Abstract
A novel strategy for the rapid detection and identification of traditional and emerging Campylobacter strains based upon Raman spectroscopy (532 nm) is presented here. A total of 200 reference strains and clinical isolates of 11 different Campylobacter species recovered from infected animals and humans from China and North America were used to establish a global Raman spectroscopy-based dendrogram model for Campylobacter identification to the species level and cross validated for its feasibility to predict Campylobacter-associated food-borne outbreaks. Bayesian probability coupled with Monte Carlo estimation was employed to validate the established Raman classification model on the basis of the selected principal components, mainly protein secondary structures, on the Campylobacter cell membrane. This Raman spectroscopy-based typing technique correlates well with multilocus sequence typing and has an average recognition rate of 97.21%. Discriminatory power for the Raman classification model had a Simpson index of diversity of 0.968. Intra- and interlaboratory reproducibility with different instrumentation yielded differentiation index values of 4.79 to 6.03 for wave numbers between 1,800 and 650 cm−1 and demonstrated the feasibility of using this spectroscopic method at different laboratories. Our Raman spectroscopy-based partial least-squares regression model could precisely discriminate and quantify the actual concentration of a specific Campylobacter strain in a bacterial mixture (regression coefficient, >0.98; residual prediction deviation, >7.88). A standard protocol for sample preparation, spectral collection, model validation, and data analyses was established for the Raman spectroscopic technique. Raman spectroscopy may have advantages over traditional genotyping methods for bacterial epidemiology, such as detection speed and accuracy of identification to the species level.
INTRODUCTION
Campylobacter species are among the predominant food-borne bacteria in the etiology of gastroenteritis globally, causing 500 million cases of human campylobacteriosis annually (66). In March 2012, the European Food Safety Authority and the European Center for Disease Prevention and Control published their annual report on zoonoses and food-borne outbreaks in the European Union for 2010. According to the report, campylobacteriosis remains the most commonly reported zoonotic infection in humans since 2005, with a total of 212,064 Campylobacter cases in humans reported in 2010, an increase for the fifth consecutive year with 7% more cases than in 2009 (16). Clinical symptoms are characterized by fever, abdominal cramps, and watery or bloody diarrhea (45). Campylobacter jejuni is the major Campylobacter species, and the ingestion of as few as 500 organisms may result in C. jejuni infection (64). While Campylobacter infection is typically self-limiting, in some cases, this infection is associated with severe enteritis, septicemia, Crohn's disease, and a higher incidence of Guillain-Barré syndrome (46, 82). C. jejuni and C. coli are confirmed human pathogens, but recent studies have validated that other Campylobacter species, e.g., C. fetus, C. concisus, C. upsaliensis, and C. sputorum, also cause gastrointestinal infections in humans (45).
Rapid identification is required if proper interventions are to be done and the source and routes of transmission are to be accurately determined. Correct typing of bacterial clinical isolates is critical to assist in epidemiological surveillance, to investigate the routes of transmission, and to understand the distribution of zoonosis and risk factors (12, 58, 72). Campylobacter spp. are widespread in the environment, members of this genus are ecologically diverse, clinical cases are sporadic, and associated outbreaks are rare. These factors make epidemiology and source tracking challenging (19, 20, 29, 69). To date, a variety of methods have been used to identify Campylobacter isolates to the species level. These include genotyping methods (76) such as traditional PCR (6, 35, 63, 75), multilocus sequence typing (MLST) (5, 13, 38, 52, 67, 71), amplified fragment length polymorphism (4, 8, 15, 40), pulsed-field gel electrophoresis (PFGE) (24, 62), loop-mediated isothermal amplification (LAMP) (80, 81), and microarray-based methods (73), as well as serotyping methods (21, 56) and mass spectrometry (17, 18, 25, 47, 78). Typically, genotyping methods are time-consuming and require highly trained personnel. Taken together, alternative molecular typing techniques would be advantageous for the detection and differentiation of Campylobacter species.
Both infrared and Raman spectroscopy methods are forms of vibrational spectroscopy, and their spectral patterns for biological samples have shown good reproducibility and high discriminatory power (41–44). In addition, these bioanalytical techniques are fast, reagentless, and easy to conduct. Thus, they provide the unique advantage of differentiating taxonomic entities at the species or subspecies level on the basis of variations in the spectral features of bacterial cells (41). Since the two groundbreaking publications in Nature about the use of infrared spectroscopy (57) and Raman spectroscopy (61) to study microorganisms, these two techniques have been extensively employed to detect and discriminate different microorganisms and have been shown to be useful as real-time typing methods in bacterial epidemiology (3, 7, 33, 34, 36, 37, 48, 49, 59, 60, 65, 77). Fourier-transformed infrared (FT-IR) spectroscopy, in combination with multivariate analyses, has been used to identify and discriminate C. jejuni and C. coli (54, 55). Recently, complementary infrared and Raman spectral features of C. jejuni planktonic cells, sessile cells in biofilm, and biofilm extracellular polymeric substance were characterized by our lab (42, 44). The biochemical compositions (i.e., carbohydrates, lipids, proteins, and nucleic acids) of Campylobacter cells were determined by using confocal micro-Raman spectroscopy on the basis of whole-organism fingerprinting (42).
Due to minor variations within the raw vibrational spectral features of different microbiological samples, the interpretation of spectra requires advanced chemometric tools (2, 41). The use of pattern recognition can unmask relationships and cluster constituents on the basis of their perceived closeness. Among the spectroscopy-based pattern recognition methods, unsupervised principal-component (PC) analysis (PCA), hierarchical cluster analysis (HCA), and supervised discriminant function analysis (DFA) are three major types, providing either cluster plots or dendrogram structures for segregation and discrimination (31, 32). Recently, soft independent modeling of class analog (SIMCA) has been extensively employed to study bacterial identification to the species level (42). In addition, Bayesian probability of vibrational spectral feature significance has been employed to validate PCs selected by PCA for classification model construction (23) and the stability of the derived supervised and/or unsupervised chemometric models could be determined by using Monte Carlo estimations (14, 68).
Here we report a fast, nondestructive, and reliable analytical approach for the identification and discrimination of Campylobacter species, including emerging taxa, by combining a micro-Raman spectroscopic analysis with a chemometric data classification approach. This technique shows great potential as a method for the classification of Campylobacter species.
MATERIALS AND METHODS
Strains.
Two hundred Campylobacter strains were included in this study. These strains represented 11 Campylobacter species, including: C. jejuni, C. coli, C. lari, C. fetus, C. concisus, C. curvus, C. helveticus, C. hyointestinalis, C. mucosalis, C. sputorum, and C. upsaliensis (see Table S1 in the supplemental material). Additionally, the strain sets for three species included members of both described subspecies, i.e., C. fetus subsp. fetus and venerealis, C. lari subsp. lari and concheus, and C. hyointestinalis subsp. hyointestinalis and lawsonii. The strains were obtained from four different laboratories in the United States and China. RM strain numbers are designations of strains from the Produce Safety and Microbiology Research Unit strain collection at the United States Department of Agriculture (USDA). MEK strain numbers are designations of strains from the Campylobacter Research Lab strain collection at Washington State University. EIQB strain numbers are designations of strains from the Chinese Entry-Exit Inspection and Quarantine Bureau strain collection at Jiangsu and Tianjin. All strains were isolated from animal, clinical, or food samples. RM strains were typed by MLST. All strains were stored frozen (−80°C) in Mueller-Hinton (MH) broth containing 12% glycerol and 75% citrated bovine blood. The bacterial strains were cultured routinely on MH agar plates supplemented with 5% citrated bovine blood (MHB) at 37°C under microaerobic conditions (10% CO2, 85% N2, 5% O2, 5% H2) during the experiment.
MLST.
MLST was performed as previously described under the conditions and with the primer sets of Miller et al. (52, 53). MLST amplifications were performed on a Tetrad thermocycler (Bio-Rad, Hercules, CA). Amplicons were purified on a BioRobot 8000 workstation (Qiagen, Valencia, CA). Cycle sequencing reactions were performed on a Tetrad thermocycler by using the ABI PRISM BigDye Terminator cycle sequencing kit (version 3.1; Applied Biosystems, Foster City, CA) and standard protocols. Cycle sequencing extension products were purified by using BigDye X-Terminator (Applied Biosystems). DNA sequencing was performed on an ABI PRISM 3730 DNA Analyzer (Applied Biosystems). Sequences were trimmed, assembled, and analyzed in SeqMan (v 9.1; DNASTAR, Madison, WI).
Preparation of samples for spectrum collection.
Campylobacter strains were cultivated in MH broth at 37°C for 24 h under microaerobic conditions (10% CO2, 85% N2, 5% O2, and 5% H2). One-hundred-microliter samples of bacterial cultures were streaked onto MHB and incubated at 37°C under microaerobic conditions for 24 to 72 h. For sample preparation, a calibrated 1-μl loop was filled with Campylobacter biomass on MHB and suspended in 100 μl of sterile deionized water. After centrifugation for 5 min at 15,000 × g, the supernatant was discarded and the bacterial pellet was transferred to a glass microarray slide coated with a thin film of gold (Thermo Scientific Inc., Waltham, MA). This gold-coated microarray slide has low fluorescence, providing a high signal-to-noise ratio, and is highly compatible with green laser (532 nm) biophotonic applications. Bacterial samples were partially dried on the gold-coated microarray slide for 30 min at 22°C. For an overview of the bacterial sample preparation procedure and the confocal micro-Raman system for spectral collection, see Fig. S1 in the supplemental material.
Raman instrumentation.
Two different confocal Raman instrumentation systems were employed in this study. The first Raman system was set up in the United States and used to collect the spectral features of Campylobacter isolates from the United States. This Raman spectroscopic analysis was performed by using a WITec alpha300 Raman microscope (WITec, Ulm, Germany) equipped with a UHTS-300 spectrometer. The spectrometer has an entrance slit of 50 μm and a focal length of 300 mm and is equipped with a 600-line/mm grating with a 532.5-nm laser power of 2 mW of incident light on the bacterial sample used. The Raman-scattered light was detected by using a 1,600- by 200-pixel charge-coupled device (CCD) array detector. The size of each pixel was 16 by 16 μm. A Nikon 20× objective focused the laser light onto the bacterial samples. An integration time of 60 s (3-s integration time with 20 signal averages) was used for bacterial spectral collection. The z displacement was controlled by a piezoelectric transducer on the objective. The WITec Control v1.5 software (WITec, Ulm, Germany) was employed for instrumental control and data collection. Collection of Raman spectra was performed over a simultaneous wavenumber shift range of 3,700 to 200 cm−1 in an extended mode.
The second Raman system was operated in China and used to collect the spectral features of Campylobacter isolates from China. A Renishaw inVia Raman microscope system (Renishaw plc, Gloucestershire, United Kingdom) equipped with a Leica microscope (Leica Biosystems, Wetzlar, Germany) and a 514.5-nm green diode laser source was used in this study. Rayleigh scattering was eliminated by the filters. Raman scattered light was collected and dispersed by a diffraction grating, and finally the Raman shift signal was recorded as a spectrum by a 576- by 384-pixel CCD array detector. Gold-coated microarray chips covered with Campylobacter samples were mounted on a standard stage of an Olympus microscope, focused under the collection assembly, and Raman spectra were collected by using a 20× objective with a detection range of 4,000 to 100 cm−1. The measurement was conducted over a 60-s exposure time (3-s integration time by 20 accumulations) with approximately 2 mW of incident laser power. The WiRE 3.0 software was used to control the Raman system and collect spectral features.
Spectral preprocessing.
Raw micro-Raman spectra contain several types of spectral interferences, including the fluorescence background of the biological sample, CCD background noise, Gaussian noise, and cosmic noise (2). The background correction was first performed by using a polynomial background fit described by Lieber et al. (39). This procedure can minimize the effect of different background profiles caused by fluorescence of the microbiological samples on gold-coated microarray slides and the thermal fluctuations on the CCD detector. Spectral smoothing was subsequently done by using a (9-point) Savitzky-Golay algorithm. Because the focal volume of the biological analyte (i.e., bacterial samples) is significantly fluctuated, subsequent normalization is necessary to further process the micro-Raman spectra for quantitative analyses. In this study, the Raman spectra were normalized on the basis of the intensity of the C-H peak in the wavenumber region of 3,100 to 2,900 cm−1 because this signal represents the total biomass of the Campylobacter cells. According to Rösch et al. (65) and our preliminary analysis, using the above-mentioned C-H vibrations for normalization yields the best results for baseline correction (data not shown).
Spectral reproducibility.
Vibrational spectrum reproducibility is a critical parameter to determine intralaboratory reliability and to subsequently establish robust and reliable spectroscopy-based chemometric models. There are several factors (parameters) that may affect vibrational spectrum reproducibility, including cell culture age, nutrient availability, cultivation temperature, and spectral wavenumber selection (42). In addition, vibrational spectral variability was due mainly to biological features and to a lesser extent to instrumental sources (55). This statement was further validated in this study by using two different Raman instruments. Spectral reproducibility (intragroup [within-strain] variation) was investigated by calculating the differentiation index (Dy1y2) value (42, 43) as follows:
The lower the Dy1y2 value, the better the reproducibility of the Raman spectra for bacterial samples.
Spectral selectivity.
Spectral selectivity is critical to the detection and discrimination of different types of Campylobacter in a mixture by using Raman spectroscopy combined with chemometrics. Factorization was employed on averaged spectra of the selected Campylobacter species. Factor analysis extracts high-dimensional Raman scattered spectra into several PCs and relevant scores. Spectral distance (SD) was subsequently calculated on the basis of this relevant score, and selectivity (S) was subsequently calculated as the ratio of SD to the sum of the threshold values of the cluster radius scores T1 and T2, according to our previous publications (42, 43). S values of >1 were considered to be significant for detection and segregation of selected Campylobacter types from a mixture.
Discriminatory power.
To determine the discriminatory powers of Raman typing, the numerical index of discrimination (D) was calculated (30). This parameter is based on the probability that two unrelated bacterial strains will be assigned to different typing groups and can be calculated by using Simpson's index of diversity, as follows:
In this equation, N is the total number of Campylobacter strains in the sample population used for the chemometric model, S is the total number of Campylobacter types involved in this model, and nj is the number of the strains belonging to the jth type. A D value of >0.9 is required for a highly discriminatory typing method, with segregation results interpreted with confidence (72, 77).
Classification chemometric models.
Two different types of segregation chemometric models were employed for Campylobacter identification to the species level. PCA and HCA are unsupervised classification methods that illustrate similarity relationships between Raman spectra without a priori knowledge about the bacteria investigated (28). DFA is a supervised classification method that constructs a dendrogram structure to segregate bacteria according to their known bacterial characterization (i.e., type) (31, 32).
A classification of constituents is required to be made before analysis by the supervised DFA model. This classification procedure maximizes the variance between groups and minimizes the variance within the group. The Mahalanobis distance was calculated and is defined as follows: M1,2 = [(x1 − x2)S−1](x1 − x2), where S is the pooled estimate of the within-group covariance matrix and x1 and x2 are mean vectors for the two groups. Thus, M1,2 is the distance between groups in units of within-group standard deviations (10).
In this study, several different classification models were established and validated, including: a PCA-based segregation model to determine spectral reproducibility on the basis of different cultivation times, an HCA-based dendrogram model to study the concordance between Raman typing and MLST for selected Campylobacter strains, a DFA-based dendrogram model to classify 11 different species of Campylobacter, and a DFA-based dendrogram global model to differentiate C. jejuni and C. coli isolates from different continents.
A Bayesian probability approach was employed to validate the PCs selected by PCA for a DFA-based dendrogram model to classify 11 different species of Campylobacter. The principle of using a Bayesian probability approach to feature significance for infrared spectra of bacteria has been extensively illustrated by others (23) on the basis that a factor with a large variance has a higher probability for model construction than a factor with a small variance. Additionally, the stability of this model was determined by using Monte Carlo estimation (14). Briefly, this estimation was employed to construct random models and calculate the variability of the intracluster distances, resulting in the determination of the intracluster geometry. The inverse of the average variance was expressed as stability (68).
To further evaluate the performance and reliability of the DFA-based global chemometric model in differentiating C. jejuni and C. coli isolates from different continents, SIMCA was continuously employed to determine the recognition rate of the spectral features for Campylobacter identification to the species level. SIMCA is a supervised chemometric model describing a plane (for two PCs), and the mean orthogonal distance of training data from this specific plane is calculated as residual standard deviation and subsequently employed to determine a critical distance (on the basis of F distribution with a 95% confidence interval) for the identification of an analyte (i.e., bacteria) to the species level (11). Prediction data were subsequently projected into each PC model, and the residual distances were calculated (whether below the statistical limit for a specific class or not) to determine the class to which the prediction data belong.
Identification of mixed Campylobacter strains.
We selected C. jejuni RM1221, C. coli RM1051, C. concisus RM3271, C. curvus RM3269, C. fetus RM1558, C. helveticus RM3228, C. hyointestinalis RM2101, C. lari RM1887, C. mucosalis RM3233, C. sputorum RM3237, and C. upsaliensis RM1488 and mixed these strains in equal biomasses, forming a cocktail. C. jejuni strain MEKF38011, C. coli strain MEK2, C. upsaliensis strain RM3776, or C. fetus strain RM2087 was added individually to the 11-strain cocktail at concentrations ranging from 5 to 100% by biomass to form a new mixture. The spectral features of the Campylobacter mixture were determined by using a confocal micro-Raman spectroscopic system.
A supervised partial least-squares regression (PLSR) model was established, and leave-one-out cross validation was applied to challenge the reliability of this chemometric model by removing one standard from the data set at a time and calibrating the remaining standards (1). The error of leave-one-out cross validation is an unbiased estimate of the actual classification error probability, while the traditionally used holdout method (which employs 70% of the data for model establishment and 30% for model validation) results in a higher estimate of classification error probability (65). This PLSR model generates a linear regression model by projecting the predicted variables (here, Raman spectral features of a new Campylobacter mixture, i.e., the 11-strain cocktail mixture plus 1 additional strain at various levels) and the observable variables (here, the relative concentration of each Campylobacter strain in a new mixture) to a new space on the basis of a causal network of confirmed latent variables (1, 41). The suitability of the developed models was assessed by determining the regression coefficient (R), latent variables, the root mean square error (RMSE) of calibration, and the RMSE of cross validation, while the overall suitability of the models for predicting the concentration of a specific Campylobacter strain in the mixture was evaluated from the residual prediction deviation (RPD) (22, 41, 79).
Statistical analysis.
The experiment was performed in three independent replicate trails. The results are expressed as the mean of three independent replicates ± the standard deviation. The significance of differences (P < 0.05) was determined by one-way analysis of variance following the t test in Matlab.
RESULTS
Raman spectral reproducibility and influence of selected wavenumber regions.
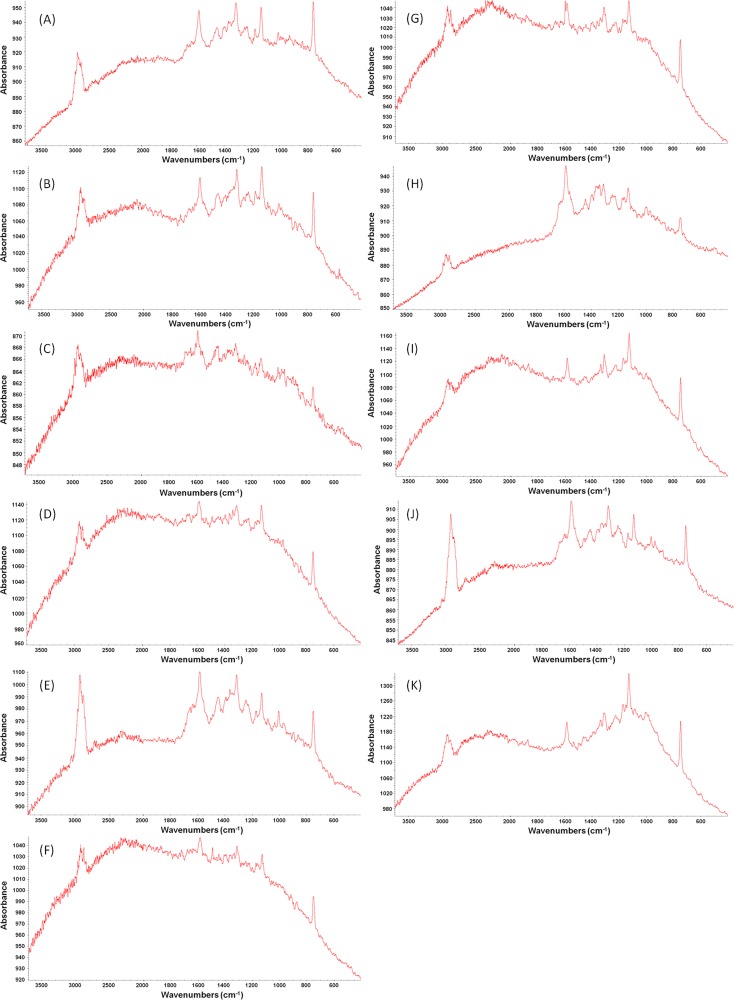
We previously analyzed C. jejuni by confocal Raman spectroscopy and made detailed band assignments (42). Here we analyzed the Raman spectral features of 200 strains from 11 Campylobacter species (Fig. 1). Five different wavenumber regions were selected to investigate their relationship to Raman spectral reproducibility, including the wavenumber region between 3,100 and 2,800 cm−1 (designated w1), which is related to the total bacterial biomass; the wavenumber region between 1,800 and 1,500 cm−1 (designated w2), which provides constituent information about proteins and peptides; the wavenumber region between 1,500 and 1,200 cm−1 (designated w3), a mixed region of proteins and fatty acids; the wavenumber region between 1,200 and 900 cm−1 (designated w4), a polysaccharide region; and the wavenumber region between 900 and 650 cm−1 (designated w5), which is often defined as the “fingerprint” region because of specific spectral patterns. The Dy1y2 values were calculated, by using the equations described in Materials and Methods, for each wavenumber region and combinations of wavenumber regions on the basis of the cultivation time for strains of each species and then summarized. As noted below, some different species required different cultivation times to reach a targeted biomass. The highest Dy1y2 values were obtained by using the spectral regions designated w1 (18.38 to 24.51) and w4 (20.63 to 26.05), while lower Dy1y2 values were derived from w2 (9.30 to 13.71), w3 (11.09 to 14.26), and w5 (7.88 to 12.45). A combination of regions w2, w3, w4, and w5 gave the lowest Dy1y2 values (4.79 to 6.03). The addition of w1 to a previous combination of wavenumber regions significantly increased the Dy1y2 values to >20. The lower the Dy1y2 value, the better the spectral reproducibility (42, 43). Therefore, we selected the wavenumber combination of w2, w3, w4, and w5, the wavenumber region between 1,800 and 650 cm−1, for further chemometric model evaluation.
Fig 1.
Different species of Campylobacter bacteria show unique Raman spectral patterns. Shown are the raw Raman spectral features of C. jejuni (A), C. coli (B), C. concisus (C), C. fetus (D), C. helveticus (E), C. hyointestinalis (F), C. curvus (G), C. lari (H), C. mucosalis (I), C. sputorum (J), and C. upsaliensis (K).
Raman spectral reproducibility and influence of cell cultivation time.
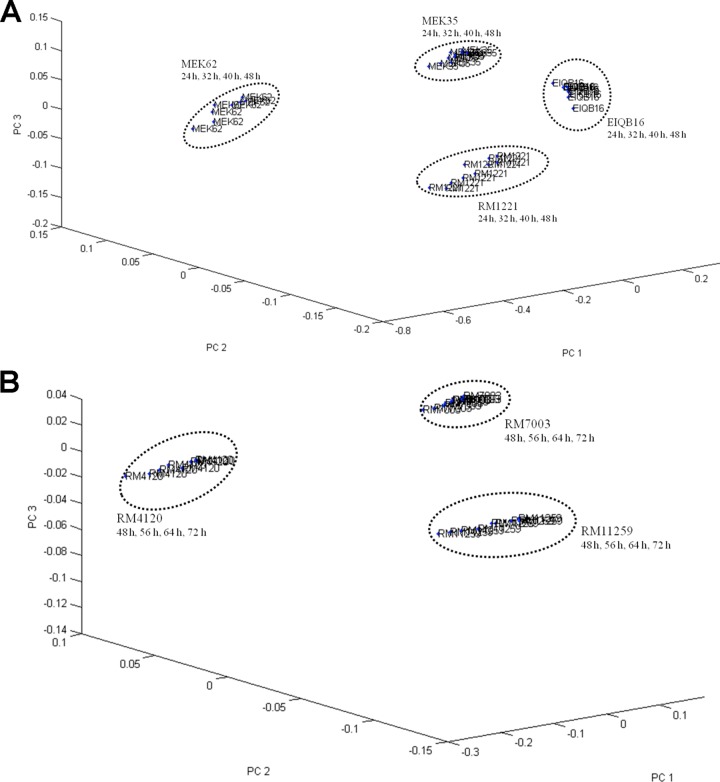
We determined whether cell cultivation time was a critical factor affecting Raman spectral reproducibility. Of the 11 different species of Campylobacter used in this study, some required a shorter cultivation time for microcolony formation under microaerobic conditions (i.e., C. jejuni, C. fetus, C. coli, C. lari, and C. hyointestinalis, 24 to 48 h), while others needed a longer cultivation time to reach a certain predetermined biomass (i.e., C. upsaliensis, C. sputorum, C. curvus, C. mucosalis, C. concisus, and C. helveticus, 48 to 72 h). PCA was employed to investigate how Raman spectral reproducibility is influenced by culturing time (Fig. 2). We randomly selected four different strains of C. jejuni (Fig. 2A) and three different strains of C. sputorum (Fig. 2B) obtained from three different laboratories (RM, EIQB, and MEK) as representative strains to individually establish two-dimensional cluster models (n = 20). As shown in Fig. 2, tight clusters were formed for each strain on the basis of different cultivation times (24 to 48 h for rapidly growing species and 48 to 72 h for slowly growing species). Calculation of the interclass distance between every two strains of the same species resulted in values ranging from 23.06 to 43.19, based upon Mahalanobis distance measurements computed between the centroids of the classes. Classes with interclass distance values of >3 are considered to be significantly different from each other (10). On the basis of these calculations, we selected 24 h as the cultivation time for the rapidly growing species and 48 h as the cultivation time for the slowly growing species.
Fig 2.
(A) Reproducibility of Raman spectra with different bacterial cultivation times on blood agar. Shown are the results of PCA of the repeated measurements of the four C. jejuni strains obtained from different laboratories. (B) Reproducibility of Raman spectra with different bacterial cultivation times on blood agar. Shown are the results of PCA of the repeated measurements of the three C. sputorum strains obtained from different laboratories.
Raman spectral reproducibility and concordance with MLST.
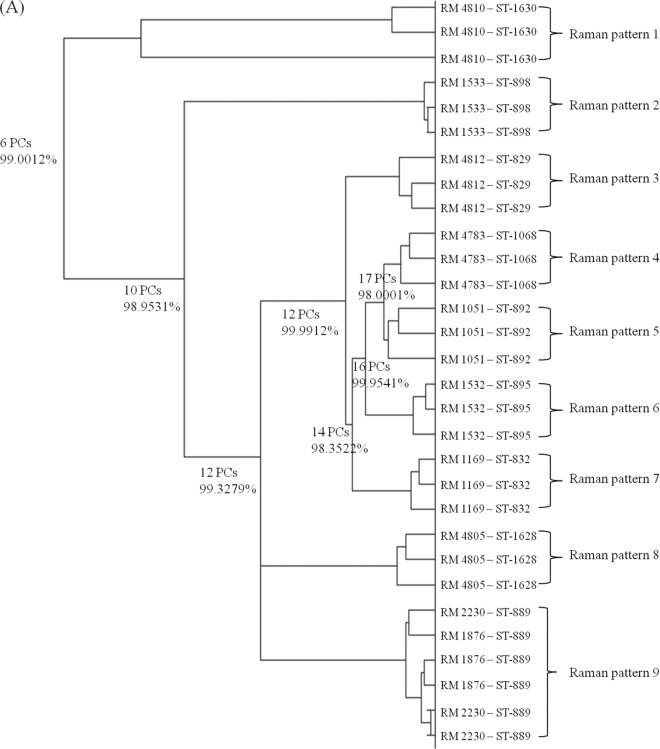
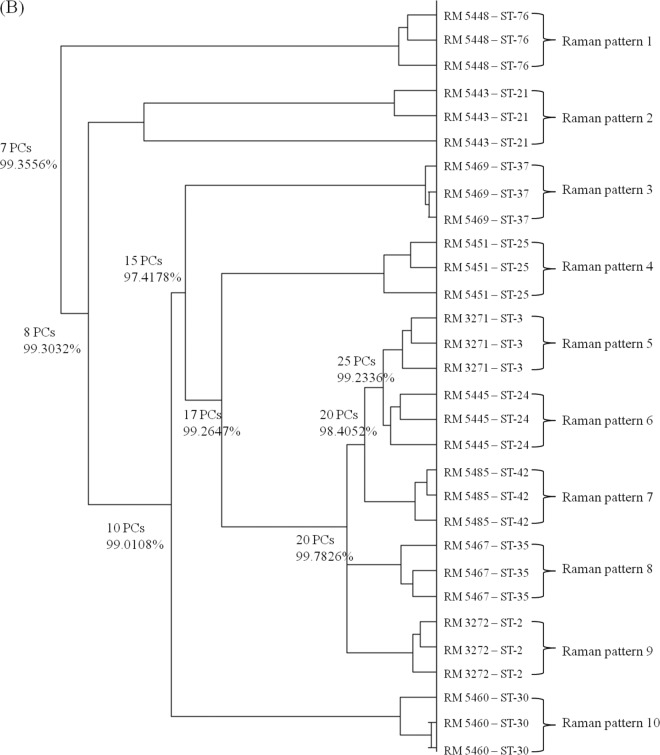
The correlation of Raman spectral reproducibility with MLST when unsupervised HCA is used is shown in Fig. 3. C. coli (Fig. 3A) and C. concisus (Fig. 3B) were selected as representative Campylobacter species. As shown in Fig. 3A, Raman patterns matched MLST profiles. In one case, C. coli RM2230 and RM1876 are both sequence type 889, and these strains cannot be distinguished by using the Raman typing method. However, for the other eight C. coli strains with different MLST profiles, each profile forms a distinct cluster based upon Raman patterns. This correlation between the MLST and Raman typing methods was also observed in C. concisus (Fig. 3B) and the other nine Campylobacter species (data not shown). Taken together, confocal micro-Raman spectroscopy could provide an alternative to MLST typing for Campylobacter classification, since the results from both classification schemes correlate well.
Fig 3.
Hierarchical cluster analysis of C. coli (A) and C. concisus (B) isolates. Full details of the numbers of PCs used at each level and the associated percentages of explained variance are provided. This dendrogram shows the reproducibility and concordance of Raman typing with MLST.
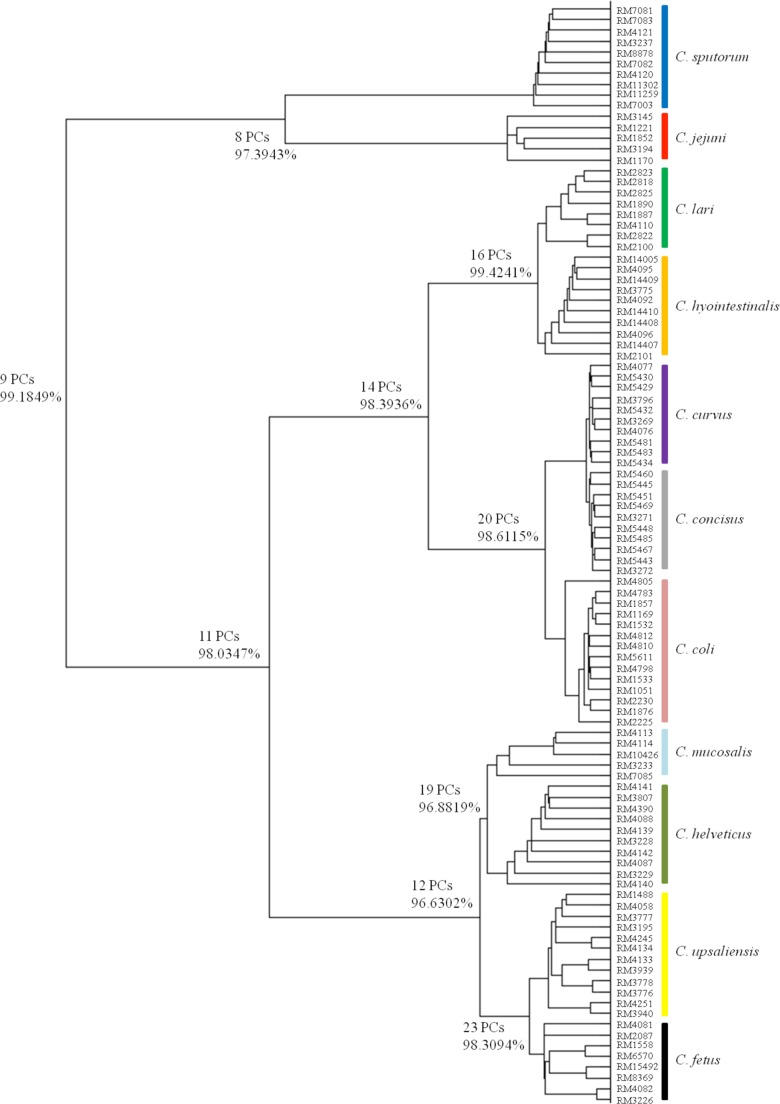
Evaluation of Raman spectroscopy for Campylobacter identification to the species level.
A total of 102 Campylobacter strains representing 11 Campylobacter species and 16 Campylobacter taxa (see Table S1a in the supplemental material) were provided by the USDA and used in a supervised DFA to determine if Raman spectroscopy can unambiguously identify Campylobacter isolates to the species level, and a clear segregation of each Campylobacter species was observed (Fig. 4). The spectral feature of each strain in the dendrogram is an average of 18 spectra collected from three independent experiments; thus, this chemometric DFA model incorporated data from a total of 1,836 Raman spectra. The corresponding Mahalanobis distances between groups in discriminant analysis were calculated and are summarized in Table 1. These Mahalanobis distances further validate the discrimination between various Campylobacter species. Additionally, the numerical index of discrimination (D), calculated for the Raman typing method by using Simpson's index of diversity, was determined to be 0.968, which is a high score and suitable for the differentiation of bacterial strains (30).
Fig 4.
Composite dendrogram derived from Raman spectroscopy-based discriminant function analysis of different species of traditional and emerging Campylobacter species.
Table 1.
Mahalanobis distances between groups in discriminant analysis
| Species | Mahalanobis distance from: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C. jejuni | C. coli | C. concisus | C. curvus | C. fetus | C. helveticus | C. hyointestinalis | C. lari | C. mucosalis | C. sputorum | C. upsaliensis | |
| C. jejuni | 0.00 | ||||||||||
| C. coli | 94.82 | 0.00 | |||||||||
| C. concisus | 115.76 | 55.38 | 0.00 | ||||||||
| C. curvus | 107.21 | 46.09 | 19.32 | 0.00 | |||||||
| C. fetus | 89.06 | 68.23 | 60.41 | 81.97 | 0.00 | ||||||
| C. helveticus | 135.66 | 73.07 | 49.29 | 72.55 | 32.07 | 0.00 | |||||
| C. hyointestinalis | 85.40 | 37.62 | 38.43 | 13.68 | 63.36 | 67.42 | 0.00 | ||||
| C. lari | 13.86 | 47.53 | 52.19 | 60.25 | 87.84 | 72.50 | 7.95 | 0.00 | |||
| C. mucosalis | 155.62 | 36.89 | 64.01 | 58.20 | 28.31 | 15.06 | 58.14 | 43.02 | 0.00 | ||
| C. sputorum | 9.11 | 98.02 | 101.03 | 81.37 | 120.38 | 83.63 | 80.33 | 95.61 | 128.30 | 0.00 | |
| C. upsaliensis | 163.48 | 89.68 | 55.38 | 63.02 | 10.42 | 27.48 | 43.43 | 56.20 | 32.17 | 175.57 | 0.00 |
We employed a Bayesian probability analysis to compare the top 25 features with PCs determined by PCA and found good agreement between the two approaches. The stability of the DFA model was determined by using Monte Carlo estimation. The 25 most significant features, 25 least significant features, and all features were selected and compared on the basis of model stability. The highest DFA model stability was derived from the use of the 25 most significant features (0.45 ± 0.07), and the lowest stability was derived from the use of the 25 least significant features (0.03 ± 0.01). The use of all features resulted in a stability similar to but slightly lower (0.41 ± 0.08) than that obtained by the use of the 25 most significant features. This may be because of interference and noise from nonsignificant features. Taken together, these findings validated the correct choice and use of the selected PCs for the DFA model.
Loading plots were determined to investigate the specific biochemical components most significant for classification and subsequent construction of the DFA dendrogram models for Campylobacter strain classification (Fig. 5). The following coding was programmed into Matlab to perform this loading plot analysis:
“figure
plot(z,coeff(:,1),′r′);
latent = diag(latent);
totalvar = sum(latent);
explained = 100*latent/totalvar;”
Fig 5.
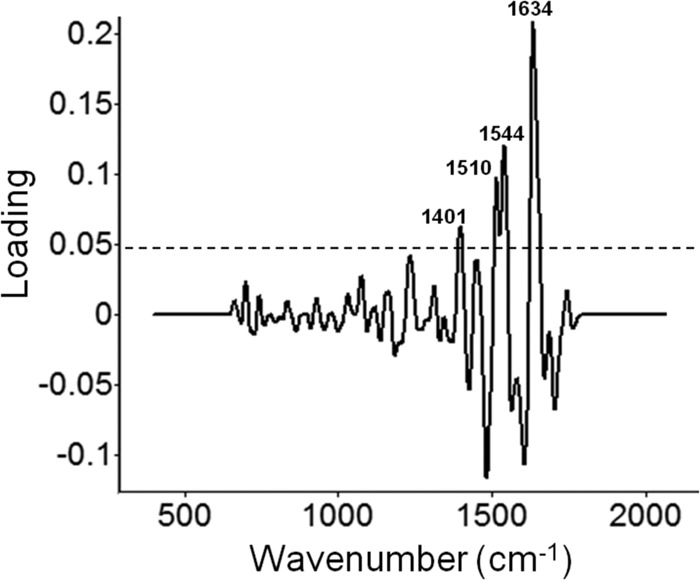
Loading plot of the first PC obtained from the DFA dendrogram in Fig. 4 to explain the classification of 11 different species of traditional and emerging Campylobacter bacteria. The loading plot explains the contributions of chemical constituents of the bacterial cell membrane to the classification model. Peaks: 1,634 cm−1, amide I; 1,544 cm−1, amide II; 1,510 cm−1, ring breathing modes in the DNA bases; 1,401 cm−1, bending modes of methyl groups of proteins.
Only the significant loadings (P < 0.05, represented by the dotted line in Fig. 5) were considered. The band at 1,634 cm−1 is assigned to amide I (9), and the band at 1,544 cm−1 is assigned to amide II (9). The band at 1,510 cm−1 is derived from ring breathing modes in the DNA bases (41). The band at 1,401 cm−1 is assigned to the bending modes of methyl groups of proteins (9). Thus, the classification of Campylobacter species was based mainly upon the secondary structural features of proteins in the Campylobacter cell membrane.
Raman spectral selectivity and sensitivity of Campylobacter detection within a mixture.
To determine the detection sensitivity and selectivity of Raman spectroscopy for a particular Campylobacter species within a mixture, four different targeted Campylobacter strains (i.e., C. jejuni MEKF38011, C. coli MEK2, C. upsaliensis RM3776, and C. fetus RM2087) were individually mixed with a prepared Campylobacter cocktail (details in Materials and Methods) at concentrations ranging from 5 to 100%, forming a composite. The Raman spectra were collected for each of these new, concentration-defined mixtures, and the selectivity values, calculated at a 95% confidence interval, for the targeted Campylobacter strain in the composite were determined. Selectivity values of >1 were considered significant for the detection and differentiation of the targeted analyte(s), in this case, the strain at various concentrations in the prepared composite (42, 43). Otherwise, overlapping clusters occurred, i.e., samples not significantly different from the spectra of the 11-strain cocktail. In this study, the selectivity value was higher than 1 for all four Campylobacter strains tested, indicating high selectivity (data not shown).
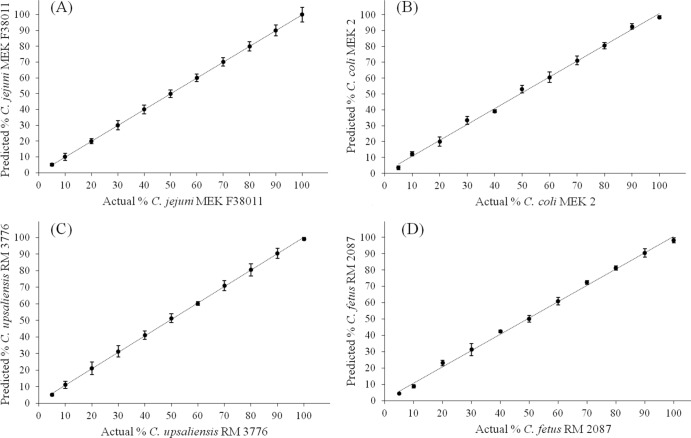
After spectral selectivity was tested, a PLSR model was subsequently established and cross validated by using the leave-one-out method. A reliable linear correlation between the targeted bacterial concentration and its corresponding Raman spectral features was observed, as shown in Fig. 6. A summary of all of the PLSR-associated parameters is shown in Table 2. These PLSR models have high R (>0.98) and RPD (>7.88) values and a low standard error for both calibrated and cross-validated models. The RMSE of calibration and cross validation were <0.61 and 0.72, respectively. These results meet the criteria that a good PLSR model should have high R (>0.95) and RPD (>5) values and a low RMSE (<1) for calibration and validation (22).
Fig 6.
Representative PLSR for actual specific Campylobacter concentration prediction in a mixture. C. jejuni (A), C. coli (B), C. upsaliensis (C), and C. fetus (D) cells were suspended in different proportions ranging from 5 to 100% of the total, and the correlation between Raman spectral features and the percentages of specific Campylobacter spp. in the mixture was determined.
Table 2.
PLSR models for prediction of specific Campylobacter concentrations in a mixture
| Species | Concn range (%) | No. of samples | No. of latent variables | Calibration |
CVa |
||||
|---|---|---|---|---|---|---|---|---|---|
| R | RMSE | RPD | R | RMSE | RPD | ||||
| C. jejuni | 5–100 | 198 | 8 | ≥0.99 | ≤0.43 | ≥14.32 | ≥0.99 | ≤0.51 | ≥7.88 |
| C. coli | 5–100 | 198 | 9 | ≥0.99 | ≤0.61 | ≥11.63 | ≥0.98 | ≤0.68 | ≥8.21 |
| C. upsaliensis | 5–100 | 198 | 9 | ≥0.99 | ≤0.38 | ≥16.57 | ≥0.99 | ≤0.49 | ≥11.05 |
| C. fetus | 5–100 | 198 | 8 | ≥0.99 | ≤0.53 | ≥13.91 | ≥0.98 | ≤0.72 | ≥9.27 |
CV, cross validation.
Global chemometric models for identification of Campylobacter isolates to the species level.
C. jejuni and C. coli account for the majority of Campylobacter-related food-borne enteritis. In this study, we used C. jejuni and C. coli strains (RM, MEK, and EIQB) from four different laboratories in the United States and China to create a composite global chemometric dendrogram model for the identification of C. coli and C. jejuni isolates to the species level and evaluation of strain variability on the basis of an expanded sample size (Fig. 7). From among them, 20 C. coli RM strains and 60 C. jejuni MEK strains were selected for the calibration set (Fig. 7, labeled in black) and another 20 C. jejuni MEK strains and 20 C. jejuni EIQB strains were selected for the validation set (Fig. 7, labeled in red). C. jejuni and C. coli were correctly classified, and strain similarity was observed for C. jejuni strains from the two different countries. We employed SIMCA to calculate the recognition rate for strains in the validation set. An average recognition rate of 97.21% was received for clinical C. jejuni strains that originated from both North America and China (Table 3), providing an additional and easy-to-use model for the classification of species by the use of Raman spectroscopy.
Fig 7.
Composite dendrogram representing the global chemometric model to identify C. jejuni and C. coli strains from North America and Asia. Entries in black represent the training data, and those in red denote the validation data. The model was established and validated as described in the text.
Table 3.
Recognition rates for Raman spectra of C. jejuni clinical strains from China and the United States determined by SIMCA
| Strain or parameter | Total no. of spectra | No. of strain spectra incorrectly classified | % of strain spectra correctly classified |
|---|---|---|---|
| EIQB1 | 9 | 0 | 100.0 |
| EIQB2 | 9 | 0 | 100.0 |
| EIQB3 | 9 | 0 | 100.0 |
| EIQB4 | 9 | 1 | 88.9 |
| EIQB5 | 9 | 0 | 100.0 |
| EIQB6 | 9 | 0 | 100.0 |
| EIQB7 | 9 | 1 | 88.9 |
| EIQB8 | 9 | 1 | 88.9 |
| EIQB9 | 9 | 0 | 100.0 |
| EIQB10 | 9 | 0 | 100.0 |
| EIQB11 | 9 | 0 | 100.0 |
| EIQB12 | 9 | 1 | 88.9 |
| EIQB13 | 9 | 0 | 100.0 |
| EIQB14 | 9 | 0 | 100.0 |
| EIQB15 | 9 | 0 | 100.0 |
| EIQB16 | 9 | 1 | 88.9 |
| EIQB17 | 9 | 0 | 100.0 |
| EIQB18 | 9 | 0 | 100.0 |
| EIQB19 | 9 | 0 | 100.0 |
| EIQB20 | 9 | 0 | 100.0 |
| MEK61 | 18 | 2 | 88.9 |
| MEK62 | 18 | 1 | 94.4 |
| MEK63 | 18 | 0 | 100.0 |
| MEK64 | 18 | 0 | 100.0 |
| MEK65 | 18 | 0 | 100.0 |
| MEK66 | 18 | 1 | 94.4 |
| MEK67 | 18 | 0 | 100.0 |
| MEK68 | 18 | 0 | 100.0 |
| MEK69 | 18 | 0 | 100.0 |
| MEK70 | 18 | 1 | 94.4 |
| MEK71 | 18 | 1 | 94.4 |
| MEK72 | 18 | 1 | 94.4 |
| MEK73 | 18 | 0 | 100.0 |
| MEK74 | 18 | 0 | 100.0 |
| MEK75 | 18 | 0 | 100.0 |
| MEK76 | 18 | 2 | 88.9 |
| MEK77 | 18 | 0 | 100.0 |
| MEK78 | 18 | 0 | 100.0 |
| MEK79 | 18 | 1 | 94.4 |
| MEK80 | 18 | 0 | 100.0 |
| Avg recognition rate (%) | 97.21 |
DISCUSSION
Previous to this study, there was no single typing method that possessed all of the advantages (e.g., accuracy, speed, etc.) sought for Campylobacter identification to the species level. As emerging Campylobacter species are discovered and their clinical importance is recognized (45), the ability to properly identify and classify these strains becomes more important. In the present study, we employed confocal micro-Raman spectroscopic typing as a tool to demonstrate the feasibility of its use to complement or substitute for MLST, which could provide an alternative method to improve Campylobacter epidemiological surveillance.
Raman spectroscopy is a noninvasive method that provides a biochemical profile of the bacterial cell wall and cell membrane (41). The sample preparation procedure is easy, spectral collection is fast, and detection is accurate (28). All of these factors are advantages of Raman spectroscopy as a tool for clinical microbiology. Confocal micro-Raman spectroscopy has been recently used to detect hospital-acquired Staphylococcus aureus-associated infections (77), pathogenic endospores (70), clean-room-relevant microbiological contamination (65), and Candida species (48). As a complementary counterpart, infrared spectroscopy, especially FT-IR spectroscopy, has been widely used to identify and differentiate various types of bacteria (3, 34, 36, 37, 49, 59, 60). Mouwen and colleagues applied FT-IR spectroscopy coupled with HCA (55) or an artificial neural network (54) to detect and differentiate C. jejuni and C. coli. However, the sample size was relatively small (fewer than 30 strains in each study). We used about 200 clinical Campylobacter isolates representing 11 different species from four different laboratories in the United States and China (see Table S1 in the supplemental material) and collected about 3,500 Raman spectra (Fig. 1) as the basis to establish various chemometric models for identification to the species level.
Reproducibility drives the reliability of a specific classification method. In the case of Raman spectroscopy, there are several factors that may affect spectral reproducibility, including bacterial cultivation time, growth temperature, medium use, and wavenumber selection (42, 43, 55). We calculated the Dy1y2 values and demonstrated that use of the wavenumber region of 1,800 to 650 cm−1 provided the lowest Dy1y2 value, indicating that the highest reproducibility of Raman spectra would be within this wavelength range (55). Fortunately, this spectral range includes important features for proteins, polysaccharides, nucleic acids, and lipids. In addition, PCA was employed to show that a cultivation time of 24 h for rapidly growing Campylobacter strains and 48 h for more slowly growing Campylobacter strains resulted in high reproducibility of Raman spectra (Fig. 2), which was further validated by calculating the interclass distance by using the Mahalanobis distance. Previous studies demonstrated that the shape of the Campylobacter cell (coccoid-spiral forms) and its chemical composition vary as the culture ages (26, 27) and that these physical parameters could possibly affect the spectral features obtained if not properly controlled (42). Additionally, the carbohydrate composition of Campylobacter cells is influenced by metabolic activity and cell membrane structure, which may also be reflected in vibrational spectral features (wavenumber region w4, 1,200 to 900 cm−1) (55). We used MH broth or MHB as the medium for Campylobacter cultivation under the same microaerobic conditions for the cultivation of all of the strains tested. In summary, this appeared to have provided a suitable protocol for bacterial sample preparation that was able to ensure high reproducibility of Raman spectra.
MLST profiles correlated well with the Raman spectral features of Campylobacter strains obtained by using unsupervised HCA (Fig. 3). However, subspecies could not be discerned on the basis of this method. Three of the species tested contained two different subspecies (i.e., C. fetus subsp. fetus and venerealis, C. lari subsp. lari and concheus, and C. hyointestinalis subsp. hyointestinalis and lawsonii), but these strains were indistinguishable. Previous studies that used FT-IR spectroscopy for C. jejuni and C. coli identification to the species level also showed a good correlation of spectral features relative to PCR typing (55). In other studies, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry, another method of bacterial typing, was employed to classify Campylobacter bacteria and showed a good correlation with genotyping techniques. Different Campylobacter species could be classified on the basis of biomarker ions, specifically from proteins (18, 47, 78). However, these MALDI-TOF mass spectroscopic methods require exact extraction protocols for the recovery of an analyte(s) from bacterial cell lysates that are more complicated than that required for the Raman spectroscopic method (17). In addition, the sample matrix is inhomogeneous and the selection of “hot” spots that optimize ion formation is required. This subsequently resulted in poor spectral reproducibility that could be compensated for, in part, by coadding individual spectra. Similar spectral reproducibility issues were also observed when using surface-enhanced Raman scattering spectroscopy for bacterial analysis (31, 32). This is the major reason that we used traditional confocal micro-Raman spectroscopy for this study.
On the basis of the use of 102 strains representing 11 Campylobacter species, a comprehensive Raman spectroscopy-based dendrogram was constructed (Fig. 4). This dendrogram was generated by using supervised DFA, and the corresponding Mahalanobis distances were calculated (Table 1) to further validate the reliability of this model for the prediction of potential Campylobacter clinical isolates in the future. Bayesian probability was employed to confirm the correct selection of PCs by PCA, with a further validation by Monte Carlo estimation on the basis of determination of model stability. A PC DFA-based loading plot was determined to evaluate the specific chemical components (Fig. 5), and protein secondary structure dominated the classification of different species of Campylobacter bacteria. This would be anticipated on the basis of other studies of bacterial classification using Raman spectroscopy (41) or infrared spectroscopy (51), where changes to amide I and other protein features tend to be the most important for bacterial classification.
Being able to identify the composition of a bacterial mixture is important because clinical and environmental samples can be composed of several different Campylobacter species (47). The correct identification with high selectivity of a specific species in a mixture can improve the accuracy of epidemiological surveillance. We employed a Raman spectroscopy-based PLSR model to predict the actual concentrations of selected Campylobacter species (i.e., C. jejuni MEKF38011, C. coli MEK2, C. upsaliensis RM3776, and C. fetus RM2087) in a prepared Campylobacter cocktail composed of 11 different species. The prediction value was very precise, with a linear relationship between the actual bacterial concentration and corresponding Raman spectral features (Fig. 6 and Table 2).
A global classification model was established and validated to segregate C. jejuni and C. coli by DFA (Fig. 7), and a further validation was performed by using SIMCA (Table 3). Both supervised chemometric models demonstrated a high recognition rate, with a DFA dendrogram model performing a bit better than SIMCA. This usually happens because the classifier in the SIMCA model sometimes identifies samples (i.e., spectra) as members of multiple groups (11). In this study, the SIMCA model had a 97% average recognition rate, indicating the good reliability of our global Raman spectroscopy-based classification model. In addition, strain similarity was observed for C. jejuni strains from different countries and successful discrimination at the strain level was still received on the basis of a distensible sample size compared to Fig. 5.
Finally, we compared the times required for Raman typing and other current typing methods. Starting from the confirmation of a positive Campylobacter culture, classical serotyping requires at least 5 to 7 days for completion (56). For genotyping methods, flaA sequencing takes approximately 2 days (8, 50, 76) and PFGE, the “gold standard,” takes 3 to 4 days (4, 6), although a rapid PFGE method that takes 24 to 30 h was recently developed (76). The LAMP technique takes about 24 h (80, 81), and currently used MLST takes a minimum of 24 h (38, 52, 67). In contrast, our Raman spectroscopic classification method can significantly save analysis time and reduce reagent cost. For example, following cultivation, sample preparation of 30 clinical Campylobacter isolates takes about 1 h, including 30 min of partial dehydration required for the preparation of a bacterial sample for presentation to the confocal micro-Raman instrument. Another 40 min is needed to collect spectra, process the data, and predict the bacterial species by using a validated chemometric model. Thus, the diagnostic work could be finished within 2 h after a validated model has been established.
Recently, a more powerful laser was generated and its application as biophotonics coupled with a microfluidic environment forming a “lab-on-a-chip” system for bacterial identification and classification has been reported (74). The same quality (e.g., resolution) of Raman spectra for bacterial samples can be obtained with a spectral collection time of 1 s. This can significantly shorten the diagnostic time and permit the use of a continuous system, thus reducing sample handling. Here we report the sample preparation, instrument operation, and data analysis procedures for the use of this Raman typing technique to classify various Campylobacter species. It has the potential to be employed as a standard diagnostic tool in each microbiology laboratory for bacterial epidemiological surveillance, especially considering that this technique is reagentless and noninvasive and requires no labeling. Further research should be conducted to develop this assay for a microfluidic platform.
Supplementary Material
ACKNOWLEDGMENTS
We thank Emma Yee and Anna Bates at the USDA Western Regional Research Center for assistance in culture preparation and shipment of the Campylobacter strains used in this study.
This work was supported by funds awarded to S.W. by the Ministry of Science and Technology of China (2011CB512014 and 2012CB720803); funds awarded to M.E.K. by the National Institutes of Health (R56 AI088518-01A1); funds awarded to B.A.R. by the National Institute of Food and Agriculture (AFRI 2011-68003-20096) and the Agricultural Research Center, Washington State University; and funds awarded to W.G.M. by the USDA Agricultural Research Service (CRIS project 5325-42000-045).
Within the scope of the main research effort “Biophotonics and Its Application to Study Campylobacter Bacteria” supported by both China (NSF) and the United States (NIH and USDA), we have recently created a database of all of the Raman spectra acquired in our six laboratories for Campylobacter species, as well as a Matlab-based program for comparing spectral features determined for each strain represented in the database.
All of the chemometric models in this study were developed with programming written by Xiaonan Lu by using Matlab (version 2010a). Readers who are interested in the Matlab programming codes used for vibrational spectroscopy-based PCA, HCA, DFA, SIMCA, and PLSR should send direct inquiries to xiaonan_lu@wsu.edu and/or xiaonan_lu@tust.edu.cn.
Footnotes
Published ahead of print 27 June 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Alsberg BK, Kell DB, Goodacre R. 1998. Variable selection in discriminant partial least-squares analysis. Anal. Chem. 70:4126–4133 [DOI] [PubMed] [Google Scholar]
- 2. Bocklitz T, Walter A, Hartmann K, Rösch P, Popp J. 2011. How to pre-process Raman spectra for reliable and stable models? Anal. Chim. Acta 704:47–56 [DOI] [PubMed] [Google Scholar]
- 3. Bosch A, et al. 2008. Fourier transform infrared spectroscopy for rapid identification of nonfermenting gram-negative bacteria isolated from sputum samples from cystic fibrosis patients. J. Clin. Microbiol. 46:2535–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Champion OL, Best EL, Frost JA. 2002. Comparison of pulsed-field gel electrophoresis and amplified fragment length polymorphism techniques for investigating outbreaks of enteritis due to campylobacters. J. Clin. Microbiol. 40:2263–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clark CG, et al. 2012. Comparison of molecular typing methods useful for detecting clusters of Campylobacter jejuni and C. coli isolates through routine surveillance. J. Clin. Microbiol. 50:798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cornelius AJ, Gilpin B, Carter P, Nicol C, On SL. 2010. Comparison of PCR binary typing (P-BIT), a new approach to epidemiological subtyping of Campylobacter jejuni, with serotyping, pulsed-field gel electrophoresis, and multilocus sequence typing methods. Appl. Environ. Microbiol. 76:1533–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daniels JK, Caldwell TP, Christensen KA, Chumanov G. 2006. Monitoring the kinetics of Bacillus subtilis endospore germination via surface-enhanced Raman scattering spectroscopy. Anal. Chem. 78:1724–1729 [DOI] [PubMed] [Google Scholar]
- 8. de Boer P, et al. 2000. Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 38:1940–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. De Gelder J, De Gussem K, Vandenabeele P, Moens L. 2007. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 38:1133–1147 [Google Scholar]
- 10. De Maesschalck R, Jouan-Rimbaud D, Massart DL. 2000. The Mahalanobis distance. Chemom. Intell. Lab. Syst. 50:1–18 [Google Scholar]
- 11. De Maesschalck R, Candolfi A, Massart DL, Heuerding S. 1999. Decision criteria for soft independent modelling of class analogy applied to near infrared data. Chemom. Intell. Lab. Syst. 47:65–77 [Google Scholar]
- 12. Dingle KE, Colles FM, Falush D, Maiden MC. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Djordjevic SP, et al. 2007. Clonal complexes of Campylobacter jejuni identified by multilocus sequence typing are reliably predicted by restriction fragment length polymorphism analyses of the flaA gene. J. Clin. Microbiol. 45:102–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Draminski M, et al. 2008. Monte Carlo feature selection for supervised classification. Bioinformatics 24:110–117 [DOI] [PubMed] [Google Scholar]
- 15. Duim B, Wassenaar TM, Rigter A, Wagenaar J. 1999. High-resolution genotyping of Campylobacter strains isolated from poultry and humans with amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 65:2369–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eurosurveillance Editorial Team 2012. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. Euro Surveill. 17(10)pii:20113 [PubMed] [Google Scholar]
- 17. Fagerquist CK, et al. 2010. Rapid identification of protein biomarkers of Escherichia coli O157:H7 by matrix-assisted laser desorption ionization-time-of-flight-time-of-flight mass spectrometry and top-down proteomics. Anal. Chem. 82:2717–2725 [DOI] [PubMed] [Google Scholar]
- 18. Fagerquist CK, et al. 2005. Genomic and proteomic identification of a DNA-binding protein used in the “fingerprinting” of Campylobacter species and strains by MALDI-TOF-MS protein biomarker analysis. Anal. Chem. 77:4897–4907 [DOI] [PubMed] [Google Scholar]
- 19. Fitzgerald C, et al. 2001. Evaluation of methods for subtyping Campylobacter jejuni during an outbreak involving a food handler. J. Clin. Microbiol. 39:2386–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. French NP, et al. 2009. Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children's playgrounds. Appl. Environ. Microbiol. 75:779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frost JA, Oza AN, Thwaites RT, Rowe B. 1998. Serotyping scheme for Campylobacter jejuni and Campylobacter coli based on direct agglutination of heat-stable antigens. J. Clin. Microbiol. 36:335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goodacre R. 2003. Explanatory analysis of spectroscopic data using machine learning of simple, interpretable rules. Vib. Spectrosc. 32:33–45 [Google Scholar]
- 23. Hamel L, Brown CW. 2012. Bayesian probability approach to feature significance for infrared spectra of bacteria. Appl. Spectrosc. 66:48–59 [Google Scholar]
- 24. Hänninen ML, Pajarre S, Klossner ML, Rautelin H. 1998. Typing of human Campylobacter jejuni isolates in Finland by pulsed-field gel electrophoresis. J. Clin. Microbiol. 36:1787–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hannis JC, et al. 2008. High-resolution genotyping of Campylobacter species by use of PCR and high-throughput mass spectrometry. J. Clin. Microbiol. 46:1220–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hazeleger WC, et al. 1995. Temperature-dependent membrane fatty acid and cell physiology changes in coccoid forms of Campylobacter jejuni. Appl. Environ. Microbiol. 61:2713–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Höller C, Witthuhn D, Janzen-Blunck B. 1998. Effects of low temperatures on growth, structure, and metabolism of Campylobacter coli SP10. Appl. Environ. Microbiol. 64:581–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang WE, Griffiths RI, Thompson IP, Bailey MJ, Whiteley AS. 2004. Raman microscopic analysis of single microbial cells. Anal. Chem. 76:4452–4458 [DOI] [PubMed] [Google Scholar]
- 29. Hughes LA, et al. 2009. Molecular epidemiology and characterization of Campylobacter spp. isolated from wild bird populations in northern England. Appl. Environ. Microbiol. 75:3007–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jarvis RM, Goodacre R. 2004. Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal. Chem. 76:40–47 [DOI] [PubMed] [Google Scholar]
- 32. Jarvis RM, et al. 2008. Surface-enhanced Raman scattering from intracellular and extracellular bacterial locations. Anal. Chem. 80:6741–6746 [DOI] [PubMed] [Google Scholar]
- 33. Kalasinsky KS, et al. 2007. Raman chemical imaging spectroscopy reagentless detection and identification of pathogens: signature development and evaluation. Anal. Chem. 79:2658–2673 [DOI] [PubMed] [Google Scholar]
- 34. Kirschner C, et al. 2001. Classification and identification of enterococci: a comparative phenotypic, genotypic, and vibrational spectroscopic study. J. Clin. Microbiol. 39:1763–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klena JD, et al. 2004. Differentiation of Campylobacter coli, Campylobacter jejuni, Campylobacter lari, and Campylobacter upsaliensis by a multiplex PCR developed from the nucleotide sequence of the lipid A gene lpxA. J. Clin. Microbiol. 42:5549–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuhm AE, Suter D, Felleisen R, Rau J. 2009. Identification of Yersinia enterocolitica at the species and subspecies levels by Fourier transform infrared spectroscopy. Appl. Environ. Microbiol. 75:5809–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kümmerle M, Scherer S, Seiler H. 1998. Rapid and reliable identification of food-borne yeasts by Fourier-transform infrared spectroscopy. Appl. Environ. Microbiol. 64:2207–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lévesque S, Frost E, Arbeit RD, Michaud S. 2008. Multilocus sequence typing of Campylobacter jejuni isolates from humans, chickens, raw milk, and environmental water in Quebec, Canada. J. Clin. Microbiol. 46:3404–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lieber CA, Mahadevan-Jansen A. 2003. Automated method for subtraction of fluorescence from biological Raman spectra. Appl. Spectrosc. 57:1363–1367 [DOI] [PubMed] [Google Scholar]
- 40. Lindstedt BA, Heir E, Vardund T, Melby KK, Kapperud G. 2000. Comparative fingerprinting analysis of Campylobacter jejuni subsp. jejuni strains by amplified-fragment length polymorphism genotyping. J. Clin. Microbiol. 38:3379–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu X, Al-Qadiri HM, Lin M, Rasco BA. 2011. Application of mid-infrared and Raman spectroscopy to the study of bacteria. Food Bioprocess Technol. 4:919–935 [Google Scholar]
- 42. Lu X, et al. 2011. Investigating antibacterial mechanisms of garlic (Allium sativum) concentrate and garlic-derived organosulfur compounds on Campylobacter jejuni by using Fourier transform infrared spectroscopy, Raman spectroscopy, and electron microscopy. Appl. Environ. Microbiol. 77:5257–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lu X, et al. 2011. Infrared and Raman spectroscopic studies of the antimicrobial effects of garlic concentrates and diallyl constituents on foodborne pathogens. Anal. Chem. 83:4137–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lu X, Samuelson DR, Rasco BA, Konkel ME. 1 May 2012. Antimicrobial effect of diallyl sulfide on Campylobacter jejuni biofilms. J. Antimicrob. Chemother. (Epub ahead of print.) doi:10.1093/jac/DKS138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Man SM. 2011. The clinical importance of emerging Campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 8:669–685 [DOI] [PubMed] [Google Scholar]
- 46. Man SM, Kaakoush NO, Mitchell HM. 2011. The role of bacteria and pattern-recognition receptors in Crohn's disease. Nat. Rev. Gastroenterol. Hepatol. 8:152–168 [DOI] [PubMed] [Google Scholar]
- 47. Mandrell RE, et al. 2005. Speciation of Campylobacter coli, C. jejuni, C. helveticus, C. lari, C. sputorum, and C. upsaliensis by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 71:6292–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Maquelin K, Choo-Smith LP, Endtz HP, Bruining HA, Puppels GJ. 2002. Rapid identification of Candida species by confocal Raman microspectroscopy. J. Clin. Microbiol. 40:594–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maquelin K, et al. 2003. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J. Clin. Microbiol. 41:324–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mellmann A, et al. 2004. Sequence-based typing of flaB is a more stable screening tool than typing of flaA for monitoring of Campylobacter populations. J. Clin. Microbiol. 42:4840–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mello C, Ribeiro D, Novaes F, Poppi RJ. 2005. Rapid differentiation among bacteria that cause gastroenteritis by use of low-resolution Raman spectroscopy and PLS discriminant analysis. Anal. Bioanal. Chem. 383:701–706 [DOI] [PubMed] [Google Scholar]
- 52. Miller WG, et al. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43:2315–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miller WG, et al. 2012. Multilocus sequence typing methods for the emerging Campylobacter species C. hyointestinalis, C. lanienae, C. sputorum, C. concisus, and C. curvus. Front. Cell. Infect. Microbiol. 2:45 doi:10.3389/fcimb.2012.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mouwen DJM, Capita R, Alonso-Calleja C, Prieto-Gómez J, Prieto M. 2006. Artificial neural network based identification of Campylobacter species by Fourier transform infrared spectroscopy. J. Microbiol. Methods 67:131–140 [DOI] [PubMed] [Google Scholar]
- 55. Mouwen DJM, Weijtens MJBM, Capita R, Alonso-Calleja C, Prieto M. 2005. Discrimination of enterobacterial repetitive intergenic consensus PCR types of Campylobacter coli and Campylobacter jejuni by Fourier transform infrared spectroscopy. Appl. Environ. Microbiol. 71:4318–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nachamkin I, Ung H, Patton CM. 1996. Analysis of HL and O serotypes of Campylobacter strains by the flagellin gene typing system. J. Clin. Microbiol. 34:277–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Naumann D, Helm D, Labischinski H. 1991. Microbiological characterizations by FT-IR spectroscopy. Nature 351:81–82 [DOI] [PubMed] [Google Scholar]
- 58. Nielsen EM, et al. 2000. Evaluation of phenotypic and genotypic methods for subtyping Campylobacter jejuni isolates from humans, poultry, and cattle. J. Clin. Microbiol. 38:3800–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Oust A, et al. 2006. Fourier transform infrared and Raman spectroscopy for characterization of Listeria monocytogenes strains. Appl. Environ. Microbiol. 72:228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Preisner O, Guiomar R, Machado J, Menezes JC, Lopes JA. 2010. Application of Fourier transform infrared spectroscopy and chemometrics for differentiation of Salmonella enterica serovar Enteritidis phage types. Appl. Environ. Microbiol. 76:3538–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Puppels GJ, et al. 1990. Studying single living cells and chromosomes by confocal Raman microspectroscopy. Nature 347:301–303 [DOI] [PubMed] [Google Scholar]
- 62. Ribot EM, Fitzgerald C, Kubota K, Swaminathan B, Barrett TJ. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ridley AM, Allen VM, Sharma M, Harris JA, Newell DG. 2008. Real-time PCR approach for detection of environmental sources of Campylobacter strains colonizing broiler flocks. Appl. Environ. Microbiol. 74:2492–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Robinson DA. 1981. Infective dose of Campylobacter jejuni in milk. Br. Med. J. (Clin. Res. Ed.) 282:1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rösch P, et al. 2005. Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contamination. Appl. Environ. Microbiol. 71:1626–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ruiz-Palacios GM. 2007. The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis. 44:701–703 [DOI] [PubMed] [Google Scholar]
- 67. Sails AD, Swaminathan B, Fields PI. 2003. Utility of multilocus sequence typing as an epidemiological tool for investigation of outbreaks of gastroenteritis caused by Campylobacter jejuni. J. Clin. Microbiol. 41:4733–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shreve J, Schneider H, Soysal O. 2011. A methodology for comparing classification methods through the assessment of model stability and validity in variable selection. Decis. Support Syst. 52:247–257 [Google Scholar]
- 69. Steinbrueckner B, Ruberg F, Kist M. 2001. Bacterial genetic fingerprinting: a reliable factor in the study of the epidemiology of human Campylobacter enteritis? J. Clin. Microbiol. 39:4155–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stöckel S, et al. 2010. Raman spectroscopy-compatible inactivation method for pathogenic endospores. Appl. Environ. Microbiol. 76:2895–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Taboada EN, et al. 2012. Development and validation of a comparative genomic fingerprinting method for high-resolution genotyping of Campylobacter jejuni. J. Clin. Microbiol. 50:788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. van Belkum A, et al. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl 3):1–46 [DOI] [PubMed] [Google Scholar]
- 73. Volokhov D, Chizhikov V, Chumakov K, Rasooly A. 2003. Microarray-based identification of thermophilic Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. J. Clin. Microbiol. 41:4071–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Walter A, März A, Schumacher W, Rösch P, Popp J. 2011. Towards a fast, high specific and reliable discrimination of bacteria on strain level by means of SERS in a microfluidic device. Lab Chip 11:1013–1021 [DOI] [PubMed] [Google Scholar]
- 75. Wang G, et al. 2002. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus. J. Clin. Microbiol. 40:4744–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wassenaar TM, Newell DGDG. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Willemse-Erix DF, et al. 2009. Optical fingerprinting in bacterial epidemiology: Raman spectroscopy as a real-time typing method. J. Clin. Microbiol. 47:652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Winkler MA, Uher J, Cepa S. 1999. Direct analysis and identification of Helicobacter and Campylobacter species by MALDI-TOF mass spectrometry. Anal. Chem. 71:3416–3419 [DOI] [PubMed] [Google Scholar]
- 79. Wolthuis R, Tjiang GCH, Puppels GJ, Schut TCB. 2006. Estimating the influence of experimental parameters on the prediction error of PLS calibration models based on Raman spectra. J. Raman Spectrosc. 37:447–466 [Google Scholar]
- 80. Yamazaki W, et al. 2008. Development and evaluation of a loop-mediated isothermal amplification assay for rapid and simple detection of Campylobacter jejuni and Campylobacter coli. J. Med. Microbiol. 57:444–451 [DOI] [PubMed] [Google Scholar]
- 81. Yamazaki W, et al. 2009. Comparison of loop-mediated isothermal amplification assay and conventional culture methods for detection of Campylobacter jejuni and Campylobacter coli in naturally contaminated chicken meat samples. Appl. Environ. Microbiol. 75:1597–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Young KT, Davis LM, Dirita VJ. 2007. Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 5:665–679 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.