Abstract
Human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2 are closely related but pathogenically distinct human retroviruses. The antisense strand of the HTLV-1 genome encodes HTLV-1 basic leucine zipper (b-ZIP) protein (HBZ), a protein that inhibits Tax-mediated viral transcription, enhances T-cell proliferation, and promotes viral persistence. Recently, an HTLV-2 antisense viral protein (APH-2) was identified. Despite its lack of a typical b-ZIP domain, APH-2, like HBZ, interacts with cyclic AMP response element binding protein (CREB) and downregulates Tax-mediated viral transcription. Here, we provide evidence that the APH-2 C-terminal LXXLL motif is important for CREB binding and Tax repression. In order to investigate the functional role of APH-2 in the HTLV-2-mediated immortalization of primary T lymphocytes in vitro and in HTLV-2 infection in vivo, we generated APH-2 mutant viruses. In cell cultures, the immortalization capacities of APH-2 mutant viruses were indistinguishable from that of wild-type HTLV-2 (wtHTLV-2), indicating that, like HBZ, APH-2 is dispensable for viral infection and cellular transformation. In vivo, rabbits inoculated with either wtHTLV-2 or APH-2 mutant viruses established a persistent infection. However, the APH-2 knockout virus displayed an increased replication rate, as measured by an increased viral antibody response and a higher proviral load. In contrast to HTLV-1 HBZ, we show that APH-2 is dispensable for the establishment of an efficient infection and persistence in a rabbit animal model. Therefore, antisense proteins of HTLV-1 and HTLV-2 have evolved different functions in vivo, and further comparative studies will provide fundamental insights into the distinct pathobiologies of these two viruses.
INTRODUCTION
Human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2 are closely related complex retroviruses that infect and immortalize T lymphocytes. HTLV-1 and HTLV-2 share a number of biological properties: a genome that encodes the typical retroviral structural and enzymatic gene products (from gag, pol, and env); the Tax and Rex positive regulatory gene products; and several accessory gene products, which are important for viral infectivity and persistence in vivo (29), require cell-cell contact for the most efficient virus transmission (15), use similar cellular surface receptors but have different requirements for efficient cell entry (25), and transform T lymphocytes in culture but favor distinct transformation targets (47). However, the clinical manifestations of infections with HTLV-1 differ from those of infections with HTLV-2. In approximately 5% of infected people, HTLV-1 causes adult T-cell leukemia (ATL) and a neurological disorder, HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) (17, 22). In contrast, HTLV-2-infected individuals demonstrate marginal lymphocytosis and sporadically develop neurological symptoms, but so far, there has been no evidence of leukemia (1, 4).
In an effort to determine the mechanisms underlying the distinct pathogeneses of HTLV-1 and HTLV-2, investigations have focused on comparing the functions of proteins encoded by the two viruses. The Tax regulatory protein encoded by both HTLV-1 and HTLV-2 is the major transactivator of viral gene expression and is essential for viral replication (14). Tax modulates the expression or activity of various cellular factors involved in growth and differentiation, disrupts cell cycle control and DNA repair processes, and displays oncogenic activity in a number of cell culture assays and animal models (19, 21, 46). Tax is also the key oncoprotein required for the HTLV-mediated transformation of primary T lymphocytes (38–40). Comparative studies of the HTLV-1 and HTLV-2 Tax proteins revealed that these proteins display many similarities but also some major differences that might account for the distinct pathogenic outcomes for HTLV-1- versus HTLV-2-infected patients (6, 13, 23, 33, 36, 43, 48, 51). However, the silencing of Tax expression in ATL patients suggests a role for additional viral gene products that likely contribute to the pathogenic process.
The HTLV-1 basic leucine zipper (b-ZIP) gene (Hbz) is located on the antisense genomic DNA strand (7, 16, 30). Both spliced (SP1) and unspliced Hbz transcripts have been detected; they encode protein isoforms that differ only in the 7 amino acids (aa) at their N termini (7, 37). Transcripts of the Hbz gene are detected in all ATL cell lines and cells freshly isolated from ATL and HAM/TSP patients (41). Further studies revealed that HBZ interacts with the cellular factors cyclic AMP-response element binding protein (CREB) and CREB binding protein (CBP/p300) through its b-ZIP domain and “LXXLL” motifs, respectively, and these interactions are responsible for the repression of Tax-mediated viral transcription (9, 31). HBZ also interacts with Jun family members, including JunB, JunD, and c-Jun, thereby modulating their transcription and regulation of viral and cellular gene expression (24, 27, 44). In addition, HBZ was reported previously to selectively suppress the classical NF-κB pathway by binding the p65 subunit (53). Although HBZ is dispensable for the HTLV-1 immortalization of T lymphocytes in culture, it was previously shown to enhance infectivity and persistence in HTLV-1-infected rabbits (2). An Hbz knockdown in HTLV-1 tumor T-cell lines correlated with a significant decrease in proliferation in cell cultures as well as tumor formation and organ infiltration in immunodeficient mice (3). Moreover, HBZ transgenic mice develop systemic inflammation and CD4+ T-cell lymphoma (42). Taken together, these data support the hypothesis that HBZ functions as a secondary oncogene and is important for the proliferation of infected CD4+ T cells, contributing to leukemogenesis and, potentially, the maintenance of the tumor cell.
Recently, an antisense HTLV-2 protein (APH-2) was indentified (20). APH-2 has less than 30% homology to HBZ. However, similar to HBZ, APH-2 has been shown to downregulate Tax-mediated viral transcription by interacting with cellular CREB (20). APH-2 expression was found to correlate with the proviral load in HTLV-2-infected carriers but did not appear to promote lymphocytosis (12). Since evidence suggests that HBZ likely contributes to HTLV-1 pathogenesis, we hypothesized that an understanding of the differences in APH-2 function would provide important insights into the distinct pathogeneses of HTLV-1 and HTLV-2.
In this study, we evaluated the functional role of APH-2 in the context of an infectious HTLV-2 molecular clone and determined its contribution to in vitro cellular immortalization and in vivo viral replication kinetics and persistence. Our findings indicate that the knockout of APH-2 and its documented repressive effect on Tax were not sufficient to disrupt the ability of the virus to immortalize primary T lymphocytes in cell cultures. In addition, rabbits infected with APH-2 mutant viruses displayed an increased antibody response to viral gene products and a higher proviral load in peripheral blood mononuclear cells (PBMCs) than did wild-type HTLV-2 (wtHTLV-2)-infected rabbits. These results suggest that unlike HBZ, which plays an important role early after infection and the establishment of viral persistence, APH-2 is dispensable for enhancing viral replication and persistent infection in vivo. In addition, our data also suggest that APH-2 could contribute to the lower virulence of HTLV-2.
MATERIALS AND METHODS
Cells.
293T cells were maintained in Dulbecco's modified Eagle's medium. The parental 729 human B-cell line and derivatives were maintained in Iscove's medium. All media were supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). Human and rabbit PBMCs were isolated by using Ficoll Hypaque (Amersham, Piscataway, NJ) and Percoll (GE Healthcare Life Sciences, Piscataway, NJ), respectively. PBL-01 and PBL-02, two newly HTLV-2-immortalized peripheral blood T-lymphocyte (PBL) lines, were cultured in RPMI 1640 medium containing 20% FBS, glutamine, and antibiotics, as described above, as well as 10 U/ml recombinant interleukin-2 (IL-2) (Roche Applied Biosciences, Indianapolis, IN). HTLV-2 infection of these two immortalized cell lines was confirmed by a p19 Gag enzyme-linked immunosorbent assay (ELISA). To generate stable 729 transfectants (virus producer cells), proviral plasmid clones containing the Neor gene were introduced into 729B cells by Nucleofection (Amaxa, Lonza, Walkersville, MD) according to the manufacturer's instructions. Stable transfectants containing the desired proviral clones were isolated and characterized as previously described (47).
Plasmids.
The plasmid containing the wtHTLV-2 proviral clone (pH6neo) was described previously (8). A schematic diagram of the deletion mutants of APH-2 is shown in Fig. 1A. The HTLV-2ΔAPH proviral clone contains a single-nucleotide change (A to C) at nucleotide (nt) 7169 of the pH6neo sequence, resulting in a stop codon at the sixth amino acid of the 183-amino-acid (aa) APH-2. The HTLV-2APHΔBD2 proviral clone has a single-nucleotide change (T to A) at nt 6655 of the pH6neo sequence, which terminates APH-2 at aa 178, deleting the C-terminal 179LXXLL183 motif referred to as binding domain 2 (BD2). Neither of these mutations altered any known HTLV-2 open reading frames (p10, p11, or Env). APH, ΔAPH, and APHΔBD2 cDNA expression plasmids (pME vector based) were generated from the pH6neo sequences. Three additional APH-2 cDNA mutants were generated by site-directed mutagenesis: in APHBD1, the central 64IXXLL68 motif (BD1) was mutated to IXXAA; in APHBD2ΔLL, APH-2 was terminated at aa 181, deleting the C-terminal 182LL183 sequence; and APHBD1ΔBD2 is a double mutant containing both BD1 and ΔBD2 mutations. All the generated mutations were confirmed by sequencing. The Tax reporter plasmid LTR-2Luc and the transfection efficiency control plasmid TK-Renilla were described previously (52).
Fig 1.
Detection of Aph-2 transcript and protein. (A) Schematic representation of the complete HTLV-2 proviral genome. LTRs are depicted with their U3, R, and U5 regions. The locations of the viral open reading frames and the opposite-strand Aph-2 are indicated. The reported wild-type APH-2 coding sequence has been expanded to show the potential functional IXXLL and LXXLL motifs and the four APH-2 mutants generated for this study (APHBD1, APHΔBD2, APHBD2ΔLL, and ΔAPH). (B) Total RNA was isolated from 729.HTLV-2 cells, HTLV-2 newly immortalized human PBLs (PBL-01 and PBL-02), and uninfected 729B negative-control cells. First-strand cDNAs were prepared by using 1 μg of total RNA and a random hexamer provided by the SuperScript first-strand synthesis system for RT-PCR in the presence (+) or absence (−) of reverse transcriptase. The 105-bp PCR product was separated on a 2% agarose gel and visualized by ethidium bromide staining. Pos C is a cDNA APH-2 expression vector plasmid. M, molecular weight ladder. (C) Cell lysates were immunoprecipitated using a rabbit anti-APH-2 polyclonal antibody and visualized by WB analysis. The p19 Gag production levels in the HTLV-2-immortalized cell lines PBL-01 and PBL-02 were 3,800 pg/ml and 9,000 pg/ml, respectively.
Transfection, reporter assays, and p19 Gag ELISA.
To assess the repressive effect of APH-2 on Tax activity, 293T cells were transfected with 1 μg wtHTLV-2 proviral plasmid in the presence or absence of variable amounts (0.2, 0.4, or 0.6 μg) of APH-2 or APH-2 mutant cDNA plasmids (ΔAPH, APHBD1, APHΔBD2, APHBD2ΔLL, and APHBD1ΔBD2), 0.1 μg of LTR-2Luc, and 0.01 μg of TK-Renilla by using Lipofectamine (Invitrogen, Carlsbad, CA). To measure Tax activity expressed from wild-type and mutant proviral clones, cells were transfected with 1 μg of proviral plasmid (HTLV-2, HTLV-2ΔAPH, and HTLV-2APHΔBD2) or empty plasmid, 0.1 μg of LTR-2Luc, and 0.01 μg of TK-Renilla. In all cases, the amount of DNA was kept constant by the addition of empty plasmid pME. After 48 h, cell supernatants were tested for the p19 Gag protein by ELISA (Zeptometrix Corporation, Buffalo, NY), and cell lysates were subjected to a dual-luciferase assay to measure reporter gene activity (Promega, Madison, WI). All experiments were performed three independent times in duplicates, and the results were normalized for transfection efficiency by using Renilla luciferase. Stable 729 transfectants containing the desired proviral clones were isolated and characterized as described previously (47, 50).
Immunoprecipitation and Western blot analysis.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.4], 1% Nonidet P-40, 150 mM NaCl, and 1 mM EDTA) with protease inhibitors (Roche, Mannheim, Germany) and subjected to immunoprecipitation (IP) using a rabbit anti-APH-2 polyclonal antibody (1:500) that we generated against the whole APH-2 protein and protein A-Sepharose beads (Thermol, Rockford, IL). Immunoprecipitates were washed, eluted, and resolved by SDS-PAGE. Western blot (WB) analysis was performed by using a rabbit anti-APH-2 polyclonal antibody (1:1,000) and a goat anti-rabbit antibody conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA).
Standard PCR and quantitative TaqMan real-time PCR.
Genomic DNA was isolated from 729 producer cells and rabbit PBMCs by using the Easy-DNA kit (Invitrogen, Carlsbad, CA). Rabbit genomic DNA (0.5 μg) was subjected to a standard 40-cycle PCR. The primer pair used for the PCR was TRE-PH-S and TRE-PH-AS (49), which amplified a 257-bp product of the HTLV-2 long terminal repeat (LTR). The HTLV-2-specific primer pair 5′-6564AAATCCAGGCCCTTCC6579-3′ and 5′-7230AAATCCTGGGAAATGG7215-3′ was used to amplify a 667-bp fragment for sequencing to confirm the presence of the APH-2 mutations in the integrated provirus or stably transfected plasmid clones. TaqMan real-time PCR (Applied Biosystems, Foster City, CA) was performed in duplicates with 0.5 μg rabbit genomic DNA and HTLV-2-specific primers GP2-S/GP2-AS and Taqman probe 8 (TMP-8) to quantitate the proviral copy number per cell in infected rabbit PBMCs as described previously (32). The absolute copy number was determined by extrapolation from a standard curve generated from log10 dilutions of plasmid DNA containing the HTLV-2 gag/pol sequence (32). The copy number per cell was generated based on the estimation that 1 μg of rabbit PBMC genomic DNA is equivalent to 134,600 cells (32).
RT-PCR.
Total RNA was extracted from 729pH6neo cells, HTLV-2-immortalized human PBLs (PBL-01 and PBL-02), and 729B cells (uninfected) by using the RNeasy kit (Qiagen, Valencia, CA). After three treatments with DNase, 1 μg total RNA was subjected to reverse transcription (RT) using a SuperScript first-strand synthesis kit (Invitrogen Corp., Carlsbad, CA). The cDNAs were subjected to standard PCR using the primer pair APH-2-S (5′-ATGGATCCCCAAACTATTTTAGG-3′) and APH-2-AS (5′-TGCTGTCCTTCTCGGTTCCT-3′), which amplified a 105-bp fragment. Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified by using primers described previously (28).
Short-term proliferation and long-term immortalization coculture assays.
Short-term microtiter proliferation assays were performed as detailed previously, with modifications (48). Briefly, freshly isolated human PBMCs were prestimulated with 2 μg/ml phytohemagglutinin (PHA) and 10 U/ml IL-2 (Roche, Indianapolis, IN) for 3 days. The 729 virus producer cells were irradiated (100 Gy) and cocultured with 104 prestimulated human PBMCs at a ratio of 1:5, respectively, with 10 U/ml IL-2 in 96-well plates. The number of virus producer cells was normalized to their p19 Gag production levels. Wells were enumerated for growth and split 1:3 weekly. For the long-term immortalization assays, irradiated virus producer cells, normalized to their p19 Gag production levels, were cocultivated at a 1:2 ratio with 2 × 106 freshly isolated human PBMCs with 10 U/ml IL-2 in 24-well culture plates (18). HTLV-2 gene expression was confirmed by the detection of the p19 Gag protein in the culture supernatant, measured weekly by an ELISA. Viable cells were counted weekly by a trypan blue exclusion test. Cells infected with wtHTLV-2 or HTLV-2 APH-2 mutants that continued to produce the p19 Gag antigen and proliferate at 12 weeks postcoculture in the presence of exogenous 10 U/ml IL-2 were identified as HTLV-2-immortalized clones.
Rabbit inoculation procedure.
Twelve-week-old pathogen-free New Zealand White rabbits (Harlan, Indianapolis, IN) were inoculated with gamma-irradiated (100 Gy) 729 virus producer cells (four rabbits for wtHTLV-2 and six rabbits each for HTLV-2ΔAPH and HTLV-2APHΔBD2) or 729 uninfected control cells (two rabbits) via the lateral ear vein. The virus-containing inocula were equilibrated based on p19 Gag production (approximately 1 × 107 total cells). At weeks 0, 2, 4, 6, 8, and 12 postinoculation, 13 ml of blood was drawn from the central auricular artery from each rabbit, and plasma and PBMCs were isolated. An HTLV WB assay (MP Diagnostics, Singapore) was used to examine plasma reactivity to specific viral antigenic determinants. Plasma samples showing reactivity to Gag (p24 or p19) and Env (gp21 or gp46) antigens was classified as positive. A commercial HTLV ELISA kit (MP Diagnostics, Singapore) was used to quantitate plasma antibody levels.
RESULTS
The Aph-2 transcript is detected in HTLV-2 stably transfected cell lines and HTLV-2-transformed cell lines.
Although several HTLV-1 Hbz transcripts have been identified, including unspliced and two spliced species, to date, only a single spliced transcript has been identified for HTLV-2 APH-2. The singly spliced Aph-2 mRNA initiates in the 3′ LTR (U5 and R regions) at multiple positions (Fig. 1A) (20). Since we made use of the infectious HTLV-2 proviral clone pH6neo in our studies, we designed the initial experiment to verify the expression of Aph-2 mRNA in our HTLV-2 producer cell line, 729.HTLV-2. This cell line produces HTLV-2 that has the capacity to infect and transform human T cells from PBLs (47, 51). Standard RT-PCR was performed on the RNA isolated from 729.HTLV-2, PBL-01, PBL-02, and 729B (uninfected negative-control) cells. We could clearly detect Aph-2 gene expression in the 729.HTLV-2 and HTLV-2-immortalized cell lines (Fig. 1B). We were unable to detect the APH-2 protein in these same cell lines by WB analysis (data not shown). However, IP using anti-APH-2 rabbit polyclonal antisera to enrich for APH-2 followed by WB analysis resulted in the detection of APH-2 in 729.HTLV-2 cells, but APH-2 protein levels in PBL-01 and PBL-02 cells were below our limit of detection. This result was not unexpected, as newly immortalized T lymphocytes have been shown to express tax mRNA but, in many cases, undetectable levels of the Tax protein.
APH-2 represses Tax-mediated viral transcription but with a lower level of activity than HTLV-1 HBZ.
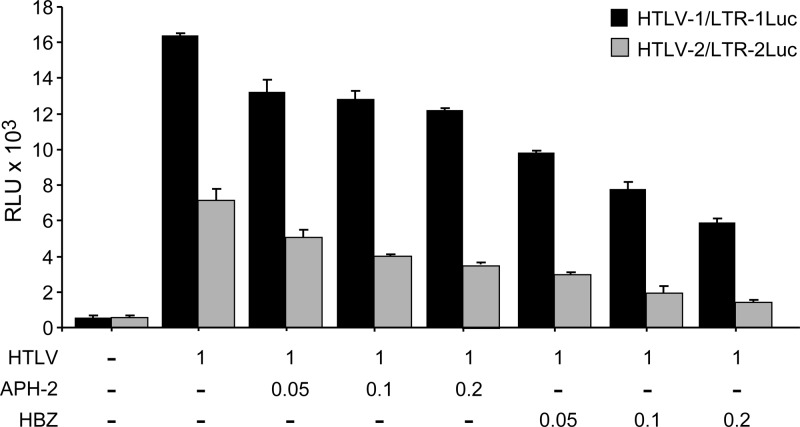
HBZ and APH-2 share a similar property of inhibiting Tax-mediated transcription (2, 20). However, the repressive functions of these two proteins have not been directly and quantitatively compared. 293T cells were transiently transfected with either the HTLV-1 or HTLV-2 proviral clone as a source of Tax and increasing amounts of HBZ or APH-2 cDNA expression vectors. Our results indicate that the repressive activity of APH-2 on Tax-mediated transcription was, on average, only 48% of that of HBZ irrespective of the HTLV-1 or HTLV-2 promoter target (Fig. 2). Thus, APH-2 is less active than HBZ in the repression of Tax-mediated viral transcription. It should be noted that these results do not take into consideration any potential differences in protein stability.
Fig 2.
Comparative functional analysis of the effects of APH-2 and HBZ on the repression of Tax-mediated transcription. 293T cells (1.5 × 105 cells) were cotransfected with 1 μg HTLV or negative-control plasmid DNA, 0.1 μg LTR-Luc reporter, 0.01 μg TK-Renilla, and various amounts (0.05, 0.1, and 0.2 μg) of the HBZ or APH-2 expression vector. All transfections were performed in duplicate and normalized to TK-Renilla to control for the transfection efficiency. Cell lysates were harvested 48 h after transfection. Black bars represent luciferase reporter activities for HTLV-1 and LTR-1-Luc, and gray bars represent luciferase reporter activities for HTLV-2 and LTR-2-Luc. RLU, relative light units.
The C-terminal LXXLL motif is critical for APH-2 activity.
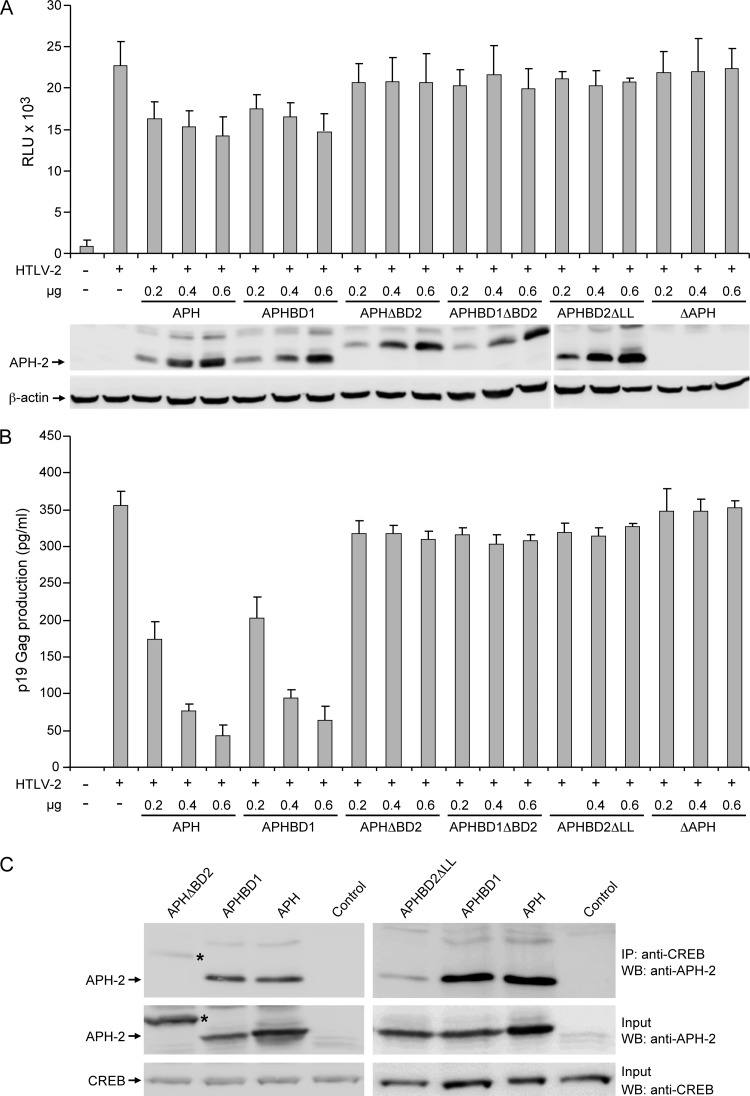
Since APH-2, like HBZ, repressed Tax-mediated viral transcription, we next analyzed the structural domains of both the proteins responsible for this function. HBZ represses Tax by interacting with CREB and CBP/p300 through its b-ZIP domain at the C terminus and LXXLL motifs in the transactivation domain at the N terminus, respectively (9, 31). In APH-2, a 179LXXLL183 motif and an LXXLL-like motif (64IXXLL68) were identified at the C terminus and central region, respectively. Based on this evidence, we generated five APH-2 mutants (Fig. 1A) to evaluate the contribution of these motifs to the repressive activity. APHΔBD2 has a deleted C-terminal LXXLL motif; APHBD1 has two leucines within the central IXXLL motif mutated to AA and was shown previously to completely abolish the function of this motif in HBZ (9). APHBD1ΔBD2 has both the BD1 and ΔBD2 mutations, APHBD2ΔLL has the last 2 amino acids of the LXXLL motif of BD2 deleted, and ΔAPH has a severely truncated APH-2 within the first 6 amino acids. The effects of exogenously expressed wild-type and mutant APH-2 proteins on Tax-mediated viral transcription were measured by using LTR-luciferase reporter assays. The results presented in Fig. 3A indicate that wtAPH-2 repressed Tax-mediated HTLV-2 transcription in a dose-dependent manner. APHBD1 also displayed a dose-dependent repressive activity on Tax at a level similar to that of wtAPH-2. However, APHΔBD2, APHBD1ΔBD2, APHBD2ΔLL, and ΔAPH lost their ability to repress Tax-mediated viral transcription. WB analysis confirmed that the amounts of wtAPH-2 and mutated APH-2 proteins correlated directly with the amount of plasmid DNA transfected, whereas the level of the control cellular protein, β-actin, remained unchanged (Fig. 3A). To our surprise, APHΔBD2 with the deleted C-terminal 5 amino acids (LXXLL motif) expressed an APH-2 protein that migrated more slowly than wtAPH-2 (Fig. 3A). However, APHBD2ΔLL displayed a protein mobility similar to that of wtAPH-2. Measurements of p19 Gag production in the cellular supernatants of these same transfected cells revealed a direct correlation with data from the transcriptional reporter assay. The overexpression of wtAPH-2 and APHBD1 resulted in a dose-dependent reduction of levels of p19 Gag production, whereas the overexpression of APHΔBD2, APHBD1ΔBD2, APHBD2ΔLL, and ΔAPH had no significant effect (Fig. 3B).
Fig 3.
The APH-2 BD2 (LXXLL) motif is required for dose-dependent repression of Tax-mediated transcription and p19 Gag production. 293T cells (1.5 × 105 cells) were cotransfected with 1 μg of the wtHTLV-2 proviral clone or negative-control DNA, 0.1 μg LTR-2-Luc, 0.01 μg TK-Renilla, and various concentrations (0.2, 0.4, and 0.6 μg) of the APH, APHBD1, APHΔBD2, APHBD1ΔBD2, APHBD2ΔLL, and ΔAPH expression vectors, as indicated. (A) Tax function was measured as firefly luciferase activity from LTR-2-Luc normalized to the Renilla luciferase activity. WB analysis was used to confirm increasing concentrations of the APH-2 proteins. β-Actin levels were measured as a loading control, as shown at the bottom. RLU, relative light units. (B) The culture supernatant was collected from cells in panel A and assayed for p19 Gag production by ELISA. (C) The APH-2 BD2 (LXXLL) motif is required for interactions with CREB. IP using an anti-CREB antibody was performed on 293T cells transfected with APHΔBD2, APHBD2ΔLL, APHBD1, APH, or control expression plasmids. Immune complexes were resolved by SDS-PAGE followed by WB analysis using an APH-2-specific antiserum. Prior to IP, 10% of lysates were saved (Input) and blotted with an anti-CREB or anti-APH-2 antibody.
APH-2 binds efficiently to CREB but very weakly to CBP/p300 (20). Since our data showed that BD2 of APH-2 is responsible for the downregulation of Tax-mediated transcription, we next evaluated whether APH-2 interacts with CREB through this domain. Since APH-2 was shown to bind very weakly to CBP/p300, we did not further evaluate the binding of APH-2 to CBP/p300. Co-IP experiments revealed that APHΔBD2 and APHBD2ΔLL (which showed wild-type protein mobility) did not efficiently interact with the transcriptional cellular cofactor CREB, whereas the functional APH and APHBD1 efficiently interacted with CREB (Fig. 3C). Taken together, these results indicate that the C-terminal LXXLL motif of APH-2 is critical for the interaction with CREB and its repression of Tax function.
Generation and characterization of HTLV-2 APH-2 mutant proviral clones.
Since we showed that the LXXLL motif in BD2 of APH-2 is responsible for binding to CREB and thereby repressing Tax function, we next evaluated this functional role of APH-2 in the context of the entire HTLV-2 proviral clone by generating two mutant proviral clones (HTLV-2ΔAPH and HTLV-2APHΔBD2). We first determined whether the APH-2 mutant proviral clones had altered Tax-mediated viral transcription. The cotransfection of either wtAPH-2 or APH-2 mutant proviral clones, as a source of Tax, and the LTR-2Luc reporter vector revealed that the APH-2 mutant proviruses display slightly higher, but statistically significant (P = 0.002 for HTLV-2ΔAPH and P = 0.004 for HTLV-2APHΔBD2), levels of Tax-mediated viral transcription than wtHTLV-2 (Fig. 4A). Consistent with data from the Tax-mediated reporter assay, cells transfected with either APH-2 mutant proviral clone also produced slightly elevated levels of p19 Gag (Fig. 4B), but this marginal increase was not found to be statistically significant. Since HTLV-2ΔAPH and HTLV-2APHΔBD2 display similar phenotypes, it reinforces our overexpression results and clearly shows that the C-terminal LXXLL motif largely determines the repressive effect of APH-2 on Tax-mediated HTLV-2 transcription. The cell lysates from the functional assay immunoprecipitated with anti-APH-2 antiserum and analyzed by WB confirmed the expression of wtAPH-2 and the slower-migrating APHΔBD2. We did not detect any protein expression from the HTLV-2ΔAPH proviral clone (Fig. 4C). These results further reiterate that in the context of the provirus, the C-terminal LXXLL motif of APH-2 plays a key role in repressing Tax-mediated transcription.
Fig 4.
Characterization of proviral clones in vitro. 293T cells (1.5 × 105 cells) were cotransfected with 1 μg wtHTLV-2, HTLV-2ΔAPH, and HTLV-2APHΔBD2 proviral clones or negative-control DNA along with 0.1 μg LTR-2-Luc and 0.01 μg TK-Renilla. All transfections were performed in duplicate and normalized to TK-Renilla to control for the transfection efficiency. Cell lysates or supernatants were harvested 48 h after transfection. Histograms present the average values from three independent experiments. (A) Measurement of Tax activity presented as relative light units (RLU). The asterisk represents the statistically significant difference in Tax activity in HTLV-2ΔAPH and HTLV-2APHΔBD2 proviral clones compared to wtHTLV-2 (P = 0.002 and P = 0.004, respectively, by a Tukey test). (B) Measurement of p19 Gag in the supernatants. (C) Detection of the APH-2 and APH-2 mutant proteins in transfected-cell lysates. The cell lysate was immunoprecipitated by a rabbit anti-APH-2 polyclonal antibody and visualized by WB analysis. β-Actin was assessed as a loading control in those cells transfected with proviral clones only. The asterisks represent a slower-migrating APH detected in both the HTLV-2APHΔBD2 proviral clone and APHΔBD2 cDNA-transfected cells.
Characterization of APH-2 mutant HTLV-2 stable viral producer cell lines.
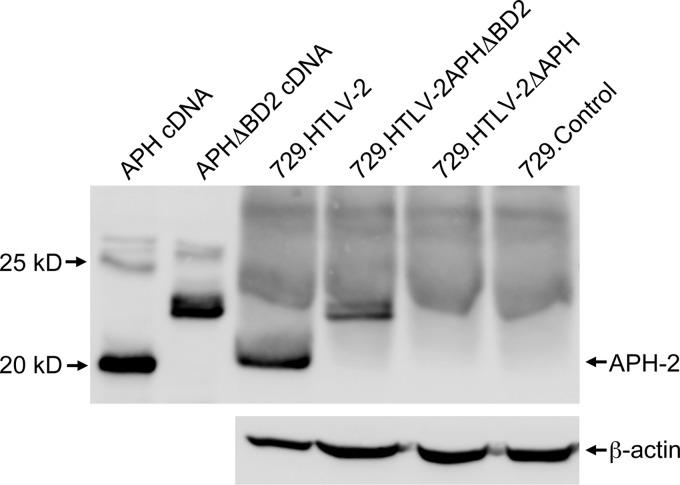
To determine the ability of APH-2 mutant HTLV-2 to replicate and induce cellular immortalization, stable 729B cell transfectants (729.HTLV-2, 729.HTLV-2ΔAPH, and 729.HTLV-2APHΔBD2) expressing the proviral clones were isolated and characterized. Each stable transfectant contained complete copies of the provirus with the expected mutations (data not shown), and the concentration of p19 Gag in the culture supernatant was quantified by ELISA as a measure of virus production (1,200 pg/ml for 729.HTLV-2, 1,400 pg/ml for 729.HTLV-2ΔAPH, and 1,600 pg/ml for 729.HTLV-2APHΔBD2). APH-2 was detected in the virus producer cell lines 729.HTLV-2 and 729.HTLV-2APHΔBD2 after IP using a rabbit anti-APH-2 polyclonal antibody (Fig. 5). However, as shown with transiently transfected cells, APHΔBD2 migrates as a slower protein species, and we did not detect any APH-2 protein expression in 729.HTLV-2ΔAPH or negative-control 729 cells.
Fig 5.
APH-2 protein expression in stably transfected cell lines. A total of 6 × 107 cells of each stably transfected cell line (729.HTLV-2, 729.HTLV-2APHΔBD2, and 729.HTLV-2ΔAPH) and the control 729 cell line were lysed and subjected to IP using a rabbit anti-APH-2 polyclonal antibody and subjected to WB analysis using an APH-2-specific antiserum. Lysates from 293T cells transfected with APH and APHΔBD2 cDNA plasmids are presented as positive controls for protein expression and mobility.
HTLV-2ΔAPH and HTLV-2APHΔBD2 promote virus-induced proliferation and immortalization of PBMCs.
We next assessed the abilities of the APH-2 mutant viruses to induce proliferation and immortalize human PBMCs in coculture assays. Freshly isolated human PBMCs were cocultured with lethally irradiated virus producer cells, 729.HTLV-2, 729.HTLV-2ΔAPH, or 729.HTLV-2APHΔBD2, in the presence of 10 U/ml of human IL-2. The number of virus producer cells used in this assay was normalized to their p19 Gag production levels. The PBMCs infected with 729.HTLV-2 cells and the two APH-2 mutant viruses showed very similar progressive growth patterns consistent with the HTLV-2 immortalization process, whereas control cells died within the first few weeks of culture (Fig. 6A). We also detected a continuous accumulation of p19 Gag in the culture supernatant, which is indicative of viral replication and virion production (Fig. 6B). In an effort to obtain a more quantitative measure of the abilities of these viruses to infect and immortalize PBMCs, a fixed number of PBMCs was cocultivated with viral producer cells in a short-term proliferation assay (48). The number of viral producer cells used in this assay was also normalized to p19 Gag production levels. Since this assay is very stringent as a result of the cultures being diluted 1:3 weekly, slowly growing or nondividing cells are eliminated very quickly, and the percentage of surviving cells is an accurate measure of the immortalization efficiency of the virus. A Kaplan-Meier plot of HTLV-2-induced T-cell proliferation or survival indicated that the percentages of wells containing proliferating lymphocytes were similar between 729.HTLV-2 cells and the two APH-2 mutant viruses (Fig. 6C). Taken together, our results demonstrate that APH-2 is not required for efficient infection or the HTLV-2-mediated immortalization of primary human T lymphocytes in vitro.
Fig 6.
HTLV-2 T-lymphocyte proliferation and immortalization assays. (A) PBMCs (2 × 106 cells) were cultured with 106 irradiated donor cells (729B, 729.HTLV-2, 729HTLV-2ΔAPH, or 729.HTLV-2APHΔBD2) in 24-well plates. A representative growth curve showing cell viability at weekly intervals is presented. (B) HTLV-2 gene expression was quantified by the detection of the p19 Gag protein in the culture supernatant using ELISA. In panels A and B, means and standard deviations of data from each time point were determined from three random independent wells. (C) A total of 104 prestimulated PBMCs were cocultured with 2 × 103 irradiated 729 stable producer cells in 96-well plates. The Kaplan-Meir plot shows the percentages of wells with proliferating cells as a function of time (weeks). The results indicated that the percentages of wells containing proliferating lymphocytes were similar between wtHTLV-2 and the HTLV-2 APH-2 mutant.
Loss of APH-2 results in enhanced viral replication in newly infected rabbits.
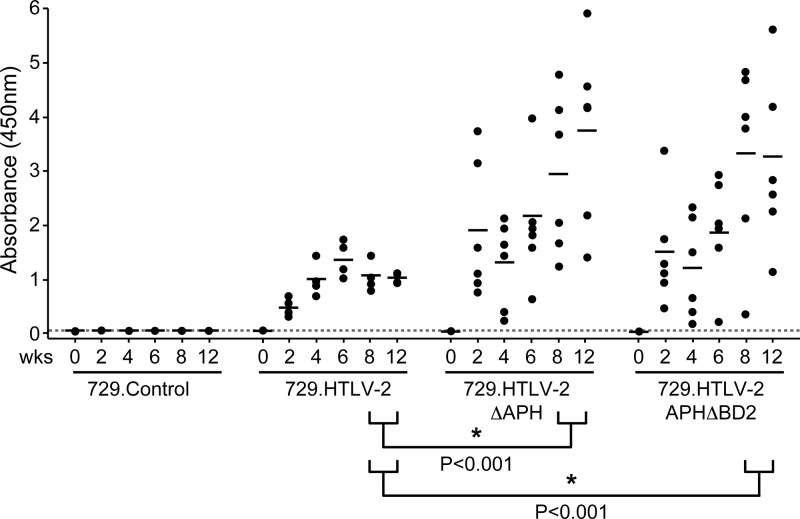
To evaluate the role of APH-2 in vivo, we compared the abilities of the 729.HTLV-2, 729.HTLV-2ΔAPH, and 729.HTLV-2APHΔBD2 cell lines to transmit virus to rabbits, which is an established model to investigate HTLV infection and persistence (10, 11). Rabbits were inoculated with lethally irradiated virus producer cells (cell inocula were equilibrated based on p19 Gag production levels), and at weeks 0, 2, 4, 6, 8, and 12, whole blood was collected and processed for the isolation of plasma and PBMCs. A WB assay was performed on the plasma collected at week 8, and the results showed clear seroconversion in all rabbits infected with wtHTLV-2 or APH-2 mutant HTLV-2 (data not shown). Moreover, a quantitative comparison of antibody responses between each rabbit was performed by using an HTLV-specific ELISA (Fig. 7). Statistical analysis revealed a significantly higher antibody response to HTLV-2 antigens in the 729.HTLV-2ΔAPH-inoculated (P < 0.001) and 729.HTLV-2APHΔBD2-inoculated (P < 0.0001) rabbits than in the 729.HTLV-2-inoculated rabbits at weeks 8 and 12 postinoculation. Consistent with our antibody data, HTLV-2 DNA sequences were detected in all 729.HTLV-2- and APH-2 mutant virus-infected rabbits (Table 1). Quantitative real-time PCR revealed that as early as 2 weeks after inoculation, proviral loads in rabbits infected with APH-2 mutant viruses were significantly higher (12- to 15-fold) than those in rabbits infected with wtHTLV-2 (Table 1). Statistical analysis revealed that the difference in proviral loads between the two APH-2 mutant virus-infected rabbit groups was not significant (Table 1). A comparison of the antibody responses and the proviral loads revealed that there was no correlation between them for any of the three infected groups at any given time point. The proviral loads showed a similar trend for all the three infected groups over time, reaching maximum levels at weeks 2 to 4 and then declining over time. By week 12, although the proviral loads in all the three infected groups had declined, they were 2.6- to 2.9-fold higher in the rabbits infected with the APH-2 mutant viruses than in wtHTLV-2-infected rabbits (Table 1). In contrast, the trends of the antibody responses were different for both mutant virus-infected rabbit groups compared to the wtHTLV-2-infected rabbits (Fig. 7). In the wtHTLV-2-infected rabbits, the antibody levels increased after week 2 postinfection and peaked at week 6, followed by a decline and stabilization by week 12. In both the APH-2 mutant virus-infected rabbit groups, the antibody levels were 2.5 to 3.9 times higher than those in the wtHTLV-2-infected rabbits at week 2, followed by a modest decline at week 4 and a subsequent rise to very high levels by weeks 8 to 12. Diagnostic DNA PCR analyses and nucleotide sequencing performed on PBMCs from rabbits 12 weeks after infection indicated that the infected cells contained the expected viral sequences and showed no indication of Aph-2 reversion (data not shown). Our results indicated that the lack of APH-2 promotes viral transcription and replication. In addition, since HTLV-2ΔAPH and HTLV-2APHΔBD2 mutants display similar phenotypes in vivo, we further conclude that the C-terminal “LXXLL” motif is critical for the functional activity of APH-2. Furthermore, the presence of APH-2 correlates with an earlier humoral response and a smaller proviral load, suggesting that APH-2 could contribute to the lower virulence of HTLV-2.
Fig 7.
Assessment of HTLV-2 infection in rabbits. The antibody response against HTLV-2 in each rabbit was measured by a commercial anti-HTLV ELISA, using both HTLV Gag and envelope proteins as antigens. Each dot represents the absorbance value for a single inoculated rabbit at 0, 2, 4, 6, 8, and 12 weeks postinoculation within each group. Inocula used for the rabbits were 729.HTLV-2 (n = 4), 729.HTLV-2ΔAPH (n = 6), 729.HTLV-2APHΔBD2 (n = 6), or 729B (n = 2) cells. The horizontal line represents the average for the rabbit group at each time point, and the dotted line represents 3 times the standard deviation of uninfected control values. Asterisks indicate that the antibody responses in rabbits inoculated with 729.HTLV-2ΔAPH cells and in rabbits inoculated with 729.HTLV-2APHΔBD2 cells are statistically different from those in rabbits inoculated with wtHTLV-2 at week 8 and week 12 postinoculation (P < 0.001 and P < 0.001, respectively). The differences between 729.HTLV-2ΔAPH and 729.HTLV-2APHΔBD2 infections were not statistically significant at any time point (P = 0.7). P values were determined by an ANOVA.
Table 1.
Detection and quantification of HTLV-2 DNA in rabbit PBMCs
| Inoculum and rabbit | PCR result (proviral copy no.) at wk after inoculationa |
|||||
|---|---|---|---|---|---|---|
| 0 | 2b | 4 | 6b | 8 | 12 | |
| 729.HTLV-2 | ||||||
| R1 | − | + (1.91) | + | + (0.32) | + | + (0.22) |
| R2 | − | + (2.29) | + | + (0.33) | + | + (0.17) |
| R3 | − | + (1.20) | + | + (0.23) | + | + (0.19) |
| R4 | − | + (1.41) | + | + (0.35) | + | + (0.19) |
| 729.HTLV-2ΔAPH-2 | ||||||
| R1 | − | + (17.72) | + | + (0.71) | + | + (0.79) |
| R2 | − | + (18.23) | + | + (1.16) | + | + (0.22) |
| R3 | − | + (16.43) | + | + (1.61) | + | + (0.70) |
| R4 | − | + (13.61) | + | + (1.04) | + | + (0.41) |
| R5 | − | + (30.74) | + | + (1.37) | + | + (0.86) |
| R6 | − | + (21.94) | + | + (2.09) | + | + (0.31) |
| 729.HTLV-2APH-2ΔBD2 | ||||||
| R1 | − | + (5.11) | + | + (0.84) | + | + (0.49) |
| R2 | − | + (23.53) | + | + (3.10) | + | + (0.65) |
| R3 | − | + (25.39) | + | + (0.82) | + | + (0.39) |
| R4 | − | + (49.01) | + | + (1.63) | + | + (0.73) |
| R5 | − | + (13.45) | + | + (1.23) | + | + (0.36) |
| R6 | − | + (33.05) | + | + (1.52) | + | + (0.33) |
| 729B | ||||||
| R1 | − | − (0.00) | − | − (0.00) | − | − (0.00) |
| R2 | − | − (0.04) | − | − (0.07) | − | − (0.00) |
−, negative PCR result; +, positive PCR result; R1, rabbit 1. Numbers in parentheses denote proviral copy numbers per 1,000 rabbit PBMCs as determined by real-time PCR.
Copy numbers in rabbits inoculated with each mutant virus at week 2 and week 6 were different from those of 729.HTLV-2 cells, as determined by an analysis of variance (ANOVA)-Tukey test (P < 0.005). There was no significant difference between each mutant virus at any time point (P > 0.5).
DISCUSSION
The antisense viral transcript has been characterized for retroviruses, including human immunodeficiency virus (HIV), feline immunodeficiency virus (FIV), and HTLV (5, 30, 45). In HTLV-1, the antisense transcript encodes the viral protein HBZ, which plays a crucial role in viral persistence and late-stage leukemogenesis (35). For HTLV-2, the antisense counterpart, APH-2, was identified recently (20). HTLV-1 and HTLV-2 are the most widely characterized strain types of HTLV. These viruses have distinct pathogenic properties: HTLV-1 is the most pathogenic virus, causing ATL, HAM/TSP, and other inflammatory disorders (17, 22), and HTLV-2 is less pathogenic and has been associated infrequently with a HAM/TSP-like neurological disorder but not with leukemia (1, 4). Differences in viral pathogenesis have led to the subsequent delineation of a number of distinct functions of the viral proteins encoded by these two viruses. Therefore, it is of great interest to determine the similarities and differences between the two antisense gene products of HTLV-1 and HTLV-2. In the present study, we showed that, like HBZ, APH-2 represses Tax-mediated viral transcription by interacting with the host cellular factor CREB through the LXXLL motif in the C terminus and is also dispensable for viral transformation in vitro. In contrast to HBZ, the loss of APH-2 increases viral replication as well as virus-induced humoral responses in infected rabbits. Our findings open new avenues for the further exploration of the distinct functional properties of these two proteins that contribute to the differential pathogeneses of HTLV-1 and HTLV-2.
Hbz is expressed in all asymptomatic carriers and patients, and the expression level of Hbz positively correlates with disease severity (34). In many ATL cases, tax is no longer expressed, and Hbz is the only viral gene that is expressed. However, the HBZ protein is scarcely detected in these patients but can be detected in HTLV-1-transformed as well as HTLV-1 stably transfected cell lines (2). Aph-2 mRNA was detected previously in HTLV-2-infected cell lines and in 27% of HTLV-2 carriers (20). Aph-2 mRNA expression correlated with HTLV-2 proviral loads in HTLV-2-infected carriers, but APH-2 expression did not appear to promote lymphocytosis in vivo or cell proliferation in vitro (12). In the present study, we further determined the presence of the APH-2 transcript and protein in HTLV-2 stably transfected B-cell lines as well as in HTLV-2-immortalized T-cell lines. Although Aph-2 mRNA was detected in both these cell types, the APH-2 protein was detected only in the HTLV-2 stably transfected B-cell lines after protein enrichment by IP using a rabbit anti-APH-2 polyclonal antibody. Thus, although Aph-2 mRNA can be detected in all cell lines, the protein is not as readily detectable as HBZ.
HBZ represses Tax-mediated viral transcription by interacting with CREB and CBP/p300 through its b-ZIP and activation domains, respectively (2, 9, 31). Moreover, Halin et al. showed previously that APH-2 represses Tax-2-mediated viral transcription both from a cDNA vector and from a proviral clone (20). Here, we compared the abilities of HBZ and APH-2 to repress Tax-mediated viral transcription from either an HTLV-1 or an HTLV-2 proviral clone. Although the HBZ and APH-2 proteins repressed Tax-mediated viral transcription, the repressive activity of APH-2 was only 48% of that of HBZ. One possible explanation for this finding is that HBZ strongly interacts with CBP/p300 coactivators through the LXXLL motifs within its transactivation domain, thereby preventing recruitment to the viral promoter. Although APH-2 can efficiently interact with CREB, the binding with CBP/p300 is weak, unlike HBZ (9, 20).
Similar to the LXXLL motifs in HBZ, APH-2 has a 179LXXLL183 motif and an LXXLL-like motif (64IXXLL68) at the C terminus and central region, respectively. To evaluate the function of these motifs in APH-2, we generated five mutants. The ectopic expression of these mutants revealed that BD2 containing the LXXLL motif at the C terminus is critical for the Tax-repressive function and the interaction with the host cellular factor CREB. This function of the LXXLL motif at the C terminus was maintained even in the context of the entire HTLV-2 proviral clone. Additionally, we also observed an unexpected shift in mobility when this motif was completely deleted. We speculate that this shift might be due to altered posttranslational modifications or an improper folding of APH-2 leading to a conformational change. Thus, both HBZ and APH-2 interact with CREB but through different motifs. Further investigation into the posttranslational modifications and even the cellular localization of the APH-2 mutant is necessary to elucidate this possibility.
We previously reported that HBZ is dispensable for the immortalization of primary T lymphocytes in cell culture (2). Others have shown that HBZ plays a role in late-stage leukemogenesis, when Tax is no longer expressed (26). Additionally, it is known that HTLV-1 causes leukemia, while HTLV-2 does not. In this study, we determined the requirement for APH-2 for the in vitro HTLV-2 immortalization of freshly isolated T lymphocytes in both long-term and stringent short-term coculture assays. The APH-2 mutant viruses immortalized freshly isolated T lymphocytes with an efficiency similar to that of wtHTLV-2. Thus, HTLV-2 APH-2, like HTLV-1 HBZ, is dispensable for the immortalization of freshly isolated T lymphocytes. However, since HTLV-2 is aleukemic, there is no in vivo evidence for a role of APH-2 in leukemogenesis.
We next evaluated the replication efficiencies of these mutant viruses in comparison with those of wtHTLV-2 along with a negative control in newly infected hosts. Since HTLV-infected individuals are chronically infected at the time of detection, the field is forced to utilize rabbits as an early-infection model to study viral replication and persistence as well as virus-triggered immune responses. In our previous study, we showed that HBZ, like other HTLV-1 and HTLV-2 accessory proteins, is important for viral persistence. Our present data indicated that unlike HBZ and other accessory proteins, APH-2 is not required for viral persistence. On the other hand, a lack of APH-2 increased the infection rate of the mutant viruses in comparison to that of wtHTLV-2 at statistically significant levels, as demonstrated by both the proviral loads and the humoral responses in the rabbits. These results suggest that APH-2 could potentially contribute to the less robust infection by HTLV-2 than by HTLV-1.
In summary, our study demonstrates that the antisense proteins in HTLV-1 and HTLV-2 confer various pathogenic properties. Although HBZ and APH-2 repress Tax-mediated viral transcription at various levels through interactions with CREB, the motifs that mediate this interaction are different. The only similar function between HBZ and APH-2 is that they are both not required for the immortalization of T cells in vitro. Unlike HBZ, which plays an important role in viral persistence, APH-2 is dispensable for enhancing viral replication and persistence. This difference, along with the increase in proviral loads when rabbits were inoculated with the APH-2 mutant viruses, provides insight into the distinctive pathogeneses of HTLV-1 and HTLV-2. Our findings warrant further investigation into the role of APH-2 in HTLV-2 pathogenesis, which could subsequently provide important insights into the use of these antisense gene products as potential therapeutic targets.
ACKNOWLEDGMENTS
We thank K. Hayes-Ozello for editorial comments on the manuscript and T. Vojt for figure preparations.
This work was supported by a grant from the National Institutes of Health (grant AI095913) to P.L.G.
Footnotes
Published ahead of print 23 May 2012
REFERENCES
- 1. Araujo A, Hall WW. 2004. Human T-lymphotropic virus type II and neurological disease. Ann. Neurol. 56:10–19 [DOI] [PubMed] [Google Scholar]
- 2. Arnold J, et al. 2006. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood 107:3976–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnold J, Zimmerman B, Li M, Lairmore MD, Green PL. 2008. Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T lymphocyte proliferation. Blood 112:3788–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartman MT, et al. 2008. Long-term increases in lymphocytes and platelets in human T-lymphotropic virus type II infection. Blood 112:3995–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Briquet S, Richardson J, Vanhee-Brossollet C, Vaquero C. 2001. Natural antisense transcripts are detected in different cell lines and tissues of cats infected with feline immunodeficiency virus. Gene 267:157–164 [DOI] [PubMed] [Google Scholar]
- 6. Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. 1999. Genetic instability and Darwinian selection in tumours. Trends Cell Biol. 9:M57–M60 doi:10.1016/S0962-8924(99)01661-X [PubMed] [Google Scholar]
- 7. Cavanagh M-H, et al. 2006. HTLV-I antisense transcripts initiating in the 3′ LTR are alternatively spliced and polyadenylated. Retrovirology 3:15 doi:10.1186/1742-4690-3-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen IS, McLaughlin J, Gasson JC, Clark SC, Golde DW. 1983. Molecular characterization of genome of a novel human T-cell leukaemia virus. Nature 305:502–505 [DOI] [PubMed] [Google Scholar]
- 9. Clerc I, et al. 2008. An interaction between the human T cell leukemia virus type 1 basic leucine zipper factor (HBZ) and the KIX domain of p300/CBP contributes to the down-regulation of tax-dependent viral transcription by HBZ. J. Biol. Chem. 283:23903–23913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cockerell GL, Rovank J, Green PL, Chen ISY. 1996. A deletion in the proximal untranslated pX region of human T-cell leukemia virus type II decreases viral replication but not infectivity in vivo. Blood 87:1030–1035 [PubMed] [Google Scholar]
- 11. Collins ND, Newbound GC, Ratner L, Lairmore MD. 1996. In vitro CD4+ lymphocyte transformation and infection in a rabbit model with a molecular clone of human T-cell lymphotropic virus type 1. J. Virol. 70:7241–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Douceron E, et al. 2012. HTLV-2 APH-2 expression is correlated with proviral load but APH-2 does not promote lymphocytosis. J. Infect. Dis. 205:82–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Endo K, et al. 2002. Human T-cell leukemia virus type 2 (HTLV-2) Tax protein transforms a rat fibroblast cell line but less efficiently than HTLV-1 Tax. J. Virol. 76:2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Felber BK, Paskalis H, Kleinman-Ewing C, Wong-Staal F, Pavlakis GN. 1985. The pX protein of HTLV-I is a transcriptional activator of its long terminal repeats. Science 229:675–679 [DOI] [PubMed] [Google Scholar]
- 15. Feuer G, Green PL. 2005. Comparative biology of human T-cell lymphotropic virus type 1 (HTLV-1) and HTLV-2. Oncogene 24:5996–6004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gaudray G, et al. 2002. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 76:12813–12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gessain A, et al. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet ii:407–410 [DOI] [PubMed] [Google Scholar]
- 18. Green PL, Ross TM, Chen ISY, Pettiford S. 1995. Human T-cell leukemia virus type II nucleotide sequences between env and the last exon of tax/rex are not required for viral replication or cellular transformation. J. Virol. 69:387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grossman WJ, et al. 1995. Development of leukemia in mice transgenic for the tax gene of human T-cell leukemia virus type I. Proc. Natl. Acad. Sci. U. S. A. 92:1057–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Halin M, et al. 2009. Human T-cell leukemia virus type 2 produces a spliced antisense transcript encoding a protein that lacks a classic bZIP domain but still inhibits Tax2-mediated transcription. Blood 114:2427–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hasegawa H, et al. 2006. Thymus-derived leukemia-lymphoma in mice transgenic for the Tax gene of human T-lymphotropic virus type I. Nat. Med. 12:466–472 [DOI] [PubMed] [Google Scholar]
- 22. Hinuma Y, et al. 1981. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc. Natl. Acad. Sci. U. S. A. 78:6476–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hirata A, et al. 2004. PDZ domain-binding motif of human T-cell leukemia virus type 1 Tax oncoprotein augments the transforming activity in a rat fibroblast cell line. Virology 318:327–336 [DOI] [PubMed] [Google Scholar]
- 24. Hivin P, et al. 2007. The HBZ-SP1 isoform of human T-cell leukemia virus type I represses JunB activity by sequestration into nuclear bodies. Retrovirology 4:14 doi:10.1186/1742-4690-4-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones KS, et al. 2006. Human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2 use different receptor complexes to enter T cells. J. Virol. 80:8291–8302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kannian P, Green PL. 2010. Human T lymphotropic virus type 1 (HTLV-1): molecular biology and oncogenesis. Viruses 2:2037–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuhlmann AS, et al. 2007. HTLV-1 HBZ cooperates with JunD to enhance transcription of the human telomerase reverse transcriptase gene (hTERT). Retrovirology 4:92 doi:10.1186/1742-4690-4-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kusuhara K, Anderson M, Pettiford SM, Green PL. 1999. Human T-cell leukemia virus type 2 Rex protein increases stability and promotes nuclear to cytoplasmic transport of gag/pol and env RNAs. J. Virol. 73:8112–8119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lairmore M, Franchini G. 2007. Human T-cell leukemia virus types 1 and 2, p 2071–2106 In Knipe DM, et al. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 30. Larocca D, Chao LA, Seto MH, Brunck TK. 1989. Human T-cell leukemia virus minus strand transcription in infected cells. Biochem. Biophys. Res. Commun. 163:1006–1013 [DOI] [PubMed] [Google Scholar]
- 31. Lemasson I, et al. 2007. Human T-cell leukemia virus type 1 (HTLV-1) bZIP protein interacts with the cellular transcription factor CREB to inhibit HTLV-1 transcription. J. Virol. 81:1543–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li M, Green PL. 2007. Detection and quantitation of HTLV-1 and HTLV-2 mRNA species by real-time RT-PCR. J. Virol. Methods 142:159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mahieux R, et al. 2000. Differences in the ability of human T-cell lymphotropic virus type 1 (HTLV-1) and HTLV-2 tax to inhibit p53 function. J. Virol. 74:6866–6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matsuoka M, Green PL. 2009. The HBZ gene, a key player in HTLV-1 pathogenesis. Retrovirology 6:71 doi:10.1186/1742-4690-6-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matsuoka M, Jeang KT. 2011. Human T-cell leukemia virus type 1 (HTLV-1) and leukemic transformation: viral infectivity, Tax, HBZ and therapy. Oncogene 30:1379–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meertens L, Chevalier S, Weil R, Gessain A, Mahieux R. 2004. A 10-amino acid domain within human T-cell leukemia virus type 1 and type 2 tax protein sequences is responsible for their divergent subcellular distribution. J. Biol. Chem. 279:43307–43320 [DOI] [PubMed] [Google Scholar]
- 37. Murata K, et al. 2006. A novel alternative splicing isoform of human T-cell leukemia virus type 1 bZIP factor (HBZ-SI) targets distinct subnuclear localization. J. Virol. 80:2495–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robek MD, Ratner L. 1999. Immortalization of CD4+ and CD8+ T lymphocytes by human T-cell leukemia virus type 1 Tax mutants expressed in a functional molecular clone. J. Virol. 73:4856–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ross TM, Narayan M, Fang Z-Y, Minella AC, Green PL. 2000. Human T-cell leukemia virus type 2 tax mutants that selectively abrogate NFκB or CREB/ATF activation fail to transform primary human T cells. J. Virol. 74:2655–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ross TM, Pettiford SM, Green PL. 1996. The tax gene of human T-cell leukemia virus type 2 is essential for transformation of human T lymphocytes. J. Virol. 70:5194–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Satou Y, Yasunaga J, Yoshida M, Matsuoka M. 2006. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Natl. Acad. Sci. U. S. A. 103:720–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Satou Y, et al. 2011. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog. 7:e1001274 doi:10.1371/journal.ppat.1001274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Semmes OJ, et al. 1996. HTLV-I and HTLV-II Tax: differences in induction of micronuclei in cells and transcriptional activation of viral LTRs. Virology 217:373–379 [DOI] [PubMed] [Google Scholar]
- 44. Thebault S, Basbous J, Hivin P, Devaux C, Mesnard JM. 2004. HBZ interacts with JunD and stimulates its transcriptional activity. FEBS Lett. 562:165–170 [DOI] [PubMed] [Google Scholar]
- 45. Vanhee-Brossollet C, et al. 1995. A natural antisense RNA derived from the HIV-1 env gene encodes a protein which is recognized by circulating antibodies of HIV+ individuals. Virology 206:196–202 [DOI] [PubMed] [Google Scholar]
- 46. Wycuff DR, Marriott SJ. 2005. The HTLV-1 Tax oncoprotein: hypertasking at the molecular level. Front. Biosci. 10:620–642 [DOI] [PubMed] [Google Scholar]
- 47. Xie L, Green PL. 2005. Envelope is a major viral determinant of the distinct in vitro cellular transformation tropism of human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2. J. Virol. 79:14536–14545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xie L, Yamamoto B, Haoudi A, Semmes OJ, Green PL. 2006. PDZ binding motif of HTLV-1 Tax promotes virus-mediated T-cell proliferation in vitro and persistence in vivo. Blood 107:1980–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yamamoto B, et al. 2008. Human T-cell leukemia virus type 2 post-transcriptional control protein p28 is required for viral infectivity and persistence in vivo. Retrovirology 5:38 doi:10.1186/1742-4690-5-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ye J, Sileverman L, Lairmore MD, Green PL. 2003. HTLV-1 Rex is required for viral spread and persistence in vivo but is dispensable for cellular immortalization in vitro. Blood 102:3963–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ye J, Xie L, Green PL. 2003. Tax and overlapping Rex sequences do not confer the distinct transformation tropisms of human T-cell leukemia virus types 1 and 2. J. Virol. 77:7728–7735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Younis I, et al. 2004. Repression of human T-cell leukemia virus type 1 and 2 replication by a viral mRNA-encoded posttranscriptional regulator. J. Virol. 78:11077–11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao T, et al. 2009. Human T-cell leukemia virus type 1 bZIP factor selectively suppresses the classical pathway of NF-kappaB. Blood 113:2755–2764 [DOI] [PubMed] [Google Scholar]