Abstract
Next-generation sequencing (NGS) has recently been used for analysis of HIV diversity, but this method is labor-intensive, costly, and requires complex protocols for data analysis. We compared diversity measures obtained using NGS data to those obtained using a diversity assay based on high-resolution melting (HRM) of DNA duplexes. The HRM diversity assay provides a single numeric score that reflects the level of diversity in the region analyzed. HIV gag and env from individuals in Rakai, Uganda, were analyzed in a previous study using NGS (n = 220 samples from 110 individuals). Three sequence-based diversity measures were calculated from the NGS sequence data (percent diversity, percent complexity, and Shannon entropy). The amplicon pools used for NGS were analyzed with the HRM diversity assay. HRM scores were significantly associated with sequence-based measures of HIV diversity for both gag and env (P < 0.001 for all measures). The level of diversity measured by the HRM diversity assay and NGS increased over time in both regions analyzed (P < 0.001 for all measures except for percent complexity in gag), and similar amounts of diversification were observed with both methods (P < 0.001 for all measures except for percent complexity in gag). Diversity measures obtained using the HRM diversity assay were significantly associated with those from NGS, and similar increases in diversity over time were detected by both methods. The HRM diversity assay is faster and less expensive than NGS, facilitating rapid analysis of large studies of HIV diversity and evolution.
INTRODUCTION
Next-generation sequencing (NGS) can provide gigabases of sequencing data at a fraction of the cost (per base sequenced) of traditional Sanger-based sequencing methods (20, 27). The sequencing depth of NGS allows detailed characterization of nucleic acid pools. In particular, this technology has been used to examine dynamic viral populations that consist of large numbers of quasispecies, such as those typically seen in HIV infection. NGS has been used to characterize viral populations in early HIV infection (6), track patterns of HIV evolution (1, 31), confirm transmission linkage between HIV infections (5), and identify the presence of superinfecting HIV strains (22).
Despite the experimental power of NGS (15), there are significant barriers to the widespread use of this technology, especially in resource-limited settings. While the cost per base pair sequenced is low, the instrument, reagent, and labor costs remain high (27). The bioinformatics requirements necessitate special training, large amounts of computing power, and well-conceived data storage methods (35). Additionally, template resampling, limited read length, and PCR error rates can complicate the use of NGS for analysis of genetic diversity, although methods are available to mitigate these effects (11, 22, 34). For these reasons, NGS-based HIV studies have typically evaluated small numbers of samples and relatively small regions of the HIV genome (12, 19, 22). Patterns of diversification may vary in different regions of the HIV genome, since different selective pressures target different viral proteins (7, 21).
Further studies of HIV diversity are required to develop a more complete understanding of HIV diversity in order to clarify the relationship between HIV diversity and pathogenesis (3, 17, 18, 33). Additionally, recent research efforts suggest that HIV diversity may be a biomarker for duration of infection that is suitable for use in HIV incidence testing (2, 13). One study demonstrated that multiregion analysis of HIV diversity may enhance the utility of HIV diversity as a biomarker for incidence analysis (2). Alternative methods for analysis of HIV diversity that are simpler and less costly may be needed to examine HIV diversification patterns in large sample sets and to analyze viral diversification across the HIV genome (16, 21). Cost savings would also be required to facilitate analysis of HIV diversity in resource-poor settings.
We recently developed a diversity assay based on high-resolution melting (HRM) technology and adapted this analytical tool for analysis of HIV (2, 9, 30). The HRM diversity assay provides a single numeric HRM score that reflects the level of HIV diversity in a specific region of the HIV genome. We have optimized the HRM diversity assay for analysis of multiple regions of the HIV genome, including regions in HIV env, gag, and pol (2). In a previous study, we demonstrated that HRM scores in a single region of gag were significantly associated with sequence-based diversity measures, based on analysis of 20 or 50 HIV clones from a small number (n = 18) of infected individuals (30). We expanded upon that study in this investigation by comparing HRM scores to sequence-based diversity measures obtained using NGS data from longitudinal samples (n = 220) for HIV gag and env.
(Portions of this work were presented at the 19th Conference on Retroviruses and Opportunistic Infections, March 2012, Seattle, WA [abstr 684]).
MATERIALS AND METHODS
Human subjects (ethics statement).
Stored samples were obtained from participants enrolled in the Rakai Community Cohort Study (RCCS). Participants provided written informed consent for sample storage and testing. The RCCS was approved by the Science and Ethics Committee of the Uganda Virus Research Institute, the Uganda National Council for Research and Technology, Western Institutional Review Board, and the Committee on Human Research at Johns Hopkins Bloomberg School of Public Health. This study was conducted according to the ethical standards set forth by these institutional review boards and the Helsinki Declaration of the World Medical Association.
Sample selection.
Samples were collected from adults enrolled in the RCCS in Rakai District, Uganda (32). Since 1994, the RCCS has interviewed participants and collected blood samples for storage and analysis (approximately 14,000 individuals from 50 villages sampled annually). Participants were followed longitudinally. In a previous study, HIV from paired, longitudinal serum samples was analyzed using NGS (env and gag regions) (23). A subset of those samples (220 paired longitudinal serum samples from 110 individuals) was selected for this study. The first of the paired samples corresponded to the first HIV-positive sample for seroconverters (median of 459 days from last negative test, interquartile range [IQR], 393 to 708 days). The second sample was collected a median of 1,106 days after the first HIV-positive sample (IQR, 679 to 1,655 days). HIV subtyping was performed previously by phylogenetic analysis of NGS data. Samples from the seroconversion time point (n = 110) included 71 subtype D (∼65%), 17 subtype A (∼15%), 12 D/A recombinants (∼11%), 4 D/C recombinants (∼4%), 1 C/A recombinant (∼1%), and 5 multisubtype dual infections (∼5%). Twelve (∼11%) of the 110 participants had infection with two distinct HIV strains at the first time point (dual infection; 5 had a mixture of subtypes A and D, and 7 had two different subtype D strains) (23).
NGS analysis.
NGS was performed in a previous study (22, 23). Briefly, HIV RNA was extracted from serum samples and amplified by reverse transcription-PCR (RT-PCR). Templates were amplified using nested PCR (gp41, E55 primer set with 14 454-bar-coded variations [MID1 to MID14]; p24, G100 primer set with 14 454-bar-coded variations [MID1 to MID14], as described by the manufacturer [Roche, Inc., Branford, CT]). Amplification was verified by gel electrophoresis. Products were purified, quantified, and diluted to 1 × 109 molecules/μl. The amplicon libraries (1 × 109 molecules/μl) were diluted to 1 × 105 molecules/μl and were added to DNA capture beads at a rate of 0.175 molecules per bead. Enriched DNA capture beads were sequenced following the manufacturer's instructions by using a 4-region gasket on the Roche 454 platform (Roche, Branford, CT). Primer failure prevented collection of a small fraction of NGS data. Primer failure occurred for two gag p24 amplicons (one from a subtype A infection and one from a D/A dual infection) and one env gp41 amplicon (D/A dual infection).
Sequence data processing.
GS Amplicon Variant Analyzer version 2.5 (Roche, Branford, CT) was used to process the resulting pool of sequence reads (median number of reads for gag, 11,394; gp41, 9,640). Read lengths for each sample were ∼390 bp for gag p24 and ∼324 bp for env gp41, not including primer sequences. Equivalent sequences were combined into single consensus sequences. Initially, the consensus sequence data were processed, removing variants that appeared fewer than 10 times. All prominent and any outlier consensus sequence populations for each sample in a given NGS run were compared by phylogenetic analysis, and contaminating sequences from that same run were removed. The remaining consensus sequences that appeared at a frequency less than 0.5% of the total read volume for that sample were also removed to normalize consensus sequence numbers between samples. After sequence processing, the median numbers of reads remaining were 3,710 for gag p24 and 5,413 for env gp41.
Calculation of sequence-based diversity measures.
Consensus sequences obtained from NGS were aligned using Clustal W (29) in MEGA (version 5.05; www.megasoftware.net), and the resulting alignments were manually edited to correct minor alignment errors. Edited alignments were then trimmed such that the region included in the alignment corresponded to the amplified region analyzed in the HRM diversity assay. The regions included in the analysis were the GAG3 HRM amplicon (GAG, ∼240 bp) and the ENV3mod HRM amplicon (ENV, ∼229 bp). The trimmed alignments were used to calculate three sequence-based diversity measures: average genetic distance between HIV genomes (percent diversity), the number of unique sequence reads/total reads × 100 (percent complexity), and the Shannon entropy, a measure that accounts for both the number of distinct reads and the proportional representation of each distinct read by summing the product of the proportional representation of each read and the log of the proportional representation of each read and dividing this sum by the log of the total number of reads according to the formula shown in Table 1.
Table 1.
Description of sequence-based diversity measures
| Diversity measure | Method of calculation |
|---|---|
| % diversity | Average pairwise genetic distance was calculated using the maximum composite likelihood model. MEGA 5.05 (www.megasoftware.net) was used to analyze gap-stripped sequence alignments. This value is reported as a percentage. |
| % complexity | [(no. of distinct variants)/(total no. of reads)] × 100 |
| Shannon entropy (S) | |
| where pi is the proportion of reads consisting of each unique sequence pattern, N is the total no. of sequences, and n is the no. of distinct sequences |
HRM diversity assay.
Diluted first-round PCR products that served as templates for the nested PCR were diluted 100-fold and analyzed in the HRM diversity assay (2). Nested PCR mixtures (10 μl) consisted of the following components: 4.6 μl H2O, 4 μl of LightScanner master mix (Idaho Technology Inc., Salt Lake City, UT), 0.2 μl each of 10 μM forward and reverse primer, and 1 μl of diluted template DNA. For nested PCR amplification of the GAG region for HRM, GAG3 forward primer G80 (ATGAGAGAACCAAGGGGAAGTGA; HXB2 positions 1471 to 1493 [26]) and GAG3 reverse primer (TTGGACCAACAAGGTTTCTGTCATCCA; HXB2 positions 1735 to 1761) were used. Both primers used to amplify the GAG amplicon were internal to primers used to prepare NGS amplicons. To amplify the ENV region for HRM, the ENV3mod forward primer ENV3F (TGCTCTGGAAARCWCATYTGC; HXB2 positions 8016 to 8036, internal to primers used to prepare NGS amplicons [2]) and ENV3mod reverse primer GP48 (TCCTACTATCATTATGAATATTTTTATATA; HXB2 positions 8265 to 8294, same as the reverse primer used for NGS [22, 26]) were used. Cycling conditions for amplification of GAG in the presence of LCGreen Plus dye (Idaho Technology Inc., Salt Lake City, UT) were as follows: 2-min hold at 95°C, 45 cycles of 94°C for 30 s and 63°C for 30 s, two sequential 30-s holds at 94°C and 28°C, and a terminal hold at 4°C. Amplification of ENV was conducted using the same methods, with the following exception: the second cycling temperature was 61°C rather than 63°C. The resulting amplicons were melted using the LightScanner Instrument (model HR 96; Idaho Technology Inc., Salt Lake City, UT), and release of the dye was quantified as a function of temperature (melting range for GAG, 68 to 98°C with a 65°C hold; melting range for ENV, 60 to 98°C with a 57°C hold). Melt data were exported from the LightScanner software package and analyzed using an R-based analytical platform called the HRM Diversity Assay Analysis Tool (DivMelt; available at cran.r-project.org/web/packages/DivMelt/index.html) to generate HRM scores. A total of 17 HRM amplification failures occurred. A single failure occurred in GAG in a subtype A infection, and 16 failures occurred in ENV (5 subtype A and 11 subtype D). Primer failure was more likely in ENV than in GAG, and primer failure was more likely in subtype A.
Statistical methods.
A linear mixed model assuming a random intercept for repeated measures from the same individual was used to assess the association between the HRM score and the sequence-based diversity measures. This approach was also used to quantify the change over time in each measure. R2, interpreted as the percent variance in each sequence-based diversity measure that was explained by the HRM score, was used to assess the strength of the prediction. A simple linear regression model was employed to examine whether a within-person change in HRM score predicted a within-person change in a sequence-based diversity measure.
Extreme values were observed for several individuals with dual infection. For this reason, all data values from individuals with dual infection were excluded from the analysis of changes in diversity over time and from the simple linear regression model. In addition, extreme values from four individuals with dual infection and one individual without dual infection were removed from the analysis of association between the HRM score and sequence-based diversity measures, as noted.
Analyses were performed using SAS software version 9.2 (Cary, NC).
RESULTS
Paired samples from 110 adults were analyzed with both NGS (23) and the HRM diversity assay. Two regions of the HIV genome were analyzed for each sample, HIV gag (GAG) and HIV env (ENV). Control amplicons were prepared from a set of plasmids containing inserts derived from subtype A and D (the set contained 4 subtype A gag, 4 subtype D gag, 4 subtype A env, and 4 subtype D env plasmids). Plasmids had a median GAG HRM score of 4.01 (range, 3.63 to 4.60) and a median ENV HRM score of 3.70 (range, 3.43 to 3.86). This is consistent with our previous studies that demonstrated that amplicons derived from plasmids usually have very low HRM scores (2). Region-specific differences in HRM scores for plasmid-derived amplicons have been noted previously and most likely reflect the differences in base composition and lengths of the amplicons (2, 24).
HIV diversity was first quantified by calculating sequence-based diversity measures from NGS data. These measures included percent diversity, percent complexity, and Shannon entropy (Tables 1 and 2). The sample amplicons used for NGS were then analyzed using the HRM diversity assay to generate HRM scores for each region (Table 2). In the analysis that used the HRM score as a predictor of sequence-based diversity measures, the HRM score was strongly associated with sequence-based measures for both GAG and ENV (P < 0.001 for all six measures) (Table 3).
Table 2.
Medians and IQRs for diversity measures
| Region | Diversity measurea | At seroconversion |
Postseroconversionb |
||
|---|---|---|---|---|---|
| Median (IQR) | nc | Median (IQR) | nc | ||
| GAG | % diversity | 0.05 (0.00–0.17) | 110 | 0.28 (0.12–0.48) | 108 |
| % complexity | 0.08 (0.04–0.15) | 110 | 0.15 (0.09–0.27) | 108 | |
| Shannon entropy | 0.06 (0.02–0.11) | 110 | 0.13 (0.09–0.18) | 108 | |
| HRM score | 4.21 (3.87–4.71) | 109 | 5.03 (4.57–5.60) | 110 | |
| ENV | % diversity | 0.20 (0.05–0.47) | 109 | 0.67 (0.33–1.23) | 110 |
| % complexity | 0.12 (0.07–0.22) | 109 | 0.31 (0.15–0.59) | 110 | |
| Shannon entropy | 0.12 (0.06–0.18) | 109 | 0.21 (0.14–0.27) | 110 | |
| HRM score | 4.27 (3.96–4.76) | 108 | 4.84 (4.09–5.69) | 96 | |
Sequence-based diversity measures included the percent diversity, percent complexity, and Shannon entropy (Table 1); the HRM score is the output from the HRM diversity assay.
Postseroconversion samples were collected approximately 1 to 8 years after the seroconversion sample.
In some cases, NGS or HRM data could not be obtained.
Table 3.
Association between HRM score and sequence-based diversity measuresa
| Region | Diversity measure | Slope (95% CI) | P value | R2 |
|---|---|---|---|---|
| GAG | % diversity | 0.19 (0.15, 0.23) | <0.001 | 0.23 |
| % complexityb | 0.10 (0.07, 0.13) | <0.001 | 0.05 | |
| Shannon entropy | 0.05 (0.04, 0.06) | <0.001 | 0.22 | |
| ENV | % diversityc | 0.51 (0.41, 0.60) | <0.001 | 0.34 |
| % complexity | 0.25 (0.18, 0.32) | <0.001 | 0.14 | |
| Shannon entropy | 0.06 (0.05, 0.06) | <0.001 | 0.40 |
Results of a linear mixed model using the HRM score to predict sequence-based diversity measures.
Two extreme values were removed for the first time point (seroconversion). One of the excluded values was from an individual who had dual infection at seroconversion (see Materials and Methods).
Three extreme values were removed for the first time point. All of these values were collected from individuals with dual infection (see Materials and Methods).
Diversification of HIV over time was examined using diversity measures derived from NGS and the HRM diversity assay (Fig. 1). Both of these analytic approaches revealed a significant increase in HIV diversity over time for all measures analyzed except for percent complexity for GAG (P < 0.001) (Table 4; Fig. 2). Furthermore, the pattern of HIV diversification observed over time was similar when using both analytic approaches for all measures analyzed except for percent complexity for GAG (Table 5).
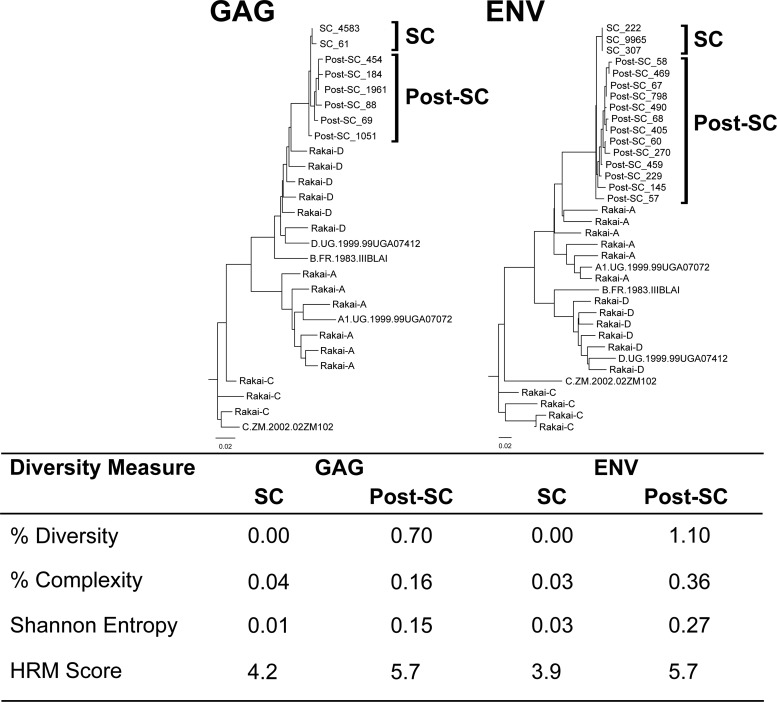
Fig 1.
Diversity data collected from a representative study participant. Neighbor-joining trees of HIV GAG and ENV next-generation consensus sequences from one individual at two time points are shown. The number of sequence reads included in each consensus sequence is shown at the end of the sequence, following an underscore. Subtype reference sequences and random sequences from unrelated individuals in Rakai are included. The diversity measures associated with both time points and regions are shown in the table below the trees. Note that this participant is infected with an A/D recombinant strain of HIV (A in env, D in gag).
Table 4.
Change in viral diversity over timea
| Region | Diversity measure | Change in measure/yr (95% CI) | P value |
|---|---|---|---|
| GAG | % diversity | 0.04 (0.03, 0.06) | <0.001 |
| % complexity | 0.02 (0.00, 0.04) | 0.053 | |
| Shannon entropy | 0.01 (0.01, 0.02) | <0.001 | |
| HRM score | 0.18 (0.15, 0.22) | <0.001 | |
| ENV | % diversity | 0.17 (0.13, 0.20) | <0.001 |
| % complexity | 0.11 (0.07, 0.15) | <0.001 | |
| Shannon entropy | 0.02 (0.01, 0.03) | <0.001 | |
| HRM score | 0.17 (0.11, 0.24) | <0.001 |
Results of a linear mixed model examining the change in viral diversity per year since seroconversion in longitudinally sampled individuals. All data were removed for individuals who had dual infection at the first time point (seroconversion).
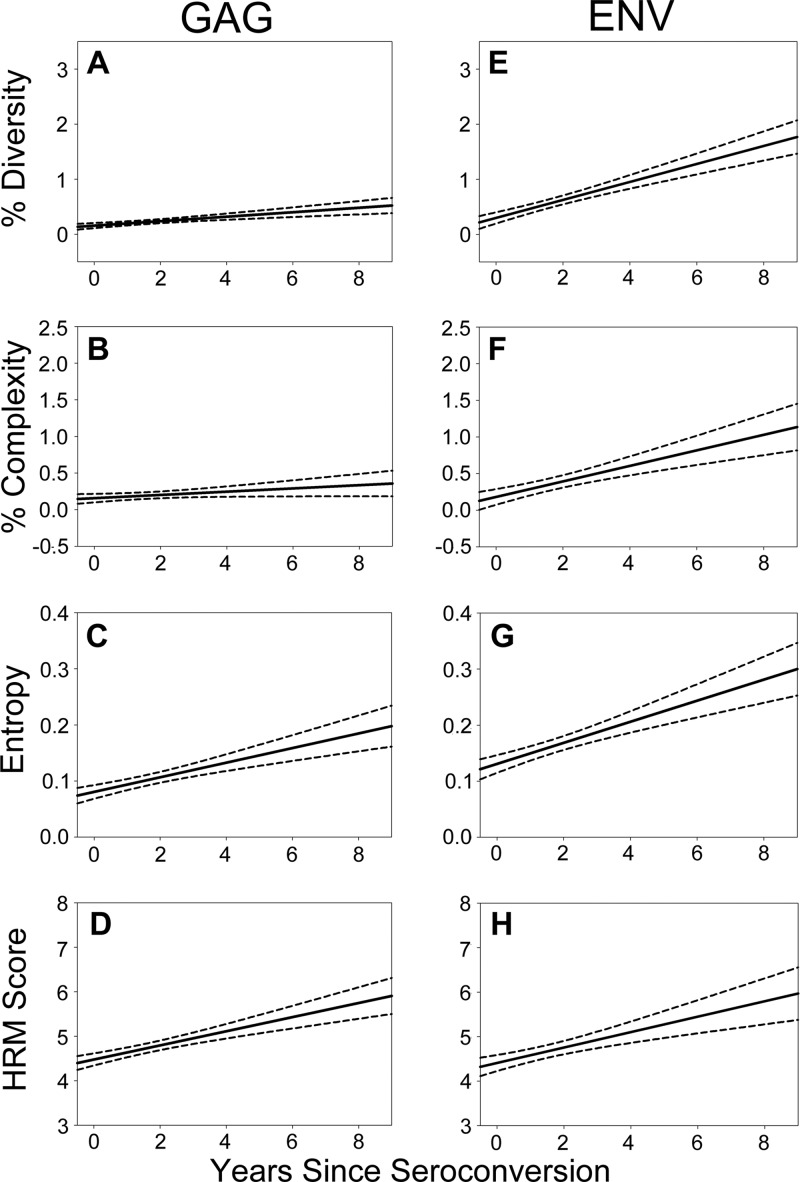
Fig 2.
HRM scores and sequence-based diversity measures increase over time after infection. HRM scores and sequence-based diversity measures (percent diversity, percent complexity, and Shannon entropy) increased for both GAG (A, B, C, and D) and ENV (E, F, G, and H) over time from seroconversion. Univariate logistic regression analysis was used to calculate the mean longitudinal change (solid line) with 95% confidence intervals (dotted lines).
Table 5.
Relationship between the change in diversity over time measured using NGS and the change in diversity over time measured using the HRM diversity assaya
| Region | Diversity measure | Change in diversity measure (95% CI) | P value |
|---|---|---|---|
| GAG | % diversity | 0.24 (0.15, 0.34) | <0.001 |
| % complexity | 0.10 (−0.01, 0.22) | 0.079 | |
| Shannon entropy | 0.06 (0.04, 0.09) | <0.001 | |
| ENV | % diversity | 0.42 (0.33, 0.52) | <0.001 |
| % complexity | 0.30 (0.15, 0.45) | <0.001 | |
| Shannon entropy | 0.05 (0.03, 0.06) | <0.001 |
Results of a simple linear regression predicting change in sequence-based diversity measures by one unit change in HRM score. All data were removed for individuals who had dual infection at the first time point (seroconversion).
DISCUSSION
This study demonstrates that quantitative measures of HIV diversity obtained using the HRM diversity assay are highly associated with sequence-based diversity measures obtained from NGS data. Specifically, HRM scores were associated with the average genetic distance between HIV genomes (percent diversity), the number of different reads/number of unique sequence reads (percent complexity), and the proportion and number of distinct reads (Shannon entropy).
HRM scores are impacted by different types of genetic variation, including single base changes and insertions and deletions (indels) (M. M. Cousins et al., submitted for publication). In contrast, the sequence-based measures analyzed in this report capture limited characteristics of the DNA region examined. For example, percent diversity is calculated using gap-stripped sequences and does not capture genetic diversity introduced by indels.
HIV diversity typically increases over time during the course of HIV infection. Rapid viral replication, frequent mutation events, and frequent recombination events generate large numbers of distinct viral variants (10, 28). Immune responses to infection, antiretroviral therapy, and other selective pressures drive the diversification and evolution of the viral population (10, 25). In this report, we observed similar increases in diversity of both the env and gag regions based on these two methods, indicating that the HRM diversity assay can be used to assess HIV diversification over time.
There are substantial differences in cost and labor needed for analysis of diversity using NGS and the HRM diversity assay. Equipment, labor, reagent, and supply costs for analysis using the HRM diversity assay are substantially lower than the costs associated with analysis using NGS. The HRM diversity assay also allows more rapid collection of diversity measures.
Despite these advantages, the HRM diversity assay is not suitable for applications where sequence data are required for phylogenetic analysis or other purposes. Also, the absence of sequence data means that the HRM diversity assay may have difficulty distinguishing between diversity acquired over a long course of infection and diversity resulting from a superinfection event. We are presently working to address this question.
The HRM diversity assay has been used to study HIV diversity in infants and adults (2, 8, 9). We are currently exploring whether the HRM diversity assay can be used alone or in combination with other biomarkers for cross-sectional HIV incidence determinations (2). In this study, HIV samples from individuals with dual HIV infection (e.g., infection with two HIV subtypes or two divergent HIV strains of the same subtype) often had very high HRM scores. We are investigating whether the HRM diversity assay can be used to screen for analysis of HIV superinfection. The HRM diversity assay may also be useful for evaluation of genetic diversity in other pathogens and other genetic systems.
ACKNOWLEDGMENTS
We thank the participants and the study team of the Rakai Community Cohort Study, which was supported by (i) the Bill and Melinda Gates Foundation (22006.03), (ii) the National Institutes of Health (NIH), Division of Allergy and Infectious Diseases (U1AI51171 and 1UO1AI075115-O1A1), (iii) the Department of the Army, U.S. Army Medical Research and Materiel Command Cooperative Agreement (DAMD17-98-2-8007), and (v) the Henry M. Jackson Foundation (5D43TW00010). This study was supported by (i) the HIV Prevention Trials Network (HPTN) sponsored by the NIAID, the National Institute on Drug Abuse (NIDA), the National Institute of Mental Health, and the Office of AIDS Research of the NIH and DHHS (U01AI068613 and UM1AI068613 to S.H.E.), (ii) NIAID (1R01-AI095068 to S.H.E.), and (iii) NIAID (UM1-AI068617 to D.D.). This study was supported in part by funding from the Division of Intramural Research, NIAID, NIH, and the Office of AIDS Research, NIH.
M.M.C. has given presentations at meetings sponsored by Idaho Technology (marketer of the LightScanner platform and reagents designed specifically for HRM analysis).
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the National Institutes of Health (NIH). Use of trade names is for identification purposes only and does not constitute endorsement by the NIH.
All authors contributed to writing the manuscript. In addition, authors had the following roles: Matthew Cousins conceived of the study, coordinated the study, optimized the HRM diversity assay for analysis of gag and env regions, designed and tested the HRM Diversity Assay Analysis Tool for analysis of data from the HRM diversity assay, generated and analyzed HRM data, analyzed sequence data, and prepared the manuscript; Stephen Porcella managed NGS data collection and analysis; San-san Ou served as data analyst for the project; Supriya Munshaw provided input related to data interpretation and presentation and assisted with data analysis; Caroline Mullis performed PCR amplification of NGS amplicons and assisted in data analysis; David Swan designed and tested the HRM Diversity Assay Analysis Tool for analysis of data from the HRM diversity assay and wrote the package for this R-based analytical tool; Craig Magaret advised on the design and guided development of the HRM Diversity Assay Analysis Tool for analysis of data from the HRM diversity assay; Dave Serwadda provided samples and longitudinal data from the RCCS; Maria Wawer provided samples and longitudinal data from the RCCS; Ron Gray provided samples and longitudinal data from the RCCS; Thomas Quinn provided input related to data interpretation and presentation; Deborah Donnell was responsible for statistical analysis for the project; Susan Eshleman served as senior investigator and was responsible for development of the HRM diversity assay, provided input related to study design, data interpretation, presentation, and analysis, and prepared the manuscript; Andrew Redd conceived of the study, coordinated the study, analyzed sequence data, and prepared the manuscript.
Footnotes
Published ahead of print 11 July 2012
REFERENCES
- 1. Buzon MJ, et al. 2011. Deep molecular characterization of HIV-1 dynamics under suppressive HAART. PLoS Pathog. 7:e1002314 doi:10.1371/joural.ppat.1002314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cousins MM, et al. 2011. Use of a high resolution melting (HRM) assay to compare gag, pol, and env diversity in adults with different stages of HIV infection. PLoS One 6:e27211 doi:10.1371/journal.pone.0027211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Delwart EL, et al. 1997. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J. Virol. 71:7498–7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reference deleted.
- 5. Eshleman SH, et al. 2011. Analysis of genetic linkage of HIV from couples enrolled in the HIV Prevention Trials Network 052 trial. J. Infect. Dis. 204:1918–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fischer W, et al. 2010. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS One 5:e12303 doi:10.1371/journal.pone.0012303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frahm N, Brander C. 2007. HIV viral diversity and escape from cellular immunity. Curr. Infect. Dis. Rep. 9:161–166 [DOI] [PubMed] [Google Scholar]
- 8. James M, et al. 2012. env, gag, and pol diversity in HIV+ children and the association of HIV diversity with age and impact of non-suppressive ART: Uganda, abstr 998. 19th Conf. Retroviruses Opportunistic Infect, CROI, Alexandria, VA [Google Scholar]
- 9. James MM, et al. 2011. Association of HIV diversity and survival in HIV-infected Ugandan infants. PLoS One 6:e18642 doi:10.1371/journal.pone.0018642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keele BF. 2010. Identifying and characterizing recently transmitted viruses. Curr. Opin. HIV AIDS 5:327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. 2011. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. U. S. A. 108:9530–9535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Knoepfel SA, Di Giallonardo F, Daumer M, Thielen A, Metzner KJ. 2011. In-depth analysis of G-to-A hypermutation rate in HIV-1 env DNA induced by endogenous APOBEC3 proteins using massively parallel sequencing. J. Virol. Methods 171:329–338 [DOI] [PubMed] [Google Scholar]
- 13. Kouyos RD, et al. 2011. Ambiguous nucleotide calls from population-based sequencing of HIV-1 are a marker for viral diversity and the age of infection. Clin. Infect. Dis. 52:532–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reference deleted.
- 15. Liang B, et al. 2011. A comparison of parallel pyrosequencing and sanger clone-based sequencing and its impact on the characterization of the genetic diversity of HIV-1. PLoS One 6:e26745 doi:10.1371/journal.pone.0026745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu S, et al. 2008. Dynamic analysis of genetic diversity of gag and env regions of HIV-1 CRF07_BC recombinant in intravenous drug users in Xinjiang Uvghur Autonomous Region, China. Arch. Virol. 153:1233–1240 [DOI] [PubMed] [Google Scholar]
- 17. Mani I, et al. 2002. Intrapatient diversity and its correlation with viral setpoint in human immunodeficiency virus type 1 CRF02_A/G-IbNG infection. J. Virol. 76:10745–10755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Markham RB, et al. 1998. Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc. Natl. Acad. Sci. U. S. A. 95:12568–12573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mild M, Hedskog C, Jernberg J, Albert J. 2011. Performance of ultra-deep pyrosequencing in analysis of HIV-1 pol gene variation. PLoS One 6:e22741 doi:10.1371/journal.pone.0022741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pacold M, et al. 2010. Comparison of methods to detect HIV dual infection. AIDS Res. Hum. Retroviruses 26:1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Piantadosi A, et al. 2009. HIV-1 evolution in gag and env is highly correlated but exhibits different relationships with viral load and the immune response. AIDS 23:579–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Redd AD, et al. 2011. Identification of HIV superinfection in seroconcordant couples in Rakai, Uganda, by use of next-generation deep sequencing. J. Clin. Microbiol. 49:2859–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Redd AD, et al. 2012. The rates of HIV superinfection and primary HIV incidence in a general population in Rakai, Uganda. J. Infect. Dis. 206:267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reed GH, Wittwer CT. 2004. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin. Chem. 50:1748–1754 [DOI] [PubMed] [Google Scholar]
- 25. Richman DD, et al. 2004. HIV evolution and escape. Trans Am. Clin. Climatol. Assoc. 115:289–303 [PMC free article] [PubMed] [Google Scholar]
- 26. Sanders-Buell E, Salminen MO, McCutchan FE. 1995. Sequencing primers for HIV-1, p 15–21 In Korber B, et al. (ed), The Human Retroviruses and AIDS 1995 Compendium. Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 27. Shendure J, Ji H. 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26:1135–1145 [DOI] [PubMed] [Google Scholar]
- 28. Tebit DM, Nankya I, Arts EJ, Gao Y. 2007. HIV diversity, recombination and disease progression: how does fitness “fit” into the puzzle? AIDS Rev. 9:75–87 [PubMed] [Google Scholar]
- 29. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Towler WI, et al. 2010. Analysis of HIV diversity using a high-resolution melting assay. AIDS Res. Hum. Retroviruses 26:913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsibris AM, et al. 2009. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One 4:e5683 doi:10.1371/journal.pone.0005683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wawer MJ, et al. 1998. A randomized, community trial of intensive sexually transmitted disease control for AIDS prevention, Rakai, Uganda. AIDS 12:1211–1225 [DOI] [PubMed] [Google Scholar]
- 33. Wolinsky SM, et al. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537–542 [DOI] [PubMed] [Google Scholar]
- 34. Zagordi O, Klein R, Daumer M, Beerenwinkel N. 2010. Error correction of next-generation sequencing data and reliable estimation of HIV quasispecies. Nucleic Acids Res. 38:7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang J, Chiodini R, Badr A, Zhang G. 2011. The impact of next-generation sequencing on genomics. J. Genet. Genomics 38:95–109 [DOI] [PMC free article] [PubMed] [Google Scholar]