Abstract
We previously described a novel genetic locus within the ULb′ region of the human cytomegalovirus (HCMV) genome that, while dispensable for replication in fibroblasts, suppresses replication in hematopoietic progenitors and augments replication in endothelial cells. This locus, referred to as the UL133-UL138 locus, encodes four proteins, pUL133, pUL135, pUL136, and pUL138. In this work, we have mapped the interactions among these proteins. An analysis of all pairwise interactions during transient expression revealed a robust interaction between pUL133 and pUL138. Potential interactions between pUL136 and both pUL133 and pUL138 were also revealed. In addition, each of the UL133-UL138 locus proteins self-associated, suggesting a potential to form higher-order homomeric complexes. As both pUL133 and pUL138 function in promoting viral latency in CD34+ hematopoietic progenitor cells (HPCs) infected in vitro, we further focused on this interaction. pUL133 and pUL138 are the predominant complex detected when all proteins are expressed together and require no other proteins in the locus for their association. During infection, the interaction between pUL133 and pUL138 or pUL136 can be detected. A recombinant virus that fails to express both pUL133 and pUL138 exhibited a latency phenotype similar to that of viruses that fail to express either pUL133 or pUL138, indicating that these proteins function cooperatively in latency and do not have independent functions that additively contribute to HCMV latency. These studies identify protein interactions among proteins encoded by the UL133-UL138 locus and demonstrate an important interaction impacting the outcome of HCMV infection.
INTRODUCTION
Human cytomegalovirus (HCMV) is a ubiquitous herpesvirus that establishes a life-long latent infection in the majority of the human population worldwide (14). The cellular and viral mechanisms that contribute to persistence of the virus and underlie the balance of replicative and latent states of infection are poorly understood. An understanding of these mechanisms is critical to designing strategies to control HCMV infection and viral pathogenesis. HCMV remains an important pathogen in immunocompromised individuals, including solid-organ and stem cell transplant patients, where reactivation poses life-threatening disease risks (1, 2). Furthermore, the persistence of the virus in immunocompetent individuals is associated with age-related pathologies, including an increased risk of atherosclerosis, immune senescence, and frailty (9, 18, 20, 21, 23).
The HCMV genome encodes nearly 200 protein-coding genes (14). Of these proteins, approximately 43 are highly conserved among other herpesvirus subfamilies and are essential for viral replication in cultured fibroblasts (4, 14). In fibroblasts, however, the majority of viral genes are dispensable for viral replication. Among these so-called nonessential genes, 20 putative open reading frames (ORFs), including UL133 to UL151, comprising the ULb′ region, are notable (3, 5, 16, 17). The ULb′ region is comprised of 15 kilobases of sequence that is uniformly lost in strains of the virus adapted to replication in cultured fibroblasts. As the genes encoded within the ULb′ region are nonessential for viral replication in fibroblasts, they are hypothesized to be important for virus dissemination, latency, and pathogenesis in the human host (7, 24). Due to the lack of a phenotype in fibroblasts, the coding potential or function has been defined for very few ULb′ genes.
Our previous work has characterized a polycistronic locus spanning UL133 to UL138 within the ULb′ region, which encodes the UL138 latency determinant, pUL138, and three additional proteins, pUL133, pUL135, and pUL136 (8, 22). Each of these proteins is membrane associated and localizes to the Golgi complex (19, 22). As expected of ULb′ sequences, the UL133-UL138 locus is dispensable for virus replication in cultured fibroblasts (22). Intriguingly, the UL133-UL138 locus is required for efficient HCMV replication in primary endothelial cells. Furthermore, we have demonstrated a requirement for the UL133-UL138 locus during a latent infection using an experimental latency model with primary CD34+ human hematopoietic progenitor cells (HPCs) (7, 19, 22). Within the locus, both pUL133 and pUL138 function in promoting a latent infection (19, 22). The existence of three cell type-dependent phenotypes suggests that the proteins within the UL133-UL138 locus differentially contribute to virus infection depending on the context of infection. The mechanism by which each protein functions in infection is not yet known.
We hypothesize that the proteins encoded by the UL133-UL138 locus function in viral infection, in part, through interactions with other viral proteins encoded within this locus. To better understand how proteins encoded in the UL133-UL138 locus cooperate in dictating the outcome of infection, we explored the possibility that these proteins physically interact and sought to map these interactions. Immunoprecipitation (IP) studies revealed a robust interaction between pUL138 and pUL133 in cells transiently expressing each protein. Potential interactions between pUL136 and both pUL133 and pUL138 were also detected in transient-overexpression experiments. Each of the proteins was also able to self-associate. Of these interactions, we detected an interaction between pUL133 and pUL138 or pUL136 during infection in fibroblasts but not in endothelial cells. As pUL133 and pUL138 are both important for the latent phenotype, we explored the possibility that pUL133 and pUL138 function cooperatively in promoting a latent infection. A mutant virus containing disruptions in both UL133 and UL138, TB40E-UL133Stop/UL138Stop, exhibited a loss of latency phenotype but did not produce higher frequencies of infectious centers than viruses containing a single disruption of UL133 or UL138. This result suggests that pUL133 and pUL138 function cooperatively to promote viral latency and do not have independent roles.
MATERIALS AND METHODS
Cells.
Human embryonic kidney 293T (HEK293T) cells (ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES, 1 mM sodium pyruvate, 2 mM l-glutamine, 0.1 mM nonessential amino acids, 100 U/ml penicillin, and 100 mg/ml streptomycin. Primary human embryonic lung fibroblasts (MRC5) (ATCC) were maintained as described previously (22). 293FT cells were cultured in DMEM supplemented with 10% normal calf serum (NCS), and 100 U/ml penicillin, and 100 mg/ml streptomycin. Primary human microvascular lung endothelial cells (HMVECs) were cultured in microvascular endothelial cell growth medium 2 (EGM-2 MV; Lonza, Walkersville, MD). All cells were maintained at 37°C with 5% CO2.
Virus strains.
Recombinant HCMV strains TB40E-WT, TB40E-UL133Stop, TB40E-UL138Stop, TB40E-UL135myc, and TB40E UL136myc were described previously (22). The TB40E-UL133Stop/UL138Stop and UL138Flag viruses were generated by using the TB40E-WT bacterial artificial chromosome (BAC) by recombination in Escherichia coli using a two-step positive-negative selection method that leaves no trace of the engineering process (22, 25, 26). Briefly, the galK cassette was substituted for UL133 in the TB40E-UL138Stop virus BAC (19) in the first step. A PCR product was generated from the UL133 coding sequence in the TB40E-UL133Stop virus genome by using primers used to create TB40E-UL133Stop (22). galK was replaced with the UL133Stop sequences by homologous recombination in the second step. Virus stocks were propagated and stored and titers were determined as described previously (19).
Expression constructs.
The construction of myc epitope-tagged expression constructs in the pCIG2-IRES-EGFP expression vector was described previously (22). To generate pCIG2-UL133HA-eGFP, UL133 was amplified by PCR from the TB40E BAC isolate by using a reverse primer with a hemagglutinin (HA) epitope tag (YPYDVPDYA) (Table 1) and cloned into the NheI and EcoRV sites of pCIG2-IRES-EGFP, as described previously (8). pCIG2-UL136HA-IRES-eGFP was generated in the same way, using a reverse primer containing the HA epitope tag. pCIG2-UL138Flag-PURO was generated by PCR amplifying UL138 from the recombinant HCMV strain TB40E-UL138Flag BAC isolate by using a reverse primer to the Flag epitope tag. The UL138Flag construct was cloned into the NheI and EcoRV sites of pCIG2-IRES-PURO, generating a 3× Flag-tagged UL138 expression construct. Lastly, pCIG2-UL138SII-EE-IRES-PURO was generated in an identical manner, using a reverse primer with a tandem StrepII (SII) (WSHPQFEK)–Glu-Glu (EE) (EYMPME) epitope tag. All constructs were confirmed by restriction digestion and sequencing.
Table 1.
Primers used for cloning into the pCIG vector and to generate recombinant viruses
| Primera | Sequence (5′–3′)b |
|---|---|
| UL133-Fwd | ccggaattcgctagcaccATGGGTTGCGACGTGCACGATCCTTCG |
| UL133HA-Rev | gggggatatcTTACGCGTAATCTGGAACATCGTATGGGTAACCGCCACCGCCCGTTCCGGTCTGATGCTGCTGC |
| T40E-UL133Stop-GalK-Fwd | CCACTGTAGGGATAAATAGTGCGATGGCGTTTGTGAGAGAACGCAGTAGCGCCTGTTGACAATTAATCATCGGCA |
| T40E-UL133Stop-GalK-Rev | GGAAGGAGATGTGGGCCAAGTCGGAAAATTCCTTATCACACCGGGGGCGGGTCAGCACTGTCCTGCTCCTT |
| T40E-UL133Stop-Fwd | CCACTGTAGGGATAAATAGTGCGATGGCGTTTGTGAGAGAACGCAGTAGCGTAAGGTTGAGACGTGCACGATCCTTCG |
| T40E-UL133Stop-Rev | GGAAGGAGATGTGGGCCAAGTCGGAAAATTCCTTATCACACCGGGGGCGGGTTACGTTCCGGTCTGATGCTGCTGCTG |
| T40E-UL138Flag-GalK-Fwd | CCGCGACGCAGTTCACCACCGTAGCCATGGTACATTATCATCAAGAATACACGCCTGTTGACAATTAATCATCGGCA |
| T40E-UL138Flag-GalK-Rev | GTCAAAACGACATTACCGCGATCCGCTCCCCTCTTTTTTCTTTTTCTCATTCAGCACTGTCCTGCTCC |
| T40E-UL138Flag-Fwd | CCGCGACGCAGTTCACCACCGTAGCCATGGTACATTATCATCAAGAATACACGGGCGGTGGCGGTGATTATAAAGATGATGATGATAAAGGGGCA |
| T40E-UL138Flag-Rev | GTCAAAACGACATTACCGCGATCCGCTCCCCTCTTTTTTCTTTTTCTCATTCATTACTTGTCGTCGTCGTCCTTGTAGT |
Fwd, forward; Rev, reverse.
Enzyme sites are in lowercase type.
Lentiviruses.
To generate lentiviruses expressing tagged protein variants, 293FT cells were transfected with pCIG2 variants encoding tagged versions of pUL133, pUL135, pUL136, or pUL138 in combination with the lentiviral packaging vectors pVSV-G, pLP1, and pLP2 (kind gifts from L. Lybarger) in a 2:1:1:1 ratio using Lipofectamine 2000 (Invitrogen, CA) according to the manufacturer's instructions. The supernatant containing lentivirus was harvested at 48 and 96 h posttransfection and pelleted at 3,220 × g for 15 min at 4°C to remove cellular debris. Lentivirus particles were pelleted for 2 h at 17,000 rpm in an SW28 rotor (Beckman Coulter, CA) at 4°C. Virus pellets (100×) were resuspended in Iscove's modified Dulbecco's medium (IMDM) plus 2% bovine serum albumin (BSA) for 1 h on ice, and aliquots were stored at −80°C until use. The titers of lentiviruses on fibroblasts were determined as the 50% tissue culture infective dose (TCID50).
Immunoprecipitation.
HEK293T cells in 100-mm dishes were transfected with Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. A total of 6.93 μg of each uniquely tagged expression construct (pUL133Flag, pUL135HA, pUL136myc, and pUL138SII-EE) was mixed in 500 μl of Opti-MEM, and 14.85 μl of Lipofectamine 2000 was mixed in 500 μl Opti-MEM. Mixtures were combined and added to cells for a 6-h transfection period before replacement with normal growth medium. At 48 h posttransfection, cells were harvested by trypsinization and pelleted at 450 × g for 6 min at 4°C. In the case of viral infection, fibroblasts were infected with recombinant viruses or transduced with lentiviruses expressing Flag- or myc-tagged pUL133, pUL135, pUL136, and pUL138 at a multiplicity of infection (MOI) of 1 for 48 h. Supernatants were aspirated, and cell pellets were resuspended in phosphate-buffered saline (PBS) plus 20 mM iodoacetamide (IAA) and pelleted a second time at 450 × g for 6 min at room temperature. Supernatants were discarded, and cell pellets were flash-frozen in liquid nitrogen and stored at −80°C.
For transfected-cell immunoprecipitations (IP), cells were lysed for 5 to 10 min on ice in 1% NP-40 IP buffer (PBS with 1% Igepal CA-630 [NP-40], 20 mM IAA, 0.2 mM phenylmethanesulfonyl fluoride [PMSF], 1× Halt protease inhibitor cocktail [Pierce], and 1 mM EDTA). Lysates were cleared of nuclear debris by centrifugation at 14,000 × g for 20 min at 4°C. Supernatants were mixed with Zysorbin (Invitrogen) at a final concentration of 0.25%, shaken for 30 min at 4°C, and pelleted at 10,000 × g for 5 min at 4°C. Final protein concentrations were determined by a bicinchoninic acid (BCA) assay (Pierce Biotechnology, Rockford, IL). One hundred micrograms of the transfected-cell lysate was mixed with 0.24, 0.975, 2.25, and 0.324 μg of rabbit antibodies specific to the myc (71D10; Cell Signaling Technology, Danvers, MA), HA (C29F4; Cell Signaling Technology), Flag (F7425; Sigma Aldrich, St. Louis, MO), and Glu-Glu (Cell Signaling Technology) epitope tags, respectively, that had been prebound to protein G plus agarose (overnight at 4°C) and incubated overnight at 4°C. The protein G immune complexes were washed 3 times for 10 min each in NP-40 wash buffer (PBS plus 0.1% Igepal CA-630 [NP-40] and 20 mM IAA). The immune complexes were collected by centrifugation at 10,000 × g for 2 min at 4°C, resuspended in nonreducing SDS sample buffer (0.0625 M Tris HCl, 2% SDS, 10% glycerol, 0.002% bromophenol blue), heated at 95°C for 5 min, and analyzed by immunoblotting.
For infected-cell IP, cells were lysed for 5 to 10 min on ice in 1% NP-40 IP buffer or radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris [pH 8.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and 1 mM PMSF plus 1× Halt protease inhibitor cocktail). Lysates were cleared of nuclear debris by centrifugation at 14,000 × g for 20 min at 4°C. Supernatants were mixed with Zysorbin at a final concentration of 0.25%, shaken for 30 min at 4°C, and pelleted at 10,000 × g for 5 min at 4°C. Final protein concentrations were determined by BCA assay. A total of 200 to 300 μg of the infected cell lysate was combined with 0.5 μg (2 μg/ml final concentration) of rabbit polyclonal antibodies specific to pUL133 or antibody-matched preimmune control sera and incubated overnight at 4°C. The immune complexes from the infected-cell lysates were bound to prewashed protein A beads (Pierce) for 1 h at 4°C. The protein A immune complexes were washed three times for 10 min each with wash buffer (10 mM Tris [pH 8.5], 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, and 1 mM PMSF) and one time for 10 min with PBS to wash away the detergent. The immune complexes were collected by centrifugation at 10,000 × g for 2 min at 4°C, resuspended in nonreducing SDS sample buffer, heated at 95°C for 5 min, and analyzed by immunoblotting.
Immunoprecipitation supernatants were concentrated by trichloroacetic acid (TCA)-acetone precipitation using 1 supernatant volume of TCA to 8 volumes of cold acetone for 3 h at −80°C. Proteins were pelleted at 17,000 × g at 4°C for 30 min before being washed once in cold acetone. Pellets were air dried for 10 min, resuspended in nonreducing sample buffer, heated at 95°C for 5 min, and analyzed by immunoblotting. For comparisons, 100 μg of each lysate was also TCA-acetone precipitated prior to gel loading.
Immunoblotting.
Immunoblotting was described previously (19). Briefly, 100 μg of immunoprecipitate or precipitated supernatant was separated on 4 to 12% NuPAGE Bis-Tris (Invitrogen, CA) or 11% Bis-Tris gels by electrophoresis and transferred onto 0.45-μm polyvinylidene difluoride (Immobilon-FL; Millipore, MA) membranes. The proteins were immunoblotted by using mouse antibodies specific to epitope tags or rabbit polyclonal antibodies directed against each protein (Open Biosystems, Rockford, IL) and detected by using fluorescently conjugated secondary antibodies and the Odyssey infrared imaging system (Li-Cor, NE). All antibodies were used as previously described (22).
Infectious-center assay.
CD34+ HPCs isolated from human cord blood were infected at an MOI of 2 for 20 h in IMDM supplemented with 10% BIT9500 serum substitute (Stem Cell Technologies, Canada), 2 mM l-glutamine, 20 ng/ml low-density lipoproteins, and 50 μM 2-mercaptoethanol. Following infection, pure populations of infected CD34+ HPCs (green fluorescent protein [GFP] positive [GFP+]) were isolated by fluorescence-activated cell sorter (FACS) analysis (FACSAria; BD Biosciences Immunocytometry Systems, San Jose, CA) by using a phycoerythrin-conjugated antibody specific to CD34 (BD Biosciences) and were cultured in transwells above an irradiated (4,000 rads) (137Cs Gammacell-40 irradiator type B; Atomic Energy of Canada Ltd., Ottawa, Canada) M2-10B4 and S1/S1 stromal cell monolayer (13) for 10 to 12 days. The frequency of production of infectious centers was measured by using a limiting-dilution assay as described previously (7). Briefly, infected HPCs were serially diluted 2-fold in long-term bone marrow culture (LTBMC) medium. An aliquot (0.05 ml) of each dilution was added to 12 wells (the first dilution corresponds to 40,000 cells per well) of 96-well tissue culture plates containing MRC5 cells. MRC5 cells were monitored for GFP expression for a period of 14 days. The frequency of infectious centers formed was calculated based on the number of GFP+ cells at each dilution using Extreme Limiting Dilution Analysis (ELDA) software (http://bioinf.wehi.edu.au/software/elda) (10).
RESULTS
Mapping pairwise interactions between proteins encoded within the UL133-UL138 locus.
The UL133-UL138 locus, while dispensable for replication in fibroblasts, is required for replication in endothelial cells and suppresses replication in hematopoietic cells, presumably for the establishment of latency (22). Previously, we demonstrated a loss of latency defect (6, 7) in recombinant viruses when the entire locus, UL133 alone (22), or UL138 alone (19, 22) was disrupted. Furthermore, each of the proteins encoded by the UL133-UL138 locus partially localizes to the Golgi complex when either transiently expressed or expressed within the context of infection (19, 22).
Given the coordinated expression and overlapping localization, we reasoned that the proteins encoded by this locus might function through direct interactions with one another. To explore the potential interactions between the proteins encoded within the UL133-UL138 locus, we cloned UL133, UL135, UL136, and UL138 into the pCIG expression vector with 3′ in-frame 3× Flag, HA, myc, and StrepII–glutamate-glutamate (EE) epitope tags, respectively. Using these tools, we examined all possible pairwise interactions between members of the UL133-UL138 locus. We transiently expressed all combinations of any two epitope-tagged proteins in HEK293T cells. At 48 h posttransfection, lysates were immunoprecipitated with rabbit antibodies specific to one epitope tag in the pair, and complexes were collected and immunoblotted with antibodies specific to the alternant epitope tag in the pair to detect the interaction. Results for each interaction pair are shown in Fig. 1.
Fig 1.
Paired protein interactions from the UL133-UL138 locus. HEK293T cells were transiently transfected with various pairs of expression vectors encoding uniquely tagged ORFs within the UL133-UL138 locus. The protein pair examined in each panel is indicated above the immunoblot. In each panel, protein lysates were assayed for protein-protein interactions by immunoprecipitation using rabbit antibodies (R) against the Flag, HA, myc, or EE epitope tag, and interacting partners were detected by immunoblotting with antibodies specific to the epitope tag of the other protein in the pair, as indicated beside the blot. Lysates incubated with protein G-Agarose+ and no antibody served as negative controls (No Ab.). The immunoprecipitations shown are representative of 3 to 4 independent experiments.
pUL133Flag and pUL135HA failed to coprecipitate when pulled down with antibody specific to either the Flag or HA epitope, suggesting that these two proteins do not interact, at least under the conditions tested (Fig. 1A). A potential interaction between pUL133Flag and pUL136myc was detected (Fig. 1B). For this immunoprecipitation pair, a small proportion of both pUL133Flag and pUL136myc was coprecipitated with antibodies specific to the epitope tag of the other protein, suggesting that this may be a transient or weak interaction. The most apparent and robust interaction occurred in cells expressing pUL133Flag and pUL138SII-EE; pUL133Flag and pUL138SII-EE were coprecipitated by using an antibody to either the EE or Flag tag, respectively (Fig. 1C). This result strongly indicates a direct interaction between pUL133 and pUL138. We detected potential interactions between pUL135HA and either pUL138SII-EE (Fig. 1D) or pUL136myc (Fig. 1E). In each case, small amounts of the interacting partner that were just above the background level were detected, similar to the pUL133Flag-pUL136myc interaction. pUL138SII-EE coprecipitated with the pUL136myc pulldown (Fig. 1F); however, pulling down in the opposite direction on pUL138SII-EE did not coprecipitate pUL136myc above background levels. The precipitation of complexes in only one direction may indicate that the SII-EE epitope may be inaccessible in complexes containing pUL136myc and pUL133SII-EE, perhaps due to the formation of higher-order complexes or interactions with other proteins. This series of immunoprecipitation experiments was conducted under a variety of buffer conditions, with no significant changes in the interactions being observed (data not shown). To ensure that the presence or absence of detectable interactions was not a result of specific epitope tag combinations, immunoprecipitations were also performed in which each protein was tagged with a different epitope or not tagged, in the case of pUL133 and pUL138, with similar results (data not shown). From our results, we consider the pUL133-pUL138 interaction to be the most robust of all possible interactions analyzed. While our data do not exclude weak or transient interactions between pUL135HA and pUL133Flag, pUL136myc, or pUL138SII-EE or between pUL136myc and pUL133Flag or pUL138SII-EE, a definitive demonstration of these interactions requires further work.
UL133-UL138 locus proteins self-associate.
Having identified potential intermolecular interactions between proteins encoded by the UL133-UL138 locus, we next asked whether these proteins had the ability to self-associate. If so, this would suggest the existence of multimeric complexes, consisting of not only multiple proteins from this locus but also multiple molecules of the same protein. We explored this possibility by immunoprecipitating complexes from lysates of HEK293T cells expressing two uniquely tagged variants of each protein. Immunoprecipitates were analyzed for the presence of both epitope tags by immunoblotting (Fig. 2). The precipitation of pUL133 with either the myc or HA antibody precipitated the other tagged form (Fig. 2A). For pUL135, the pUL135HA pulldown coprecipitated pUL135myc, but the pulldown of pUL135myc did not coprecipitate pUL135HA (Fig. 2B). Similar to pUL133, pUL136 expressed as either a myc- or HA-tagged variant coprecipitated with the alternant tagged variant (Fig. 2C). Finally, pUL138 expressed as either a myc- or a Flag-tagged variant coprecipitated the alternant tagged variant (Fig. 2D). These results suggest that each protein encoded by the UL133-UL138 locus can form higher-order complexes with itself.
Fig 2.
UL133-UL138 locus proteins self-associate. HEK293T cells were transiently transfected with sets of two uniquely tagged versions of each of the four proteins expressed from the UL133-UL138 locus and screened for their ability to self-associate by coimmunoprecipitation using rabbit antibodies specific to the myc, HA, and Flag epitope tags, followed by immunoblotting using mouse antibodies specific to the myc, HA, and Flag epitope tags, as indicated beside each blot.
pUL133 and pUL138 interact directly and specifically.
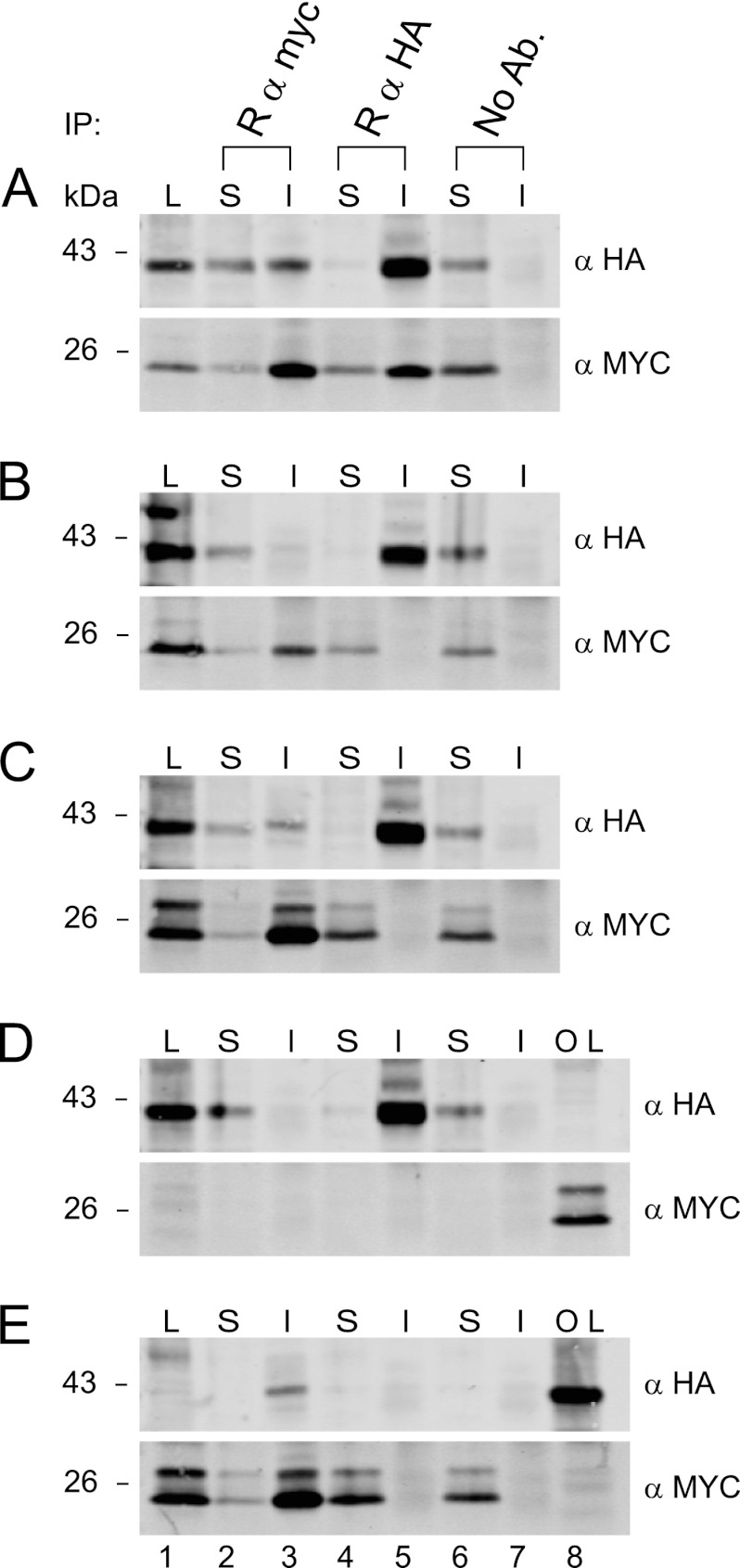
pUL133 and pUL138 demonstrated the most robust interaction in our pairwise analysis (Fig. 1). As both of these proteins are important for the latency phenotype in vitro (22), we focused on this interaction in further analyses. In an effort to better characterize this interaction, we analyzed the interaction quantitatively under various conditions by immunoprecipitation. Immunoprecipitations using rabbit antibodies specific to the myc or HA tag were performed with 100 μg of lysate derived from HEK293T cells expressing both pUL133HA and pUL138myc. Both the immunoprecipitates and the supernatants were evaluated by immunoblotting using mouse antibodies to the myc and HA epitope tags. In lysates containing pUL133HA and pUL138myc, immunoprecipitation with the anti-myc antibody nearly cleared the supernatant (Fig. 3A, bottom, lane 2) and precipitated pUL138myc (Fig. 3A, bottom, lane 3) but also coprecipitated more than 50% of pUL133HA (Fig. 3A, top, lanes 2 and 3). Similarly, the immunoprecipitation of pUL133HA with the anti-HA antibody completely cleared the supernatant (Fig. 3A, top, lane 4) and precipitated pUL133HA (Fig. 3A, top, lane 5) and also coprecipitated more than 50% of pUL138myc (Fig. 3A, bottom, lanes 4 and 5). A no-antibody immunoprecipitation control (Agarose+ beads only) failed to precipitate either protein, as all the protein remained in the supernatant (Fig. 3A, lanes 6 and 7), indicating that the proteins do not nonspecifically precipitate with the protein G-Agarose+ beads. These results demonstrate a specific interaction between pUL133myc and pUL138HA. During coprecipitation, pUL138myc and pUL133HA were nearly, but not completely, cleared from the supernatants by the pulldown of the interacting partner, suggesting that the majority, but not necessarily all, of each protein exists in a pUL133-pUL138 complex.
Fig 3.
pUL133 specifically interacts with pUL138. HEK293T cells were transiently transfected with pUL133HA and pUL138myc alone or in combination, and 100 μg of lysate was analyzed for protein interactions under various conditions by coimmunoprecipitation using rabbit antibodies to the HA and myc epitope tags, followed by immunoblotting with mouse antibodies to the HA and myc epitope tags. Both immunoprecipitates (I) and protein remaining in the supernatants (S) were examined. Lysates (L) served as a positive control for the antibody. (A) Lysate from cells expressing both pUL133myc and pUL138HA. (B) SDS (1%) was added to the lysate from cells expressing both pUL133myc and pUL138HA and boiled prior to immunoprecipitation. (C) Mixed lysates. A lysate from cells expressing only pUL133HA was combined with a lysate expressing only pUL138myc prior to immunoprecipitation. (D) Lysate from cells expressing only pUL133myc. (E) Lysate from cells expressing only pUL138myc. Opposite lysate controls (OL) in lanes 8 of panels D and E are protein expression and antibody controls for the myc and HA blots, respectively.
Importantly, immunoprecipitations from lysates containing both pUL133HA and pUL138myc, which were boiled in 1% SDS prior to coimmunoprecipitation, precipitated only the target of each antibody and failed to coprecipitate the interacting partner (Fig. 3B, lanes 3 and 5). Furthermore, immunoprecipitations from a mixture of two lysates, one containing only pUL133HA combined with one containing only pUL138myc, precipitated only the targets of each antibody and failed to precipitate the interacting partners (Fig. 3C, lanes 3 and 5), indicating that these complexes form in intact cells and do not form postlysis. To demonstrate the specificity of each immunoprecipitation, an anti-myc immunoprecipitation did not precipitate pUL133HA from lysates containing only pUL133HA (Fig. 3D, top, lane 3), whereas pUL133HA was efficiently precipitated by the pulldown with the anti-HA antibody (Fig. 3D, top, lane 5). Furthermore, the anti-HA antibody did not precipitate pUL138myc from lysates containing only pUL138myc (Fig. 3E, bottom, lane 5), whereas pUL138myc was efficiently precipitated by the pulldown with the anti-myc antibody (Fig. 3E, bottom, lane 3). Again, proteins remained in the supernatant in the no-antibody controls (Fig. 3D and E, lanes 6 and 7). Lysates containing each protein were run as a control for the antibodies used for immunoblotting (Fig. 3D and E, lane 8). Taken together, these data indicate a specific and robust interaction between pUL133 and pUL138.
pUL133 and pUL138 form the predominant UL133-UL138 locus interaction.
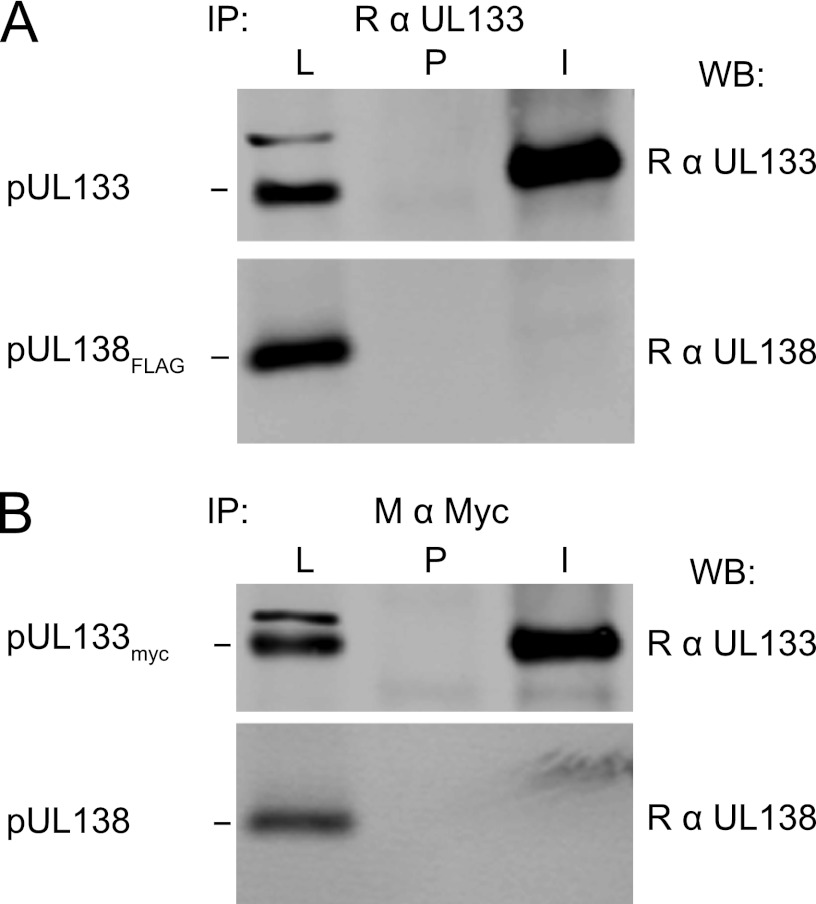
As pUL133 and pUL138 were found to weakly or transiently interact with other UL133-UL138 locus proteins (Fig. 1), we analyzed the pUL133-pUL138 interaction in cells relevant to HCMV infection that concomitantly expressed all four UL133-UL138 locus proteins. MRC5 fibroblasts were transduced with lentivirus constructs expressing myc-tagged variants of each protein. pUL136 is expressed at very low levels during infection (22), and expression from lentiviral vectors is necessary to achieve protein expression levels sufficient for immunoprecipitation. Lysates were immunoprecipitated with pUL133 polyclonal antisera and blotted with the myc antibody to detect possible myc-tagged interacting proteins. While all UL133-UL138 locus proteins were abundantly expressed, the only interacting partner to coprecipitate with pUL133 was pUL138 (Fig. 4A, lane 6), suggesting that this is the primary interaction with pUL133. This experiment did not confirm the interaction between pUL133 and pUL136 detected in Fig. 1. No proteins were precipitated with the preimmune sera (Fig. 4A, lane 5). Importantly, pUL138 was not coimmunoprecipitated with the pUL133 antibody when pUL133 was not present in the lysate (Fig. 4A, compare lanes 1 and 3). This experiment demonstrates an exclusive and specific interaction between pUL133 and pUL138 that does not include other members of the UL133-UL138 locus.
Fig 4.
The pUL133-pUL138 interaction is the predominant interaction when all UL133-UL138 proteins are expressed together. (A) MRC5 fibroblasts were transduced with lentiviruses expressing myc-tagged pUL135, pUL136, and pUL138 in the absence (lanes 1 to 3) or presence (lanes 4 to 6) of a pUL133myc-expressing lentivirus. Interactions with pUL133 were analyzed by immunoprecipitation using rabbit polyclonal antisera against pUL133, followed by immunoblotting with mouse anti-myc antibody (M α myc) to detect proteins expressed from each construct. Preimmune pUL133 antiserum was used as a control. Data for lysates (L), the preimmune serum control (P), and rabbit anti-UL133 IP (I) are shown. (B) MRC5 fibroblasts were transduced with lentiviruses expressing myc-tagged pUL133, pUL135, pUL136, and pUL138 in various combinations. Interactions with pUL133 were analyzed by immunoprecipitation as described above for panel A. Data for lysates, preimmune control IP, and rabbit anti-UL133 IP are shown for each combination. WB, Western blotting.
We further analyzed the requirement for or contribution of other UL133-UL138 locus proteins in the pUL133-pUL138 interaction; we analyzed this complex under conditions where pUL135, pUL136, or pUL138 was omitted. The interaction between pUL133 and pUL138 does not depend on nor is it altered by the absence of other proteins from this locus, since the omission of any single protein from the lysate did not alter the interaction between pUL133 and pUL138 (Fig. 4B, compare lane 3 to lanes 6 and 9). From these data, we conclude that despite possible interactions with pUL136, pUL136 is not required for the formation of the pUL133-pUL138 complex, and our data suggest that pUL136 does not enter pUL133-pUL138 complexes. When pUL138 was omitted, we did not readily detect other proteins from the locus in a complex with pUL133 (Fig. 4B, lane 12).
pUL133 interacts with pUL136 and pUL138 during infection.
To investigate the interaction between pUL133 and pUL138 in the context of a viral infection, MRC5 fibroblasts were infected with a recombinant virus, TB40E-UL138Flag, expressing a C-terminal Flag-tagged variant of pUL138 for 48 h. Immunoprecipitation using the polyclonal antisera specific to pUL133 coprecipitated a portion of pUL138Flag (Fig. 5A, lane I), demonstrating that the interaction between pUL138 and pUL133 occurs during infection. No pUL138 was detected in immunoprecipitations using preimmune sera (Fig. 5A, lane P). Immunoprecipitates from cells infected with recombinant viruses expressing myc-tagged variants of pUL135 (Fig. 5B) or pUL136 (Fig. 5C) demonstrated that the pUL133 pulldown coprecipitates a portion of pUL136 but not pUL135. These data are consistent with the findings from our one-on-one interaction studies during transient expression (Fig. 1). We also performed immunoprecipitations at 3 days postinfection and obtained similar results (data not shown).
Fig 5.
pUL133 interacts with pUL138 or pUL136 during infection of fibroblasts. Lysates derived from fibroblasts infected with TB40E-UL138Flag (A), TB40E-UL135myc (B), or TB40E-UL136myc (C) were prepared at 48 h postinfection and immunoprecipitated by using rabbit polyclonal antisera specific for pUL133. Coprecipitating proteins were detected by immunoblotting using rabbit polyclonal antibodies specific to pUL133 and polyclonal antisera against pUL138 (A) or mouse anti-myc antibody (B and C). Data for lysates (L), the preimmune serum control (P), and rabbit anti-UL133 IP (I) are shown.
Given the requirement for the UL133-UL138 locus for viral replication in endothelial cells (22), we investigated the interaction between pUL133 and pUL138 in primary human microvascular lung endothelial cells (HMVECs). These cells exhibit a defect (≥2 logs) for replication of the UL133-UL138NULL virus similar to that of a variety of other endothelial cell types, including human umbilical vein endothelial cells (HUVECs) (22). HMVECs were infected with TB40E-UL138Flag or TB40E-UL133myc. At 96 h postinfection (hpi), pUL133 or pUL133myc was immunoprecipitated from lysates by using pUL133-specific polyclonal antisera or the mouse anti-myc antibody, respectively. Despite abundant protein being present in the lysates (Fig. 6A and B, lane L), we did not detect an interaction between pUL133 and pUL138 in endothelial cells (Fig. 6A and B, bottom, lane I). These data indicate that the interactions between pUL133 and pUL138 are dependent on the cell type. While we did not detect the pUL133-pUL138 complex in endothelial cells, we cannot rule out the existence of other complexes derived from the UL133-UL138 locus that were not detected in MRC5 cells.
Fig 6.
The pUL133-pUL138 interaction is absent in infected endothelial cells. Lysates derived from HMVECs infected with TB40E-UL138Flag (A) or TB40E-UL133myc (B) were prepared at 4 days postinfection and immunoprecipitated by using rabbit polyclonal antisera specific for pUL133 or mouse monoclonal antibodies specific to the myc epitope tag. Coprecipitating proteins were detected by immunoblotting using rabbit polyclonal antibodies specific to pUL133 and pUL138. Data for lysates (L), the preimmune serum control (P), and rabbit anti-UL133 IP (I) are shown.
pUL133 and pUL138 cooperatively promote HCMV latency.
Our work has demonstrated a role for both pUL133 (22) and pUL138 (19) in suppressing viral replication in CD34+ HPCs for latency. To better understand the requirement for pUL133 and pUL138 in latency, we created a recombinant virus, TB40E-UL133Stop/UL138Stop, containing disruptions (substitutions of stop codons) in both UL133 and UL138 (Fig. 7A). Viruses containing disruptions in UL133 or UL138 consistently replicate to slightly increased yields relative to those of the wild-type (WT) virus (22). As would be expected, TB40E-UL133Stop/UL138Stop replicated similarly to the wild-type, UL133Stop, and UL138Stop viruses (Fig. 7B). We confirmed the coding sequence disruptions by immunoblotting. All recombinant viruses expressed equivalent levels of IE1 and IE2 (Fig. 7C), confirming the replication kinetics depicted in Fig. 7B. The recombinant viruses failed to express proteins targeted for disruption (Fig. 7C). The expression of pUL135 and other UL133-UL138 proteins indicates that the disruption of pUL133 or pUL138 does not alter the expression of other proteins in the locus.
Fig 7.
pUL133 and pUL138 contribute cooperatively to HCMV latency. (A) Schematic of recombinant viruses constructed for this study. (B) MRC5 cells were infected with TB40-WT, TB40-UL133Stop, TB40-UL138Stop, or TB40-UL133Stop/UL138Stop at an MOI of 0.5. Virus yields were measured as the TCID50 at the times indicated. (C) MRC5 cells infected with TB40E-WT or mutants at an MOI of 1.0 were harvested at 2 days postinfection (dpi), and lysates prepared from equal numbers of cells were analyzed by immunoblotting using either mouse anti-IE1 and anti-IE2 antibody to detect IE genes or rabbit polyclonal antibodies against each UL133-UL138 protein. α-Tubulin was used as a protein loading control. (D) CD34+ HPCs infected with TB40E viruses were maintained in LTBMC medium for 10 dpi, after which lysates from infected CD34+ HPCs were serially diluted onto MRC5 cells plated into 96-well dishes. The frequency of formation of infectious centers was determined 15 to 20 days later from the number of GFP+ wells at each dilution by using ELDA software. The statistical significance comparing each of the mutant viruses to TB40E-WT was determined by Student's t test (P = 0.002, P = 0.011, and P = 0.003 for TB40E-UL133Stop, TB40E-UL138Stop, and TB40E-UL133Stop/UL138Stop, respectively). The standard errors of the means from three independent experiments are shown.
We analyzed TB40E-UL133Stop/UL138Stop for its ability to establish and maintain a latent infection in CD34+ HPCs relative to the wild-type virus or viruses containing a disruption in only UL133 or UL138. CD34+ HPCs purified from cord blood were infected at an MOI of 2. Infected (GFP+) CD34+ cells were purified 24 h later by fluorescence-activated cell sorting and seeded into a long-term bone marrow culture. At 10 days postinfection, we analyzed the frequency of production of infectious centers using a modified TCID50 assay and Extreme Limiting Dilution Analysis (ELDA) software. Relative to TB40E-WT, TB40E-UL133Stop/UL138Stop exhibited an enhanced replicative capacity, producing a 2-fold-higher frequency of infectious centers (Fig. 7D). Infection with viruses containing single mutations in either UL133 or UL138 resulted in similar frequencies of infectious centers relative to that of the double mutant virus. In fact, Student's t tests comparing the means from infection with TB40E-UL133Stop or -UL138Stop to those from infection with the double mutant virus indicate that these data are not statistically different. This result suggests a cooperative relationship between pUL133 and pUL138 in establishing latency and that pUL133 and pUL138 do not independently or additively contribute to this phenotype.
DISCUSSION
The UL133-UL138 locus encodes four novel proteins important to cell type-dependent outcomes of HCMV infection, including latency (8, 22). We have previously shown that pUL133 and pUL138 suppress replication for a latent infection in CD34+ HPCs infected in vitro but are not required for replication in fibroblasts (19, 22). While the UL133-UL138 locus is required for efficient replication in endothelial cells, UL138 alone is dispensable (22). Given the coordinated expression and overlapping subcellular localization of pUL133, pUL135, pUL136, and pUL138, we hypothesized that these proteins cooperate to dictate the outcome of infection. In this work, we have explored possible interactions between these proteins to understand how they may cooperate in infection. The protein interactions revealed by this work are summarized by the interaction map shown in Fig. 8. Each of the proteins, pUL133, pUL135, pUL136, and pUL138, was able to self-associate, indicating the potential to form high-order homomeric complexes (Fig. 2). In addition, we detected a robust and highly specific interaction between pUL133 and pUL138 when expressed transiently (Fig. 1, 3, and 4) or during virus infection in fibroblasts (Fig. 5). Interestingly, this interaction was not detected in endothelial cells (Fig. 6), suggesting that the interaction depends on the cell type infected. The interaction between pUL133 and pUL138 is of significant interest, since both of these proteins are important for viral latency (22). The disruption of both UL133 and UL138 resulted in the same phenotype in CD34+ cells as the disruption of either gene alone, suggesting that pUL133 and pUL138 do not independently or additively contribute to latency (Fig. 7D). Interestingly, pUL133 and pUL138 are always expressed together in any context that has been tested (22), consistent with a requirement for both proteins. A demonstration of the existence of the pUL133-pUL138 interaction in CD34+ HPCs has not yet been possible due to the low levels of protein expression in these cells and the requirement for large cell numbers for immunoprecipitation and antibody detection (data not shown).
Fig 8.
Map of UL133-UL138 locus proteins. pUL133, pUL135, pUL136, and pUL138 were all demonstrated to self-associate, as indicated by a loop. Interactions demonstrated between proteins are indicated by lines connecting the proteins, and the weight of the line indicates the robustness of the interaction (i.e., detected in transfections and infections and the relative amount of protein coprecipitated). pUL133-pUL138 interacted when expressed transiently, when expressed with other proteins from the UL133-UL138 locus, and during infection. pUL133 also interacted with pUL136 when transiently expressed or during infection. All other interactions shown were detected only during transient overexpression.
pUL138 was recently shown to enhance cell surface levels of tumor necrosis factor receptor (TNFR) (12, 15). Increased cell surface TNFR levels were observed for cells transiently expressing pUL138 alone and were not observed for cells expressing pUL133 (15). These data suggest that pUL138 modulates cell surface levels of TNFR independently of its interaction with pUL133. In contrast, our data (Fig. 6) establish cooperative, rather than independent, roles for pUL133 and pUL138 in latent infection. Therefore, more work is required to understand the relationship between the pUL138 modulation of TNFR, interactions with UL133-UL138 locus proteins, and functions in latency. pUL138-mediated enhanced surface levels of TNFR correlate with increased sensitivity to NF-κB stimulation and a corresponding induction of immediate-early (IE) gene expression. We also previously reported that pUL138 expression results in slightly increased levels of major immediate-early promoter activation and IE gene expression (19). These finding are suggestive of a role for pUL138 in activating viral replication but are seemingly inconsistent with a role for these proteins in suppressing viral replication for latency in CD34+ HPCs. Elegant in vivo studies using murine cytomegalovirus (MCMV) have shown that IE gene activation does not necessarily correspond to viral reactivation (11). Therefore, while pUL138 activity results in IE gene expression, this activation may be an unintended consequence of the pUL138 function that does not result in a reactivation of viral replication. The virus may encode other functions to curtail full-blown reactivation initiated by the IE-stimulatory activity of pUL138. As many viral proteins are multifunctional, pUL138 may suppress viral replication by means separable from the effect on IE gene expression. It will be important to determine how pUL133 may influence the role of pUL138 in modulating cell surface levels of TNFR and how this impacts the outcome of infection.
In addition to the interaction between pUL133 and pUL138, we also detected interactions between pUL136 and either pUL133 or pUL138 (Fig. 1). The interaction between pUL133 and pUL136 was detectable during virus infection (Fig. 5). The role of these interactions in viral infection remains unknown. Determining the role for pUL136 in the context of the UL133-UL138 locus is complicated by the low levels of pUL136 expression during infection (22), the internal ribosomal entry site within the UL136 coding sequence important for the translation of pUL138 (8), and the multiple isoforms of pUL136 (22). When all four UL133-UL138 locus proteins were expressed, the predominant interaction recovered from the immunoprecipitation of pUL133 was the interaction with pUL138. The failure to coprecipitate pUL136 may indicate that this is a less abundant or less favorable interaction. It is intriguing to speculate how multiple interactions may exist among the proteins encoded by the UL133-UL138 locus and how the balance of these interactions in different cell types may dictate context-dependent outcomes of infection. The nature of these complexes and their individual contributions to infection will become more apparent as we probe the role of each protein in different contexts of infection.
The UL133-UL138 locus is dispensable for replication in fibroblasts, inhibitory to replication in CD34+ HPCs, and augmenting for replication in endothelial cells (22). Determining the role of the interactions between the proteins encoded by the UL133-UL138 locus and with cellular proteins is critical to an understanding of how these proteins contribute to the outcome of infection. Our work has mapped the possible interaction between proteins encoded by this locus and demonstrated the existence of a complex between pUL133 and pUL138 in fibroblasts but not endothelial cells. We are not able to directly examine protein interactions in primary CD34+ HPCs, but both proteins function to suppress viral replication for latency, and the disruption of either protein or both proteins results in a similar phenotype. This result indicates that these proteins cooperate to establish latency but do not have additive functions, as would be expected if they functioned independently. Given their ability to modulate outcomes of infection, the proteins encoded by the UL133-UL138 locus and their interactions represent pivotal targets for the control of viral infection.
ACKNOWLEDGMENTS
We gratefully acknowledge Christian Sinzger for the gift of the TB40E BAC and Tom Shenk for the gift of antibodies. We thank Paula Campbell at the AZCC/ARL Division of Biotechnology Cytometry Core Facility for FACS expertise and Evlyn Hadley for her work in procuring umbilical cord blood.
This work was supported by Public Health Service grant AI079059 from the National Institutes of Allergy and Infectious Diseases and by a Cancer Center Support Grant (CCSG) (grant CA020374). F.G. is a Pew scholar in the biomedical sciences, supported by the Pew Charitable Trusts.
The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute, the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, or the Pew Charitable Trusts.
Footnotes
Published ahead of print 6 June 2012
REFERENCES
- 1. Boeckh M, Geballe AP. 2011. Cytomegalovirus: pathogen, paradigm, and puzzle. J. Clin. Invest. 121:1673–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol. 325:417–470 [DOI] [PubMed] [Google Scholar]
- 3. Cha TA, et al. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davison AJ. 2002. Evolution of the herpesviruses. Vet. Microbiol. 86:69–88 [DOI] [PubMed] [Google Scholar]
- 5. Dolan A, et al. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301–1312 [DOI] [PubMed] [Google Scholar]
- 6. Goodrum F, Jordan CT, Terhune SS, High KP, Shenk T. 2004. Differential outcomes of human cytomegalovirus infection in primitive hematopoietic subpopulations. Blood 104:687–695 [DOI] [PubMed] [Google Scholar]
- 7. Goodrum F, Reeves M, Sinclair J, High K, Shenk T. 2007. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood 110:937–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grainger L, et al. 2010. Stress-inducible alternative translation initiation of human cytomegalovirus latency protein pUL138. J. Virol. 84:9472–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horvath R, et al. 2000. The possible role of human cytomegalovirus (HCMV) in the origin of atherosclerosis. J. Clin. Virol. 16:17–24 [DOI] [PubMed] [Google Scholar]
- 10. Hu Y, Smyth GK. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347:70–78 [DOI] [PubMed] [Google Scholar]
- 11. Kurz SK, et al. 1999. Focal transcriptional activity of murine cytomegalovirus during latency in the lungs. J. Virol. 73:482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Le VT, Trilling M, Hengel H. 2011. The cytomegaloviral protein pUL138 acts as potentiator of tumor necrosis factor (TNF) receptor 1 surface density to enhance ULb′-encoded modulation of TNF-alpha signaling. J. Virol. 85:13260–13270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miller CL, Eaves CJ. 2002. Long-term culture-initiating cell assays for human and murine cells, p 123–141 In Klug CA, Jordan CT. (ed), Hematopoietic stem cell protocols. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 14. Mocarski ES, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2710–2772 In Knipe DM, et al. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 15. Montag C, et al. 2011. The latency-associated UL138 gene product of human cytomegalovirus sensitizes cells to tumor necrosis factor alpha (TNF-alpha) signaling by upregulating TNF-alpha receptor 1 cell surface expression. J. Virol. 85:11409–11421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murphy E, Rigoutsos I, Shibuya T, Shenk TE. 2003. Reevaluation of human cytomegalovirus coding potential. Proc. Natl. Acad. Sci. U. S. A. 100:13585–13590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murphy E, et al. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 100:14976–14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pawelec G, Derhovanessian E. 2011. Role of CMV in immune senescence. Virus Res. 157:175–179 [DOI] [PubMed] [Google Scholar]
- 19. Petrucelli A, Rak M, Grainger L, Goodrum F. 2009. Characterization of a novel Golgi-localized latency determinant encoded by human cytomegalovirus. J. Virol. 83:5615–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pourgheysari B, et al. 2007. The cytomegalovirus-specific CD4+ T-cell response expands with age and markedly alters the CD4+ T-cell repertoire. J. Virol. 81:7759–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Streblow DN, Orloff SL, Nelson JA. 2001. Do pathogens accelerate atherosclerosis? J. Nutr. 131:2798S–2804S [DOI] [PubMed] [Google Scholar]
- 22. Umashankar M, et al. 2011. A novel human cytomegalovirus locus modulates cell type-specific outcomes of infection. PLoS Pathog. 7:e1002444 doi:10.1371/journal.ppat.1002444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vasto S, et al. 2007. Role of persistent CMV infection in configuring T cell immunity in the elderly. Immun. Ageing 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang W, et al. 2005. Human cytomegalovirus genes in the 15-kilobase region are required for viral replication in implanted human tissues in SCID mice. J. Virol. 79:2115–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36 doi:10.1093/nar/gni035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu D, et al. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]