Abstract
Varicella-zoster virus (VZV) is the causative agent of chickenpox and herpes zoster (shingles). After the primary infection, the virus remains latent in sensory ganglia and reactivates upon weakening of the cellular immune system due to various conditions, erupting from sensory neurons and infecting the corresponding skin tissue. The current varicella vaccine is highly attenuated in the skin and yet retains its neurovirulence and may reactivate and damage sensory neurons. The factors involved in neuronal invasion and establishment of latency are still elusive. Previously, we constructed a library of whole-gene deletion mutants carrying a bacterial artificial chromosome sequence and a luciferase marker in order to perform a comprehensive VZV genome functional analysis. Here, screening of dispensable gene deletion mutants in differentiated neuronal cells led to the identification of ORF7 as the first known, likely a main, VZV neurotropic factor. ORF7 is a virion component localized to the Golgi compartment in infected cells, whose deletion causes loss of polykaryon formation in epithelial cell culture. Interestingly, ORF7 deletion completely abolishes viral spread in human nervous tissue ex vivo and in an in vivo mouse model. This finding adds to our previous report that ORF7 is also a skin-tropic factor. The results of our investigation will not only lead to a better understanding of VZV neurotropism but could also contribute to the development of a neuroattenuated vaccine candidate against shingles or a vector for delivery of other antigens.
INTRODUCTION
Varicella-zoster virus (VZV), upon encountering a naïve host, causes a primary infection commonly known as chickenpox (varicella) (1, 4, 5). The disease is generally considered mild and self-resolving even in the absence of treatment (2), although occasionally it has severe and lethal consequences (9, 23). The virus reaches sensory nerve ganglia, where it remains latent for life, unless temporary or permanent immunosuppressive conditions within the host facilitate its reactivation as shingles or herpes zoster (HZ) affecting thoracic, cranial, or lumbosacral dermatomes. Many patients report excruciating and relentless pain during HZ episodes (16, 34, 36, 44). The reactivation is sometimes associated with postherpetic neuralgia (PHN), a severe pain along the affected sensory nerves that can linger for years even after the herpetic rash resolves (4). The drug treatments available to date against VZV-elicited diseases are useful only in alleviating some of the symptoms and in shortening the disease duration but cannot clear the virus or prevent establishment of latency (27, 30). PHN is difficult to manage, especially in the elderly, who frequently suffer from other age-related conditions, and the use of the standard PHN treatment, including tricyclic antidepressants, anticonvulsants, and opioids, can be hazardous (16, 36).
Chickenpox was a ubiquitous childhood disease before the anti-VZV vaccination was mandated in 1995 in the United States. Since then, the numbers of hospitalizations, ambulatory visits, and deaths caused by primary varicella have decreased dramatically (19, 20). However, there is a certain degree of controversy surrounding the most commonly used vaccine strain, the live-attenuated v-Oka, regarding its capacity to protect against reactivation, its mechanistically undefined attenuation, and its known neurovirulence (19–21, 25). Of special concern is the ability of v-Oka to cause HZ in vaccinated individuals and even to spread to naïve hosts (32, 49). A clearer view on this issue is delayed by the fact that the vaccinated population is now in its teens, while reactivations are commonly expected to occur at age 50 and beyond in otherwise healthy subjects, although HZ has been reported in people of all ages. Moreover, the currently available HZ vaccine is simply a higher-titer live-attenuated v-Oka inoculum, which is only 51% effective in lowering the risk of HZ in the elderly and is contraindicated during pregnancy and for those with active infections or with extremely weakened immune systems (29). A worrisome rise in shingles occurrence in young healthy adults in the postvaccination era warrants doubled efforts to prevent this potentially debilitating disease (33, 52, 53). A safer and more effective vaccine is highly desirable.
VZV has a 125-kb genome that encodes 70 open reading frames (ORFs), including factors required for efficient invasion of and egress from specific tissues during the course of natural infection. We sought to identify those VZV factors responsible for neurotropism. In our previous studies, we screened a comprehensive library of whole-gene deletion viral mutants harboring enhanced green fluorescent protein (EGFP) and luciferase markers to determine the essentiality of the individual genes for replication in cultured human skin melanoma cells (MeWo) (60). We reported that 18 ORFs (ORF1, -2, -3, -7, -8, -10, -11, -12, -13, -14, -15, -36, -47, -57, -58, -59, -64/69, and -65) are dispensable for viral replication and that their deletion mutants grow like wild-type virus in MeWo cells (60). Four of these (ORF7, -10, -14, and -47) were also deemed skin-tropic, the last three in accordance with previous publications (8, 40, 41). We further tested these mutants by transfecting viral DNA in a primary permissive cell line, the human retinal epithelial ARPE-19 cells (50), and found all 18 to be replication competent (Table 1). We hypothesized that these genes function as modulators for viral entry, assembly, or egress in different tissues. Of particular interest was whether one or more of these 18 genes dispensable for viral replication were implicated in neuronal infection.
Table 1.
Summary of the growth abilities of 18 ORF deletion mutants in vitroa
| VZV ORF | Growth in cell line: |
HSV-1 homolog | VZV ORF function | |||
|---|---|---|---|---|---|---|
| MeWob | ARPE-19b | SOCc | SH-SY5Yb | |||
| 1 | + | + | + | + | None | Transmembrane protein |
| 2 | + | + | + | + | None | Unknown |
| 3 | + | + | + | + | UL55 | Unknown |
| 7 | + | + | − | − | UL51 | Tegument protein with unknown functiond |
| 8e | + | + | + | + | UL50 | Deoxyuridine triphosphatase |
| 10 | + | + | − | + | UL48 | Tegument protein; transactivator of immediate-early genes |
| 11 | + | + | + | + | UL47 | Tegument protein with unknown functiond |
| 12 | + | + | + | + | UL46 | Tegument protein with unknown functiond |
| 13 | + | + | + | + | None | Thymidylate synthase |
| 14 | + | + | − | + | UL44 | Glycoprotein gC |
| 15 | + | + | + | + | UL43 | Integral membrane proteind |
| 36 | + | + | + | + | UL23 | Deoxypyrimidine kinase |
| 47e | + | + | − | + | UL13 | Serine-threonine kinase |
| 57 | + | + | + | + | None | Tegument protein; role in virion egress |
| 58 | + | + | + | + | UL3 | Unknown |
| 59e | + | + | + | + | UL2 | Uracil-DNA glycosylase |
| 64/69 | + | + | + | + | US10 | Tegument protein with unknown functiond |
| 65 | + | + | + | + | US9 | Virion protein involved in axonal transportd |
The growth abilities of 18 dispensable deletion mutants in MeWo cells and ARPE-19 cells compared to their growth in SH-SY5Y cells and human fetal skin organ cultures (SOC). “+” denotes normal growth and “−” denotes defective or null phenotypes for the ORF deletion mutants in the respective cell type or in SOC. The ORF7 deletion mutant was the only mutant with a severe growth defect in SH-SY5Y cells.
Results from this study.
Results from previous studies.
Putative function based on homology.
Partial ORF deletion virus study included.
In this study, all 18 deletion mutants were tested in a human neuroblastoma line, SH-SY5Y, following a similar transfection approach (58–60). We confirmed our results in cultured whole dorsal root ganglia (DRG) (24), as well as in the SCID-hu DRG xenograft mouse model established by the Arvin laboratory (55). We report here that ORF7 is the first full-length VZV gene shown to be responsible for neuroinvasion. This characteristic, along with the previously reported skin tropism, makes ORF7 an attractive target for the development of novel therapies against both chickenpox and herpes zoster.
MATERIALS AND METHODS
Viruses.
The parental strain p-Oka was used to generate bacterial artificial chromosome (BAC) luciferase VZV (VZVLuc BAC, designated WT here) as previously published (60). Briefly, WT was maintained and propagated in Escherichia coli DY380. Deletion mutants were created by electroporating PCR-amplified Kanr cassettes with homology arms to the region to be deleted into WT-containing DY380. The correct colonies were selected on kanamycin-LB plates and maintained as 10% glycerol stocks. Revertants were made by rescuing the full-length ORF back into the full deletion mutant. All primers and viruses, including the ORF7 deletion mutant (7D) and its rescue virus (7R), utilized in this study were the same as previously published (60).
Cells and growth curves.
MeWo and ARPE-19 cells were grown in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS), 50 U/ml penicillin, and 50 μg/ml streptomycin (Invitrogen). Growth curves were performed as follows: 6-well plates of confluent ARPE-19 cells were infected with 100 (for plaque characterization and measurements) or with 200 (for growth curves, as indicated in the figure legends) PFU, at a multiplicity of infection (MOI) of 8.3e−5 or 1.7e−4, respectively. Plaque diameters were directly measured using the built-in microruler of the microscope. Growth kinetics measurements were performed by recording luminescence signal (photons/s/cm2/steradian) from triplicate samples with the IVIS50 imaging system for 7 days by adding d-luciferin to a final concentration of 150 μl/ml as previously described (60). To evaluate the effects of infection treatment in time, we utilized repeated-measures analysis of variance (ANOVA). Analyses were undertaken using the program R (http://www.r-project.org/).
SH-SY5Y cells were maintained in DMEM–F-12 (1:1) with 10% FBS, 50 U/ml penicillin, and 50 μg/ml streptomycin. For differentiation, cells were seeded in 6-well plates, 1 million cells per well. Differentiation was carried out in two steps: a 5-day treatment with DMEM–F-12 (1:1), 5% FBS, 50 U/ml penicillin, 50 μg/ml streptomycin, and 50 μM retinoic acid, followed by a 7-day treatment with DMEM–F-12 with 50 U/ml penicillin, 50 μg/ml streptomycin, 50 nM brain-derived neurotrophic factor (BDNF), and 100 nM nerve growth factor (NGF) (Sigma-Aldrich) as described in the work of Encinas et al. (17). Fresh medium was added to the cultures every third day. Differentiation was determined when cells displayed extensive neurite growth. Cells were infected at the completion of the differentiation treatment (day 12) with 100 PFU/well (MOI, 8.3e−5). Human embryonic stem cell (hESC)-derived neurospheres, containing a mixture of neurons and neural and glial progenitors, were generated by coculturing undifferentiated hESCs with murine PA6 stromal cells. This stromal cell-derived inducing action on hESCs was first described in the work of Pomp et al. (45). Five or six neurospheres, each containing 1,000 to 5,000 cells, were seeded in 24-well culture dishes on polylysine-laminin-coated glass coverslips in differentiation medium consisting of DMEM–F-12 (1:1), B27 supplement (1:50), 2 mM glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, 10 ng/ml NGF, 5 ng/ml BDNF, and 10 ng/ml neurotrophin 3 (NT3). The medium was changed every 3 days. hESC-derived neurons were infected with 150 μl cell-free viral suspension (375 PFU/well; MOI, 0.01 to 0.06) 2 weeks after neurospheres showed extensive neurite outgrowth, indicating differentiation into neurons.
Cell-associated viruses.
Cells seeded in 6-well culture plates were transfected with 1.5 μg purified viral DNA per well using FuGene6 (Roche) according to the manufacturer's protocol. These cells were propagated, harvested, titrated, and maintained in liquid nitrogen as cell-associated viral stocks in FBS with 10% dimethyl sulfoxide (DMSO) (Sigma-Aldrich).
Cell-free viruses.
Cell-free VZVs were provided by Beijing Wantai Co., Ltd. Briefly, VZV-infected MRC-5 cells were washed with phosphate-buffered saline (PBS), resuspended in cryoprotective solution (Beijing Wantai Co., Ltd.), and stored at −80°C. During thawing, frozen cells were broken by shaking them vigorously with 1-mm glass beads. The viral samples obtained were frozen for 2 h at −40°C, lyophilized for 16 h at −25°C, and then brought to 25°C at a speed of 10°C per hour and allowed to dry at room temperature for 6 h. Titers were determined by infectious focus assay.
Cell-free inactivated virus purification.
MRC-5 cells were infected by VZV at an MOI of 0.01 and grown at 35°C in DMEM with 2% FBS. The infected cells were harvested, sonically disrupted, and clarified by centrifugation at 4°C, 3,000 rpm, for 10 min. The virus suspension was inactivated with 1:4,000 β-propionolactone solution at 4°C for 24 h and concentrated by filtration through a 300-kDa ultrafiltration membrane. The concentrated solution was chromatographed at a speed of 5 ml/min over a 60-ml Q Sepharose HP (Pharmacia) column (2.6 by 25 cm) equilibrated with 10 mM PBS, pH 7.5, on an Akta Explorer 100 fast protein liquid chromatography (FPLC) system (Amersham Pharmacia). The virus was eluted with 10 mM PBS (pH 7.5), 350 mM NaCl, and with 10 mM PBS (pH 7.5), 1 M NaCl. The second eluate was collected and chromatographed through a Sepharose 6FF (Pharmacia) column (2.6 by 95 cm) equilibrated in 10 mM PBS (pH 7.5), 150 mM NaCl, operated at a pump speed of 5 ml/min. The first fraction was collected and analyzed by SDS-PAGE and Western blotting.
Ex vivo DRG cultures and growth curves.
Human fetal DRG (1- to 2-mm3) cells were dissected from 18- to 20-week-old spinal cord specimens, obtained in accordance with approved protocols from Zhongshan Hospital (Xiamen, China). DRG were rinsed in PBS, cleansed of connective tissue, and maintained at 37°C in 12-well culture plates with 250 μl DMEM–F-12 (1:1) and 2% FBS overnight (48). Next day, infection was carried out using cell-free viral suspensions at 100 PFU/well for 24 h. For daily luciferase activity measurements, fresh medium containing 150 μg/ml d-luciferin was added to the samples and incubated for 10 min at 37°C. The luminescence signal was recorded using the IVIS200 imaging system and LiveImage software (Xenogen). Fresh medium was added to the wells before their return to the incubator.
SCID-hu xenografts and growth curves.
The SCID-hu DRG xenograft model for studying VZV pathogenesis has been established and extensively characterized by Arvin et al. (3, 46, 55–57). DRG obtained as described above were implanted under the kidney capsule of 4- to 6-week-old male C.B-17scid/scid mice. Visual inspection at 4 weeks postxenotransplantation revealed a small (2- to 3-mm3) well-vascularized graft attached to the mouse kidney. After 4 weeks, the implants were surgically exposed and injected with cell-free viral suspensions at 100 PFU. Mice were injected intraperitoneally with 3 mg d-luciferin before each measurement. Luciferase activity within the implants was measured every 48 h for 10 days using the IVIS200 imaging system and LiveImage software.
Western blots.
Cell-free virus pellets or infected whole-cell lysates were boiled in lysis buffer with 0.5% beta-mercaptoethanol (Bio-Rad) and subjected to 12% SDS-PAGE. The proteins were transferred to a polyvinylidene difluoride (PVDF) membrane and probed with mouse monoclonal anti-gE (9H8; Abcam), anti-EGFP (a gift from David Dubnau), anti-ORF7 mouse monoclonal antibody (8H3; T. Cheng and H. Zhu, unpublished data), or antiactin mouse monoclonal antibody (sc-8432; Santa Cruz Biotechnology) and then with horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibodies (Sigma-Aldrich).
IFAs.
For the ORF7 colocalization assay, cells were grown on round glass coverslips (Corning Glass Inc.) in 24-well plates (Falcon; Becton, Dickinson Labware). After viral infection with 100 PFU/well (MOI, 2.49e−6), cells were fixed in 1% paraformaldehyde (PFA; 10 min at room temperature) and permeabilized in 0.2% Triton (20 min on ice) by sequential incubation with primary mouse monoclonal anti-ORF7 antibody and rabbit antigiantin (Abcam) or human antimitochondrial antibody (Abcam) and Texas Red (TR)- or fluorescein isothiocyanate (FITC)-labeled secondary antibodies (Vector Laboratories) for 30 min each (all solutions in PBS). Finally, cells were equilibrated in PBS, stained for DNA with Hoechst 33258 (0.5 μg/ml), and mounted in Fluoromount-G (Fisher Scientific). For all other immunofluorescence assays (IFAs), confluent cells seeded in 6-well plates and infected with 200 PFU per well (MOI, 1.7e−4) were fixed at 7 days postinfection (dpi) and incubated with anti-gE mouse monoclonal antibody (9H8; Abcam). Antibody detection was performed with Alexa Fluor 594-conjugated anti-mouse secondary antibodies (Sigma-Aldrich), and antibodies were mounted in Vectashield mounting solution with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories) and visualized using AxioVision software (Carl Zeiss MicroImaging).
For live-cell imaging and plasma membrane staining, confluent ARPE-19 cells grown on glass coverslips in 6-well plates were infected with 100 PFU (MOI, 8.3e−5) of WT and 7D VZV, respectively. After 7 days, the cells were washed twice with PBS and then incubated for 10 min at 37°C in PBS containing 5 μg/ml Alexa Fluor 594-wheat germ agglutinin (WGA) and 2 μM Hoechst 33342 (Image-iT live plasma membrane and nuclear labeling kit; Invitrogen). After incubation, the cells were washed twice with PBS and mounted on microscope slides for visualization using an Olympus IX50 inverted fluorescence microscope and images were captured with a Jenoptik ProgRes C14 camera.
Immunocytochemistry.
DRG xenografts were harvested from SCID mice and immediately fixed in 4% formalin in phosphate buffer (0.1 M, pH 7.4) and embedded in paraffin. Subsequent treatment for immunohistochemical analysis was performed as described above. Three-micrometer sections of DRG were stained with mouse anti-VZV gE monoclonal antibody. WT-infected and 7D-infected DRG at day 10 postinfection were stained with mouse anti-VZV gE monoclonal antibody (Abcam Co., Ltd.), and secondary antibody was biotin-labeled anti-mouse antibody, detected by using HRP-labeled streptavidin (brown staining) (catalog no. KIT-9702; Fuzhou Maixin Biotechnology Development Co., Ltd.). Sections were counterstained with hematoxylin (blue staining).
FISH.
For fluorescence in situ hybridization (FISH) detection of viral (VZV) DNA in the paraffinized tissue section slides, the slides were first deparaffinized with xylene, rehydrated in an ethanol series (100%, 95%, 70%, and 50%), and rinsed with water. Then, the section was treated for 1 h at 37°C with RNase (Roche; 100 μg/ml in PBS) to remove RNA. After refixing in 4% paraformaldehyde (10 min at room temperature), samples were equilibrated in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), dehydrated in ethanol (70, 80, and 100% ethanol for 3 min each at 20°C), air dried, and incubated overnight at 37°C with the biotinylated hybridization probe that was prepared by nick translation using WT VZVLuc BAC DNA as the template based on the procedure described previously (51). For DNA detection, probe and cells were simultaneously heated at 94°C for 4 min to denature DNA. After hybridization, samples were washed at 37°C with 55% formamide in 2× SSC (twice, for 15 min each), 2× SSC (10 min), and 0.25× SSC (twice, for 5 min each). Hybridized probes were labeled with FITC-avidin (Vector Laboratories; 1:500 in 4× SSC plus 0.5% bovine serum albumin [BSA]), and signals were amplified by using biotinylated antiavidin (Vector Laboratories; 1:500). Finally, tissue sections were equilibrated in PBS, stained for DNA with Hoechst 33258 (0.5 μg/ml), and mounted in Fluoromount-G.
Microscopy.
ARPE-19 and SH-SY5Y immunofluorescence was detected using an Olympus IX50 inverted fluorescence microscope and processed with ImageReady software. The hESC preparations were viewed with an Olympus IX70 microscope, photographed using a cooled digital camera (Jenoptiks), and processed using ImageJ software. Images were enhanced using ImageJ and Paint Shop Pro software with all changes in the images (i.e., contrast, brightness, gamma, and sharpening) made evenly across the entire field, and no features were removed or added digitally. For intracellular ORF7 localization assays, cells were examined at ×100 magnification with a Leica TCS SPII confocal laser scanning system. Digital images obtained were cropped and adjusted for contrast with Photoshop.
For transmission electron microscopy (TEM), DRG xenografts were fixed in 2% PFA and 2% glutaraldehyde in phosphate buffer (0.1 M, pH 7.4), postfixed with 1% osmium tetroxide, and incubated in 1% aqueous uranyl acetate overnight. The samples were dehydrated in a series of increasing ethanol concentrations followed by a final propylene oxide step. The samples were embedded in Embed812. Ultrathin sections (50 to 80 nm) were stained with 3.5% aqueous uranyl acetate for 5 min and with 0.2% lead citrate for 3 min. The sections were analyzed using a JEM2100HC transmission electron microscope.
RESULTS
Screening for VZV neurotropic factors.
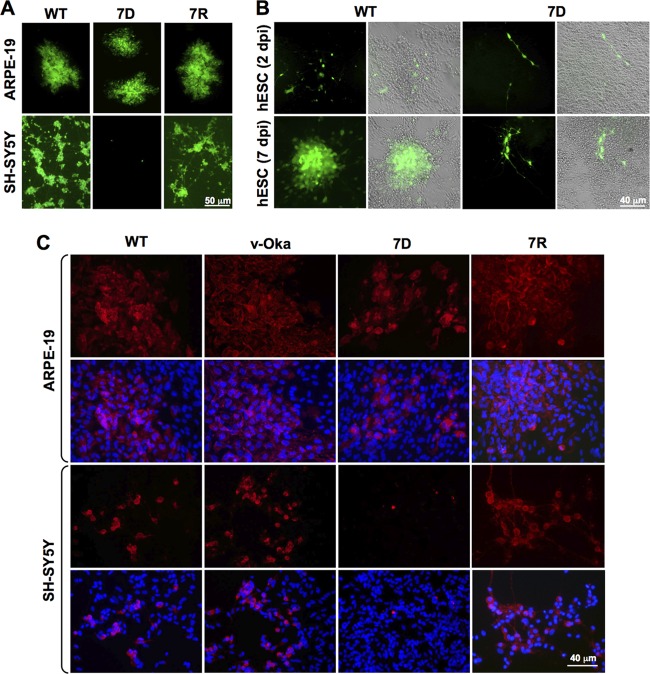
Our previous genome-wide mutagenesis studies showed that VZV contained 26 nonessential ORFs, among which eight ORFs displayed a severe growth defect in MeWo cells and skin organ cultures, while 18 ORFs were fully dispensable in MeWo cells and their growth kinetics were similar to those of WT (60). Among the latter group, four ORFs encoded skin-tropic factors, including the newly identified ORF7 (60). In order to determine whether any of the 18 dispensable ORFs encodes neurotropic factors, we transfected differentiated SH-SY5Y neuroblastoma cells with each of these deletion BAC DNA mutants. As a control, human retinal pigment epithelial (ARPE-19) cells were transfected in parallel. Every experiment was performed in triplicate. The ORF7 deletion mutant (7D) was the only one unable to form viral plaques in differentiated neuroblastoma cells, while the virus grew and spread in MeWo and ARPE-19 cells (60) (Fig. 1A; Table 1).
Fig 1.
ORF7 is required for VZV replication in human neurons in vitro. (A) ARPE-19 and SH-SY5Y cells were plated on glass coverslips in 6-well tissue culture plates and infected with 100 PFU of cell-free VZV particles (MOI, 8.3e−5). SH-SY5Y cells were fully differentiated at the time of infection. At 7 dpi, viral plaques were detected by EGFP expression (green) in WT-, 7D-, and 7R-infected ARPE-19 cells, as well as in WT- and 7R-infected SH-SY5Y cells. Although some isolated EGFP-positive cells are seen in 7D-infected SH-SY5Y cells, no plaques were detected 7 dpi. (B) hESC-derived neuronal cells, prepared as described in Materials and Methods, were infected with cell-free WT and 7D VZV, and live cultures were visualized at 2 and 7 dpi. Green-fluorescent plaques were detected in WT but not in 7D infections at the experimental endpoint, despite the presence of sparse EGFP-positive cells. Right panels in each set represent merged visible and fluorescence images of the left panels. (C) ARPE-19 and SH-SY5Y cells seeded on glass coverslips in 6-well plates were infected with 100 PFU of WT, v-Oka, 7D, and 7R cell-free particles and then fixed and probed with mouse monoclonal antibodies against the viral envelope glycoprotein gE (red, upper panels). Nuclei were detected using DAPI (blue) in the merged lower panels. Plaques were visualized at 7 dpi. Widespread expression of gE in plaque patterns indicates productive viral replication in all samples except for 7D-infected SH-SY5Y cells, where only a few isolated cells stain positive for gE. Notably, WT, 7R, and v-Oka showed robust proliferation in SH-SY5Y cells.
In a previous report, we determined that the 7D phenotype was reversible by rescuing the wild-type ORF7 sequence back into the 7D mutant genome (rescuant 7R). Construction of 7R was described in detail in the work of Zhang et al. (60). In this study, we used the same 7D and 7R mutant viruses characterized before and found that 7D had a null phenotype in neuronal cells whereas 7R grew as well as did the WT (Fig. 1A).
To confirm that ORF7 is required for efficient viral production in neuronal cells, we infected human embryonic stem cell (hESC)-derived neurons (37) with WT and 7D cell-free viral particles (Fig. 1B). Two days postinfection, live cultures were microscopically examined and numbers of EGFP-positive cells were counted and averaged from 10 fields. Isolated green-fluorescing cells were seen in both WT (7 EGFP-positive cells per field) and 7D (3 EGFP-positive cells per field) infections (Fig. 1B, upper panels). After 7 days, however, WT formed large green-fluorescing plaques, while 7D showed only isolated green-fluorescing cells in numbers similar to those on day 2 (>5 EGFP-positive cells per field), indicating that the spread efficiency of 7D was dramatically diminished (Fig. 1B, lower panels). To conclude, no evidence of plaque formation was detected in the case of 7D at any point postinfection either in differentiated human neuroblastoma or in genetically normal human neurons derived from hESC, in contrast to the robust proliferation of WT virus in these cells.
In order to determine whether 7D would be a safer vaccine candidate against VZV than v-Oka, it was of interest to compare the phenotype of 7D with that of v-Oka and WT in neurons. To this end, we performed an immunofluorescence assay of WT-, v-Oka-, 7D-, and 7R-infected ARPE-19 and SH-SY5Y cells using antibodies against the most abundant viral envelope glycoprotein, gE (Fig. 1C). Cells seeded to confluence in 6-well plates were infected with 100 PFU (MOI, 8.3e−5) of each virus and visualized 7 days postinfection. The results indicate that efficient virion production is achieved only by v-Oka and WT in differentiated neuroblastoma, but not by 7D, and that the defect was rescued in the 7R infection. Although a very few, isolated cells expressing EGFP or gE were observed in the 7D-infected neuronal cells 7 days postinfection (Fig. 1A to C), they did not evolve into viral plaques even 14 days after infection (data not shown). ARPE-19 infection was performed as a control to verify that 7D does not harbor a defect in gE expression. These results were confirmed by Western blotting and PCR (data not shown).
Characterization of ORF7.
We used bioinformatic and homology-based predictions as starting points in an effort to determine the function of VZV ORF7. Since 7D is severely impaired in neurons, it was impractical to study it in this system. We had noticed that 7D had a phenotype distinct from that of WT in epithelial cells, so we sought to acquire clues about the role of ORF7 by observing infection characteristics in ARPE-19 cells and extrapolating our findings to neurons.
To date, no information is available on VZV ORF7 other than the homology-based prediction that it encodes a nonessential tegument protein possibly involved in viral packaging and/or egress. VZV ORF7 is homologous to UL51 of the other human alphaherpesviruses, herpes simplex virus 1 (HSV-1) and HSV-2, and orthologous to several animal herpesviruses, including the suidherpesvirus 1 (PrV-1 or SHV-1). The gene is 780 bp long with a predicted size of 29 kDa (14). Bioinformatic analysis of VZV ORF7 resulted in the following predictions: (a) globular protein with a structured N terminus consisting of random coil/alpha-helix-rich regions interspaced by extended sheets and a flexible, disordered proline-rich C terminus (12, 35); (b) globular, soluble protein with no predicted transmembrane motifs (18, 28, 38); (c) four predicted phosphorylation sites but none conserved in the other human alphaherpesviruses or in PrV-1, suggesting a potentially different pattern or functional significance of ORF7 phosphorylation, if it indeed occurs (6, 7); (d) conserved cysteine at position 9, possible palmitoylation site (47) but no predicted myristoylation or prenylation (farnesyl/geranyl) sites (39). The HSV homolog UL51 is a tegument phosphoprotein associated with the trans-Golgi network (TGN) and whose deletion caused a decrease in titer and in plaque size (13, 42, 43). UL51 of PrV-1 is also a TGN phosphoprotein, suspected to be required for efficient viral secondary envelopment and egress and whose truncation results in a small-plaque-size phenotype (31).
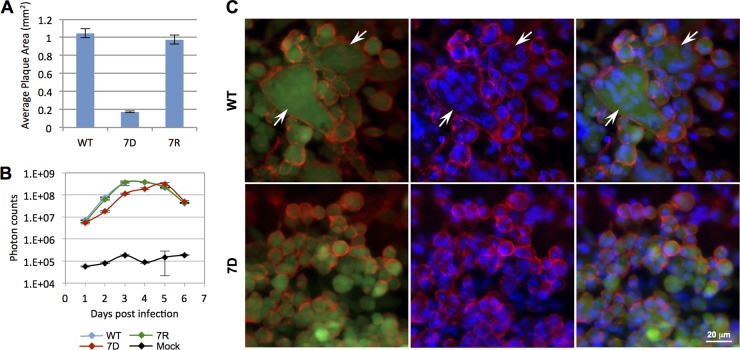
As previously reported for UL51 deletion mutants, 7D also showed a drastic decrease in plaque size in infected ARPE-19 cells, to approximately 16% of that of the WT, based on averaged measurements of over 40 plaques (Fig. 2A). We also performed a growth curve assay by infecting ARPE-19 cells with 200 PFU of cell-free WT, 7R, 7D, and mock, respectively (Fig. 2B). Our results showed that 7D has a significant growth delay compared to the WT and 7R viruses, correlating with the observed decrease in plaque size. In contrast, WT and 7R have comparable plaque sizes (Fig. 2A) and similar growth kinetics (Fig. 2B, P < 3.95e−09, ANOVA).
Fig 2.
ORF7 is required for syncytium formation. (A) ARPE-19 cells were infected with 100 PFU of cell-free WT, 7R, or 7D (MOI, 8.3e−5) and visualized at 7 dpi. Plaque sizes were averaged from a minimum of 40 plaques per virus. Error bars represent standard errors. Notably, the 7D mean plaque size was about 16% of that of WT, while WT and 7R plaque sizes were similar. (B) ARPE-19 cells were seeded in 6-well plates and infected with 200 PFU of WT, 7D, or 7R cell-free VZV or mock infected (PBS). Viral replication was measured daily for 8 days by adding d-luciferin to the wells and recording the bioluminescence signal with the IVIS50 imaging system. The total photon count (photons/s/cm2/steradian) was averaged from 3 samples per virus. Error bars represent standard deviations. The growth kinetic difference between 7D and WT was statistically significant (P < 3.95e−09, ANOVA); however, the presence of 7D plaques suggests that ARPE-19 cells have a compensatory mechanism allowing viral spread in the absence of ORF7. (C) Live ARPE-19 cells infected with WT (upper panels) or 7D (lower panels) cell-free virus and visualized 7 dpi. Left panels show live images of green-fluorescent viral plaques stained with cell membrane WGA-Alexa Fluor 594 probe (red). Middle panels show cell nuclei stained with DAPI (blue) and cell membrane probe (red). Right panels are merged images of left and middle panels. The white arrows indicate multinucleated cells (syncytia) in the WT infection, but no indication of syncytia is seen in 7D. Similar results were obtained using cell-associated viruses.
Live epithelial cells infected with cell-free viruses were visualized at day 7 postinfection (Fig. 2C). After staining for visualization of plasma membrane (red) and nuclei (blue) (detailed in Materials and Methods), syncytium formation, a hallmark of VZV infection, was readily evident in WT infection (indicated by white arrows), demonstrating that WT induced cell-to-cell fusion and formation of multinucleated cellular aggregates. 7D infection caused discrete changes in morphology (cell rounding); however, no evidence of polykarya was observed, suggesting that ORF7 may be involved in the yet-obscure mechanism of plasma membrane fusion by VZV virions (11). The results shown here were from experiments performed with cell-free viruses; comparable results were obtained from cell-associated infections (data not shown).
ORF7 is predicted to be incorporated into the virion as a putative tegument protein (14). To test this prediction, we generated a mouse monoclonal antibody (8H3) against ORF7 protein, whose size is 29 kDa (Fig. 3A), and compared purified virions with infected cell lysates on a Western blot (Fig. 3B). Mouse monoclonal antibody against the VZV envelope glycoprotein gE (82 to 95 kDa) was used as virion component positive control. Interestingly, cell-free virion gE was consistently a faster-migrating species than was its cell-associated virion counterpart, possibly due to partial digestion of envelope components during purification, although infectivity was not hindered. Anti-EGFP (27-kDa) antibody served as a negative control because EGFP, although present in the recombinant viral genome, is a self-standing gene under the control of the simian virus 40 (SV40) promoter; therefore, it only is expressed in infected cells but is not packaged into the viral particle (58, 60). Actin served as an additional control to show that cell-free VZV was free of cellular products. The results indicate that ORF7 protein is indeed a virion component.
Fig 3.

ORF7 is a viral component that localizes to the Golgi network in infected cells. (A) Cell lysates from WT- and 7D-infected cells were subjected to a Western blot assay and probed with mouse monoclonal antibody against ORF7 and HRP-conjugated goat anti-mouse secondary antibodies. Probing with anti-gE antibody (left) served as a positive control. One distinct band of the anticipated size (29 kDa) corresponding to ORF7 product was detected in the WT sample (right). (B) WT cell-free purified particles (V) and WT-infected cell lysates (C) were subjected to a Western blot assay using anti-ORF7 (panel 1), anti-gE (panel 2), anti-EGFP (panel 3), and antiactin (panel 4) mouse monoclonal antibodies. As expected, gE, being a viral envelope constituent, was present in both samples, whereas EGFP and actin were present only in the cell lysates, since they are not packaged into the virion. ORF7 was detected in the cell-free purified VZV, indicating that it is likely an integral component of the virion, as predicted. (C) In order to determine subcellular localization, GFP-negative WT-infected ARPE-19 cells were stained with mouse anti-ORF7 (green). ORF7 was detected in a discrete punctate pattern in the perinuclear region. The Golgi network was labeled with rabbit antigiantin antibodies (red, left), and mitochondria were labeled with human antimitochondrial antibodies (red, right). Examples of infected (I) and uninfected (U) cells are shown with white arrows. Nuclei were visualized using Hoechst 33258 (blue). These images indicate that ORF7 colocalizes with the Golgi network but not with the mitochondria within infected cells. (D) Mock infected ARPE-19 cells (left), as well as WT-infected ARPE-19 cells (middle) and SH-SY5Y cells (right), were fixed and stained with anti-ORF7 antibodies (red). ORF7 has similar distributions in the two cell types.
We sought to determine which cellular compartment is populated by ORF7 protein in infected cells. Figure 3C shows that ORF7 colocalizes with the Golgi network and not with mitochondria in an immunofluorescence assay using anti-ORF7 (green) and anti-Golgi antibodies (red, left) or antimitochondrial antibodies (red, right). This finding is consistent with the localization of ORF7 homologs in HSV-1 and PrV-1, both of which were reported to have a distinct association with the Golgi network in infected cells (31, 42, 43). The Golgi localization appears consistent in both epithelial cells (Fig. 3D, left) and differentiated neuroblastoma cells (right), hinting at the possible role of ORF7 in virion formation, sorting, or egress.
ORF7 neurotropism in human tissue.
To further test ORF7 neurotropism in more physiologically relevant systems, ex vivo and in vivo infections of dorsal root ganglia (DRG) were performed. In the ex vivo experiment, human fetal DRG were infected by coincubating DRG and cell-free WT and 7D suspensions (Fig. 4A) (48). The luminescence signal was recorded for 20 days postinfection as an indirect measure of viral replication within the tissue samples; 7D had a significantly impaired growth in comparison to the WT. The same defect was recapitulated in the SCID-hu DRG xenograft mouse model established by the Arvin laboratory (3, 55, 56). Human fetal DRG xenografts were surgically exposed and directly injected with 100 PFU of WT, 7D, and 7R live virus (Fig. 4B). The luminescence signal was recorded for 10 days. As observed ex vivo, no luminescent signal was detected in the 7D-infected SCID-hu DRG xenograft. Importantly, 7R grew at the same rate as did the WT, demonstrating that the defective phenotype of 7D is fully restored by rescuing ORF7.
Fig 4.
ORF7 is essential for viral replication in human neurons ex vivo and in vivo. (A) Human fetal DRG cultured on NetWell inserts were infected with cell-free WT and 7D viruses at 100 PFU/well. Over a period of 20 days, the bioluminescence signal was recorded every 48 h with the IVIS system after adding d-luciferin to the culture medium. The growth curve generated represents total photon counts (photons/s/cm2/steradian) averaged from 4 samples per virus, and the error bars represent the standard deviations. (B) Human fetal DRG implanted under the kidney capsule of SCID mice were infected by surgical exposure and direct injection of DRG with 100 PFU of cell-free WT, 7R, and 7D VZV, respectively. Luciferase activity in the infected tissue was detected every other day after administration of d-luciferin intraperitoneally for 10 dpi. The growth curve was generated from 5 mice per virus as described for panel A. (C) At 10 dpi, WT- and 7D-infected DRG were harvested from SCID mice and fixed. Sections were stained for gE (brown) and counterstained with hematoxylin (blue). Lower panels represent higher-magnification pictures of selected areas of the upper panels. Red arrowheads indicate neurons. Vacuolization and possible neuron-satellite cell aggregates (black arrows) were observed in the WT-infected sample, correlating with productive replication and widespread presence of viral gE antigen (brown), while 7D samples retained normal DRG morphology and no gE was detected. (D) FISH staining of the samples described in panel A was performed using VZV genomic DNA probes (green). In WT-infected tissue (left panels), intranuclear signal was detected from nuclei of both neurons (red arrowheads) and adjacent satellite cells (white arrowheads), indicative of active viral DNA replication within intranuclear replication centers. In contrast, 7D DNA was not detected in any of the samples tested at the experimental endpoint.
The infected DRG grafts were harvested and fixed for histochemical analysis and immunostaining (Fig. 4C). Large amounts of viral gE antigen were detected in WT- but not in 7D-infected neurons or surrounding satellite cells. For WT, the pervasive presence of gE antigen indicates active viral replication. Vacuolar structures might indicate formation of neuron-satellite cell complexes or satellite cell polykarya (black arrows). In contrast, 7D was unable to propagate within the implant as suggested by the absence of gE antigen; therefore, the appearance of the DRG is that of normal tissue.
As seen in Fig. 4D, fluorescence in situ hybridization (FISH) of DRG xenografts using whole-VZV-genome fluorescently labeled probes revealed the widespread presence of viral DNA in the nuclei of WT-infected neurons (red arrowheads) and satellite cells (white arrowheads). Clustering of viral DNA in specific intranuclear replication centers is an indication of productive replication (bright green fluorescence). In contrast, no viral DNA was detected in the 7D-infected sample, indicating that deletion of ORF7 halted viral spread in the DRG tissue. This also correlates with the immunohistochemistry analysis (Fig. 4C). These results confirm that the absence of ORF7 from the viral genome causes VZV to lose its ability to spread in human DRG tissue.
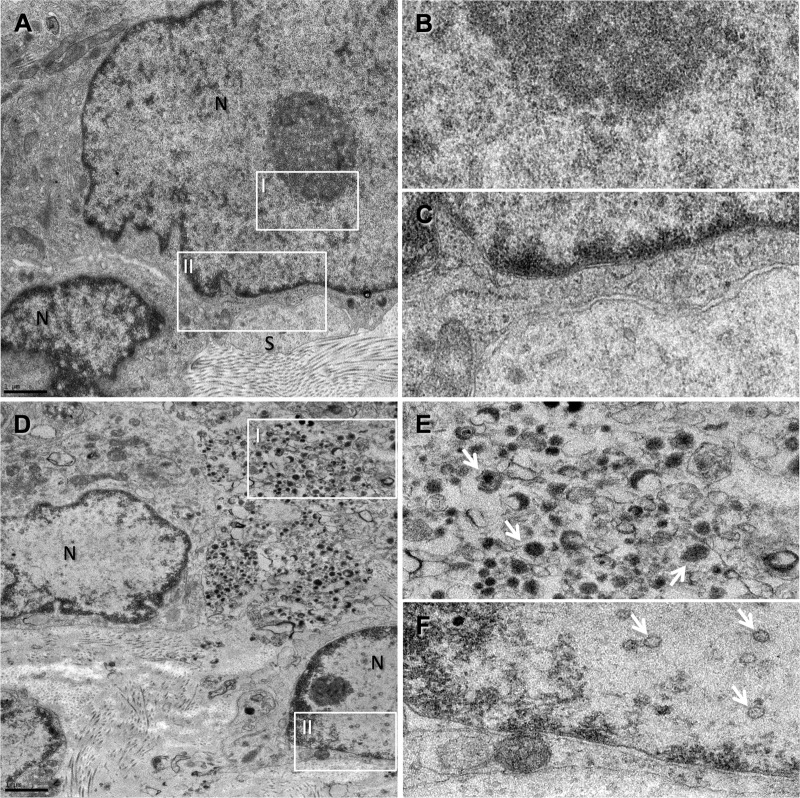
TEM analysis of the WT- and 7D-infected samples further confirmed the mutant's inability to replicate in DRG as indicated by the complete absence of virions or viral capsids (Fig. 5A to C). Active replication of WT VZV (Fig. 5D) was indicated by the detection of clusters of typical VZV polymorphic virions within the cytoplasm of neurons and intercellular regions (Fig. 5E) and the presence of intranuclear capsids (Fig. 5F).
Fig 5.
TEM analysis of infected DRG xenografts shows no evidence of virion production in the 7D samples. TEM images taken from 7D (A to C)- and WT (D to F)-infected DRG samples. The nuclei of sensory neurons (N) and surrounding satellite cells (S) are labeled. In the 7D sample, no evidence of intranuclear capsids (B; box I in panel A) or of cytoplasmic virions (C; box II in panel A) and no tissue damage were observed, whereas the WT-infected DRG sample shows some vacuolization in the plasma membrane region (D), accompanied by the presence of polymorphic virions (white arrows) in the cytoplasm of the infected neuron (E; box I in panel D), as well as intranuclear capsids (F; box II in panel D).
DISCUSSION
VZV is fundamentally different from the simplex viruses and unique in the herpes paradigm. For instance, VZV has a strict cellular association in cultured cells and is almost entirely human host restricted. These qualities posed many challenges in the efforts to characterize its genes and set it apart from the other alphaherpesviruses, whose host promiscuity and capacity to generate sufficient amounts of cell-free particles allowed for a much earlier understanding of their replication and pathogenesis. In order to circumvent some of the problems in the study of VZV, we utilized a VZV carrying a bacterial artificial chromosome sequence and a luciferase marker to generate a library of full and partial gene deletion mutants, which we screened in melanoma cells and skin organ cultures (60), in an effort to reveal all the genes involved in skin tropism. Here, we addressed the question whether VZV encodes factors which render neuronal specificity. To this end, we selected all 18 VZV genes which were dispensable in MeWo cells and screened them in differentiated neuron cultures. We found ORF7 to be the only gene whose absence completely halted viral spread not only in culture but also in ex vivo DRG and in the SCID-hu DRG xenograft model.
The SCID-hu DRG xenograft model has been established and characterized by Arvin's group (3, 56). The model has proven an excellent tool for the study of VZV pathogenesis and latency in human neurons. Moreover, factors implicated in neuropathogenesis are being brought to light by employing this model. Zerboni et al. have also demonstrated that the live-attenuated VZV vaccine strain has replication properties similar to those of the clinical VZV isolates, which correlates with our results in differentiated neuroblastoma cells (54, 56).
All alphaherpesviruses are known for their ability to infect, remain latent, and reactivate from sensory ganglion neurons. However, the mechanisms involved in these processes are largely unknown. While some studies found no evidence of cell-free infectious particles being released from these neurons (10), others suggest that VZV can be shed from neurons during productive infection (24, 37). Gowrishankar et al. (24) demonstrated a ratio of extracellular to intracellular infectious units in intact DRG of 1:16,000 or less, possibly indicating that both cell-to-cell and virus-to-cell spread mechanisms are in effect. Although seemingly intact extracellular virions have been observed not only in neurons (37) but also in skin cell culture by electron microscopy (26), it is unclear why these particles are so rarely infectious in culture, whereas it is known that cell-free virus is produced and shed for efficient transmission in the vesicular rashes associated with the natural infection. In contrast, herpes simplex viruses produce copious amounts of cell-free infectious particles in most cultured cell types tested, including nonhuman lines.
Another striking VZV feature is its divergent requirement for certain glycoproteins compared to homologs in herpes simplex viruses (15). It is, therefore, not surprising that the VZV homologs do not necessarily have conserved function across the alphaherpesviruses, as we see here in the case of ORF7. The VZV ORF7 gene product has the following characteristics in common with its homologs: (a) perinuclear/cytoplasmic/Golgi localization, (b) smaller plaques than WT and altered plaque morphology, (c) dispensability for replication in certain cell lines, (d) similar protein size (∼29 kDa), and (e) a conserved cysteine at position 9—a potential palmitoylation site for Golgi targeting. However, VZV ORF7 is only 65% similar and 47% identical in sequence to HSV-1 UL51 and 78% similar and 63% identical to PrV-1 UL51, which may account for two major differences observed. First, the HSV-1 UL51 deletion mutant showed drastically decreased titers in Vero kidney epithelial cells, whereas 7D was found to have growth properties similar to those of WT in MeWo cells (60) but decreased replication activity in ARPE-19 cells (this study). Second, the PrV UL51 deletion mutant is not essential for neuroinvasion, displaying the same growth curve and killing effect as those of WT in a mouse model (31), while 7D is unable to grow in differentiated neuronal cells in culture or in DRG ex vivo and in vivo.
Observations on the 7D phenotype in cultured ARPE-19 cells lead to the conclusion that the ORF7 gene product is involved in syncytium formation, which might explain its requirement for efficient VZV replication in skin (60). Moreover, ORF7 protein is a virion component that localizes to the Golgi compartment within infected cells, suggesting a possible role in efficient viral packaging and egress.
ORF7 appears to have a determining function in cell-to-cell fusion, and it is likely to work in conjunction with other viral or cellular proteins to achieve this effect. Another possible role of ORF7 is capsid stabilization, but we believe that this is unlikely, because 7D should have a uniform phenotype regardless of cell type. Another possibility might be that ORF7 is important in the process of viral entry into cells, specifically in fusing the viral envelope with the cell membrane. However, it has been speculated that VZV does not favor this mechanism as a mode of entry, particularly in epithelial cells, which explains its almost exclusive cell association in tissue culture (22). VZV, unlike all other alphaherpesviruses, is confined to the cell body instead of being released into the culture medium by cell lysis, which is an indication that VZV has evolved alternative modes of spread within the host, favoring cell-to-cell rather than virus-to-cell spread. It was suggested by Gershon et al. (22) that the cation-independent mannose-6-phosphate receptor (MPRci) present on most VZV-permissive cells may confine VZV to a cell-associated status by rerouting nascent virions to the late endosome. This pathway is averted in suprabasal epidermis keratinocytes, which lose their MPRci receptors as they mature, thus allowing cell-free particle release for transmission from one host to another (22). It is not inconceivable that the cell-associated spread within the host could be a means to evade the immune system during natural infection in order to maintain latency (11), in contrast with the cell-free mode of transmission from host to host.
Finally, our results suggest that the most likely role for ORF7 is efficient packaging, i.e., targeting to sites of secondary envelopment and/or recruitment of other virion components at these sites (which may differ according to cell type) and consequently facilitation of viral egress. As we noted earlier, we observed isolated green-fluorescing cells (Fig. 1A and B), which also express the viral antigen gE (Fig. 1C) in the differentiated neuronal cells infected with 7D. Although we cannot at this time exclude the possibility that spurious EGFP expression in the absence of effective viral gene expression could occur, the alternative explanation could be that the viral mutant entered the cells (albeit with low efficiency), viral genome replication took place, and structural genes were expressed (gE); however, viral egress was severely impaired. This hypothesis is supported by the fact that both PrV-1 and HSV-1 UL51 mutants seem to be defective in secondary envelopment and accumulate in the perinuclear space after the first envelopment event (31, 42, 43). Further studies are needed to ascertain whether this hypothesis is correct.
Syncytium formation may be critical for neuroinvasion; thus, a polykaryon-defective VZV mutant, such as 7D, would be unable to efficiently establish infection of neural tissue. It has been suggested that, within human DRG, VZV infects neurons via fusion of infected satellite cells with the neuronal soma, forming neuron-satellite cell complexes (46, 56). Since viral spread within nervous tissue is dramatically impaired, absence of ORF7 may trigger loss of infectivity specifically due to the incapacity of the virus to fuse potentially infected satellite cells with sensory neurons; however, further evidence is needed to validate this mechanism. An alternative possibility is that although in epithelial cells ORF7 works to sort virions to the Golgi network after the first envelopment event, the situation is different in neurons. Here, secondary envelopment may occur at a different site, as is the case with HSV-1, which acquires its secondary envelope from varicosities and growth cones at the tip of the axons or at axonal mid-distal regions (48). In this case, ORF7 may be essential for sorting to this secondary envelopment site, which remains to be determined.
In summary, we report here the discovery and preliminary characterization of VZV ORF7, the first and, for now, the only known VZV gene required for viral spread in human neurons. These findings could open multiple avenues for future investigations into the mechanism of VZV neuroinvasion, which will have an important impact upon VZV prevention and therapy. 7D, unlike the commonly used vaccine strain v-Oka, is a pure and genetically defined strain lacking the ability to infect human skin and nervous tissue in vivo, thus meeting crucial safety and genetic stability criteria for a next-generation chickenpox and shingles vaccine. 7D could also be used as a safer viral vector for gene therapy and for immunization against other viral or even bacterial antigens.
ACKNOWLEDGMENTS
We thank Ann Arvin and Leigh Zerboni for their kind advice on the SCID-hu DRG xenograft model preparation. We thank Scott Carver for statistical analysis and Frederick Coffman, Drew Boileau, and Min Lu for critically reading the manuscript.
This work was supported by a pilot grant from the Research Center for Minority Institutes (RCMI) Program (2G12RR003050-24), American Cancer Society grant RSG-090289-01-MPC, and NIH/NCRR U54RR022762.
Footnotes
Published ahead of print 6 June 2012
REFERENCES
- 1. Abendroth A, Arvin A. 1999. Varicella-zoster virus immune evasion. Immunol. Rev. 168:143–156 [DOI] [PubMed] [Google Scholar]
- 2. Arvin AM. 1995. Aspects of the host response to varicella-zoster virus: a review of recent observations. Neurology 45:S36–S37 [DOI] [PubMed] [Google Scholar]
- 3. Arvin AM. 2006. Investigations of the pathogenesis of varicella zoster virus infection in the SCIDhu mouse model. Herpes 13:75–80 [PubMed] [Google Scholar]
- 4. Arvin AM. 2001. Varicella-zoster virus, p 2731–2767 In Knipe DM, et al. (ed), Fields virology, 4th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 5. Arvin AM. 2001. Varicella-zoster virus: molecular virology and virus-host interactions. Curr. Opin. Microbiol. 4:442–449 [DOI] [PubMed] [Google Scholar]
- 6. Blom N, Gammeltoft S, Brunak S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351–1362 [DOI] [PubMed] [Google Scholar]
- 7. Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633–1649 [DOI] [PubMed] [Google Scholar]
- 8. Che X, Zerboni L, Sommer MH, Arvin AM. 2006. Varicella-zoster virus open reading frame 10 is a virulence determinant in skin cells but not in T cells in vivo. J. Virol. 80:3238–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choo PW, Donahue JG, Manson JE, Platt R. 1995. The epidemiology of varicella and its complications. J. Infect. Dis. 172:706–712 [DOI] [PubMed] [Google Scholar]
- 10. Cohen JI. 2007. VZV: molecular basis of persistence (latency and reactivation), p 689–699 In Arvin A, et al. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 11. Cole NL, Grose C. 2003. Membrane fusion mediated by herpesvirus glycoproteins: the paradigm of varicella-zoster virus. Rev. Med. Virol. 13:207–222 [DOI] [PubMed] [Google Scholar]
- 12. Combet C, Blanchet C, Geourjon C, Deleage G. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147–150 [DOI] [PubMed] [Google Scholar]
- 13. Daikoku T, Ikenoya K, Yamada H, Goshima F, Nishiyama Y. 1998. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J. Gen. Virol. 79:3027–3031 [DOI] [PubMed] [Google Scholar]
- 14. Davison AJ, Scott JE. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759–1816 [DOI] [PubMed] [Google Scholar]
- 15. Dingwell KS, Doering LC, Johnson DC. 1995. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J. Virol. 69:7087–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drolet M, et al. 2010. The impact of herpes zoster and postherpetic neuralgia on health-related quality of life: a prospective study. CMAJ 182:1731–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Encinas M, et al. 2000. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J. Neurochem. 75:991–1003 [DOI] [PubMed] [Google Scholar]
- 18. Frottin F, et al. 2006. The proteomics of N-terminal methionine cleavage. Mol. Cell. Proteomics 5:2336–2349 [DOI] [PubMed] [Google Scholar]
- 19. Galea SA, et al. 2008. The safety profile of varicella vaccine: a 10-year review. J. Infect. Dis. 197(Suppl 2):S165–S169 [DOI] [PubMed] [Google Scholar]
- 20. Gershon AA, Arvin AM, Levin MJ, Seward JF, Schmid DS. 2008. Varicella vaccine in the United States: a decade of prevention and the way forward. J. Infect. Dis. 197(Suppl 2):S39–S40 [DOI] [PubMed] [Google Scholar]
- 21. Gershon AA, Katz SL. 2008. Perspective on live varicella vaccine. J. Infect. Dis. 197(Suppl 2):S242–S245 [DOI] [PubMed] [Google Scholar]
- 22. Gershon MD, Gershon AA. 2010. VZV infection of keratinocytes: production of cell-free infectious virions in vivo. Curr. Top. Microbiol. Immunol. 342:173–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gilden DH, Kleinschmidt-DeMasters BK, LaGuardia JJ, Mahalingam R, Cohrs RJ. 2000. Neurologic complications of the reactivation of varicella-zoster virus. N. Engl. J. Med. 342:635–645 [DOI] [PubMed] [Google Scholar]
- 24. Gowrishankar K, et al. 2007. Productive varicella-zoster virus infection of cultured intact human ganglia. J. Virol. 81:6752–6756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hambleton S, Steinberg SP, Larussa PS, Shapiro ED, Gershon AA. 2008. Risk of herpes zoster in adults immunized with varicella vaccine. J. Infect. Dis. 197(Suppl 2):S196–S199 [DOI] [PubMed] [Google Scholar]
- 26. Harson R, Grose C. 1995. Egress of varicella-zoster virus from the melanoma cell: a tropism for the melanocyte. J. Virol. 69:4994–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hatchette T, et al. 2008. Foscarnet salvage therapy for acyclovir-resistant varicella zoster: report of a novel thymidine kinase mutation and review of the literature. Pediatr. Infect. Dis. J. 27:75–77 [DOI] [PubMed] [Google Scholar]
- 28. Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 29. Kimberlin DW, Whitley RJ. 2007. Varicella-zoster vaccine for the prevention of herpes zoster. N. Engl. J. Med. 356:1338–1343 [DOI] [PubMed] [Google Scholar]
- 30. Klassen TP, Hartling L, Wiebe N, Belseck EM. 2005. Acyclovir for treating varicella in otherwise healthy children and adolescents. Cochrane Database Syst. Rev. 2005(4):CD002980 doi:10.1002/14651858.CD002980.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klupp BG, et al. 2005. Functional analysis of the pseudorabies virus UL51 protein. J. Virol. 79:3831–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. LaRussa P, Steinberg S, Meurice F, Gershon A. 1997. Transmission of vaccine strain varicella-zoster virus from a healthy adult with vaccine-associated rash to susceptible household contacts. J. Infect. Dis. 176:1072–1075 [DOI] [PubMed] [Google Scholar]
- 33. Leung J, Harpaz R, Molinari NA, Jumaan A, Zhou F. 2011. Herpes zoster incidence among insured persons in the United States, 1993–2006: evaluation of impact of varicella vaccination. Clin. Infect. Dis. 52:332–340 [DOI] [PubMed] [Google Scholar]
- 34. Liesegang TJ. 2004. Herpes zoster virus infection. Curr. Opin. Ophthalmol. 15:531–536 [DOI] [PubMed] [Google Scholar]
- 35. Linding R, Russell RB, Neduva V, Gibson TJ. 2003. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31:3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lydick E, Epstein RS, Himmelberger D, White CJ. 1995. Herpes zoster and quality of life: a self-limited disease with severe impact. Neurology 45:S52–S53 [DOI] [PubMed] [Google Scholar]
- 37. Markus A, et al. 2011. Varicella zoster virus infection of neurons derived from human embryonic stem cells: direct demonstration of axonal infection, transport of VZV and productive neuronal infection. J. Virol. 85:6220–6233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martinez A, et al. 2008. Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics 8:2809–2831 [DOI] [PubMed] [Google Scholar]
- 39. Maurer-Stroh S, Eisenhaber F. 2005. Refinement and prediction of protein prenylation motifs. Genome Biol. 6:R55 doi:10.1186/gb-2005-6-6-r55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moffat JF, et al. 1998. Attenuation of the vaccine Oka strain of varicella-zoster virus and role of glycoprotein C in alphaherpesvirus virulence demonstrated in the SCID-hu mouse. J. Virol. 72:965–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moffat JF, et al. 1998. The ORF47 and ORF66 putative protein kinases of varicella-zoster virus determine tropism for human T cells and skin in the SCID-hu mouse. Proc. Natl. Acad. Sci. U. S. A. 95:11969–11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nozawa N, et al. 2003. Subcellular localization of herpes simplex virus type 1 UL51 protein and role of palmitoylation in Golgi apparatus targeting. J. Virol. 77:3204–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nozawa N, et al. 2005. Herpes simplex virus type 1 UL51 protein is involved in maturation and egress of virus particles. J. Virol. 79:6947–6956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Opstelten W, et al. 2002. Herpes zoster and postherpetic neuralgia: incidence and risk indicators using a general practice research database. Fam. Pract. 19:471–475 [DOI] [PubMed] [Google Scholar]
- 45. Pomp O, et al. 2008. PA6-induced human embryonic stem cell-derived neurospheres: a new source of human peripheral sensory neurons and neural crest cells. Brain Res. 1230:50–60 [DOI] [PubMed] [Google Scholar]
- 46. Reichelt M, Zerboni L, Arvin AM. 2008. Mechanisms of varicella-zoster virus neuropathogenesis in human dorsal root ganglia. J. Virol. 82:3971–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ren J, et al. 2008. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 21:639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saksena MM, et al. 2006. Herpes simplex virus type 1 accumulation, envelopment, and exit in growth cones and varicosities in mid-distal regions of axons. J. Virol. 80:3592–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schmid DS, Jumaan AO. 2010. Impact of varicella vaccine on varicella-zoster virus dynamics. Clin. Microbiol. Rev. 23:202–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schmidt-Chanasit J, et al. 2008. In vitro replication of varicella-zoster virus in human retinal pigment epithelial cells. J. Clin. Microbiol. 46:2122–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tang Q, Bell P, Tegtmeyer P, Maul GG. 2000. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J. Virol. 74:9694–9700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Whitley RJ. 2005. Changing dynamics of varicella-zoster virus infections in the 21st century: the impact of vaccination. J. Infect. Dis. 191:1999–2001 [DOI] [PubMed] [Google Scholar]
- 53. Yih WK, et al. 2005. The incidence of varicella and herpes zoster in Massachusetts as measured by the Behavioral Risk Factor Surveillance System (BRFSS) during a period of increasing varicella vaccine coverage, 1998–2003. BMC Public Health 5:68 doi:10.1186/1471-2458-5-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zerboni L, et al. 2011. Varicella-zoster virus glycoprotein E is a critical determinant of virulence in the SCID mouse-human model of neuropathogenesis. J. Virol. 85:98–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zerboni L, Ku CC, Jones CD, Zehnder JL, Arvin AM. 2005. Varicella-zoster virus infection of human dorsal root ganglia in vivo. Proc. Natl. Acad. Sci. U. S. A. 102:6490–6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zerboni L, Reichelt M, Arvin A. 2010. Varicella-zoster virus neurotropism in SCID mouse-human dorsal root ganglia xenografts. Curr. Top. Microbiol. Immunol. 342:255–276 [DOI] [PubMed] [Google Scholar]
- 57. Zerboni L, et al. 2007. Aberrant infection and persistence of varicella-zoster virus in human dorsal root ganglia in vivo absence of glycoprotein I. Proc. Natl. Acad. Sci. U. S. A. 104:14086–14091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang Z, Huang Y, Zhu H. 2008. A highly efficient protocol of generating and analyzing VZV ORF deletion mutants based on a newly developed luciferase VZV BAC system. J. Virol. Methods 148:197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Z, et al. 2007. Genetic analysis of varicella-zoster virus ORF0 to ORF4 by use of a novel luciferase bacterial artificial chromosome system. J. Virol. 81:9024–9033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang Z, et al. 2010. Genome-wide mutagenesis reveals that ORF7 is a novel VZV skin-tropic factor. PLoS Pathog. 6:e1000971 doi:10.1371/journal.ppat.1000971 [DOI] [PMC free article] [PubMed] [Google Scholar]