and vary markedly among patients infected with different hepatitis C virus (HCV) genotypes (GTs). Patients infected with GT3 viruses consistently respond better to IFN treatment than do patients infected with GT1 viruses. The mechanisms underlying this difference are not well understood. In this study, we sought to determine the effects of HCV NS5A proteins from different genotypes on IFN signaling. We found that the overexpression of either GT1 or GT3 NS5A proteins significantly inhibited IFN-induced IFN-stimulated response element (ISRE) signaling, phosphorylated STAT1 (P-STAT1) levels, and IFN-stimulated gene (ISG) expression compared to controls. GT1 NS5A protein expression exhibited stronger inhibitory effects on IFN signaling than did GT3 NS5A protein expression. Furthermore, GT1 NS5A bound to STAT1 with a higher affinity than did GT3 NS5A. Domain mapping revealed that the C-terminal region of NS5A conferred these inhibitory effects on IFN signaling. The overexpression of HCV NS5A increased HCV replication levels in JFH1-infected cells through the further reduction of levels of P-STAT1, ISRE signaling, and downstream ISG responses. We demonstrated that the overexpression of GT1 NS5A proteins resulted in less IFN responsiveness than did the expression of GT3 NS5A proteins through stronger binding to STAT1. We confirmed that GT1 NS5A proteins exerted stronger IFN signaling inhibition than did GT3 NS5A proteins in an infectious recombinant JFH1 virus. The potent antiviral NS5A inhibitor BMS-790052 did not block NS5A-mediated IFN signaling suppression in an overexpression model, suggesting that NS5A's contributions to replication are independent of its subversive action on IFN. We propose a model in which the binding of the C-terminal region of NS5A to STAT1 leads to decreased levels of P-STAT1, ISRE signaling, and ISG transcription and, ultimately, to preferential GT1 resistance to IFN treatment.
INTRODUCTION
More than 170 million individuals are infected with hepatitis C virus (HCV) worldwide, and the disease often progresses to chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (1, 33). Alpha interferon (IFN-α) has been the backbone of therapy for hepatitis C virus infection. Unfortunately, a number of patients do not respond well to therapy (9, 15, 20, 40). The response rates to IFN therapy are also markedly different among patients infected with different HCV genotypes (GTs). Patients infected with HCV GT1 demonstrate sustained virological response (SVR) rates of 38 to 52%, whereas those infected with GT3 achieve higher SVR rates of 66 to 88% (5, 15, 34). However, the precise mechanisms for HCV GT1 hyporesponsiveness are still not fully understood (38, 40). Several HCV proteins, including core and NS5A, have been demonstrated to contribute to the lack of a response of patients with HCV infection to IFN treatment (35, 38, 40, 47). We have previously reported that HCV replication and core protein expression decrease phosphorylated STAT1 (P-STAT1) accumulation (28, 29). HCV NS5A was also shown to inhibit IFN signaling through the suppression of STAT1 phosphorylation (14, 24, 27). However, the molecular mechanisms by which HCV NS5A antagonizes IFN activity are not well characterized. Furthermore, HCV NS5A genotype-mediated IFN resistance is still controversial. In general, viral infection stimulates the type I IFN (IFN-α/β) pathway. The binding of IFNs to their cellular receptors activates an intracellular signaling cascade. The activated Jak1 and Tyk2 kinases further phosphorylate STAT1 and STAT2. The subsequent phosphorylation of the receptor-recruited STATs promotes the formation of heterodimers between STAT1 and STAT2, which further bind to a third protein, IFN-regulatory factor 9 (IRF9) or P48, to form the IFN-stimulated gene factor 3 (ISGF3) complex. This complex translocates into the nucleus and binds the IFN-stimulated response element (ISRE) in IFN-stimulated gene (ISG) promoters, leading to the upregulation of IFN effector proteins such as the double-stranded RNA-activated protein kinase (PKR), the 2′,5′-oligoadenylate synthetase (OAS), and the MxA (myxovirus resistance A) proteins (4, 13, 19, 44). The level of IFN-induced P-STAT1 has been shown to correlate with the global induction of ISGs and was significantly higher in sustained HCV virological responders than in nonresponders (3, 17). It was described previously that the NS5A protein plays a role in the inhibition of IFN activity via the interaction of its PKR binding domain (PKRBD) (amino acid [aa] positions 2209 to 2274) with the antiviral protein PKR, which blocks PKR activity (6, 11). This interaction is presumed to allow viral protein synthesis to occur during IFN treatment. In addition, a number of recent studies have demonstrated the inhibitory effects of the NS5A protein on the IFN-induced Jak-STAT signaling pathway (14, 24, 27, 43). However, the molecular mechanisms by which HCV NS5A interferes with P-STAT1 and IFN signaling have not been fully characterized. In order to characterize the mechanism of HCV resistance to IFN therapy among NS5A proteins from different genotypes, we investigated the effects of GT1 and GT3 HCV NS5A proteins on the IFN-α-induced Jak-STAT signaling pathway in overexpression and infectious recombinant virus (JFH1) models (39). NS5A is an essential component of the HCV replication complex (21, 41). The compound BMS-790052 is a potent NS5A inhibitor (8). We also tested the effects of BMS-790052 (12) on the NS5A-mediated suppression of IFN signaling.
MATERIALS AND METHODS
Cell cultures and infectious viruses.
The human hepatocellular carcinoma cell line Huh7.5.1 (50) and HCV JFH1-infected Huh7.5.1 cells were grown at 37°C in a 5% CO2 atmosphere in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics (0.1 mg/ml streptomycin and 100 U/ml penicillin). The Huh7.5.1 cells were infected with HCV JFH1 as previously described (30, 31, 45). The infectious GT1 and GT3 NS5A recombinant viruses were produced from recombinant DNA constructs, as previously reported (39). The infectious GT1 and GT3 NS5A recombinant viruses were developed by replacing the complete NS5A of the J6/JFH1 recombinant virus with NS5A from HCV GT1 isolate H77C (GT1a) or GT3 isolate S52 (GT3a) (2, 32, 39). An NS5A-specific inhibitor, BMS-790052 (12) (Selleck Chemicals LLC, Houston, TX), was tested for its effects on HCV JFH1 replication, ISRE signaling, and NS5A protein expression.
Plasmid constructs and transfection.
Genotype 1 and 3 HCV strains are the most prevalent forms of infection in Thailand (16). Our project was initiated in Thailand several years ago, before the JFH1 virus was available. The GT1 and GT3 NS5A constructs used in this study were generated from HCV-infected patients in Thailand (26). Full-length cDNAs encoding HCV NS5A were prepared based on the consensus genotypes 1a, 1b, 3a, and 3b isolated from patient sera by reverse transcription (RT)-PCR. Primer pairs used for the amplification of the NS5A gene for each genotype are shown in Table 1. A mammalian expression vector, pCAGGS.MCS (37), was modified in this study to generate pCAGGS-V5/His. Different cDNA fragments of the NS5A gene were cloned separately into the KpnI and NotI endonuclease restriction sites of pCAGGS-V5/His. The target genes in the recombinant plasmids were then confirmed by DNA sequence analysis. The NS4B (GT1) construct was described previously (22). Huh7.5.1 cells at a concentration of 4 × 105 cells/well were seeded into 6-well plates for 24 h and transfected with 2 μg/well of each plasmid by using FuGene HD transfection reagent (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's protocol. RNA or protein lysates were harvested after 48 h of transfection.
Table 1.
Primers used for real-time PCR and generation of NS5A constructsa
| Gene | Primer (5′–3′) |
|
|---|---|---|
| Forward | Reverse | |
| GAPDH | ACAGTCCATGCCATCACTGCC | GCCTGCTTCACCACCTTCTTG |
| JFH1 | TCTGCGGAACCGGTGAGTA | TCAGGCAGTACCACAAGGC |
| PKR | CCAGTGATGATTCTCTTGAGAGC | CCCCAAAGCGTAGAGGTCCA |
| OAS | GGTGGTAAAGGGTGGCTCCTC | TCTGCAGGTAGGTGCACTCC |
| MxA | CCCTTCCCAGAGGCAGCGGG | CTGATTGCCCACAGCCACTC |
| NS5A GT1a | TTGGTACCATGTCCGGYTCCTGGYTRAGG | TTAAGCGGCCGCAGCACACGACRTCCTC |
| NS5A GT1b | TTGGTACCATTATGTCCGGCTCGTGGCTAAGG | TTAAGCGGCCGCAGCAGACGACGTCCTC |
| NS5A GT3a | TTGGTACCATGAGCGGTGACTGGCTGCGTACC | TTAAGCGGCCGCAGCAGACCACGCTCTGCTC |
| NS5A GT3b | TTGGTACCATGAACGGTGATTGGTTACATG | TTAAGCGGCCGCAGCAGACCACGCTCTGCTC |
| NS5A GT1b N terminus | TTGGTACCATTATGTCCGGCTCGTGGCTAAGG | TTAAGCGGCCGCCCGCAGACAACTGGCTAGC |
| NS5A GT1b C terminus | TTGGTACCATGCCTTCCTTGAGGGCAAC | TTAAGCGGCCGCAGCAGACGACGTCCTC |
| NS5A GT3a N terminus | TTGGTACCATGAGCGGTGACTGGCTGCGTACC | AATTGCGGCCGCCAGCCGATAGTTGGCTGG |
| NS5A GT3a C terminus | AAGGTACCATGCCGTCGTTGAAGGCCAC | TTAAGCGGCCGCAGCAGACCACGCTCTGCTC |
The underlined nucleotides indicate restriction sites, GGTACC for KpnI and GCGGCCGC for NotI.
ISRE-luciferase reporter assay.
IFN-induced ISRE signaling was monitored as previously described (28, 29). Briefly, plasmids pISRE-luc, expressing firefly luciferase, and pRL-TK, expressing Renilla luciferase, were cotransfected with selected NS5A plasmids by using Fugene HD transfection reagent (Roche, Indianapolis, IN) according to the manufacturer's protocol. ISRE-mediated transcription was investigated in the presence of 100 IU/ml pegylated IFN-α (Peg-interferon alfa-2b; Schering Co.) for 24 h. Relative luciferase activity was then assessed by the Promega dual-luciferase reporter assay system (Promega, Madison, WI). To validate the inhibitory effect of infectious HCV on IFN-induced ISRE signaling, we cotransfected plasmids pISRE-luc and pRL-TK into JFH1-infected Huh7.5.1 cells for 24 h. Cells were then treated with IFN-α at different doses for another 24 h.
Quantitative real-time PCR.
Total cellular RNAs from Huh7.5.1 cells transfected with a plasmid encoding the HCV NS5A protein, and JFH1-infected cells were harvested by using the QIA Shredder and RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Total cDNA was synthesized by reverse transcription using Applied Biosystems high-capacity cDNA reverse transcription kits (Invitrogen, Carlsbad, CA) with random primers. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), HCV JFH1, PKR, OAS, and MxA mRNA levels were quantified by real-time PCR using the Bio-Rad IQ5 system (Bio-Rad Laboratories) and Finnzymes SYBR green I dye (New England BioLabs, Ipswich, MA). GAPDH was used as a control for basal RNA levels. The primer sequences used are listed in Table 1. The reaction mixture was first heated at 95°C for 3 min, and 45 cycles of PCR amplification were then performed, as follows: 94°C for 20 s, 60°C for 30 s, and 72°C for 20 s. The mRNA level of each gene was normalized to GAPDH levels to obtain mRNA arbitrary units (folds).
Protein sample preparation.
At the time of harvesting, cells were washed with phosphate-buffered saline (PBS) and lysed by using radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 5 mM EDTA) containing a protease inhibitor cocktail (Sigma Life Science and Biochemicals, St. Louis, MO). Whole-cell protein lysates were subsequently sonicated and boiled at 95°C for 5 min in SDS-PAGE sample buffer and chilled on ice for another 5 min before being subjected to a Western blot (WB) assay.
Western blotting.
While WB may not be precisely quantitative, it does provide semiquantitative information about protein levels. We also loaded equal quantities of protein (20 μg) into each lane. Protein samples were separated by SDS-PAGE with NuPAGE Novex precast 4-to-12% gradient Bis-Tris gels (Invitrogen, Carlsbad, CA) and blotted onto nitrocellulose membranes, and the membranes were blocked with 5% bovine serum albumin (BSA). The targeted proteins were detected with specific primary antibodies, including rabbit anti-STAT1 (1:1,000), rabbit anti-phospho-STAT1 (1:1,000), rabbit anti-STAT2 (1:1,000), rabbit anti-phospho-STAT2 (1:1,000) (Cell Signaling Technology, Inc., Beverly, MA), mouse anti-HCV core C7-50 (1:2,000) (Affinity BioReagents, Inc., Golden, CO), mouse anti-V5 (1:5,000) (Invitrogen Life Technologies, Carlsbad, CA), mouse anti-NS4B (1:1,000), mouse anti-NS5A (1:1,000) (Virogen, Watertown, MA), and mouse anti-actin (1:10,000) (Sigma Life Science and Biochemicals, St. Louis, MO) antibodies. The secondary antibodies used were horseradish peroxidase (HRP)-conjugated ECL donkey anti-rabbit IgG and HRP-conjugated ECL sheep anti-mouse IgG (Amersham Biosciences, Piscataway, NJ). The blots were detected by chemiluminescence using the ECL Western blotting detection kit (Amersham Biosciences, Piscataway, NJ). Relative protein levels were quantified by using Image J software (NIH) to measure HCV NS5A and actin densitometry in Western blots. NS5A/actin protein arbitrary units (NAPAU) were calculated from the corresponding densitometry.
Immunoprecipitation assay.
Huh7.5.1 cells were transfected with 2 μg/well of NS5A constructs (tagged with V5/His) in a 6-well plate. Forty-eight hours after transfection, cells were lysed, and whole-cell lysates were collected. We added 20 μg of protein to each well to ensure equal protein loading in each lane. Furthermore, 100 μg (1 μg/μl) of protein lysate was incubated with 10 μg (1 μg/μl) of antibody in each coimmunoprecipitation (co-IP) experiment. Twenty microliters of each immunoprecipitation (IP) lysate was loaded onto the WB. STAT1 immunoprecipitation was performed by using the Roche immunoprecipitation kit (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions. The STAT1-NS5A protein complex (20 μl IP lysate) was then analyzed by Western blotting. Anti-V5 antibody (Invitrogen Life Technologies, Carlsbad, CA) was used as the primary antibody, and HRP-conjugated ECL sheep anti-mouse IgG (Amersham Biosciences, Piscataway, NJ) was used as the secondary antibody.
Statistical analysis.
Data analysis was performed by using a 2-tailed Student t test. Data are expressed as means ± standard deviations (SD) of data from at least three sample replicates, unless stated otherwise.
RESULTS
Both HCV JFH1 infection and NS5A expression inhibit IFN-α-induced ISRE signaling.
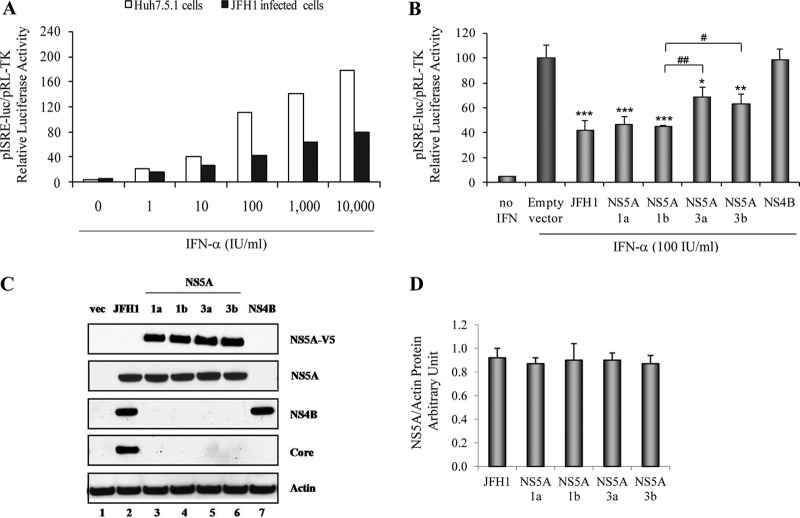
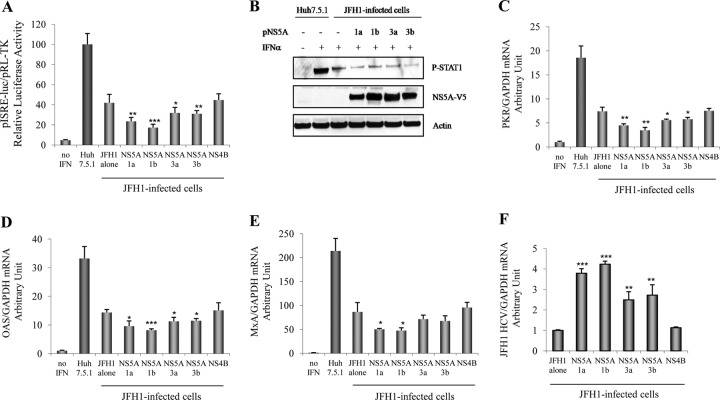
We first observed that IFN-α could induce ISRE signaling in Huh7.5.1 cells in a dose-dependent manner (from 1 to 10,000 IU/ml), as expected (Fig. 1A). We confirmed that HCV strongly decreased IFN-induced ISRE signaling in JFH1-infected Huh7.5.1 cells (Fig. 1A). Interestingly, we found that the GT1a, GT1b, GT3a, and GT3b NS5A proteins each significantly inhibited IFN-induced ISRE signaling by 52.9% ± 6.3% (P = 0.001), 55.0% ± 1.0% (P < 0.001), 31.5% ± 8.3% (P = 0.016), and 36.5% ± 7.7% (P = 0.009), respectively, compared to the empty vector control (Fig. 1B). In contrast, NS4B (GT1) did not block ISRE signaling. Between genotypes, we noted that GT1 NS5A proteins inhibited IFN signaling significantly more strongly than did GT3 NS5A proteins (P < 0.05) (Fig. 1B). Western blot results confirmed the expression of each of the NS5A, NS4B, and core viral proteins in transfected and JFH1-infected Huh7.5.1 cells (Fig. 1C). Western blotting and densitometry verified equivalent levels of NS5A protein expression corresponding to HCV JFH1, GT1a, GT1b, GT3a, and GT3b (Fig. 1D).
Fig 1.
JFH1 replication and overexpression of GT1a, GT1b, GT3a, and GT3b NS5A proteins inhibit IFN-α-induced ISRE promoter activity. (A) Huh7.5.1 cells and JFH1-infected Huh7.5.1 cells were cotransfected with plasmid pISRE-luc (expressing firefly luciferase) and pRL-TK (expressing Renilla luciferase). After 24 h, cells were treated with different doses of IFN-α: 1, 10, 100, 1,000, and 10,000 IU/ml. ISRE-mediated IFN signaling was monitored by a dual-luciferase reporter assay system at 24 h after IFN treatment. The multiplicity of infection of JFH1 in our experiments was 0.2. We found that JFH1-infected Huh7.5.1 cells were about 50% infected at 48 h and at least 95% infected at day 6 by HCV core immunofluorescence staining both in this study and in our previous report (30). (B) Full-length GT1a, GT1b, GT3a, or GT3b NS5A; NS4B constructs; or the empty vector control, pCAGGS-V5/His, was cotransfected with pISRE-luc and pRL-TK into Huh7.5.1 cells. The transfected cells were treated with 100 IU/ml of IFN-α for 24 h. Empty-vector-transfected cells were used as a negative control, and JFH1-infected Huh7.5.1 cells were used as a positive replication control. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with the empty vector control cells). #, P < 0.05; ##, P < 0.01 (compared between GT1 and GT3 NS5A proteins). (C) Western blot analysis of viral protein expression. Lane 1, pCAGGS-V5/His; lane 2, JFH1; lane 3, GT1a NS5A; lane 4, GT1b NS5A; lane 5, GT3a NS5A; lane 6, GT3b NS5A; lane 7, NS4B. (D) NS5A/actin protein arbitrary units (NAPAU) for JFH1, GT1a, GT1b, GT3a, and GT3b from the corresponding densitometry. The NAPAU (from three independent WB images) were 0.92 ± 0.08 (JFH1), 0.87 ± 0.05 (GT1a), 0.90 ± 0.14 (GT1b), 0.90 ± 0.06 (GT3a), and 0.87 ± 0.07 (GT3b), respectively. These data indicated relative equivalent NS5A protein expression levels for HCV JFH1, GT1a, GT1b, GT3a, and GT3b.
Both HCV JFH1 infection and NS5A expression block IFN-α-induced STAT1 phosphorylation.
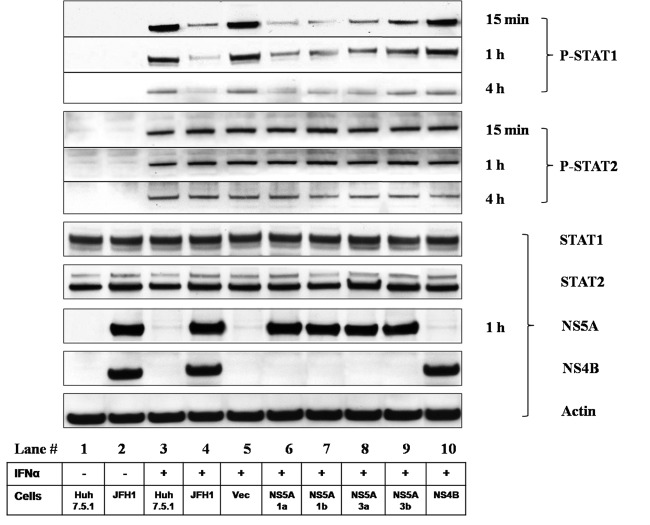
To elucidate the effects of the NS5A protein on IFN-α-induced STAT1 phosphorylation (P-STAT1), we performed a P-STAT1 Western blot analysis with Huh7.5.1 cells expressing the NS5A protein. We confirmed the activation of P-STAT1 and P-STAT2 in IFN-α-treated Huh7.5.1 cells (Fig. 2). We observed an inhibitory effect of NS5A on STAT1 phosphorylation at 15 min, 1 h, and 4 h of IFN-α treatment. However, this effect was selective, since the overexpression of NS5A did not affect STAT2 phosphorylation at 15 min, 1 h, and 4 h of IFN-α treatment (Fig. 2). JFH1 infection as well as the overexpression of the NS5A proteins of GT1a, GT1b, GT3a, and GT3b strongly decreased P-STAT1 but not P-STAT2 levels compared to those of IFN-α-treated Huh7.5.1 cells without JFH1 infection. In contrast, the empty vector and NS4B overexpression had no effect on P-STAT1 levels (Fig. 2). Furthermore, GT1 (GT1a and GT1b) NS5A expression exhibited a stronger suppression of P-STAT1 levels than did GT3 (GT3a and GT3b) NS5A expression. These data suggest that the inhibitory effect of JFH1 infection and NS5A expression on IFN signaling is attributable to the reduction in the level of STAT1 phosphorylation.
Fig 2.
HCV NS5A reduces STAT1 phosphorylation. The inhibitory effect of NS5A on IFN-α-induced STAT1 phosphorylation was assessed in cells expressing the NS5A protein. Huh7.5.1 cells were transfected with NS5A constructs for 48 h. Huh7.5.1, JFH1-infected Huh7.5.1, or construct-transfected Huh7.5.1 cells were incubated with IFN-α (100 IU/ml) for 15 min, 1 h, or 4 h. Whole-cell lysates were analyzed by Western blot analysis for P-STAT1, P-STAT2, STAT1, STAT2, NS5A, NS4B, and actin protein levels. IFN-α activated P-STAT1 and P-STAT2 levels in Huh7.5.1 cells (lane 3). JFH1-HCV replication (lane 4) as well as the overexpression of the GT1a, GT1b, GT3a, and GT3b NS5A proteins (lanes 6, 7, 8, and 9, respectively) strongly decreased P-STAT1 levels. The empty vector and NS4B overexpression had no effect on P-STAT1 levels (lanes 5 and 10, respectively). JFH1 infection or NS5A overexpression had no effect on P-STAT2 levels. GT1a and GT1b NS5A (lanes 6 and 7, respectively) expression exhibited a stronger suppression of P-STAT1 levels than did GT3a and GT3b NS5A (lanes 8 and 9, respectively) expression. Lanes: 1 and 3, Huh7.5.1 cells; 2 and 4, JFH1-infected Huh7.5.1 cells; 5, pCAGGS-V5/His; 6, GT1a NS5A; 7, GT1b NS5A; 8, GT3a NS5A; 9, GT3b NS5A; 10, NS4B. IFN-α treatment is shown in lanes 3 to 10.
Both HCV JFH1 infection and NS5A expression inhibit IFN-α-induced ISGs.
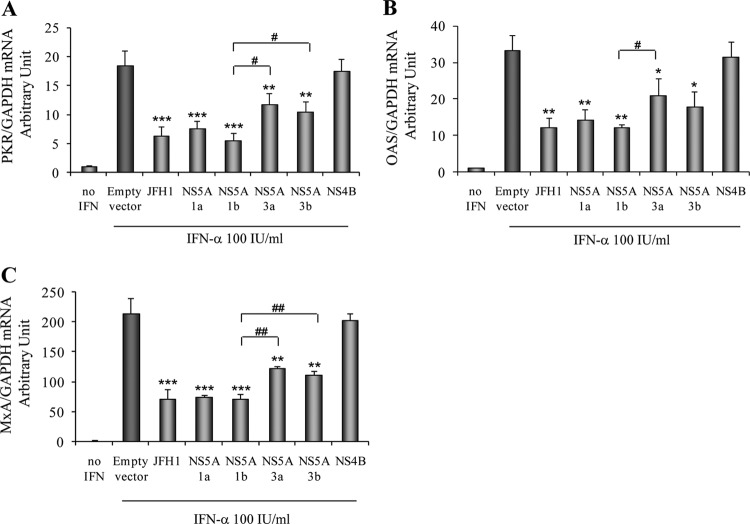
To further determine the effects of NS5A on the IFN-α-induced downstream ISG response, we monitored PKR, OAS, and MxA mRNA levels in JFH1-infected and NS5A-overexpressing Huh7.5.1 cells. We confirmed that IFN-α activated PKR, OAS, and MxA mRNA expressions in Huh7.5.1 cells transfected with the empty vector (Fig. 3). On the other hand, we found that HCV infection strongly decreased PKR, OAS, and MxA mRNA levels in JFH1-infected Huh7.5.1 cells (Fig. 3). The overexpression of the GT1a, GT1b, GT3a, and GT3b NS5A proteins significantly inhibited PKR mRNA levels by 59.1% ± 6.7% (P < 0.001), 69.9% ± 6.6% (P < 0.001), 37% ± 10.8% (P = 0.004), and 43.6% ± 9.7% (P = 0.002), respectively, compared to the empty vector control. In contrast, NS4B had no effect on PKR expression (Fig. 3A). Moreover, the overexpression of the GT1a, GT1b, GT3a, and GT3b NS5A proteins also significantly decreased OAS mRNA levels by 56.6% ± 8.3% (P = 0.005), 63% ± 2.2% (P = 0.002), 36.7% ± 14.1% (P = 0.03), and 46.3% ± 12.8% (P = 0.01), respectively, and MxA mRNA levels by 64.7% ± 1.2% (P = 0.001), 66.6% ± 4.1% (P = 0.001), 42.2% ± 3.5% (P = 0.007), and 47.3% ± 2.7% (P = 0.004), respectively, compared to the empty vector control (Fig. 3B and C). We found that the GT1 NS5A proteins (GT1a and GT1b) induced a stronger reduction of the ISG expression level than did the GT3 NS5A proteins (GT3a and GT3b). These data imply that HCV replication or NS5A overexpression impairs ISG expression through an inhibition of STAT1 phosphorylation.
Fig 3.
Overexpression of NS5A reduces ISG mRNA levels. Huh7.5.1 cells were infected with HCV JFH1 or transfected with GT1a, GT1b, GT3a, or GT3b NS5A or NS4B constructs. Twenty-four hours after transfection, cells were treated with 100 IU/ml of IFN-α for another 24 h. Total RNA was harvested and reverse transcribed. The levels of ISGs mRNA were determined by quantitative real-time PCR. Each ISG mRNA level was normalized to the GAPDH level to obtain arbitrary units. (A) Overexpression of NS5A reduced PKR mRNA levels. (B) Overexpression of NS5A inhibited OAS mRNA expression levels. (C) Overexpression of NS5A reduced MxA mRNA levels. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (for comparisons between NS5A overexpressions and the empty vector). #, P < 0.05; ##, P < 0.01 (for comparisons between GT1 and GT3 NS5A proteins).
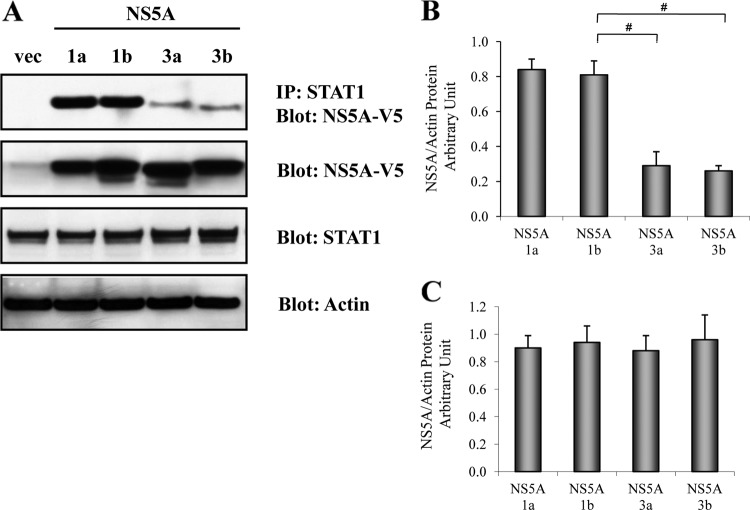
GT1 NS5A binds to STAT1 with a higher affinity than GT3 NS5A.
To further investigate whether the HCV NS5A protein physically associates with STAT1, we performed STAT1 immunoprecipitation followed by anti-V5 Western blotting. We found that the GT1a, GT1b, GT3a, and GT3b NS5A proteins each bind to the STAT1 protein (Fig. 4). Interestingly, we observed that the GT1 NS5A proteins (GT1a and GT1b) bound to STAT1 with a higher affinity than the GT3 NS5A proteins (GT3a and GT3b). Western blotting confirmed NS5A, STAT1, and actin protein expressions (Fig. 4). These data suggest a direct physical interaction between the NS5A and STAT1 proteins. Taken together, we speculate that the binding of NS5A to the STAT1 protein impairs STAT1 phosphorylation, ISRE signaling, and downstream ISG responses. Furthermore, we found that the GT1a and GT1b NS5A proteins suppress IFN responsiveness more strongly than the GT3a and GT3b NS5A proteins in conjunction with stronger binding to STAT1. Moreover, the WB-IP assay and densitometry revealed that the STAT1-NS5A interaction differences were more pronounced between HCV GT1 and GT3 (Fig. 4) than were differences in ISRE signaling (Fig. 1B) or downstream ISG mRNA expression (Fig. 3) between the two genotypes. This disparity suggests the possibility that GT1 and GT3 exert other differential effects at points downstream of STAT1 binding (10, 18, 23).
Fig 4.
GT1 NS5A has stronger binding to STAT1 than GT3 NS5A. The plasmid DNA constructs encoding HCV NS5A of GT1a, GT1b, GT3a, or GT3b were transfected into Huh7.5.1 cells. Cell lysates were harvested at 48 h after transfection. STAT1 immunoprecipitation was performed by using the Roche immunoprecipitation (IP) kit. IP cell lysates were separated by SDS-PAGE and transferred onto nitrocellulose membranes, followed by anti-V5 immunoblotting to determine interactions between NS5A and STAT1. (A) GT1a and GT1b NS5A proteins have stronger binding affinity for STAT1 than GT3a and GT3b NS5A proteins (top). Total cell lysates were used for Western blotting to monitor the expressions of the NS5A (anti-V5), STAT1, and actin proteins. (B) NAPAU for STAT1-IP lysates of GT1a, GT1b, GT3a, and GT3b from the corresponding densitometry. The NAPAU (from three independent WB images) were 0.84 ± 0.06 (GT1a), 0.81 ± 0.08 (GT1b), 0.29 ± 0.08 (GT3a), and 0.26 ± 0.23 (GT3b). These densitometry data confirmed that the GT1 (GT1a and GT1b) NS5A proteins had stronger binding to STAT1 than did the GT3 (GT3a and GT3b) NS5A proteins. (C) NAPAU for whole-cell lysates of GT1a, GT1b, GT3a, and GT3b from the corresponding densitometry. The NAPAU (from three independent WB images) were 0.90 ± 0.09 (GT1a), 0.94 ± 0.12 (GT1b), 0.88 ± 0.11 (GT3a), and 0.96 ± 0.18 (GT3b). These densitometry data verified the relative equal NS5A protein expression levels between GT1 (GT1a and GT1b) and GT3 (GT3a and GT3b).
Overexpression of NS5A in JFH1-infected Huh7.5.1 cells is permissive for HCV replication.
GT1 and GT3 NS5A proteins may exhibit different degrees of suppression of the Jak-STAT antiviral pathway and the downstream ISG response, which may result in a reduction of the IFN-induced antiviral response. We next examined the functional effects of the overexpression of the GT1a, GT1b, GT3a, and GT3b NS5A proteins on IFN signaling, ISG expression, and JFH1 replication. Huh7.5.1 cells were first transfected with NS5A constructs, followed by HCV JFH1 infection. The cells were then treated with or without IFN-α (100 IU/ml). We again confirmed that IFN-induced ISRE signaling was strongly inhibited in JFH1-infected Huh7.5.1 cells. Interestingly, the overexpression of the GT1a, GT1b, GT3a, and GT3b NS5A proteins in JFH1-infected Huh7.5.1 cells produced a stronger reduction in the level of ISRE signaling than did JFH1 infection alone. The overexpression of the GT1a and G1b NS5A proteins exhibited a stronger reduction of the level of ISRE signaling than did the overexpression of the GT3a and GT3b NS5A proteins (Fig. 5A). In contrast, NS4B overexpression led to no further decrease in the level of signaling. Furthermore, NS5A overexpression in combination with JFH1 infection further decreased P-STAT1 levels compared to levels found with JFH1 infection alone (Fig. 5B). We found that the overexpression of NS5A further decreased mRNA levels of PKR, OAS, and MxA in JFH1-infected Huh7.5.1 cells compared to cells infected with JFH1 alone. The overexpression of the GT1a and GT1b NS5A proteins displayed a more substantial reduction of ISG mRNA levels than did the overexpression of the GT3a and GT3b NS5A proteins (Fig. 5C to E). In contrast, the overexpression of NS4B led to no further decrease in the level of ISG expression. We also observed that the overexpression of NS5A in JFH1-infected Huh7.5.1 cells further increased HCV replication levels compared to those in cells infected with JFH1 alone, whereas NS4B had no effect on HCV replication in JFH1-infected Huh7.5.1 cells. The overexpression of GT1a or GT1b NS5A had greater permissive effects on HCV replication level than did the overexpression of GT3a or GT3b NS5A (Fig. 5F). Taken together, these data indicate that the overexpression of NS5A functionally increases HCV replication levels through further reductions in levels of P-STAT1, ISRE signaling, and downstream ISG responses.
Fig 5.
Overexpression of NS5A further increases HCV replication in JFH1-infected Huh7.5.1 cells through stronger suppression of IFN signaling. GT1a, GT1b, GT3a, and GT3b NS5A and NS4B plasmids were cotransfected with pISRE-luc and pRL-TK into Huh7.5.1 cells for 18 h, followed by JFH1 infection for 6 h, and cells were then treated with IFN-α (100 IU/ml) for 24 h. Samples were harvested to measure the relative luciferase activity, protein for Western blot analysis, or mRNA for qPCR. (A) Cell lysates were harvested to measure the relative luciferase activity. The GT1a, GT1b, GT3a, and GT3b NS5A proteins exhibited stronger reductions of ISRE signaling than did JFH1 infection alone. NS4B overexpression showed no further decrease in signaling. The GT1a and GT1b NS5A proteins exhibited stronger reductions of ISRE signaling than did the GT3a and GT3b NS5A proteins. (B) Protein lysates were harvested for Western blotting to measure P-STAT1, NS5A-V5, and actin protein levels. The overexpression of NS5A in JFH1-infected Huh7.5.1 cells further reduced P-STAT1 levels compared to levels in cells infected with JFH1 alone. (C to E) Lysates were harvested to monitor PKR (C), OAS (D), and MxA (E) mRNA levels. The overexpression of NS5A further reduced PKR, OAS, and MxA mRNA levels in JFH1-infected Huh7.5.1 cells compared to levels in cells infected with JFH1 alone. The GT1a and GT1b NS5A proteins caused a stronger inhibition of the ISG mRNA levels than did the GT3a and GT3b NS5A proteins. In contrast, the overexpression of NS4B led to no further reduction in the expression levels of ISGs. (F) NS5A overexpression in JFH1-infected Huh7.5.1 cells further increased HCV replication levels compared to those in cells infected with JFH1 alone, whereas NS4B had no positive effect on HCV replication. The GT1a and GT1b NS5A proteins showed higher increases in HCV replication levels than did the GT3a and GT3b NS5A proteins. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (for comparisons between the overexpression of GT1a, GT1b, GT3a, or GT3b NS5A and JFH1 infection alone).
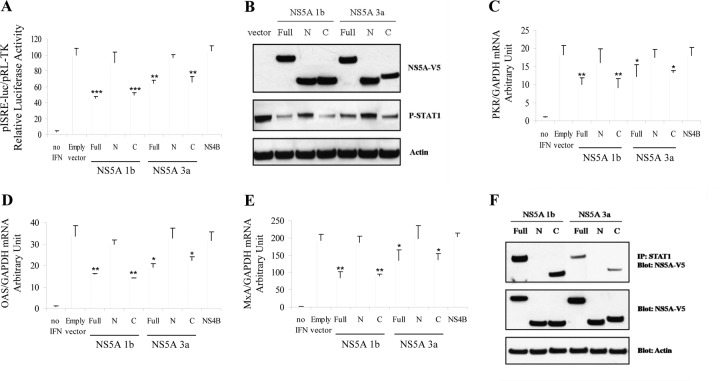
The NS5A C terminus physically interacts with STAT1 and inhibits IFN signaling.
To determine the physical domain of HCV NS5A responsible for the disruption of IFN signaling and the reduction of P-STAT1 levels, we constructed and transfected plasmids expressing full-length NS5A (aa residues 1 to 447), the N-terminal region of NS5A (N-terminal NS5A) (aa residues 1 to 236), and C-terminal NS5A (aa residues 237 to 447) into Huh7.5.1 cells. We found that the overexpression of C-terminal NS5A produced a degree of ISRE signaling inhibition comparable to that of full-length NS5A. In contrast, the overexpression of N-terminal NS5A or NS4B did not block ISRE signaling. A similar result was seen for both GT1 and GT3 (Fig. 6A). In addition, the overexpression of full-length NS5A as well as C-terminal NS5A (in both GT1b and GT3a) was associated with a reduction of P-STAT1 levels, whereas N-terminal NS5A had no effect on P-STAT1 levels (Fig. 6B). We also found that the overexpression of C-terminal NS5A (in both GT1b and GT3a) decreased ISG (including PKR, OAS, and MxA) mRNA levels similarly to full-length NS5A, whereas N-terminal NS5A had no effect on ISG expression (Fig. 6C to E). To investigate whether C-terminal NS5A is responsible for the physical association with STAT1, we performed STAT1 immunoprecipitation followed by NS5A-V5 immunoblotting with cells transfected with the full-length, N-terminal, or C-terminal NS5A construct. We found that both full-length NS5A and the C terminus of NS5A (both GT1b and GT3a) bind to STAT1 in Huh7.5.1 cells (Fig. 6F). In contrast, N-terminal NS5A did not bind to STAT1. These data indicate that the STAT1 interaction domain is located in the C terminus of NS5A (aa residues 237 to 447) and that this domain is responsible for the inhibition of type I IFN signaling and the reduction in levels of P-STAT1 and the downstream ISG response.
Fig 6.
C-terminal NS5A displays inhibitory effects on IFN signaling through binding to STAT1. Full-length HCV NS5A (aa 1 to 447) was also classified into domain I (aa 1 to 213), domain II (aa 250 to 342), and domain III (aa 356 to 447) in a previous report (42). To identify the NS5A domain responsible for the disruption of IFN signaling and the reduction of P-STAT1 levels, we constructed and transfected full-length NS5A (amino acids 1 to 447), N-terminal NS5A (aa 1 to 236) (domain I), and C-terminal NS5A (aa 237 to 447) (domains II and III) plasmids into Huh7.5.1 cells. Samples were harvested to measure the relative luciferase activity, protein for Western blotting, or mRNA for qPCR. (A) Full-length, N-terminal, or C-terminal NS5A plasmids or NS4B plasmids were cotransfected with pISRE-luc and pRL-TK into Huh7.5.1 cells for 24 h, and cells were then treated with IFN-α (100 IU/ml) for 24 h. Cell lysates were harvested to measure the dual-luciferase activity. The full-length and C-terminal NS5A proteins produced comparable degrees of ISRE signaling inhibition in both the GT1 and GT3 NS5A proteins. However, N-terminal NS5A or NS4B did not reduce ISRE signaling. (B) Full-length, N-terminal, or C-terminal NS5A plasmids were transfected into Huh7.5.1 cells for 48 h. Cells were then treated with IFN-α (100 IU/ml) for 1 h. Cell lysates were collected for a protein assay. Western blot analysis for NS5A-V5 confirmed the expression of full-length, C-terminal, or N-terminal NS5A (top). C-terminal and full-length NS5A overexpression (for both GT1b and GT3a) reduced P-STAT1 protein levels. In contrast, N-terminal NS5A had no effect on the P-STAT1 level (middle). Actin protein levels are shown in the bottom panel. (C to E) The empty vector or full-length NS5A, N-terminal or C-terminal NS5A, or NS4B plasmids were transfected into Huh7.5.1 cells. Cells were then treated with IFN-α (100 IU/ml) for 24 h. Cell lysates were harvested to measure mRNA levels. Full-length or C-terminal NS5A produced comparable levels of reduction of PKR (C), OAS (D), and MxA (E) mRNA levels compared to the empty vector for both GT1b and GT3a. N-terminal NS5A or NS4B overexpression did not reduce PKR, OAS, and MxA mRNA expression levels. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (for comparisons between NS5A expressions and the vector control). (F) Full-length, N-terminal, or C-terminal NS5A constructs were transfected into Huh7.5.1 cells. Protein lysates were collected for STAT1 immunoprecipitation by using the Roche IP kit. Protein levels in the IP lysate (top) or whole-cell lysate (middle) were assessed by anti-V5 Western blotting. Full-length NS5A or the C terminus of NS5A (both GT1b and GT3a) bound to STAT1. In contrast, N-terminal NS5A did not bind to STAT1.
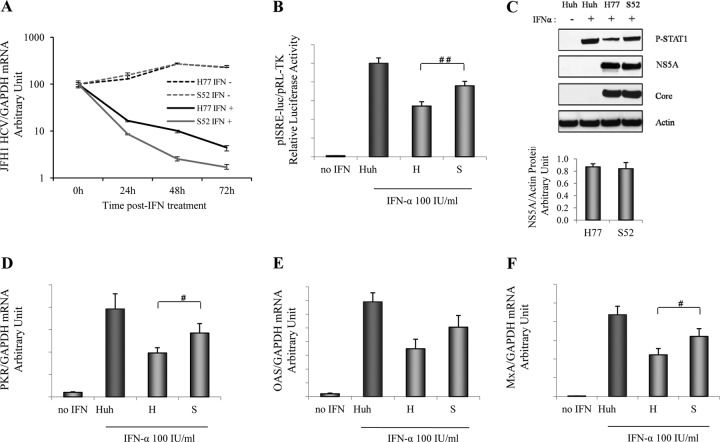
GT1 NS5A exhibits higher resistance to IFN than GT3 NS5A in an infectious replication model.
To test the effects of the GT1 and GT3 NS5A proteins on IFN resistance in the context of an infectious HCV replication model, we examined the influence of an alteration of the NS5A genotype on sensitivity to IFN-α by using H77 (GT1a NS5A)/JFH1 (H77) and S52 (GT3a NS5A)/JFH1 (S52) recombinant viruses (39). In these constructs, full-length NS5A in J6/JFH1 was replaced with GT1a or GT3a NS5A to generate H77 or S52 infectious recombinant virus, as previously reported (39). It was reported previously that NS5A has distinct cis- and trans-acting functions in HCV RNA replication. HCV NS5A's cis-acting function may occur as part of the HCV replication complex through basally phosphorylated NS5A, while the trans-acting function may occur outside the replication complex and requires hyperphosphorylation (7). We found that both the H77 and S52 viruses replicated equivalently by analyzing both RNA and protein levels (NS5A and core) without IFN treatment (Fig. 7A and C). Although we did not monitor NS5A adaptive mutations in the H77 and S52 viruses, our findings and our previously reported description (39) indicate that H77, S52, and J6/JFH1 are comparably replicative viruses. In addition, HCV RNA was markedly inhibited by IFN treatment in a time-dependent manner (Fig. 7A). Interestingly, HCV H77 (GT1a NS5A) replication demonstrated a weaker response to IFN than did HCV S52 (GT3a NS5A) replication (Fig. 7A). We also found that the H77 (GT1a NS5A) recombinant virus exhibited a more substantial reduction in IFN-induced ISRE signaling (Fig. 7B) and P-STAT1 levels (Fig. 7C) than did the S52 (GT3a NS5A) virus. We also observed that infection with the H77 (GT1a NS5A) recombinant virus led to a stronger reduction of the ISG expression level, including PKR, OAS, and MxA mRNA levels, than did infection with the S52 (GT3a NS5A) virus (Fig. 7D to F). These results confirm that GT1 NS5A has greater inhibitory effects on IFN signaling than does GT3 NS5A when inserted into an infectious replication model. It would be of interest to study NS5A sequences corresponding to the selection of GT3 NS5A-harboring clones (S52) that have acquired the phenotype associated with GT1 NS5A-harboring clones (H77).
Fig 7.
GT1 NS5A proteins exhibit higher levels of resistance to IFN than GT3 NS5A proteins in NS5A/JFH1 recombinant virus. To determine GT1 and 3 NS5A resistances to IFN in the replication model, we used the H77 (GT1a NS5A)/JFH1 (H77) and S52 (GT3a NS5A)/JFH1 (S52) recombinant viruses. Full-length NS5A in J6/JFH1 was replaced with GT1a or GT3a NS5A to generate the infectious recombinant virus H77 or S52, as previously reported (39). Huh7.5.1 cells were infected with the H77 or S52 recombinant virus. (A) HCV H77 and S52 respond to IFN treatment in a time-dependent manner. Twenty-four hours after infection, cells were treated with IFN-α (100 IU/ml) or without IFN for another 24, 48, and 72 h. We confirmed the relative equal HCV RNA levels between the H77 and S52 viruses without IFN treatment. We found that IFN treatments reduced HCV RNA levels in a time-dependent manner. S52 (GT3 NS5A) was more sensitive to IFN treatment than H77 (GT1 NS5A). (B) HCV H77 exhibited a stronger inhibition of ISRE signaling than HCV S52. Plasmids pISRE-luc and pRL-TK were cotransfected into H77- or S52-infected Huh7.5.1 cells for 24 h. Cells were treated with IFN-α (100 IU/ml) for another 24 h. The relative luciferase activity was finally assessed. We found that the H77 (GT1 NS5A) recombinant virus exhibited a greater reduction of IFN-induced ISRE signaling did HCV S52 (GT3 NS5A). ##, P < 0.01 (for comparisons between H77 and S52). (C) HCV H77 exhibited a greater reduction of the P-STAT1 level. Huh7.5.1 cell were infected with HCV H77 or S52 for 48 h. The cells were treated with IFN-α (100 IU/ml) for 1 h. IFN-α induced P-STAT1 accumulation in Huh7.5.1 cells. HCV H77 reduced the P-STAT1 protein level more than did HCV S52. The NAPAU for H77 and S52 were 0.87 ± 0.05 and 0.84 ± 0.10, respectively. HCV core and NS5A Western blotting confirmed the relative equal HCV replication levels between H77 and S52. (D to F) HCV H77 inhibited ISG expression more than HCV S52. Huh7.5.1 cells were infected with HCV H77 or S52 for 24 h. The cells were treated with IFN-α (100 IU/ml) for 24 h. Cell lysates were harvested to measure mRNA levels. H77 showed a stronger suppression of ISG expression levels, including PKR (D), OAS (E), and MxA (F) mRNA levels, than S52. #, P < 0.05 (for comparisons between H77 and S52).
HCV NS5A ISDR variation may contribute to GT1 and GT3 sensitivity to IFN.
NS5A interferon sensitivity-determining-region (ISDR) substitutions have been associated with the IFN response and HCV viral load (25, 46). To assess the possible correlation between the NS5A ISDR and IFN efficacy, we compared ISDR sequences (amino acids 2209 to 2248) between the GT1a, GT1b, GT3a, and GT3b NS5A proteins. We found five amino acid differences between GT1 and GT3, located at positions 2216, 2220, 2221, 2239, and 2248 (Table 2). We speculate that these amino acid variations may be associated with GT3 viruses being more responsive to IFN than GT1 viruses, and we will perform these studies as a logical follow-up to the present study.
Table 2.
Comparison of amino acid sequences of NS5A ISDRs between GT1a, GT1b, GT3a, and GT3b
| Protein | Amino acid sequence at residues 2209–2248a |
|---|---|
| GT1a NS5A | PSLKATCTTNHDSPDAELIEANLLWRQEMGGNITRVESEN |
| GT1b NS5A | PSLRATCTTRHDSPDADLIEANLLWRQEMGGNITRVESEN |
| GT3a NS5A | PSLKATCQTHRPHPDAELVEANLLWRQEMGSNITRVESET |
| GT3b NS5A | PSLKATCQTHGPHPDAELIDANLLWRQEMGSNITRVESET |
The underlined sequences indicate variant amino acids between GT1 and GT3 NS5A which are at positions 2216, 2220, 2221, 2239, and 2248.
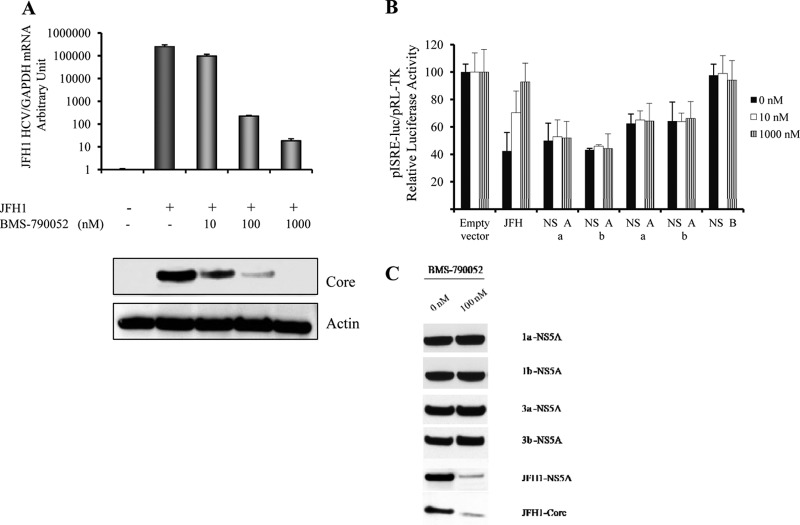
BMS-790052 does not affect NS5A-STAT1 interactions.
To evaluate the effect of a pharmacological NS5A inhibitor on HCV replication, we monitored HCV RNA and core protein levels in JFH1-infected cells treated with the NS5A inhibitor BMS-790052. As expected, we found that BMS-790052 inhibited HCV RNA levels in JFH1-infected cells in a dose-dependent manner (Fig. 8A). Western blotting for the core protein confirmed the proportional reduction of HCV core protein and RNA levels (Fig. 8A, bottom). We also found that BMS-790052 rescued the HCV-mediated inhibition of ISRE signaling in the JFH1 replication model in a dose-dependent manner (Fig. 8B). However, BMS-790052 had no effect on the NS5A overexpression-mediated inhibition of ISRE signaling (Fig. 8B). Western blotting demonstrated that BMS-790052 reduced the HCV core level in JFH1-infected cells. However, BMS-790052 had no effect on the overexpression of the NS5A protein (Fig. 8C). We also found that BMS-790052 neither rescued NS5A overexpression-mediated P-STAT1 reduction nor interrupted NS5A-STAT1 interactions (data not shown). These results indicate that the NS5A inhibitor BMS-790052 reduces HCV RNA and core levels by blocking HCV replication but does not disrupt the NS5A-STAT1 interaction.
Fig 8.
BMS-790052 inhibits JFH1-HCV replication but has no effect on the overexpression of the NS5A protein. (A) BMS-790052 reduced HCV JFH1 RNA levels in a dose-dependent manner. JFH1-infected Huh7.5.1 cells were treated with the NS5A inhibitor BMS-790052 at 0, 10, 100, or 1,000 nM for an additional 24 h. HCV RNA and core protein levels were monitored to determine HCV replication levels. (Top) BMS-790052 reduced HCV RNA levels in JFH1-infected cells. (Bottom) HCV core Western blotting confirmed the reduction of HCV replication levels. (B) BMS-790052 had no effect on NS5A overexpression-mediated inhibition of ISRE signaling. Plasmids pISRE-luc and pRL-TK were cotransfected into JFH1-infected Huh7.5.1 cells or cotransfected with the empty vector; the GT1a, GT1b, GT3a, or GT3b NS5A plasmid; or the NS4B plasmid into Huh7.5.1 cells for 12 h. Cells were incubated with BMS-790052 at 0, 10, or 1,000 nM for an additional 12 h. Cells were then treated IFN-α (100 IU/ml) for 24 h. We found that HCV JFH1 replication or NS5A overexpression inhibited IFN-induced ISRE signaling. BMS-790052 rescued the HCV-mediated inhibition of ISRE signaling in the JFH1 replication model in a dose-dependent manner. However, BMS-790052 had no effect on the NS5A overexpression-mediated inhibition of ISRE signaling. (C) BMS-790052 had no effect on overexpressed NS5A proteins. JFH1-infected Huh7.5.1 cells or Huh7.5.1 cells transfected with the GT1a, GT1b, GT3a, or GT3b NS5A construct were treated with BMS-790052 at 0 nM or 100 nM for 24 h. Protein lysates were harvested for anti-NS5A-V5, NS5A, or HCV core Western blot analysis. BMS-790052 reduced HCV NS5A and core levels in JFH1-infected Huh7.5.1 cells. However, BMS-790052 did not affect NS5A overexpression protein levels.
DISCUSSION
IFN-α has been the mainstay of therapy for chronic hepatitis C. Unfortunately, many patients do not respond well to therapy. The response rates to IFN treatment vary among patients infected with different HCV genotypes. Patients infected with genotype 3 viruses respond much more effectively to IFN treatment than patients infected with genotype 1 viruses (5, 15, 34, 48). However, the mechanisms underlying this difference are not well understood. In the present study, we used both HCV NS5A overexpression and recombinant NS5A/JFH1 virus replication models to dissect the effects of HCV NS5A proteins corresponding to specific genotypes (GT1a, GT1b, GT3a, and GT3b) on the IFN-α-induced Jak-STAT1 signaling pathway, including IFN-induced ISRE promoter activity, P-STAT1 levels, and downstream ISG expression. We demonstrated that the overexpression of the GT1a, GT1b, GT3a, and GT3b NS5A proteins significantly inhibited IFN-induced ISRE signaling, P-STAT1 levels, and ISG expression. However, the overexpression of the GT1a and GT1b NS5A proteins had stronger inhibitory effects on IFN signaling than did the GT3a and GT3b NS5A proteins. It was reported previously that GT1 NS5A displays more resistance to IFN than does GT2 NS5A (24, 36, 43). Our studies also demonstrate that the overexpression of GT1 NS5A has stronger inhibitory effects on IFN than does the overexpression of GT3 NS5A. We further tested the NS5A genotype-inhibitory effects on IFN signaling in an NS5A/JFH1 recombinant virus replication model (39). Our results demonstrated that the H77 (GT1a NS5A)/JFH1 recombinant virus conferred more resistance to IFN treatment than did S52 (GT3a NS5A)/JFH1 through a stronger reduction of IFN-induced ISRE signaling, P-STAT1 levels, and ISG expression levels. However, in a previous study (39), it was reported that an NS5A (GT1 to GT7)/JFH1 recombinant virus had a similar sensitivity to IFN-α treatment. We speculate that the discrepancy between our findings and those of the previous report is attributable to the HCV detection methods used. We monitored HCV replication in the presence of IFN-α by measuring both HCV RNA levels by quantitative PCR (qPCR) and HCV NS5A and core protein levels by Western blotting, compared to the previous report, in which HCV infectivity was measured by calculating the percentage of HCV NS3-positive cells by immunostaining. We also found that NS5A proteins bind to STAT1 and that GT1 NS5A proteins exhibit stronger binding to STAT1 than do GT3 NS5A proteins, suggesting a basis for these differential functional effects. It was reported previously that the C-terminal region of NS5A is required for IFN antagonism (43). Our results reveal, for the first time, that the C terminus of NS5A (both GT1 and GT3) but not the N terminus of NS5A exhibits inhibitory effects on IFN signaling through binding to the STAT1 protein. In a previous report, we demonstrated that IFN-λ1, IFN-λ2, and IFN-λ3 (interleukin-29 [IL-29], IL28A, and IL28B, respectively) inhibited HCV JFH1 replication through the Jak-STAT signaling pathway. We found that IFN-λ3 induced ISG expression through P-STAT1/P-STAT2 activation and the IFN-induced ISRE signaling pathway (49). A further characterization of the mechanism of HCV NS5A's response to lambda interferon-induced Jak-STAT signaling and ISGs is warranted. We found that the NS5A inhibitor BMS-790052 rescued the HCV-mediated suppression of IFN-induced ISRE signaling through its strong inhibition of HCV replication in JFH1-infected cells. However, BMS-790052 exhibited no significant effect on the activity of the overexpressed NS5A protein or on the NS5A-STAT1 interaction, implying a functional dissociation between NS5A's replication and IFN subversion functions.
It was identified previously that the inhibitory activity of BMS-790052 maps to the first 100 amino acids of HCV NS5A (N-terminal NS5A) (12). In this study, we demonstrated that C-terminal NS5A confers a blockage of IFN signaling pathway through binding to STAT1. It is likely that the existence of distinct binding sites on NS5A for BMS-790052 and STAT1 explains the finding that BMS-790052 does not affect the overexpression of NS5A-mediated IFN antagonism. We therefore conclude that NS5A impairs type I IFN signaling through the inhibition of STAT1 phosphorylation in both HCV replication and NS5A expression models. GT1 NS5A suppresses IFN responsiveness more strongly than GT3 NS5A in conjunction with stronger binding to STAT1. We propose a model in which HCV mediates its antagonistic effects on IFN treatment through C-terminal NS5A binding to STAT1, which results in reduced P-STAT1, ISRE signaling, and ISG expression levels. These observations provide new insights into the mechanisms by which HCV NS5A proteins from different genotypes influence IFN signaling and may account for genotype-related differences in IFN treatment responses. The results further suggest a potential target for the development of novel antiviral strategies. Further analyses of NS5A's qualitative and quantitative effects at more physiologic levels will take steps toward an understanding of HCV evasion of host control in a native context.
ACKNOWLEDGMENTS
This work was supported in part by NIH grants AI069939 (R.T.C.) and AI082630 (R.T.C.), an NIH-MGH Center for Human Immunology pilot/feasibility study grant (W.L.), the Royal Golden Jubilee Ph.D. Program of Thailand Research Fund (K.K.), and German Research Foundation (Deutsche Forschungsgemeinschaft) grant Ji 145/1-1 (N.J.).
We are grateful to the following investigators for supplying reagents: Francis Chisari for the Huh7.5.1 cell line, Takaji Wakita for the infectious HCV JFH1 DNA construct, and Jens Bukh for infectious HCV NS5A/JFH1 recombinant virus DNA constructs (H77 and S52).
Footnotes
Published ahead of print 6 June 2012
REFERENCES
- 1. Barth H, Liang TJ, Baumert TF. 2006. Hepatitis C virus entry: molecular biology and clinical implications. Hepatology 44:527–535 [DOI] [PubMed] [Google Scholar]
- 2. Cai Z, et al. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 79:13963–13973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen L, et al. 2005. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology 128:1437–1444 [DOI] [PubMed] [Google Scholar]
- 4. Darnell JE, Jr, Kerr IM, Stark GR. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415–1421 [DOI] [PubMed] [Google Scholar]
- 5. Ferenci P, et al. 2005. Predicting sustained virological responses in chronic hepatitis C patients treated with peginterferon alfa-2a (40 KD)/ribavirin. J. Hepatol. 43:425–433 [DOI] [PubMed] [Google Scholar]
- 6. Francois C, et al. 2000. Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis. J. Virol. 74:5587–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fridell RA, et al. 2011. Distinct functions of NS5A in hepatitis C virus RNA replication uncovered by studies with the NS5A inhibitor BMS-790052. J. Virol. 85:7312–7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fridell RA, Qiu D, Wang C, Valera L, Gao M. 2010. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob. Agents Chemother. 54:3641–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fried MW, et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982 [DOI] [PubMed] [Google Scholar]
- 10. Gale M, Jr, et al. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 18:5208–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gale MJ, Jr, et al. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217–227 [DOI] [PubMed] [Google Scholar]
- 12. Gao M, et al. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garcia-Sastre A, Biron CA. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312:879–882 [DOI] [PubMed] [Google Scholar]
- 14. Gong GZ, Cao J, Jiang YF, Zhou Y, Liu B. 2007. Hepatitis C virus non-structural 5A abrogates signal transducer and activator of transcription-1 nuclear translocation induced by IFN-alpha through dephosphorylation. World J. Gastroenterol. 13:4080–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hadziyannis SJ, et al. 2004. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140:346–355 [DOI] [PubMed] [Google Scholar]
- 16. Hansurabhanon T, et al. 2002. Infection with hepatitis C virus among intravenous-drug users: prevalence, genotypes and risk-factor-associated behaviour patterns in Thailand. Ann. Trop. Med. Parasitol. 96:615–625 [DOI] [PubMed] [Google Scholar]
- 17. He XS, et al. 2006. Global transcriptional response to interferon is a determinant of HCV treatment outcome and is modified by race. Hepatology 44:352–359 [DOI] [PubMed] [Google Scholar]
- 18. He Y, et al. 2002. Subversion of cell signaling pathways by hepatitis C virus nonstructural 5A protein via interaction with Grb2 and P85 phosphatidylinositol 3-kinase. J. Virol. 76:9207–9217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Honda K, Yanai H, Takaoka A, Taniguchi T. 2005. Regulation of the type I IFN induction: a current view. Int. Immunol. 17:1367–1378 [DOI] [PubMed] [Google Scholar]
- 20. Hoofnagle JH, et al. 2003. Maintenance therapy with ribavirin in patients with chronic hepatitis C who fail to respond to combination therapy with interferon alfa and ribavirin. Hepatology 38:66–74 [DOI] [PubMed] [Google Scholar]
- 21. Huang Y, Staschke K, De Francesco R, Tan SL. 2007. Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology 364:1–9 [DOI] [PubMed] [Google Scholar]
- 22. Hugle T, et al. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70–81 [DOI] [PubMed] [Google Scholar]
- 23. Ide Y, Tanimoto A, Sasaguri Y, Padmanabhan R. 1997. Hepatitis C virus NS5A protein is phosphorylated in vitro by a stably bound protein kinase from HeLa cells and by cAMP-dependent protein kinase A-alpha catalytic subunit. Gene 201:151–158 [DOI] [PubMed] [Google Scholar]
- 24. Kang SM, Won SJ, Lee GH, Lim YS, Hwang SB. 2010. Modulation of interferon signaling by hepatitis C virus non-structural 5A protein: implication of genotypic difference in interferon treatment. FEBS Lett. 584:4069–4076 [DOI] [PubMed] [Google Scholar]
- 25. Kohashi T, et al. 2006. Site-specific mutation of the interferon sensitivity-determining region (ISDR) modulates hepatitis C virus replication. J. Viral Hepat. 13:582–590 [DOI] [PubMed] [Google Scholar]
- 26. Kumthip K, et al. 2011. Correlation between mutations in the core and NS5A genes of hepatitis C virus genotypes 1a, 1b, 3a, 3b, 6f and the response to pegylated interferon and ribavirin combination therapy. J. Viral Hepat. 18:e117–e125 doi:10.1111/j.1365-2893.2010.01379.x [DOI] [PubMed] [Google Scholar]
- 27. Lan KH, et al. 2007. HCV NS5A inhibits interferon-alpha signaling through suppression of STAT1 phosphorylation in hepatocyte-derived cell lines. J. Hepatol. 46:759–767 [DOI] [PubMed] [Google Scholar]
- 28. Lin W, et al. 2005. Hepatitis C virus expression suppresses interferon signaling by degrading STAT1. Gastroenterology 128:1034–1041 [DOI] [PubMed] [Google Scholar]
- 29. Lin W, et al. 2006. Hepatitis C virus core protein blocks interferon signaling by interaction with the STAT1 SH2 domain. J. Virol. 80:9226–9235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin W, et al. 2010. Hepatitis C virus regulates transforming growth factor beta1 production through the generation of reactive oxygen species in a nuclear factor kappaB-dependent manner. Gastroenterology 138:2509–2518, 2518.e1 doi:10.1053/j.gastro.2010.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin W, et al. 2011. HIV and HCV cooperatively promote hepatic fibrogenesis via induction of reactive oxygen species and NFkappaB. J. Biol. Chem. 286:2665–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lindenbach BD, et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 33. Machida K, et al. 2010. c-Jun mediates hepatitis C virus hepatocarcinogenesis through signal transducer and activator of transcription 3 and nitric oxide-dependent impairment of oxidative DNA repair. Hepatology 52:480–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mangia A, et al. 2005. Peginterferon alfa-2b and ribavirin for 12 vs. 24 weeks in HCV genotype 2 or 3. N. Engl. J. Med. 352:2609–2617 [DOI] [PubMed] [Google Scholar]
- 35. Mbow ML, Sarisky RT. 2004. What is disrupting IFN-alpha's antiviral activity? Trends Biotechnol. 22:395–399 [DOI] [PubMed] [Google Scholar]
- 36. Neumann AU, et al. 2000. Differences in viral dynamics between genotypes 1 and 2 of hepatitis C virus. J. Infect. Dis. 182:28–35 [DOI] [PubMed] [Google Scholar]
- 37. Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199 [DOI] [PubMed] [Google Scholar]
- 38. Pawlotsky JM. 2003. Mechanisms of antiviral treatment efficacy and failure in chronic hepatitis C. Antiviral Res. 59:1–11 [DOI] [PubMed] [Google Scholar]
- 39. Scheel TK, Gottwein JM, Mikkelsen LS, Jensen TB, Bukh J. 2011. Recombinant HCV variants with NS5A from genotypes 1–7 have different sensitivities to an NS5A inhibitor but not interferon-alpha. Gastroenterology 140:1032–1042 [DOI] [PubMed] [Google Scholar]
- 40. Taylor DR, Shi ST, Lai MM. 2000. Hepatitis C virus and interferon resistance. Microbes Infect. 2:1743–1756 [DOI] [PubMed] [Google Scholar]
- 41. Tellinghuisen TL, Foss KL, Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032 doi:10.1371/journal.ppat.1000032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tellinghuisen TL, Marcotrigiano J, Gorbalenya AE, Rice CM. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576–48587 [DOI] [PubMed] [Google Scholar]
- 43. Tsai YH, et al. 2008. The non-structural 5A protein of hepatitis C virus exhibits genotypic differences in interferon antagonism. J. Hepatol. 49:899–907 [DOI] [PubMed] [Google Scholar]
- 44. Velazquez L, Fellous M, Stark GR, Pellegrini S. 1992. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell 70:313–322 [DOI] [PubMed] [Google Scholar]
- 45. Wakita T, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Watanabe H, et al. 2001. Number and position of mutations in the interferon (IFN) sensitivity-determining region of the gene for nonstructural protein 5A correlate with IFN efficacy in hepatitis C virus genotype 1b infection. J. Infect. Dis. 183:1195–1203 [DOI] [PubMed] [Google Scholar]
- 47. Wohnsland A, Hofmann WP, Sarrazin C. 2007. Viral determinants of resistance to treatment in patients with hepatitis C. Clin. Microbiol. Rev. 20:23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zeuzem S, et al. 2004. Peginterferon alfa-2b plus ribavirin for treatment of chronic hepatitis C in previously untreated patients infected with HCV genotypes 2 or 3. J. Hepatol. 40:993–999 [DOI] [PubMed] [Google Scholar]
- 49. Zhang L, et al. 2011. IL28B inhibits hepatitis C virus replication through the JAK-STAT pathway. J. Hepatol. 55:289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhong J, et al. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]