Abstract
The first yellow-grain fungal mycetoma, in a 60-year-old man from Central Sudan, is reported. Morphological and phylogenetic analysis of the ribosomal small subunit (SSU), large subunit (LSU), internal transcribed spacer (ITS), β-tubulin (BT2), actin (ACT1), and elongation factor (TEF1) genes revealed that the isolate deviated from any known agent of mycetoma; it clustered in the genus Pleurostoma (anamorph genus, Pleurostomophora) in the order Calosphaeriales. The novel species, here named Pleurostomophora ochracea, is characterized by phenotypic features. The species proved to be highly susceptible to itraconazole, ketoconazole, posaconazole, and voriconazole, but not to fluconazole. The fungus was inhibited by caspofungin at 8 μg/ml, while no inhibition was found with 5-flucytosine (MIC > 64 μg/ml). Compared to other members of the genus Pleurostomophora, P. ochracea is slow growing, with a relatively high optimum growth temperature (36 to 37°C). This is the first case of a yellow-grain fungal mycetoma; yellow grains are otherwise of bacterial nature. Our case emphasizes that identification of mycetoma agents by the color of the grain only is not sufficient and may lead to inappropriate therapy.
INTRODUCTION
Mycetoma is a chronic granulomatous, progressive, subcutaneous inflammatory infection (12). It is caused by true fungi or by bacteria, and hence, it is classified as eumycetoma or actinomycetoma, respectively (1, 3). The triad of a painless subcutaneous mass, multiple sinuses, and seropurulent discharge containing grains is pathognomic of mycetoma (5, 14). Eumycetoma is more common than actinomycetoma throughout the world, and in Sudan, it accounts for 70% of all mycetoma cases (5). A variety of fungi are implicated in causing eumycetoma (Table 1). In Sudan, eumycetoma is usually caused by the fungus Madurella mycetomatis, which forms black grains in human tissue (1, 15). The most common fungal pathogens causing eumycetoma produce either black grains or white grains (5, 10). While yellow grains have been encountered in cases of actinomycetoma, they have never been reported before in eumycetoma. The available diagnostic tools for mycetoma are limited and are mostly of a phenotypic nature (1). A first indication is usually obtained from the color of the grain: black grains are produced by fungi and yellow grains by bacteria, while white grains can be of bacterial or fungal nature. Culture media used for diagnosis are based upon this first indication. Here, we present a case of yellow-grain mycetoma, which was initially misdiagnosed as actinomycetoma based on the color of the grain and on cytology. In this report, we show that cultural and molecular techniques, in combination with an updated taxonomy, can be successfully used to identify the yellow-grain-producing fungus Pleurostomophora ochracea as a new causative agent of human mycetoma.
Table 1.
Agents of mycetoma in mammals and their phylogenetic positionsa
| Name | Order | Grain |
|---|---|---|
| Acremonium kiliense | Hypocreales | White |
| Acremonium potronii | Hypocreales | White |
| Acremonium recifei | Hypocreales | White |
| Cylindrocarpon cyanescens | Hypocreales | White |
| Cylindrocarpon destructans | Hypocreales | White |
| Fusarium falciforme | Hypocreales | White |
| Fusarium verticillioides | Hypocreales | White |
| Fusarium solani | Hypocreales | White |
| Phialemonium obovatum | Hypocreales | White |
| Aspergillus flavus | Eurotiales | White |
| Aspergillus hollandicus | Eurotiales | White |
| Aspergillus nidulans | Eurotiales | White |
| Bipolaris spicifera | Pleosporales | Black |
| Corynespora cassiicola | Pleosporales | Black |
| Curvularia geniculata | Pleosporales | Black |
| Curvularia lunata | Pleosporales | Black |
| Leptosphaeria senegalensis | Pleosporales | Black |
| Leptosphaeria thompkinsii | Pleosporales | Black |
| Madurella grisea | Pleosporales | Black |
| Pseudochaetosphaeronema larense | Pleosporales | Black |
| Pyrenochaeta mackinnonii | Pleosporales | Black |
| Pyrenochaeta romeroi | Pleosporales | Black |
| Neotestudina rosatii | Pleosporales | White/black |
| Madurella mycetomatis | Sordariales | Black |
| Madurella pseudomycetomatis | Sordariales | Black |
| Madurella fahalii | Sordariales | Black |
| Madurella tropicana | Sordariales | Black |
| Phaeoacremonium parasiticum | Diaporthales | White |
| Phaeoacremonium krajdenii | Diaporthales | White |
| Exophiala jeanselmei | Chaetothyriales | Black |
| Pseudallescheria boydii | Microascales | White |
| Trichophyton sp. | Onygenales | White |
| Microsporum canis | Onygenales | White |
Adapted from de Hoog et al. (10).
CASE REPORT
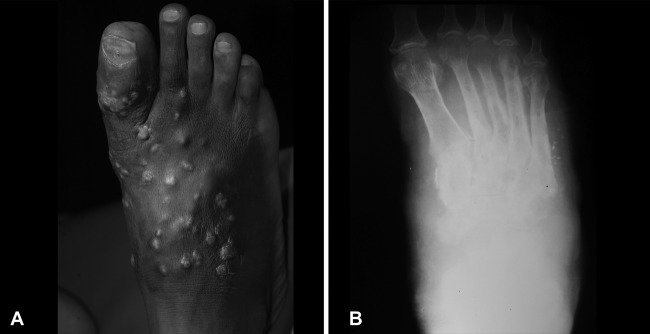
The patient was a 60-year-old male farmer from central Sudan. In 2006, he presented to the Mycetoma Clinic at the Mycetoma Research Centre of the University of Khartoum, Khartoum, Sudan, with a massive, recurrent right-foot swelling. His condition started 20 years prior to presentation with a small, painless right-foot swelling, which had gradual onset and progress and was painful. He developed multiple sinuses, and the seropurulent discharge contained yellow grains. The patient underwent four surgical excisions under general and spinal anesthesia done elsewhere. He had neither a history of local trauma nor a family history of mycetoma. The social and geographic history and drug use were not contributory to his problem. On examination, he looked well, not pale and neither jaundiced nor cyanosed. He had a normal pulse rate (75/min), respiratory rate (16/min), blood pressure (120/80), and temperature (37°C). Also, his renal and hepatic function tests, as well as his full blood count, were within normal limits. Local examination revealed a painless massive mass involving the dorsal and plantar parts of the right foot. There were multiple active and healed sinuses, and the seropurulent discharge contained yellow grains (Fig. 1). The foot X-ray confirmed a massive soft tissue swelling and revealed a periosteal reaction and bone destruction (Fig. 1). Fine-needle aspiration for cytology was taken from the lesion and showed an intense inflammatory infiltrate with grains typical of actinomycetes. The patient was started on streptomycin (1 g per day) and cotrimoxazole (945 mg twice a day) and was followed up regularly in the outpatient clinic. However, there was no clinical response to this treatment. He was then switched to amikacin sulfate (15 mg/kg of body weight twice a day) and cotrimaxozole (945 mg twice a day) with no clinical improvement. He underwent surgical debulking to improve the response to medical treatment. Numerous yellow grains were collected from discharging sinuses, which were hard in consistency and irregular in size. Culturing of the grains revealed fungal growth, indicating that the patient was suffering from a eumycetoma rather than an actinomycetoma. Unfortunately, changing the treatment accordingly was not possible; the patient was lost at follow-up and was never seen again.
Fig 1.
Clinical presentation of the patient. (A) The patient had a mycetoma on the right foot, with massive, recurrent foot swelling. (B) On the foot X-ray, a massive soft tissue swelling, a periosteal reaction, and bone destruction are seen.
MATERIALS AND METHODS
Clinical specimen.
A clinical specimen was collected from the patient when he presented with yellow-grain mycetoma at the Mycetoma Research Centre, University of Khartoum, Khartoum, Sudan, on 27 October 2008. After obtaining written consent from the patient, an aspirate with visible yellow grains was taken from the sinuses. The yellow grains were collected for further identification.
Fungal isolation.
Grains collected from the sinus were washed twice in physiological saline and inoculated into Lowenstein-Jensen (LJ) medium (Hi Media Laboratories, Mumbai, India). The grain was cultured for 3 to 4 weeks at 37°C. This initial inoculation on LJ medium was guided by the color of the grain: yellow grains were considered to be of bacterial origin. After a week, the color of the grains converted from yellow to black, and the grain was subcultured on Sabouraud's dextrose agar (SDA) (Hi Media). The culture was identified morphologically as a fungal species. For identification, the isolate was shipped to the Department of Medical Microbiology and Infectious Diseases, Erasmus MC, Rotterdam, Netherlands, and to the Centraalbureau voor Schimmelcultures (CBS) KNAW Fungal Biodiversity Centre, Utrecht, Netherlands, and deposited in the CBS collection under number CBS 131321.
Morphology and physiology.
Colony characteristics and growth morphology were studied by inoculating the isolate onto plates of malt extract agar (MEA) (Oxoid, United Kingdom), oat meal agar (OA) (homemade at CBS, Netherlands), corn meal agar (CMA) (homemade at CBS, Netherlands), and potato dextrose agar (PDA) (Oxoid, United Kingdom) and incubating the plates in the dark at 30°C for up to 4 weeks. Microscopic mounts in lactic acid with cotton blue were made from cultures grown on a PDA plate and slide cultures after 2 weeks of incubation at 30°C. The slides were examined and measured with a light microscope (Nikon Eclipse 80i), and pictures were taken using a camera attached to the microscope (Nikon; digital-sight DS-5 M). A minimum of 10 measurements per structure were taken after processing in Adobe Photoshop CS3, and the average was calculated. Cardinal growth temperatures were determined on PDA, and the cultures were incubated in the dark for 4 weeks at temperatures ranging from 15°C to 40°C at 3°C intervals. The ability of the isolate to assimilate carbohydrate sources was determined with the API 20C AUX system (bioMérieux, Marcy l'Etoile, France). Prior to the carbohydrate assimilation test, the homogenized fungal suspension was prepared by ultrasonic treatment as published previously (4, 36) and then was adjusted to a 2 McFarland standard with the medium provided. Finally, 100 μl of this inoculum was used to fill the cupules of the test strips as directed by the manufacturer. The ability of the isolate to hydrolyze casein, gelatin, and starch was determined by culturing the strains on selective plates according to the manufacturer's instructions. The ability to produce urease was determined by culturing on urea agar (urea base concentrate [Difco] with 1.5% [wt/vol] agar). Sodium chloride tolerance was determined by comparing the growth after 2 weeks on SDA slants containing 0, 0.5, 5, 10, and 30% NaCl.
DNA extraction, amplification, and sequencing.
Genomic DNA was extracted by scraping material off Sabouraud plates, freezing it in liquid nitrogen, and grinding it with a mortar and pestle. DNA was extracted from the resulting pulp with the Promega Wizard Kit (Promega) according to the manufacturer's instructions. The ribosomal DNA (rDNA) internal transcribed spacer (ITS) gene and partial sequences of the actin (ACT1), β-tubulin (BT2), and elongation factor 1α (TEF1) genes, as well as the 18S rDNA gene (small subunit [SSU]) and 28S rDNA gene (large subunit [LSU]), were amplified and sequenced. The primer pairs for the genes were ITS4 and ITS5 (39), ACT-512F and ACT-783R (9), T1 and BT2b (18, 28), EF2 and EF728F (9, 23), NS1 and NS24 (17, 39), and LRoR and LR5 (38), respectively. Additional primers used for sequencing of the SSU included NS2, NS3, NS6, and NS7 (39).
Alignment and phylogenetic analysis.
A consensus sequence was computed from the forward and reverse sequences with SeqMan from the Lasergene package (DNAstar, Madison, WI). Sequences retrieved from GenBank are listed in Fig. 2 and in Table 2. Sequences were aligned with Multiple Sequence Comparison by Log-Expectation (MUSCLE) using the EMBL-EBI Web server (http://www.ebi.ac.uk/Tools/msa/muscle/). An alignment was constructed for the complete ITS (ITS1-5.8S-ITS2), including 30 strains representing 12 species. A combined 28S LSU and 18S SSU alignment for 39 strains from 36 fungal species was constructed. To investigate the phylogenetic relationship of the newly isolated fungus and related taxa, maximum-likelihood, maximum-parsimony, and Bayesian analyses were used for both alignments. Maximum likelihood with 500 bootstraps using the Tamura-Nei model and maximum-parsimony analysis were conducted in MEGA v. 5.05 (33, 34). Bayesian analysis was done using MrBayes v. 3.1.2 software. All trees were constructed by the outgroup method and edited in MEGA v. 5.05.
Fig 2.
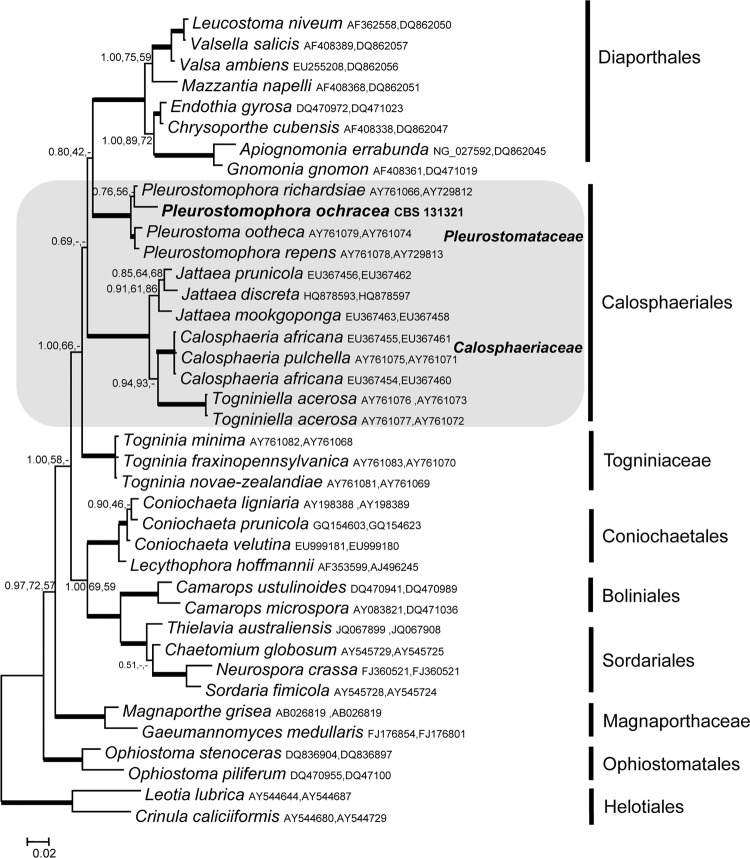
Phylogram of two loci (SSU and LSU) obtained by Bayesian analysis, maximum likelihood, and maximum parsimony (values of >0.8 for Bayesian probability and >80% for maximum likelihood and maximum parsimony are shown with thick branches). Leotia lubrica and Crinula caliciiformis were used as the outgroup. The portion of the phylogram relating to Calosphaeriales is shaded. The names of families within the order and the new isolate are shown in bold.
Table 2.
Calosphaeriales strains
| Strain no. | Name | ITS | Source |
|---|---|---|---|
| STE-U 6181 | Calosphaeria africana | EU367445 | Plant |
| CBS 120870 | Calosphaeria africana | EU367444 | Plant |
| CBS 115999 | Calosphaeria pulchella | EU367451 | Plant |
| SM05 | Calosphaeria pulchella | HM237300 | Plant |
| LM06 | Calosphaeria pulchella | HM237298 | Plant |
| DC04 | Calosphaeria pulchella | HM237299 | Plant |
| SS07 | Calosphaeria pulchella | HM237297 | Plant |
| CBS 127681 | Jattaea discreta | HQ878587 | Plant |
| CBS 119343 | Jattaea leucospermi | EU552127 | Plant |
| STE-U 6401 | Jattaea mookgoponga | EU367450 | Plant |
| CBS 120867 | Jattaea mookgoponga | EU367449 | Plant |
| CBS 120871 | Jattaea prunicola | EU367446 | Plant |
| STE-U 6400 | Jattaea prunicola | EU367448 | Plant |
| STE-U 6399 | Jattaea prunicola | EU367447 | Plant |
| CBS 294.39 | Pleurostomophora repens | AF083195 | Plant |
| CBS 131321 | Pleurostomophora ochracea | (JX073270) | Human |
| CBS 115329 | Pleurostoma ootheca | HQ878590 | Plant |
| IFM5 54325 | Pleurostomophora richardsiae | AB364704 | Human |
| CBS 302.62 | Pleurostomophora richardsiae | AB364698 | Plant |
| CBS 506.90 | Pleurostomophora richardsiae | AB364702 | Human |
| CBS 295.39 | Pleurostomophora richardsiae | AB364697 | Plant |
| IFM 4926 | Pleurostomophora richardsiae | AB364694 | Human |
| PC1 | Pleurostomophora richardsiae | AB364693 | Human |
| CBS 483.80 | Pleurostomophora richardsiae | AB364701 | Human |
| IFM 41579 | Pleurostomophora richardsiae | AB364695 | Human |
| CBS 270.33 | Pleurostomophora richardsiae | AY729811 | Human |
| CBS 406.93 | Pleurostomophora richardsiae | AB364703 | Plant |
| CBS 631.94 | Phaeoacremonium aleophilum | AF266647 | Plant |
| CBS 222.95 | Phaeoacremonium inflatipes | AF266655 | Plant |
| STE-U 5963 | Togninia minima | EU128019 | Plant |
| CBS 113648 | Togniniella acerosa | EU367453 | Plant |
| CBS 113726 | Togniniella acerosa | EU367452 | Plant |
Antifungal susceptibility.
Antifungal susceptibilities for eight antifungal drugs were determined in triplicate by using the colorimetric Sensititre YeastOne method (Trek Diagnostic Systems, East Grinstead, United Kingdom) as described previously (36). In short, the isolate was cultured for 10 days in RPMI 1640 medium supplemented with l-glutamine (0.3 g/liter) and 20 mM morpholinepropanesulfonic acid at 37°C. Mycelia were harvested by 5 min of centrifugation at 2,158 × g and washed once with sterile saline. After sonication (20 s at 28-μm maximum power; Soniprep, Beun de Ronde, Netherlands) of the hyphal suspension, Tween 60 was added at 0.05% (vol/vol), and the transmissions were adjusted to 70% at 660 nm (Novaspec II; Pharmacia Biotech). The inoculated plates were incubated at 37°C for 7 days. The drug concentrations used ranged from 0.016 μg/ml to 8 μg/ml for amphotericin B, itraconazole, ketoconazole, and voriconazole; from 0.25 μg/ml to 128 μg/ml for fluconazole; and from 0.125 μg/ml to 64 μg/ml for 5-flucytosine.
Nucleotide sequence accession numbers.
The GenBank accession numbers for isolate CBS 131321 amplified genes are as follows: 28S rRNA, JX073274; 18S rRNA, JX073269; β-tubulin, JX073271; actin, JX073275; and elongation factor, JX097097.
MycoBank accession number.
The MycoBank accession number for P. ochracea is MB800514.
RESULTS
Phylogeny.
A BLAST search with the ITS, 18S SSU, and 28S LSU sequences did not yield identity with any known fungus. Therefore, the sequences were used to determine the higher-order phylogeny for our isolate. The alignment of LSU and SSU consisted of 1,909 characters, of which 1,520 were constant, 62 were parsimony uninformative, and 327 were parsimony informative. Introns were found in SSUs of three strains from the data set, and they were deleted from the alignment. Maximum-parsimony analysis of the combined data set resulted in 8 most parsimonious trees (length of the tree = 1,109; consistency index [CI] = 0.500451; retention index [RI] = 0.759444; rescaled consistency index [RCI] = 0.380065). The SSU and LSU phylogenetic analyses showed that the reported pathogenic isolate was closely related to the family Pleurostomataceae, with high bootstrap support with all algorithms used (1.00, 100, and 100) (Fig. 2) in the order Calosphaeriales.
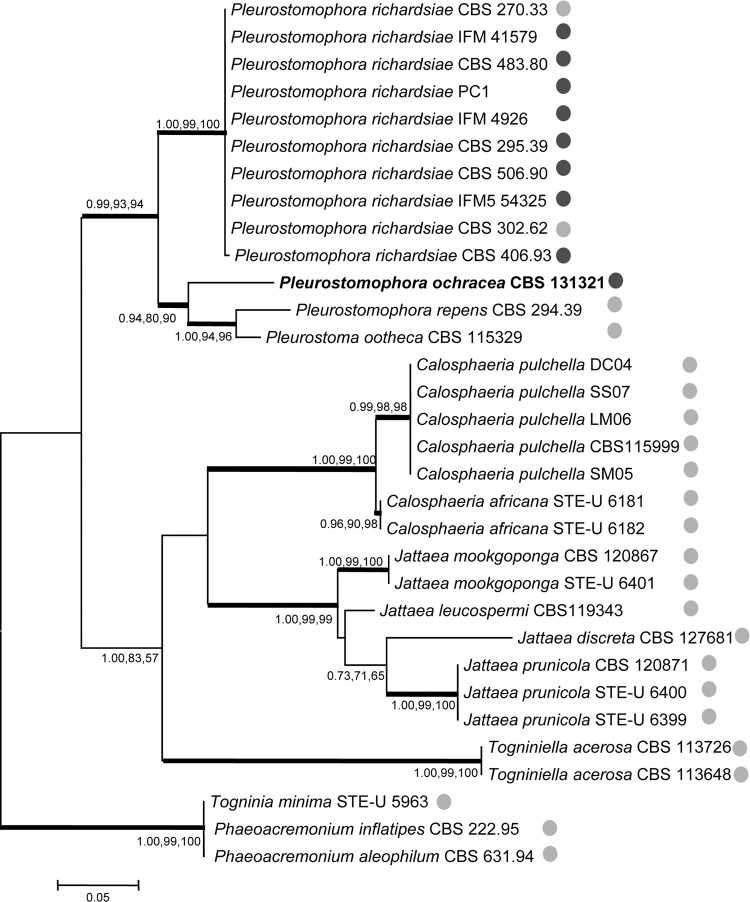
The ITS alignment consisted of 600 characters, of which 342 were constant, 242 were parsimony informative, and 16 were parsimony uninformative. Maximum-parsimony analysis resulted in 217 most parsimonious trees (length = 356; CI = 0.688202; RI = 0.904475; RCI = 0.622462). ITS analysis confirmed that the species belonged to the Pleurostomataceae, in which it formed a well-supported clade with the genus Pleurostoma (0.99, 93, and 94) (Fig. 3). Moreover, analysis of the actin, β-tubulin, and elongation factor of the newly isolated strain and a representative isolate from each species currently attributed to the Pleurostomataceae confirmed that the isolate was appropriately placed in this family (data not shown) as a member of the anamorph genus Pleurostomophora.
Fig 3.
Phylogenetic tree resulting from Bayesian analysis, maximum likelihood, and maximum parsimony for the ITS gene (values of >0.8 for Bayesian probability and >80% for maximum likelihood and maximum parsimony are shown with thick branches). Togninia minima, Phaeoacremonium inflatipes, and Phaeoacremonium aleophilum were used as the outgroup. Light-gray circles, strains isolated from plants; dark-gray circles, human-pathogenic strains.
Physiology.
The minimum growth temperature for the isolate was 15°C, and the maximum was above 40°C, with an optimum at 36 to 37°C (Fig. 4). Measurements were also taken after 8 days of incubation on PDA to compare the growth pattern of the isolate with those of other Pleurostomophora species. The colony diameter reached only 1 to 2 mm when grown at 20°C. The isolate decomposed casein in the first week. Starch was hydrolyzed; some plates showed a narrow clear zone around the colony, and in others, a zone extending up to 2.4 cm was observed. Gelatin hydrolysis was positive using both the tube and plate techniques. Urease was strongly positive within 3 days. The isolate showed some sensitivity to cycloheximide and was sensitive to salt, exhibiting some inhibition with 0.5%, poor or no growth with 5%, and no growth with 10% and 30% NaCl.
Fig 4.
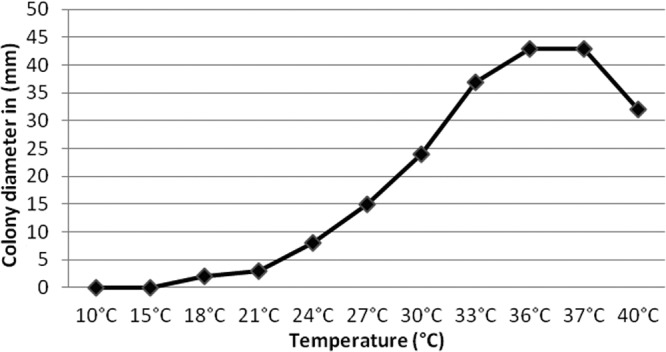
Colony diameters after 1 month of incubation at various temperatures ranging from 15 to 36°C at 3°C intervals, including 10°C, 37°C, and 40°C.
Antifungal susceptibility.
The MICs of the strain tested are shown in Table 3. Our strain was highly susceptible to the azoles itraconazole, ketoconazole, posaconazole, and voriconazole. Low MICs were also found for amphotericin B. The only azole for which a high MIC was found (128 μg/ml) was fluconazole. The fungus was inhibited by caspofungin at 8 μg/ml, but not by 5-flucytosine (MIC > 64 μg/ml).
Table 3.
Antifungal susceptibility of P. ochracea
| Antifungal agent | MIC (μg/ml) |
|---|---|
| Amphotericin B | 1 |
| Itraconazole | 0.25 |
| Ketoconazole | 0.25 |
| Fluconazole | 128 |
| Voriconazole | 0.5 |
| Posaconazole | 1 |
| 5-Flucytosin | >64 |
| Caspofungin | 8 |
Taxonomy.
Pleurostomophora ochracea Mhmoud, Abdalla Ahmed, Fahal, de Hoog, van de Sande, sp. nov. MycoBank accession number MB800514. Etymology: named after its formation of yellow grains in human tissue.
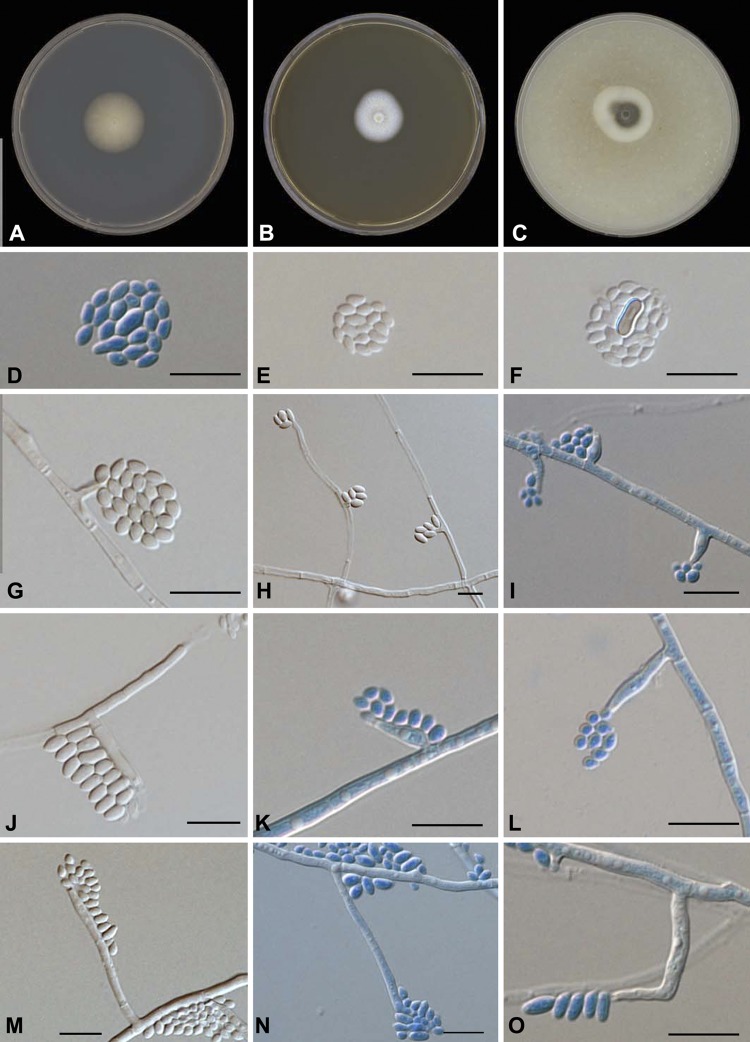
After 1 week of growth on all media tested (PDA, MEA, OA, and CMA), the colony surface and reverse were typically creamy white to pale yellow and felty, initially with some mycelial tufts. On OA and CMA, colonies turned dark olivaceous brown to nearly black after 1 month of incubation. No diffusible pigment was produced in any medium (Fig. 5). Vegetative hyphae were branched, septate, and hyaline to pale brown, turning dark brown after longer incubation. Hyphae were 1.2 to 3.2 μm wide. Hyphal walls were smooth or verruculose. Conidiophores were absent. Conidiogenous cells were variable and poorly differentiated, and phialidic. Three distinct types of phialides were observed. Type I phialides were short adelophialides without basal septa, occasionally wider at the base, and were (0.4) 1.0 to 5.0 (6.5) µm (where the parenthetical values are the minimum and maximum sizes, respectively) by (1) 1.0 to 2.0 (3) μm. Type II phialides were elongate-ampulliform, swollen at the base and tapered toward the apex, (6.2) 7 to 8.6 (8.7) by (1.3) 1.0 to 2.0 (2.5) μm. Type III phialides were (10.7) 11.6 to 17.9 (20.0) by (1.3) 1.7 to 2.4 (2.6) μm, (sub)cylindrical to elongate-ampulliform (Fig. 5). Conidia were smooth-walled and aggregate at the tip of the phialides; most conidia were hyaline, and some were brown. Two types were observed: either small and subspherical to ellipsoidal, (2.0) 2.2 to 3.0 (3.5) by (1.3) 1.6 to 2.0 (2.3) μm, or larger and allantoid, (3.5) 4 to 6 (6.2) by (1.2) 1.7 to 2.5 (2.6) μm (Fig. 5). Teleomorph unknown. Holotype: dried culture in CBS Herbarium H-20972; ex-type strain CBS 131321, isolated by N. A. Mhmoud and A. H. Fahal from human yellow-grain mycetoma, Khartoum, Sudan.
Fig. 5.
P. ochracea growth and morphology. Shown are colonies after 2 weeks of incubation at 37°C. (A) PDA. (B) MEA. (C) OA. (D, E, and F) Different-shape conidia. (G, H, and I) Type I phialides. (J, K, and L) Type II phialides. (M, N, and O) Type III phialides. All scale bars, 10 μm.
DISCUSSION
Mycetoma is a chronic progressive disease characterized by suppurative, swollen lesions and sinuses and can be caused by both bacteria and fungi (2, 15). Based on the color of the grain, fungal mycetomata can be divided into two large groups: those causing black or white grains. Black-grain mycetoma is mainly associated with M. mycetomatis, Madurella grisea, Leptosphaeria senegalensis, Pyrenochaeta romeroi, and Exophiala jeanselmei; white-grain mycetoma is usually caused by Pseudallescheria boydii, Acremonium kiliense, and other, occasional agents (5) (Table 1). Yellow-grain eumycetoma, to our knowledge, has never been reported. In areas of high endemicity, like the Sudan, 70% of all mycetoma cases are eumycetoma, caused by M. mycetomatis, a species producing black grains (5, 15). The remaining 30% of mycetoma cases are of bacterial origin, and many of these are characterized by the presence of yellow grains. With the limited availability of diagnostic techniques in routine laboratories in areas of high endemicity, the color of grains has decisive diagnostic value (15). In this communication, we report on the first authenticated case of human eumycetoma infection caused by a yellow-grain-producing fungus.
We identified the causative agent by multigene phylogenetic analysis. The novel pathogenic isolate clustered in the ascomycete order Calosphaeriales. The small order Calosphaeriales comprises only four genera separated into two families: the family Calosphaeriaceae contains three genera, viz., Calosphaeria, Jattaea, and Togniniella, while our fungus clustered in the Pleurostomataceae, containing only a single teleomorph genus, Pleurostoma, and a single anamorph genus, Pleurostomophora (30). Our fungus was paraphyletic to Pleurostoma ootheca and Pleurostomophora repens (Fig. 3). Because of the absence of sexual sporulation, we prefer to classify the new species in the anamorph genus Pleurostomophora.
The majority of causative agents of mycetoma belong to the orders Sordariales (particularly the genus Madurella) and Pleosporales (particularly coelomycetes) (Table 1). The order Calosphaeriales is a new addition to the list of orders harboring potential agents. The order was described by Barr in 1983 (6, 7); recent taxonomic revision showed that the Togniniaceae telemorph of Phaeoacremonium had some affinity to Diaporthales (27), but in our study, the family was found as a separate cluster (Fig. 2), which is in agreement with the results of Réblová (29). Based on SSU and LSU data, and unlike previous publications (21, 29), the Pleurostomataceae showed affinity to the Diaporthales, whereas the Calosphaeriaceae formed a separate, well-supported clade (Fig. 2). The order Calosphaeriales, comprising only 10 species, partly at large mutual phylogenetic distances, is still poorly defined. Supposedly related fungi, such as Phaeoacremonium, were found to be unalignable for the species-level genes used in this study, i.e., with degrees of variability equal to or larger than that of the ITS.
Most members of the cluster Pleurostoma-Pleurostomophora are wood-inhabiting fungi (8, 37). Two cases of phaeohyphomycosis caused by P. repens have been reported (20, 26). However, reexamination of both pathogenic strains revealed misidentifications, with the correct name being Phaeoacremonium krajdenii (26). Pleurostomophora richardsiae is the only confirmed human-pathogenic species in the order Calosphaeriales. It is known as an environmental fungus but has been reported as the causative agent of subcutaneous granuloma, phaeohyphomycosis, a chromoblastomycosis-like infection, and a bone infection (19, 22, 24, 32, 35).
Pleurostomophora ochracea and P. richardsiae both produce different types of phialides and conidia, but in P. richardsiae it is noticeably more pronounced (37). In P. ochracea, all three types of phialides are able to produce differently shaped conidia. All species of Pleurostomophora and the teleomorph genus Pleurostoma grow at temperatures ranging between 10 and 40°C, but P. ochracea has a relatively high optimum growth temperature (36°C versus 30°C) and a low growth rate. This is expressed after 8 days at 20°C, where colony diameters of P. ochracea are only 1 to 2 mm, while P. richardsiae reaches 18 to 19 mm, P. ootheca 30 to 32 mm, and P. repens 59 to 61 mm (37).
The clinical isolate of P. ochracea in this study appeared to be highly susceptible, in vitro, to ketoconazole and itraconazole, compounds that are commonly used in the treatment of subcutaneous infections, including mycetoma by pigmented fungi (16, 21, 25, 31). Of all azole antifungals tested, fluconazole was the least effective. Once it was established that the causative agent was a fungus, our patient was advised to take ketoconazole at 400 mg/day, according to the guidelines of the Mycetoma Research Centre in Khartoum, Sudan. However, the patient refused treatment and did not return to the center, and follow-up was impossible.
As noted in most cases of mycetoma, it is difficult to explain how our patient acquired his infection. He did not report any type of injury. Since the patient is a farmer by occupation, he is in daily contact with soil, thorns, wood, and other trauma-causing objects; even minor trauma might have facilitated the introduction of the fungus into his foot. The present case report shows that proper species identification in the diagnosis of mycetoma remains troublesome and that identification based exclusively on the color of the grains may be erratic (11, 13). Molecular identification is recommended to ascertain appropriate therapy.
ACKNOWLEDGMENT
Wendy W. J. van de Sande is supported by a postdoctoral fellowship from the Netherlands Organization for Scientific Research (NWO, VENI-Grant 916.11.178).
Footnotes
Published ahead of print 3 July 2012
REFERENCES
- 1. Ahmed A, et al. 2002. Environmental occurrence of Madurella mycetomatis, the major agent of human eumycetoma in Sudan. J. Clin. Microbiol. 40:1031–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahmed A, van de Sande W, Verbrugh H, Fahal A, van Belkum A. 2003. Madurella mycetomatis strains from mycetoma lesions in Sudanese patients are clonal. J. Clin. Microbiol. 41:4537–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahmed A, et al. 2007. Management of mycetoma: major challenge in tropical mycoses with limited international recognition. Curr. Opin. Infect. Dis. 20:146–151 [DOI] [PubMed] [Google Scholar]
- 4. Ahmed AO, et al. 2004. In vitro susceptibilities of Madurella mycetomatis to itraconazole and amphotericin B assessed by a modified NCCLS method and a viability-based 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) assay. Antimicrob. Agents Chemother. 48:2742–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahmed AO, et al. 2004. Mycetoma caused by Madurella mycetomatis: a neglected infectious burden. Lancet Infect. Dis. 4:566–574 [DOI] [PubMed] [Google Scholar]
- 6. Barr ME. 1983. The ascomycete connection. Mycologia 75:1–13 [Google Scholar]
- 7. Barr ME. 1985. Notes on the Calosphaeriales. Mycologia 77:509–565 [Google Scholar]
- 8. Barr ME. 1990. Prodromus to nonlichenized pyrenomycetous members of the class Hymenoascomycetes. Mycotaxon 39:43–184 [Google Scholar]
- 9. Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556 [Google Scholar]
- 10. de Hoog GS, Adelmann D, Ahmed AO, van Belkum A. 2004. Phylogeny and typification of Madurella mycetomatis, with a comparison of other agents of eumycetoma. Mycoses 47:121–130 [DOI] [PubMed] [Google Scholar]
- 11. de Hoog GS, et al. 1993. Diagnostic problems with imported cases of mycetoma in The Netherlands. Mycoses 36:81–87 [DOI] [PubMed] [Google Scholar]
- 12. Fahal AH. 2010. Management of mycetoma. Expert Rev. Dermatol. 5:87–93 [Google Scholar]
- 13. Fahal AH. 2006. Mycetoma, clinicopathological monograph, 1st ed Khartoum University Press, Khartoum, Sudan [Google Scholar]
- 14. Fahal AH. 2004. Mycetoma: a thorn in the flesh. Trans. R. Soc. Trop. Med. Hyg. 98:3–11 [DOI] [PubMed] [Google Scholar]
- 15. Fahal AH, Hassan MA. 1992. Mycetoma. Br. J. Surg. 79:1138–1141 [DOI] [PubMed] [Google Scholar]
- 16. Fahal AH, Rahman IA, El-Hassan AM, Rahman ME, Zijlstra EE. 2011. The safety and efficacy of itraconazole for the treatment of patients with eumycetoma due to Madurella mycetomatis. Trans. R. Soc. Trop. Med. Hyg. 105:127–132 [DOI] [PubMed] [Google Scholar]
- 17. Gargas A, Taylor JW. 1992. Polymerase chain reaction (PCR) primers for amplifying and sequencing 18S rDNA from lichenized fungi. Mycologia 84:589–592 [Google Scholar]
- 18. Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61:1323–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gueho E, Bonnefoy A, Luboinski J, Petit JC, de Hoog GS. 1989. Subcutaneous granuloma caused by Phialophora richardsiae: case report and review of the literature. Mycoses 32:219–223 [DOI] [PubMed] [Google Scholar]
- 20. Hironaga M, Nakano K, Yokoyama I, Kitajima J. 1989. Phialophora repens, an emerging agent of subcutaneous phaeohyphomycosis in humans. J. Clin. Microbiol. 27:394–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hood SV, Moore CB, Cheesbrough JS, Mene A, Denning DW. 1997. Atypical eumycetoma caused by Phialophora parasitica successfully treated with itraconazole and flucytosine. Br. J. Dermatol. 136:953–956 [PubMed] [Google Scholar]
- 22. Ikai K, Tomono H, Watanabe S. 1988. Phaeohyphomycosis caused by Phialophora richardsiae. J. Am. Acad. Dermatol. 19:478–481 [DOI] [PubMed] [Google Scholar]
- 23. Jacobs K, et al. 2004. Leptographium wingfieldii introduced into North America and found associated with exotic Tomicus piniperda and native bark beetles. Mycol. Res. 108:411–418 [DOI] [PubMed] [Google Scholar]
- 24. Lieb DF, Smiddy WE, Miller D, Cooperman EW. 2003. Case report: fungal endophthalmitis caused by Phialophora richardsiae. Retina 23:406–407 [DOI] [PubMed] [Google Scholar]
- 25. Mahgoub ES, Gumaa SA. 1984. Ketoconazole in the treatment of eumycetoma due to Madurella mycetomii. Trans. R. Soc. Trop. Med. Hyg. 78:376–379 [DOI] [PubMed] [Google Scholar]
- 26. Meyers WM, Dooley JR, Kwon-Chung KJ. 1975. Mycotic granuloma caused by Phialophora repens. Am. J. Clin. Pathol. 64:549–555 [DOI] [PubMed] [Google Scholar]
- 27. Mostert L, Groenewald JZ, Summerbell RC, Gams W, Crous PW. 2006. Taxonomy and pathology of Togninia (Diaporthales) and its Phaeoacremonium anamorphs. Studies Mycol. 54:1–113 [Google Scholar]
- 28. O'Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet Evol. 7:103–116 [DOI] [PubMed] [Google Scholar]
- 29. Réblová M. 2011. New insights into the systematics and phylogeny of the genus Jattaea and similar fungi of the Calosphaeriales. Fungal Diversity 49:167–198 [Google Scholar]
- 30. Réblová M, Mostert L, Gams W, Crous PW. 2004. New genera in Calosphaeriales: Togniniella and its anamorph Phaeocrella, and Calosphaeriophora as anamorph of Calosphaeria. Studies Mycol. 50:533–550 [Google Scholar]
- 31. Richardson MD, Kokki M. 2001. Therapeutic guidelines in systemic fungal infections. Current Medical Literature Ltd, London, United Kingdom [Google Scholar]
- 32. Son YM, et al. 2010. Chromoblastomycosis caused by Phialophora richardsiae. Ann. Dermatol. 22:362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512–526 [DOI] [PubMed] [Google Scholar]
- 34. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Torstrick RF, Harrison K, Heckman JD, Johnson JE. 1979. Chronic bursitis caused by Phialophora richardsiae. A case report. J. Bone Joint Surg. Am. 61:772–774 [PubMed] [Google Scholar]
- 36. van de Sande WWJ, Luijendijk A, Ahmed AO, Bakker-Woudenberg IA, van Belkum A. 2005. Testing of the in vitro susceptibilities of Madurella mycetomatis to six antifungal agents by using the Sensititre system in comparison with a viability-based 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) assay and a modified NCCLS method. Antimicrob. Agents Chemother. 49:1364–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vijaykrishna D, et al. 2004. Pleurostomophora, an anamorph of Pleurostoma (Calosphaeriales), a new anamorph genus morphologically similar to Phialophora. Studies Mycol. 50:387–395 [Google Scholar]
- 38. Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172:4238–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322 In Innis MA, Gelfand DH, Sninsky JJ, White TJ. (ed), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, NY [Google Scholar]