Abstract
Simian immunodeficiency virus (SIV) infection of rhesus macaques has become an important surrogate model for evaluating HIV vaccine strategies. The extreme resistance to neutralizing antibody (NAb) of many commonly used strains, such as SIVmac251/239 and SIVsmE543-3, limits their potential relevance for evaluating the role of NAb in vaccine protection. In contrast, SIVsmE660 is an uncloned virus that appears to be more sensitive to neutralizing antibody. To evaluate the role of NAb in this model, we generated full-length neutralization-sensitive molecular clones of SIVsmE660 and evaluated two of these by intravenous inoculation of rhesus macaques. All animals became infected and maintained persistent viremia that was accompanied by a decline in memory CD4+ T cells in blood and bronchoalveolar lavage fluid. High titers of autologous NAb developed by 4 weeks postinoculation but were not associated with control of viremia, and neutralization escape variants were detected concurrently with the generation of NAb. Neutralization escape was associated with substitutions and insertion/deletion polymorphisms in the V1 and V4 domains of envelope. Analysis of representative variants revealed that escape variants also induced NAbs within a few weeks of their appearance in plasma, in a pattern that is reminiscent of the escape of human immunodeficiency virus type 1 (HIV-1) isolates in humans. Although early variants maintained a neutralization-sensitive phenotype, viruses obtained later in infection were significantly less sensitive to neutralization than the parental viruses. These results indicate that NAbs exert selective pressure that drives the evolution of the SIV envelope and that this model will be useful for evaluating the role of NAb in vaccine-mediated protection.
INTRODUCTION
Neutralizing antibodies (NAbs) are key components mediating protective immunity against infectious pathogens. For many viruses, such as influenza, smallpox, and poliovirus, the appearance of NAbs correlates with virus clearance and protection against reinfection (69). The induction of NAbs is used as the gold standard for evaluating vaccine efficacy against these viruses. For human immunodeficiency virus type 1 (HIV-1), there is still no effective vaccine available, and vaccines currently undergoing clinical trials induced only weak NAbs against primary isolates (33, 64). In contrast, autologous NAbs can be detected from HIV-1-infected patients within a few weeks postinfection and drive virus escape from neutralization (19, 38, 51, 52, 63). Cross-neutralizing antibodies are also detectable in approximately 30% of chronically HIV-1-infected patients (15, 54, 59). Studies in nonhuman primates using passively administered antibodies have shown that preexisting NAbs, when given at the appropriate dose and time, are effective in preventing HIV, simian immunodeficiency virus (SIV), and simian-human immunodeficiency virus (SHIV) infection (1, 4, 16, 34, 44, 48, 58, 61). Further evidence for the role of antibody in disease progression is implicated by a subsequent decline of autologous NAb production and significant increase in the post-acute-phase plasma viral load in SIV-infected macaques depleted of B cells by treatment with anti-CD20 antibody (35, 57). Similarly, B cell depletion as a part of clinical treatment for lymphocytic B cell lymphoma resulted in increased viremia in an HIV-1-infected patient (24). Clinical studies on vertical transmission also indicate that the presence of cross-neutralizing antibodies may lower the risk of mother-to-child HIV-1 transmission (14, 56). Overall, these studies indicate that NAbs play an important role in limiting HIV-1 virus replication and suggest that induction of NAbs, especially broadly cross-neutralizing antibodies, is a promising goal for development of an effective vaccine against HIV-1 infection.
The effect of NAbs on HIV-1 virus disease progression and clinical outcome has not been well elucidated. The appearance of NAbs during HIV-1 infection is not associated with clearance of virus replication or protection from disease progression (5, 17, 46). Viruses can quickly escape from neutralization by sequential substitution and insertion/deletion changes in their envelope, and such viruses persist into chronic infection without attenuation or decrease of replicative ability (6, 32, 51, 60, 63). The role of NAbs in long-term nonprogressors (LTNP) is controversial. Some groups report that NAb responses correlated with protection from disease (11, 36, 47), while other groups found that NAb responses were low in LTNP cohorts (15, 45). Clinical studies on individuals with broadly cross-neutralizing Abs showed that these individuals had higher viral loads and that the breadth of NAbs did not affect disease progression (12, 15, 46), confounding a clear role for NAbs in preventing viral pathogenesis. Indeed, escape from neutralization was also observed in individuals from whom highly potent broadly cross-neutralizing antibodies were derived (7, 8, 65). Since clinical outcome and disease progression in HIV infection are affected by many other factors, such as host genetics, diversity of the infecting virus, and difference in routes of infection, it is difficult to evaluate the effect of NAbs on HIV-1 disease progression in such clinical studies.
Simian immunodeficiency virus (SIV)-infected rhesus macaques provide a good surrogate model for AIDS vaccine development. Studies on SIV/SHIV-infected macaques have shown an important role of NAbs in preventing HIV infection and transmission, as described above and as reviewed in more detail by Lifson and Haigwood (31). Several studies conducted to investigate the NAb response in macaques infected with SIV found that NAbs appear to exert selective pressure on virus envelope sequence variation (9, 10, 42, 43, 53, 55). For pigtail macaques infected with the less-pathogenic SIVmneCL8, this is associated with increased virulence and escape from NAb (53). However, SIVmac239 and SIVmac251 are relatively resistant to neutralization in vitro, and the autologous NAb response in SIVmac-infected macaques was weaker and later than generally observed in HIV-infected patients (66). In this present study, we investigated the NAb response in macaques infected with molecular clones derived from the SIVsmE660 virus stock. SIVsmE660 virus is an uncloned pathogenic virus isolate that is widely used as a heterologous challenge virus in vaccine studies. It is more sensitive to neutralization than SIVmac239 and SIVmac251 (39) and therefore might be more comparable to primary HIV-1 isolates. We generated two full-length infectious molecular clones from the uncloned SIVsmE660 virus stock and characterized their pathogenesis in rhesus macaques and the evolution of neutralizing antibody in vivo.
MATERIALS AND METHODS
Viruses.
SIVsmE660 virus was isolated terminally from the spleen of a rhesus macaque (RhE660) following three passages of a primary SIVsm strain in rhesus macaques (23). The neutralization-resistant SIVsmE543-3 clone was derived from a terminal isolate from RhE543 (21); RhE660 was inoculated with blood collected at 1 year postinfection of RhE543. Thus, these are closely related viruses. The uncloned SIVsmE660 virus stock was generated by growth in pig-tailed macaque peripheral blood mononuclear cells (PBMCs) as previously described (18).
Construction of SIVsmE660 molecular clones.
The strategy to construct SIVsmE660 molecular clones was similar to that used previously to clone virus from a rapid-progressor macaque (29). In brief, virus RNA was isolated from cell-free SIVsmE660 virus stock with the QIAamp viral RNA kit (Qiagen, Germany). Reverse transcriptase PCR (RT-PCR) was performed using SuperScript III (Invitrogen, Carlsbad, CA) and Platinum Taq high-fidelity polymerase (Invitrogen) according to the manufacturer's instructions. Two RT-PCR products were amplified using the following pairs of primers: R-F (5′-CAGTCG CTC TGC GGA GAG GCT GG-3′) and Gag-R (5′-TGA CGC AGA CAG TAT TAT AAA GGC TC-3′) or Bgl-F (5′-GGC AGC AGA TCT TGG CCT TGG C-3′) and R-R (5′-TGC TTA CTT CTA AAT GGC AGC TTT-3′). These two PCR products shared the R region of the long-terminal-repeat region (LTR) and contained the U3 and U5 regions of the LTR in separate products. The complete LTR was obtained by an overlapping PCR with these two PCR products as templates and Bgl-F and Gag-R as primers. The LTR PCR product was cloned into PCR4-Topo vector (Invitrogen) and sequenced. The 5′ terminal LTR was subcloned into PUC19 plasmid by restriction digestion with NdeI and NarI. The plasmid was cut with EcoRI and SmaI and ligated with the 3′ terminal LTR, which was cut out with EcoRI and NarI, and the 3′ terminus was blunted with Klenow fragment. The resultant vector, designated pUC-2LTR, has a 5′ LTR up to the NarI site and a 3′ fragment from the BglII site to the 3′ end of the SIV genome. The region spanning the NarI to BglII sites was amplified from SIVsmE660 virus stock by RT-PCR with the primers Nar-F (5′-GGTTGGCGC CCG AAC AGG GAC TT-3′) and Bgl-R (5′-GGC CAA GAT CTG CTG CCA CCT CTG TC-3′). The PCR product was digested with NarI and BglII and directly cloned into PUC-2LTR to obtain full-length SIVsmE660 clones. Several clones were obtained by transformation into STBL2 chemically competent cells (Invitrogen) with ampicillin selection. Plasmids were prepared with the HiSpeed plasmid midi kit (Qiagen, Germany) and sequenced using an Applied Biosystems 3130XL genetic analyzer as previously described (29).
Preparation of virus stocks.
293T cells were maintained in Gibco GlutaMAX Dulbecco's modified Eagle medium (DMEM) plus 10% fetal calf serum (FCS), 100 U/ml penicillin, and 100 μg/ml streptomycin and transfected with 10 μg plasmid using FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN). Virus stocks were collected from the supernatant of transfected cells after 48 h and filtered with a 0.22-μm filter. The 50% tissue culture infectivity doses (TCID50) of virus stocks were tested on TZM-bl cells (62) and calculated by the Reed and Muench method (49).
Macaque PBMC infection.
Rhesus peripheral blood mononuclear cells (PBMCs) were cultured in complete RPMI 1640 medium containing 10% interleukin-2 (IL-2) and stimulated with 2 μg/ml phytohemagglutinin (PHA) for 72 h. After washing, 106 activated PBMCs were infected with 103 TCID50 of the viruses at 37°C for 60 min. The infected PBMCs were washed and cultured in complete RPMI 1640 medium containing 10% IL-2. Virus production was monitored by the reverse transcriptase (RT) activity of supernatant collected at 3-day intervals. RT values were quantified with a phosphorimaging plate (FujiFilm, Japan).
Animal study.
Simian T cell lymphotropic virus (STLV)-, simian retrovirus (SRV)-, and SIV-seronegative, colony-bred rhesus macaques of Indian origin (Macaca mulatta) were maintained in accordance with the guidelines of the Committee on the Care and Use of Laboratory Animals under a NIAID-approved animal study protocol and were housed in a biosafety level 2 (BSL2) facility using BSL3 practices. The TRIM5α genotypes of rhesus macaques were determined as previously described (25), and major histocompatibility complex class I (MHC-I) genotypes were determined by the Rhesus Macaque MHC Typing Core facility at the University of Wisconsin (Table 1). Four rhesus macaques heterozygous for TRIM5TFP/Q were intravenously inoculated with 103 TCID50 of the cloned viruses. Blood, plasma, bronchoalveolar lavage (BAL) fluids, peripheral lymph nodes, and sera were collected sequentially to monitor the viral load, CD4+ T cell subsets, and SIV-specific NAbs.
Table 1.
MHC genotypes of the studied macaquesa
| Macaque | MHC-I genotype | Inoculated virus |
|---|---|---|
| Rh805 | B17 | SIVsmE660-FL6 |
| Rh806 | A01, B01 | SIVsmE660-FL6 |
| Rh807 | A01, A08 | SIVsmE660-FL14 |
| Rh808 | A01, A02 | SIVsmE660-FL14 |
All the macaques have the TRIM5TFP/Q genotype.
Quantification of virus in plasma.
The viral RNA levels in plasma were determined by quantitative reverse transcriptase PCR (RT-PCR) using a Prism 7700 sequence detector (Applied Biosystems, Foster City, CA) with a detection limit of 100 copies/ml, as described previously (22). The primers and probe were as follows: forward primer, 5′-CATCTAGTGGTGGAAACAGGAACA-3′; reverse primer, 5′-AATTTCCTCCTCTGCCACTAGGT-3′; and probe, 5′-ATGCCAGCAACAAGCAGACCAACAGC-3′.
SIV-specific in situ hybridization.
Formalin-fixed, paraffin-embedded lymph node tissues were assayed for SIV viral RNA expression by in situ hybridization (ISH) with SIV-specific RNA probes as previously described (21).
Flow cytometric analysis.
Blood and BAL fluid samples were stained with combinations of the following fluorochrome-conjugated monoclonal antibodies: anti-human CD3-Alexa 700, anti-human CD20 phycoerythrin (PE)-Cy7, anti-human CD4 PE-Cy5.5, anti-human CD8 Pacific Blue, anti-human CD95 PE-Cy5, anti-human CD28 PE-Texas Red, and anti-CCR5 PE. All the antibodies were purchased from BD and used according to the manufacturer's instructions. Labeled cells were fixed with 0.5% paraformaldehyde and analyzed with the fluorescence-activated cell sorter (FACS) LSRFortessa (BD Biosciences). Data analysis was performed with FlowJo software (v7.6.1; TreeStar).
Analysis of Env regions in plasma viral RNA.
Viral RNA was isolated from plasma collected at different time points using the QIAamp viral RNA kit. Reverse transcription was performed using SuperScript III with primer Env-R (5′-CATCATCCACATCATCCATG-3′). The Env region was amplified with primers Env-S (5′-GGTGTTGCTATCATTGTCAGC-3′) and Env-R. The PCR products were cloned into PCR4-Topo vector with the TOPO-TA cloning kit and sequenced. Env sequences were aligned using ClustalW and compared with the parental SIVsmE660 clones. The Env sequence alignments were analyzed by FindModel, and the Hasegawa-Kishino-Yano plus gamma with invariant sites (HKY+G+I) model was selected as the best model to generate phylogenetic trees. Maximum likelihood phylogenetic trees were constructed by MEGA5 with parental SIVsmE660 clones as roots. Nonsynonymous and synonymous nucleotide substitutions were calculated using the Highlighter for Nucleotide Sequences v2.1.1 program. Potential N-linked glycosylation sites were predicted using the N-GlycoSite program (68). (The FindModel, Highlighter, and N-GlycoSite programs are available at http://www.hiv.lanl.gov/.) Env variants were digested with BsmI and BglII and subcloned into the backbone of the parental SIVsmE660-FL6 clone to make chimeric virus variants. Chimeric viruses were made by transfection of 293T cells for neutralization assays.
Neutralization assay.
SIV-specific NAbs were evaluated using the TZM-bl neutralization assay as previously described (40). Briefly, serially diluted, heat-inactivated serum samples were mixed with 50 TCID50 of the viruses and incubated at 37°C for 1 h. After incubation, 104 TZM-bl cells were added to each well with DEAE-dextran at a final concentration of 12.5 μg/ml to enhance virus infectivity. After 40 h of culture at 5% CO2 and 37°C, luciferase activity was measured with the luciferase assay kit (Promega) and read on Mithras LB940 (Berthold Technologies). Average relative luminescence units (RLU) of cell controls were subtracted as backgrounds. The 50% inhibitory dose (ID50) was calculated with nonlinear regression by PRISM5 and expressed as the highest dilution of sera which resulted in a 50% reduction of RLU compared with the virus control.
Nucleotide sequence accession numbers.
The complete sequences of the SIVsmE660 molecular clones have been deposited in GenBank under accession numbers JQ864084 to JQ864087. The Env sequences are deposited in GenBank under accession numbers JQ864088 to JQ864180.
RESULTS
Construction of neutralization-sensitive and replication-competent SIVsmE660 molecular clones.
We obtained four molecular clones from the SIVsmE660 virus stock by RT-PCR which were designated SIVsmE660-FL6, SIVsmE660-FL8, SIVsmE660-FL10, and SIVsmE660-FL14. The genomic organization of these clones was mapped according to the sequence of the molecular clone SIVsmE543-3. All of these clones encoded intact gag, pol, env, vif, vpx, vpr, tat, rev, and nef genes. The sequences were overall more than 99.5% identical to each other. The Env proteins encoded by these clones were 99% identical to one another, with a total of eight amino acid substitutions, mainly within the V1/V2 and V4 regions, and one insertion/deletion polymorphism in V1. In contrast, the E660 clones were 92.6% and 81.3% identical to SIVsmE543-3 and SIVmac239, respectively. Most of the differences were located in the V1/V2 region, which is believed to be a main neutralizing epitope in the SIV envelope (43, 55, 66).
Since there are reports indicating that TRIM5α genotypes affect SIVsmE660 acquisition in repeated mucosal challenge models (50, 67), we examined the capsid sequence of Gag of the SIVsmE660 clones. Compared with SIVsmE543-3, substitutions were observed in the cyclophilin A binding loop of capsid in three of the clones (LPA224-226 to PPA224-226 in SIVsmE660-FL8 and FL10, LPA224-226 to LPP224-226 in SIVsmE660-FL14). We therefore evaluated the restriction sensitivity of these clones by a single-cycle infection assay on feline cell lines that stably express different macaque TRIM5α alleles as previously described (25). The three clones with substitutions in the cyclophilin A binding loop had escaped from restriction of TRIM5-cyclophilin A chimera (TRIM5CypA), whereas the other clone was restricted by both TRIM5CypA and TRIM5TFP (SIVsmE660-FL6) (data not shown).
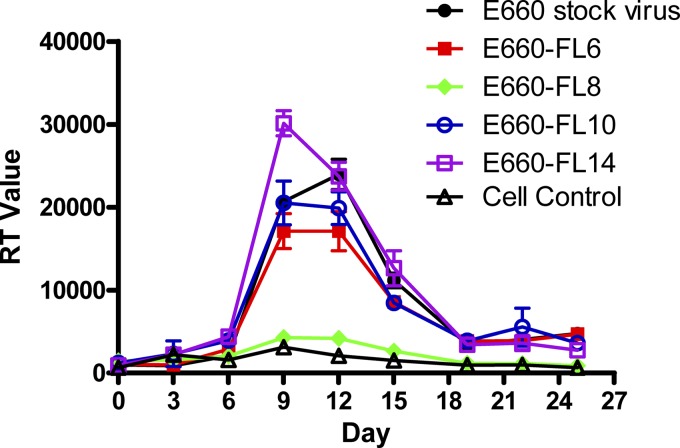
Transfection of 293T cells with SIVsmE660-FL6, -FL10, and -FL14 clones produced high titers (104 TCID50) of virus, whereas the virus yield following transfection of FL8 was 10-fold lower (data not shown). The replication of these clones was evaluated on PHA-stimulated rhesus PBMCs using an equivalent input of virus based on the TCID50. As shown in Fig. 1, the molecular clones SIVsmE660-FL6, -FL10, and -FL14 replicated well in rhesus PBMCs, with kinetics similar to those of the uncloned SIVsmE660 stock virus. Virus production peaked at day 9 postinfection. In contrast, the molecular clone SIVsmE660-FL8 did not replicate efficiently in macaque PBMCs and was not evaluated further.
Fig 1.
Peripheral blood mononuclear cell (PBMC) infection with SIVsmE660 clones. PBMCs were stimulated with 10% IL-2 for 3 days and infected with SIVsmE660 clones at a multiplicity of infection (MOI) of 0.001. Noninfected PBMCs were used as negative controls, and PBMCs infected with SIVsmE660 stock virus were used as positive controls. Virus production was quantified and shown as RT values in supernatants collected at 3-day intervals.
We chose to evaluate two of the three clones in vivo, SIVsmE660-FL6 and SIVsmE660-FL14 (Table 1). The sensitivity to neutralization of these two clones was tested in TZM-bl neutralization assays using a panel of serum samples, including six from SIVsmE660-infected macaques and four from SIVsmE543-3-infected macaques (detailed in Table 2). All of the sera were collected from macaques infected for more than 1 year. SIVsmE543-3 was tested in parallel as a neutralization-resistant control. The ID50 titers of sera are shown in Table 2. SIVsmE660-FL6 and SIVsmE660-FL14 clones were neutralized not only by the homologous sera from SIVsmE660-infected macaques (Rh422, Rh456, RhDC6B, RhDBPX, RhDCM6, and RhDC5M) but also by the heterologous sera from SIVsmE543-3-infected macaques (Rh703, Rh719, Rh735, and Rh777). The ID50 titers of sera against SIVsmE660-FL6 and SIVsmE660-FL14 varied from 6.7 × 103 to 7.7 × 105 ID50. In contrast, the neutralization-resistant virus SIVsmE543-3 was only weakly neutralized by serum samples from two SIVsmE543-3-infected macaques with low ID50 titers (Rh735 and Rh777; ID50 titers of 55 and 36 ID50, respectively). Thus, both of the SIVsmE660 clones were very sensitive to neutralization. This phenotype is not reflective of the more intermediate phenotype of the uncloned SIVsmE660 stock (39), suggesting that the uncloned stock may be a mixture of sensitive and resistant variants.
Table 2.
Neutralization antibody ID50 titers against Env clones from 24-w.p.i plasmaa
| Macaque and virus | ID50 of: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SIVsmE660-infected macaque |
SIVsmE543-3-infected macaque |
|||||||||
| Rh422 | Rh456 | RhDC6B | RhDBPX | RhDCM6 | RhDC5M | Rh703 | Rh719 | Rh735 | Rh777 | |
| Rh805 | ||||||||||
| H805-24w-2 | <20 | 61 | 600 | <20 | <20 | 240 | <20 | <20 | <20 | 29 |
| H805-24w-3 | <20 | 139 | 829 | <20 | <20 | 140 | <20 | <20 | <20 | <20 |
| H805-24w-8 | 29 | 137 | 290 | <20 | <20 | 138 | <20 | <20 | <20 | 28 |
| Rh806 | ||||||||||
| H806-24w-2 | 40,710 | 128,840 | 49,741 | 2,287 | 127,602 | 107,176 | 10,316 | 133,639 | 189,488 | 3,218 |
| H806-24w-3 | 45,229 | 119,214 | 96,190 | 3,516 | 128,497 | 91,130 | 13,470 | 132,642 | 192,350 | 5,874 |
| H806-24w-5 | 88,897 | 140,696 | 210,280 | 5,782 | 172,721 | 124,344 | 22,481 | 277,093 | 276,044 | 8,662 |
| Rh807 | ||||||||||
| H807-24w-4 | <20 | <20 | 326 | <20 | <20 | 73 | <20 | <20 | <20 | <20 |
| H807-24w-5 | <20 | 154 | 342 | <20 | 164 | 140 | <20 | <20 | <20 | <20 |
| H807-24w-7 | <20 | 26 | 261 | <20 | <20 | 359 | <20 | <20 | <20 | <20 |
| Rh808 | ||||||||||
| H808-24w-1 | <20 | 729 | 434 | <20 | <20 | 91 | <20 | <20 | <20 | 25 |
| H808-24w-5 | 23,680 | 65,088 | 65,807 | 5,205 | 133,679 | 91,290 | 8,038 | 137,877 | 167,798 | 3,416 |
| H808-24w-10 | 232 | 331 | 402 | <20 | <20 | 112 | <20 | <20 | <20 | <20 |
| Controls | ||||||||||
| SIVsmE660-FL6 | 53,658 | 167,359 | 82,676 | 7,327 | 118,644 | 73,426 | 14,675 | 109,311 | 243,486 | 6,908 |
| SIVsmE660-FL14 | 88,656 | 775,565 | 294,326 | 22,535 | 313,521 | 507,644 | 13,155 | 251,673 | 430,011 | 6,784 |
| SIVsmE543-3 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | 55 | 36 |
Neutralization antibody ID50 titers against Env clones from 24-w.p.i plasma were determined by a TZM-bl neutralization assay. Sera from six SIVsmE660-infected rhesus macaques and four SIVsmE543-3-infected macaques were used. Control viruses include the neutralization-sensitive virus clones SIVsmE660-FL6 and SIVsmE660-FL14 and the neutralization-resistant virus SIVsmE543-3.
Replication of SIVsmE660 clones in rhesus macaques.
The in vivo replication of SIVsmE660-FL6 and SIVsmE660-FL14 was evaluated in rhesus macaques. We selected four rhesus macaques with the same moderately susceptible TRIMTFP/Q genotype to minimize the potential effect of TRIM5α in this study. The MHC genotypes of these macaques were also examined and listed in Table 1. Two of the macaques (Rh805 and Rh806) were intravenously inoculated with 103 TCID50 of SIVsmE660-FL6, and two (Rh807 and Rh808) were inoculated with the same amount of SIVsmE660-FL14. Infection was established in all four macaques, as indicated by the presence of viral RNA in plasma (Fig. 2A), SIV-infected cells in lymph nodes by ISH staining (Fig. 2B), and seroconversion by Western blot analysis (data not shown). Plasma viral load peaked by 2 weeks postinfection (w.p.i.) in Rh805, Rh807, and Rh808 and varied from 7 × 106 to 1 × 107 copies/ml. The viral loads in these three macaques were subsequently maintained between 104 and 105 during chronic infection. Plasma viremia at both peak and chronic stages of infection in Rh806 was 10-fold lower than that in the other three macaques. A decline of peripheral CD4+ T cells, which was due to the depletion of both naïve (CD28hi CD95lo) (Fig. 2D) and memory (CD28hi CD95hi) (Fig. 2E) subsets of CD4+ T cells, was observed in all of the infected macaques (Fig. 2C). The decrease in the CD4+ T cell percentage in BAL fluids was also observed in all the infected macaques (Fig. 2F), indicative of the depletion of CD4+ T cells at mucosal sites. The macaque with the lowest viral load (Rh806) retained significantly more peripheral memory CD4+ T cells and BAL fluid CD4 T cells than the other infected macaques, consistent with the role of virus in depletion of these cells. The control of viremia in this animal was not explained by trivial factors such as MHC-I haplotype, since all of these animals expressed at least one restrictive MHC-I allele. One macaque (H805) was euthanized at 92 weeks postinfection with clinical AIDS (disseminated Mycobacterium avium infection and lymphoma), confirming the pathogenic nature of these cloned viruses.
Fig 2.
Replication of SIVsmE660 clones in rhesus macaques. (A) Viral loads were quantified and shown as numbers of RNA copies per ml in plasma samples. (B) SIV RNA-positive cells (black spots) were detected by ISH staining from lymph nodes collected at day 10 from macaque RH805. (C to E) Peripheral CD4+ T cells (C), peripheral naïve CD4+ T cells (CD4+ CD28+ CD95−) (D), and peripheral memory CD4+ T cells (CD4+ CD28+ CD95+) (E) were quantified by FACS and shown as the absolute numbers per microliter of blood. (F) Percentages of CD4+ cells in BAL fluid were quantified by FACS.
Neutralizing antibody response.
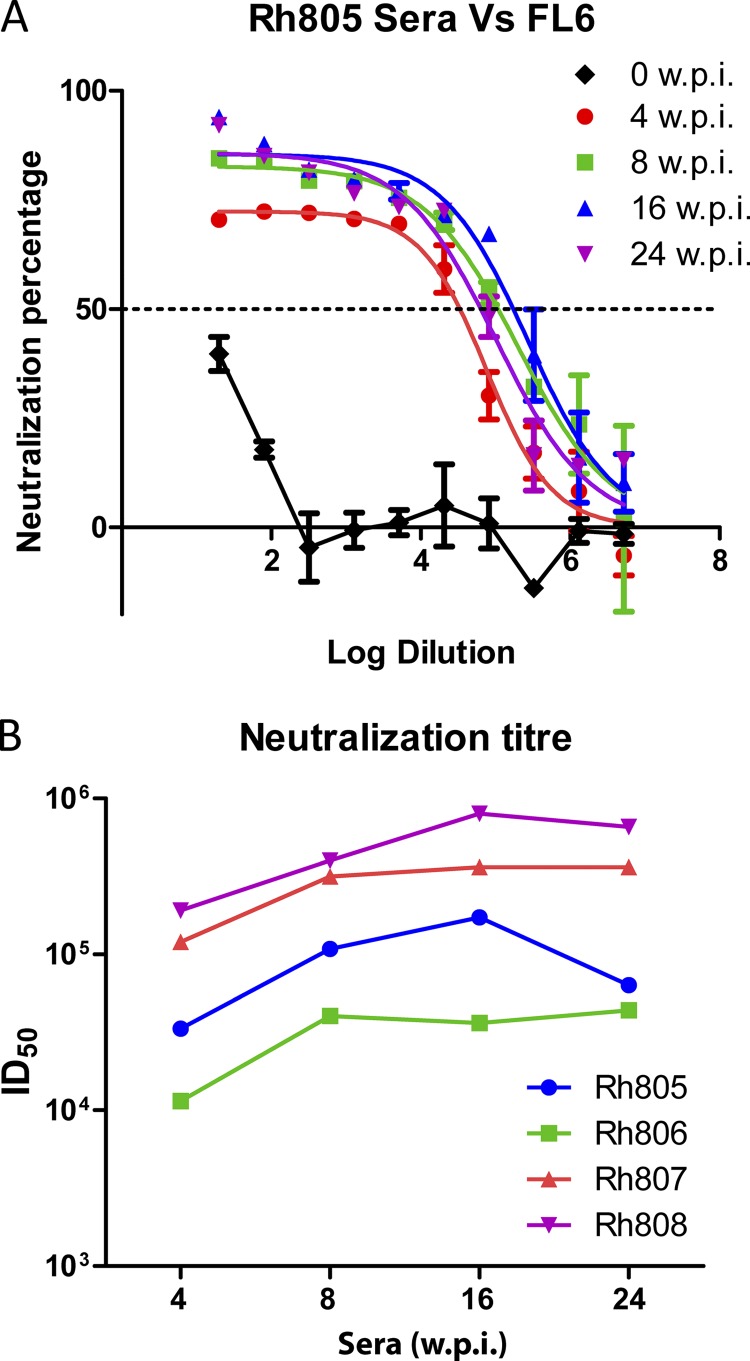
To evaluate the neutralizing antibody response during SIV infection, we collected serum samples at 4, 8, 16, and 24 w.p.i. and tested neutralization against parental E660-FL6 and E660-FL14 by the TZM-bl neutralization assay. As shown in Fig. 3, autologous NAbs were detected as early as 4 w.p.i. in all the macaques. The median reciprocal NAb titer at 4 w.p.i. was 7.7 × 104 per ml, with a range of 1.14 × 103 to 1.92 × 105. The titers of the NAbs reached peak levels by 16 w.p.i. with a median reciprocal titer of 2.68 × 105 per ml (range, 3.63 × 104 to 8 × 105), which was maintained throughout the subsequent infection. NAb titers in the macaque with lower viral load (Rh806) were also lower than those in the other three macaques.
Fig 3.
ID50 titers of sera against parental SIVsmE660 clones. Serum samples were collected at 0, 4, 8, 16, and 24 w.p.i and tested against the parental SIVsmE660 clones FL6 and FL14 by a TZM-bl neutralization assay. (A) Serum samples collected from Rh805 were tested against SIVsmE660-FL6. ID50 titers were calculated with nonlinear regression by PRISM5 and expressed as the reciprocals of the highest dilutions of sera which mount a 50% reduction in RLU from that of the virus control. (B) ID50 titers of sera from infected macaques against the inoculated parental clones.
Env diversity.
Virus isolated from PBMCs of these animals sequentially revealed that the later isolates were resistant to neutralization of contemporaneous serum samples (data not shown). Therefore, to elucidate the mechanisms underlying this apparent neutralization escape, we examined the Env variation during infection in representative animals infected with each of the two E660 clones. The Env region was amplified from Rh805 (inoculated with E660-FL-6) and Rh807 (inoculated with FL-14), and plasma samples were collected at 2, 4, 8, 16, and 24 w.p.i. The Env sequences were compared pairwise with parental SIVsmE660-FL6 and SIVsmE660-FL14 clones, respectively. The diversity of Env sequences was analyzed by bootstrap maximum likelihood trees. Synonymous and nonsynonymous mutations were visualized by Highlighter analysis. As shown in Fig. 4, substitutions in Env were infrequent in clones derived from acute-infection samples but accumulated during infection with more-divergent env variants found in plasma samples during the chronic phase (16 and 24 w.p.i.). Common mutations were not observed in Env sequences cloned from acute-phase plasma samples of Rh805 and Rh807. In contrast, clones derived from chronic-phase samples exhibited a concentration of mutations in the V1 and V4 domains. Common mutations were not observed in gp41. Phylogenetic analysis of these clones (Fig. 4, right panel) showed sequential evolution of env variants over time that tended to cluster by time point.
Fig 4.
Diversity of Env sequences. Env sequences from Rh805 (A) and Rh807 (B) at 2, 4, 8, 16, and 24 w.p.i. were aligned with Env sequences from SIVsmE660-FL6 or SIVsmE660-FL14. Phylogenetic trees (right) were constructed by maximum likelihood methods using the Hasegawa-Kishino-Yano plus gamma with invariant sites (HKY+G+I) model. The left panels show nucleotide mutations in Env regions as silent or synonymous changes (green), nonsynonymous or nonsilent changes (red), and deletions (gray) in a Highlighter plot.
To extend our analysis to all four infected animals, we also examined Env regions from 24-w.p.i. plasma samples of Rh806 and Rh808. The sequences of the V1/V2 and V4 regions of Env gp120 regions from the four macaques are shown in Fig. 5. Similar types of mutations, including insertion/deletion polymorphisms in the V1 region and substitutions in the V4 region, were observed in all of the infected macaques. Deletions in the V1 region were observed as early as 4 weeks postinfection, with scattered substitutions distinct to each animal appearing later. Substitutions in the V4 region were also observed by 16 w.p.i., with a two-amino-acid deletion observed in one animal (Rh805). As in the V1 region, the specific substitutions in V4 varied in each of the four animals but clustered in the region encompassing amino acids (aa) 419 to 435. The diversity in these variable regions, as well as the high conservation in other regions, suggested that these Env variants might have been selected to escape antibody neutralization, as previously reported in studies of SIV infection (28, 55, 66).
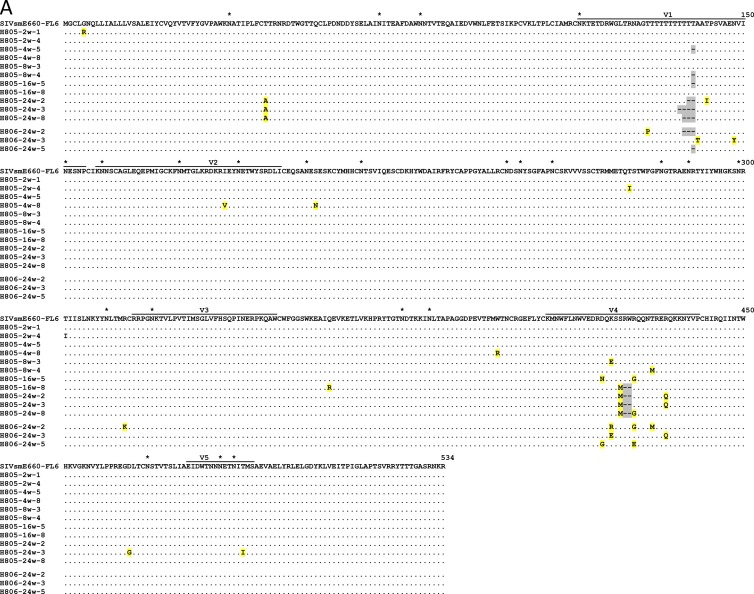
Fig 5.
Alignment of Env gp120 amino acid sequences. The gp120 amino acids of typical Env variants from Rh805 and Rh806 (A) and Rh807 and Rh808 (B) were aligned to parental SIVsmE660-FL6 (A) and SIVsmE660-FL14 (B). Identical amino acids are shown as dots (.), and deletions are shown as dashes (-) and highlighted with gray. Amino acid substitutions are shown and highlighted with yellow. Potential glycosylation sites are indicated with asterisks (*) in SIVsmE660-FL6 and -FL14, and mutations resulting in a change of potential glycosylation sites are highlighted with red in the Env variants.
Alterations in potential N-linked glycosylation sites in the gp120 protein were also mapped, as shown in Fig. 5. The two SIVsmE660 clones differ in terms of potential N-linked glycosylation sites; FL14 has an N70S substitution that results in the loss of a potential N-linked glycosylation site in the C1 domain. All the clones from chronic-phase plasma samples of Rh807 and Rh808 had the reverse S70N mutation, suggesting an important role of this glycosylation site for virus replication or neutralization escape. A K420N mutation, which results in the addition of a glycosylation site in the V4 domain, was observed in a few clones from 24-w.p.i. plasma of Rh807 and Rh808. In contrast, no alterations in potential N-linked glycosylation sites were observed in the SIVsmE660-FL6-infected macaques Rh805 and Rh806.
Neutralization escape.
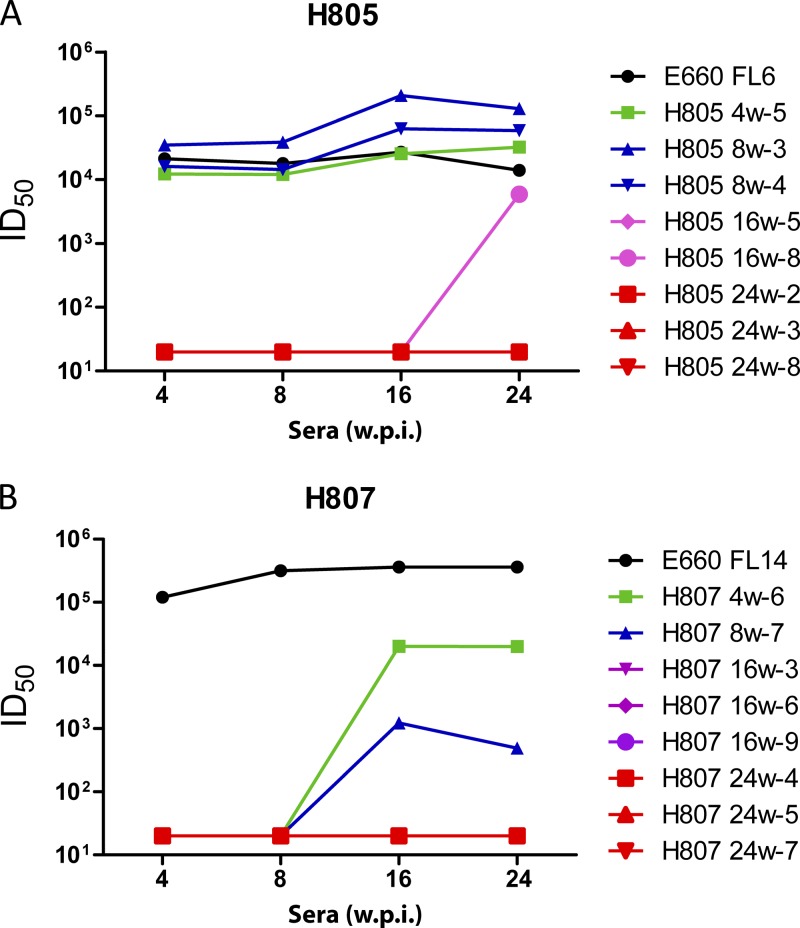
To confirm whether the Env variants were the result of selection by NAbs, we tested the ability of autologous sera to neutralize different Env variants. Representative chimeric viruses were constructed by subcloning Env variants into the SIVsmE660-FL6 backbone. Virus stocks were generated from transfection of 293T cells and evaluated for neutralization sensitivity in TZM-bl neutralization assays. In macaque Rh805 (Fig. 6A), Env variants obtained from acute-phase plasma were still neutralized by sera collected at all time points. The ID50 values were similar or slightly higher than those of the parental E660-FL6 clone. In contrast, Env variants from 16 w.p.i and 24 w.p.i. had completely escaped neutralization (ID50, <20). One Env variant from 16-w.p.i plasma (H805 16w-8) was neutralized by sera at 24 w.p.i. (ID50 titer, 5,901 ID50). In Rh807 (Fig. 6B), all the clones from 16 w.p.i. and 24 w.p.i. had escaped neutralization by serum samples collected from 4 w.p.i. to 24 w.p.i. H807 4w-6 and H807 8w-7, the clones from 4 w.p.i. and 8 w.p.i., respectively, were not neutralized by sera from 4 w.p.i and 8 w.p.i but were still neutralized by sera from 16 w.p.i. and 24 w.p.i., with 10- to 100-fold reduced ID50 titers.
Fig 6.
Neutralization of sera against clones from different time points. Serum samples from Rh805 (A) and Rh807 (B) were tested against selected clones from the same macaque at different time points by the TZM-bl neutralization assay. ID50 titers were calculated and are shown. Viruses tested are identified in the legend by the rhesus ID, the plasma sample that was the source of the envelope clone, and a clone number (e.g., 805 24w-2).
Several 24-w.p.i. clones from the remaining two study animals, Rh806 and Rh808, were also tested with autologous sera. Two clones in Rh808, H808 24w-1 and H808 24w-10, behaved similarly to clones from Rh805 and Rh807 which completely escaped neutralization by the tested serum samples (ID50, <20). H808 24w-5 was still neutralized but with reduced ID50 titers. All the clones from Rh806, the animal that maintained good control of viremia, were still neutralized by the tested sera, although with reduced ID50 titers.
We next investigated the sensitivity to neutralization of Env variants from 24-w.p.i. plasma samples using the panel of sera from chronically infected rhesus macaques (listed in Table 2). Compared with E660-FL6 and E660-FL14, the sensitivity to neutralization was greatly reduced for all the Env variants from Rh805 and Rh807 plasma samples collected at 24 w.p.i. Weak neutralization was observed when sera from macaques infected with SIVsmE660 stock virus (Rh456, RhDC6B, and RhDC5M) were used. The ID50 titers against these Env variants were more than 100-fold lower than those against E660-FL6 clones and E660-FL14 clones. In contrast, all the clones from Rh806 had sensitivity similar to that of the E660-FL6 and E660-FL14 clones. Two clones from Rh808 (Rh808 24w-1 and Rh808 24w-10) were resistant to neutralization, while the other clone, Rh808 24w-5, was still neutralized by all the tested sera, including heterologous sera from macaques infected with SIVsmE543-3.
De novo antibody responses against the escape mutants.
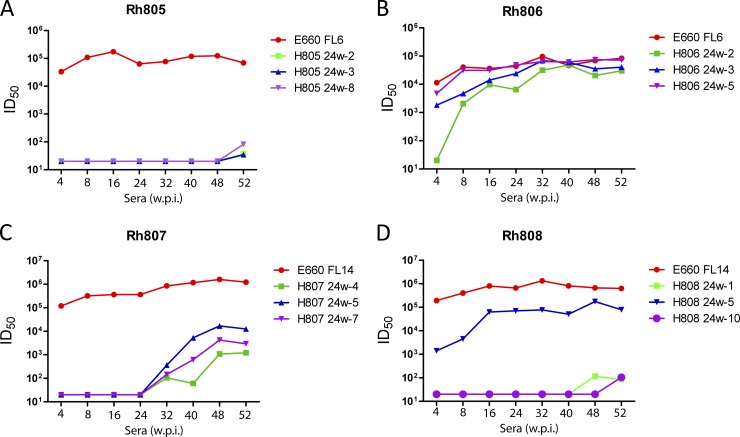
The neutralization assay results indicated that virus variants that acquired resistance to neutralization were responsible for maintaining infection in SIVsmE660-inoculated macaques. We also examined whether the macaques were able to develop NAbs against these neutralization-resistant variants. Sera from later time points (32, 40, 48, and 52 w.p.i.) were tested to evaluate the neutralization properties of the escape variants, using the parental E660-FL6 and E660-FL14 clones as a control. As shown in Fig. 7, the titers of NAbs against the E660-FL6 and E660-FL14 clones remained stable and high in all four macaques. Sera from Rh807 displayed neutralization to the escape clones (H807 24w-4, H807 24w-5, and H807 24w-7), and ID50 titers increased with time. However, the ID50 titers against the escape variants were more than 100-fold lower than those against the E660-FL14 clone. In Rh805 and Rh808, neutralization to resistant clones was not detected until 48 and 52 w.p.i., more than 6 months after the resistant clones were detected. The observed neutralization was extremely weak, with low ID50 titers (34 to 105 ID50).
Fig 7.
Neutralization of late-stage sera against Env escape variants. Serum samples from Rh805, Rh806, Rh807, and Rh808 at a late stage were tested against selected escape Env variants. ID50 titers were calculated and are shown. Viruses tested are identified in the legend by the rhesus ID, the plasma sample that was the source of the envelope clone, and a clone number (e.g., 805 24w-2).
DISCUSSION
In the present study, we evaluated two infectious molecular clones from the SIVsmE660 virus stock and studied their pathogenesis in rhesus macaques. We focused on characterization of the neutralizing antibody response and virus evolution to escape neutralization, providing fundamental information for future studies. Despite their sensitivity to neutralizing antibodies and a rapid autologous neutralizing antibody response, these two clones were able to maintain persistent plasma viremia when inoculated intravenously into rhesus macaques. We restricted our studies to macaques with a moderately sensitive TRIM5 genotype, and therefore, this study may not reflect the full variability among macaques with restrictive or permissive genotypes. However, virus loads were comparable to those in macaques infected with the uncloned SIVsmE660 stock virus (40). The continual decline of CD4+ T cells in the infected macaques is consistent with disease progression, and one animal was recently euthanized due to opportunistic infections (data not shown). Although limited by the small study size, the pathogenicity appears to be less than that observed in macaques inoculated with neutralization-resistant molecular clones, such as SIVmac239 and SIVsmE543-3. These neutralization-sensitive clones will be useful for investigating the role of NAbs in disease progression and vaccine protection. A number of recent studies have suggested that envelope is a critical component of vaccines in the SIV/macaque model (2, 30, 40), and envelope antibodies correlate with protection in the recent RV144 trial (20). However, vaccine studies with SIVmac239 are limited for use in the evaluation of neutralizing antibody response in protection since they are so resistant to neutralization. In addition to the utility of these NAb-sensitive E660 clones, viruses with a neutralizing phenotype intermediate between SIVmac251/239 and SIVsmE660-FL6/FL14, such as these later-stage SIVsmE660 env clones, are ideal for evaluating potential vaccine strategies since they are more representative of the phenotype of primary HIV-1 isolates.
There is still controversy regarding the role of autologous NAb responses in the initial downregulation of primary viremia in HIV-1 infection (41). Since we observed high titers of autologous NAb by 4 weeks, it is possible that these responses might have developed earlier. However, NAb was not detected in plasma samples collected at 2 and 3 weeks (data not shown); this result will need confirmation using serum samples since anticoagulants are known to interfere with neutralizing assays. The appearance of autologous NAbs in macaques in the present study was much earlier than that in SIVmac251-infected macaques (66). Autologous NAbs against parental SIVsmE660 clones were detected by 4 weeks postinfection, more similar to the kinetics in HIV-infected patients (51). The temporal association of neutralization escape mutants and evolution of an autologous NAb response in this study, as well as in SIVmac251-infected animals (66), is entirely consistent with a role of NAbs in the pathogenesis of SIV infection of macaques.
Despite the similarities in kinetics of autologous neutralization, NAb titers in SIVsmE660-infected macaques were higher by orders of magnitude than what have been observed in studies on HIV-1-infected patients (3, 51, 63). This difference may be due to the sensitivity of the E660 clones compared to that of the transmitted/founder virus HIV-1 Envs used for assays to evaluate HIV-1 antibody responses. Measuring NAbs against the inoculated SIV clone should be equivalent to measuring NAbs against the transmitted/founder Env since most of the Env sequences cloned from plasma at the peak of infection are similar to Env sequences of the inoculated SIV clone. But this is also is an explanation for the very high titers in these animals, since the transmitted/founder viruses used for equivalent HIV-1 studies would be significantly more resistant to neutralization, and thus, the titers would be much lower and more difficult to measure. While titers against subtype B HIV-1 Envs tend to remain at moderate levels, there are subtype differences, since early autologous neutralizing antibody responses are quite high in subtype C HIV-1 infection (37). Interestingly, several studies have recently demonstrated that NAb responses are very potent in HIV-2-infected cohorts, with NAb titers ranging from 104 to 106 per ml (13, 26), levels which are similar to titers in the SIVsmE660-infected macaques in this study. While some studies suggest that HIV-2 does not escape NAb as effectively as HIV-1, a study by de Silva et al. demonstrated a similar pattern of envelope evolution and the development of neutralization resistance (13). Compared with HIV-1-infected cohorts, disease progression during HIV-2 infection is usually slower and plasma viral loads are lower, suggesting that the potent NAb response may contribute to the delayed disease progression. The SIVsmE660-infected macaques in the present study should provide a good model to test this hypothesis in vivo. Future studies will compare the disease progressions of macaques infected with SIVsmE660 clones and their more-resistant derivatives that arose during in vivo passage to further understand the relationship between NAb response and disease progression.
In this study, we found that the neutralization-sensitive SIVsmE660 clones maintained persistent infection in the presence of high titers of NAbs by rapidly escaping from neutralizing antibody responses, as previously described in HIV-1 studies (51, 63). Sequence analysis of Env sequences from Rh805 and Rh807 showed that substitutions and deletions in the V1 and V4 domains accumulated over time, suggesting that these domains may be the target of NAbs or that changes in these regions influence the overall conformation of the envelope glycoprotein. Most Env clones from 4 and 8 w.p.i. had mutations only in either the V1 or V4 domain, whereas clones from later time points had mutations in both domains. Neutralization assays also showed that early clones from Rh807 escaped neutralization of concurrent serum samples but were still neutralized by late-phase serum samples (Fig. 6). Env variants from 16 w.p.i. and 24 w.p.i. completely escaped neutralization of serum samples collected before 24 w.p.i. These results revealed that escape mutants gradually acquired neutralization resistance and that the resistant Env variants are selected by NAbs.
In contrast with studies in HIV-infected patients (52, 63), we did not observe consistent changes in glycosylation sites in E660 envelopes during neutralizing antibody escape. Although uncommon in escape variants, we did observe mutations in some Env variants from 24-w.p.i plasma of Rh807 and Rh808 that resulted in the addition of a glycosylation site in the V4 domain. We observed a common S70N mutation, which results in the addition of a glycosylation site in the C1 domain, in all the clones from chronic-stage plasma of Rh807 and Rh808. Interestingly, this glycosylation site is very conserved among SIVmac and SIVsm viruses (27). Whether this glycosylation in the C1 domain contributes to neutralization escape will be investigated in the future.
De novo antibody responses developed against SIVsmE660 escape variants, a result which has not been observed in SIVmac239-infected macaques (55). NAbs against the escape variants from 24-w.p.i. plasma can be detected a few weeks later in all the macaques. These results also suggest that the escape variants are capable of being neutralized. Future studies will be required to identify the neutralizing epitopes recognized by the de novo antibodies. The NAb responses against the escape Env variants were much weaker than those against the parental clones. In Rh805 and Rh808, NAbs against the escape variants were not detected until 6 to 7 months later (48 to 52 w.p.i.). The weak and delayed NAb responses against escape variants may explain why virus replication is not controlled by the potent neutralizing antibody response. The sensitivity to neutralization of these escape variants was also tested with the panel of sera from macaques infected with uncloned SIVsmE660 stock virus or SIVsmE543-3. The escape mutants are more resistant to both homologous and heterologous NAbs than the parental SIVsmE660-FL6 and -FL14 clones, while they are more sensitive to neutralization than SIVsmE543-3. Such variants, with intermediate sensitivity to neutralization, may be a useful addition to challenge virus stocks for vaccine evaluation in the SIV macaque model, used either alone or in combination with their neutralization-sensitive parents.
In summary, we generated two pathogenic SIVsmE660 molecular clones in the present study. These clones are sensitive to neutralization and induce a more potent NAb response in rhesus macaques than neutralization-resistant SIVmac and SIVsmE543-3 viruses and thus can be used as a model to study the role of NAb in disease progression. We also observed virus evolution to escape from neutralization and obtained several escape mutants with intermediate sensitivity to neutralization compared to that of SIVsmE660-FL6, SIVsmE660-FL14, and SIVsmE543-3 clones. SIV clones with intermediate sensitivity to neutralization will be a useful adjunct to vaccine and challenge strains for vaccine development using the SIV model.
ACKNOWLEDGMENTS
We thank Heather Cronise-Santis, Joanne Swerczek, and Richard Herbert in NIHAC for excellent care of the study animals.
This work was supported by the intramural research program of NIAID, NIH.
Footnotes
Published ahead of print 13 June 2012
REFERENCES
- 1. Baba TW, et al. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200–206 [DOI] [PubMed] [Google Scholar]
- 2. Barouch DH, et al. 2012. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature 482:89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Binley JM, et al. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bottiger D, Putkonen P, Oberg B. 1992. Prevention of HIV-2 and SIV infections in cynomolgus macaques by prophylactic treatment with 3′-fluorothymidine. AIDS Res. Hum. Retroviruses 8:1235–1238 [DOI] [PubMed] [Google Scholar]
- 5. Bunnik EM, et al. 2010. Adaptation of HIV-1 envelope gp120 to humoral immunity at a population level. Nat. Med. 16:995–997 [DOI] [PubMed] [Google Scholar]
- 6. Bunnik EM, Lobbrecht MS, van Nuenen AC, Schuitemaker H. 2010. Escape from autologous humoral immunity of HIV-1 is not associated with a decrease in replicative capacity. Virology 397:224–230 [DOI] [PubMed] [Google Scholar]
- 7. Bunnik EM, et al. 2010. Emergence of monoclonal antibody b12-resistant human immunodeficiency virus type 1 variants during natural infection in the absence of humoral or cellular immune pressure. J. Gen. Virol. 91:1354–1364 [DOI] [PubMed] [Google Scholar]
- 8. Bunnik EM, et al. 2009. Changing sensitivity to broadly neutralizing antibodies b12, 2G12, 2F5, and 4E10 of primary subtype B human immunodeficiency virus type 1 variants in the natural course of infection. Virology 390:348–355 [DOI] [PubMed] [Google Scholar]
- 9. Burns DP, Collignon C, Desrosiers RC. 1993. Simian immunodeficiency virus mutants resistant to serum neutralization arise during persistent infection of rhesus monkeys. J. Virol. 67:4104–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burns DP, Desrosiers RC. 1991. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J. Virol. 65:1843–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cao Y, Qin L, Zhang L, Safrit J, Ho DD. 1996. Characterization of long-term survivors of human immunodeficiency virus type 1 infection. Immunol. Lett. 51:7–13 [DOI] [PubMed] [Google Scholar]
- 12. Deeks SG, et al. 2006. Neutralizing antibody responses against autologous and heterologous viruses in acute versus chronic human immunodeficiency virus (HIV) infection: evidence for a constraint on the ability of HIV to completely evade neutralizing antibody responses. J. Virol. 80:6155–6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Silva TI, et al. 2012. Potent autologous and heterologous neutralizing antibody responses occur in HIV-2 infection across a broad range of infection outcomes. J. Virol. 86:930–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dickover R, et al. 2006. Role of maternal autologous neutralizing antibody in selective perinatal transmission of human immunodeficiency virus type 1 escape variants. J. Virol. 80:6525–6533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doria-Rose NA, et al. 2009. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J. Virol. 83:188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emini EA, et al. 1992. Prevention of HIV-1 infection in chimpanzees by gp120 V3 domain-specific monoclonal antibody. Nature 355:728–730 [DOI] [PubMed] [Google Scholar]
- 17. Euler Z, et al. 2010. Cross-reactive neutralizing humoral immunity does not protect from HIV type 1 disease progression. J. Infect. Dis. 201:1045–1053 [DOI] [PubMed] [Google Scholar]
- 18. Goldstein S, et al. 1994. Immunization with whole inactivated vaccine protects from infection by SIV grown in human but not macaque cells. J. Med. Primatol. 23:75–82 [DOI] [PubMed] [Google Scholar]
- 19. Gray ES, et al. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81:6187–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haynes BF, et al. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hirsch V, et al. 1997. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J. Virol. 71:1608–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hirsch VM, et al. 1996. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J. Virol. 70:3741–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hirsch VM, Zack PM, Johnson PR. 1990. Molecular characterization of SIV in tissues from experimentally infected macaques. J. Med. Primatol. 19:287–294 [PubMed] [Google Scholar]
- 24. Huang KH, et al. 2010. B-cell depletion reveals a role for antibodies in the control of chronic HIV-1 infection. Nat. Commun. 1:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kirmaier A, et al. 2010. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 8:e1000462 doi:10.1371/journal.pbio.1000462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kong R, et al. 2012. Broad and potent neutralizing antibody responses elicited in natural HIV-2 infection. J. Virol. 86:947–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuiken C, et al. (ed). 2011. HIV sequence compendium. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, NM [Google Scholar]
- 28. Kuwata T, et al. 2007. A rapid progressor-specific variant clone of simian immunodeficiency virus replicates efficiently in vivo only in the absence of immune responses. J. Virol. 81:8891–8904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuwata T, et al. 2006. Infectious molecular clones from a simian immunodeficiency virus-infected rapid-progressor (RP) macaque: evidence of differential selection of RP-specific envelope mutations in vitro and in vivo. J. Virol. 80:1463–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lai L, et al. 2011. Prevention of infection by a granulocyte-macrophage colony-stimulating factor co-expressing DNA/modified vaccinia Ankara simian immunodeficiency virus vaccine. J. Infect. Dis. 204:164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lifson JD, Haigwood NL. 2011. Lessons in nonhuman primate models for AIDS vaccine research: from minefields to milestones. Cold Spring Harb. Perspect. Med. 2:a007310 doi:10.1101/cshperspect.a007310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mahalanabis M, et al. 2009. Continuous viral escape and selection by autologous neutralizing antibodies in drug-naive human immunodeficiency virus controllers. J. Virol. 83:662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mascola JR, et al. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340–348 [DOI] [PubMed] [Google Scholar]
- 34. Mascola JR, et al. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207–210 [DOI] [PubMed] [Google Scholar]
- 35. Miller CJ, et al. 2007. Antiviral antibodies are necessary for control of simian immunodeficiency virus replication. J. Virol. 81:5024–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Montefiori DC, et al. 1996. Neutralizing and infection-enhancing antibody responses to human immunodeficiency virus type 1 in long-term nonprogressors. J. Infect. Dis. 173:60–67 [DOI] [PubMed] [Google Scholar]
- 37. Moore PL, et al. 2011. Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. J. Virol. 85:3128–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moore PL, et al. 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 5:e1000598 doi:10.1371/journal.ppat.1000598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ourmanov I, Bilska M, Hirsch VM, Montefiori DC. 2000. Recombinant modified vaccinia virus Ankara expressing the surface gp120 of simian immunodeficiency virus (SIV) primes for a rapid neutralizing antibody response to SIV infection in macaques. J. Virol. 74:2960–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ourmanov I, et al. 2009. Improved survival in rhesus macaques immunized with modified vaccinia virus Ankara recombinants expressing simian immunodeficiency virus envelope correlates with reduction in memory CD4+ T-cell loss and higher titers of neutralizing antibody. J. Virol. 83:5388–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Overbaugh J, Morris L. 2012. The antibody response against HIV-1. Cold Spring Harb. Perspect. Med. 2:a007039 doi:10.1101/cshperspect.a007039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Overbaugh J, Rudensey LM. 1992. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J. Virol. 66:5937–5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Overbaugh J, Rudensey LM, Papenhausen MD, Benveniste RE, Morton WR. 1991. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J. Virol. 65:7025–7031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Parren PW, et al. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pereyra F, et al. 2008. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J. Infect. Dis. 197:563–571 [DOI] [PubMed] [Google Scholar]
- 46. Piantadosi A, et al. 2009. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J. Virol. 83:10269–10274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pilgrim AK, et al. 1997. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term-nonprogressive infection. J. Infect. Dis. 176:924–932 [DOI] [PubMed] [Google Scholar]
- 48. Prince AM, et al. 1991. Prevention of HIV infection by passive immunization with HIV immunoglobulin. AIDS Res. Hum. Retroviruses 7:971–973 [DOI] [PubMed] [Google Scholar]
- 49. Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 50. Reynolds MR, et al. 2011. The TRIM5α genotype of rhesus macaques affects acquisition of simian immunodeficiency virus SIVsmE660 infection after repeated limiting-dose intrarectal challenge. J. Virol. 85:9637–9640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rong R, et al. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 5:e1000594 doi:10.1371/journal.ppat.1000594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rudensey LM, Kimata JT, Long EM, Chackerian B, Overbaugh J. 1998. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J. Virol. 72:209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sather DN, et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sato S, et al. 2008. Potent antibody-mediated neutralization and evolution of antigenic escape variants of simian immunodeficiency virus strain SIVmac239 in vivo. J. Virol. 82:9739–9752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Scarlatti G, et al. 1993. Mother-to-child transmission of human immunodeficiency virus type 1: correlation with neutralizing antibodies against primary isolates. J. Infect. Dis. 168:207–210 [DOI] [PubMed] [Google Scholar]
- 57. Schmitz JE, et al. 2003. Effect of humoral immune responses on controlling viremia during primary infection of rhesus monkeys with simian immunodeficiency virus. J. Virol. 77:2165–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shibata R, et al. 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 5:204–210 [DOI] [PubMed] [Google Scholar]
- 59. Simek MD, et al. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. van Gils MJ, et al. 2010. Rapid escape from preserved cross-reactive neutralizing humoral immunity without loss of viral fitness in HIV-1-infected progressors and long-term nonprogressors. J. Virol. 84:3576–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang C, et al. 2009. Ecosystem services assessment of two watersheds of Lancang River in Yunnan, China with a decision tree approach. Ambio 38:47–54 [DOI] [PubMed] [Google Scholar]
- 62. Wei X, et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wei X, et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 64. Wrin T, Nunberg JH. 1994. HIV-1MN recombinant gp120 vaccine serum, which fails to neutralize primary isolates of HIV-1, does not antagonize neutralization by antibodies from infected individuals. AIDS 8:1622–1623 [DOI] [PubMed] [Google Scholar]
- 65. Wu X, et al. 2012. Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site. J. Virol. 86:5844–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yeh WW, et al. 2010. Autologous neutralizing antibodies to the transmitted/founder viruses emerge late after simian immunodeficiency virus SIVmac251 infection of rhesus monkeys. J. Virol. 84:6018–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yeh WW, et al. 2011. The TRIM5 gene modulates penile mucosal acquisition of simian immunodeficiency virus in rhesus monkeys. J. Virol. 85:10389–10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang M, et al. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246 [DOI] [PubMed] [Google Scholar]
- 69. Zinkernagel RM. 2003. On natural and artificial vaccinations. Annu. Rev. Immunol. 21:515–546 [DOI] [PubMed] [Google Scholar]