Abstract
A variable-number tandem-repeat (VNTR) typing assay for the differentiation of Mycobacterium abscessus strains was developed. This assay showed complete reproducibility, locus stability, and a discriminatory power (Hunter-Gaston discriminatory index [HGDI] of 0.9563) that is superior to that of multilocus sequencing. It is a promising tool for the investigation of Mycobacterium abscessus epidemiology and nosocomial outbreaks.
TEXT
Mycobacterium abscessus is a rapidly growing Mycobacterium species that has been increasingly recognized as an opportunistic human pathogen (6, 8) associated with both community-acquired (3) and nosocomial infections (5, 9). This species, belonging to the Mycobacterium chelonae-M. abscessus complex, is known to comprise many phenotypically and genotypically closely related members that can be distinguished by various molecular identification methods. A division into Mycobacterium abscessus sensu stricto, Mycobacterium massiliense, and Mycobacterium bolletii was suggested (1, 2), but phylogenetic studies using rpoB and multilocus sequencing did not support their separation into 3 different species (13). Recently, Leão et al. recommended the creation of two subspecies, Mycobacterium abscessus subspecies bolletii comb. nov. to comprise the combination of Mycobacterium massiliense and Mycobacterium bolletii and Mycobacterium abscessus subspecies abscessus subspecies nov. to replace Mycobacterium abscessus sensu stricto (12).
Variable-number tandem-repeat (VNTR) analysis has been well established for the differentiation of Mycobacterium tuberculosis strains (14) but has not been used as a genotyping tool for epidemiological studies on Mycobacterium abscessus. Recently, Choi et al. used two tandem-repeat loci to distinguish Mycobacterium abscessus from other mycobacterial species and discriminated Mycobacterium abscessus, Mycobacterium massiliense, and Mycobacterium bolletii by different copy numbers of the two VNTR targets (7). In this article, we describe the development and evaluation of a new VNTR assay for the molecular typing of Mycobacterium abscessus strains.
Thirty-eight clinical isolates of acid-fast bacilli from sputum and bronchoalveolar lavage specimens were collected from the Microbiology Diagnostic Laboratory of University Malaya Medical Centre, Kuala Lumpur, from 2009 to 2010. These isolates were identified as Mycobacterium abscessus by a PCR restriction assay as previously described (16) as well as by a reverse line probe hybridization assay (GenoType Mycobacterium CM; Hain Lifescience GmbH, Germany). All strains were kept in Middlebrook 7H9 broth with 15% glycerol at −80°C until required for further testing. Although all of the patients from whom the isolates were obtained were symptomatic with productive cough, only 7 met the minimum criteria set by the American Thoracic Society (11) for the diagnosis of nontuberculous mycobacterial lung disease: 4 with chest radiographic changes and positive growth in their bronchoalveolar lavage specimen and 3 with positive growth in at least 2 different sputum samples collected on separate occasions. None had clinical features of cystic fibrosis, which is a disease rarely diagnosed in Malaysia.
For the development of the Mycobacterium abscessus VNTR assay (MaVA), the genome sequence of M. abscessus ATCC 19977 (GenBank accession no. CU458896.1) was retrieved from the NCBI GenBank database. Tandem repeat (TR) regions in the genome were identified using Tandem Repeat Finder software (4). From 201 TRs found, 18 were chosen based on several criteria: copy number of ≥2, period size of ≥25 bp, and sequence identity of ≥95% (Table 1). PCR primers targeting the regions flanking these 18 TRs were designed using Primer3 software (15).
Table 1.
Primers used in PCR amplification of 18 Mycobacterium abscessus tandem-repeat loci
| Locus | Primer sequence (5′→3′) |
Period size (bp)a | |
|---|---|---|---|
| Forward | Reverse | ||
| TR2 | AATGGGTTCTTACGCAGGTT | GAGGGCACACACCAAAGG | 33 |
| TR28 | GAGACCGAACAACGACTGCT | CCGGTAATGAATTGGGTTGA | 27 |
| TR45 | CGAACTGCCTCGTGATCG | CACTCTCCTGACGCCAGAC | 32 |
| TR86 | GCGCGTATCTTGAACCAATC | GGCGTACTCGTCGTAAAAGG | 33 |
| TR101 | CCAGTGAACGACGCGATAC | ACAGCTTCAGTTGGCATGTG | 33 |
| TR109 | GCGTGTGGGCATATCAATTA | CAATCTCGAGGTGGATGTGA | 32 |
| TR116 | GAACACCTCAACCGCAGTG | ATTAGCGCGATAGGCTCACC | 33 |
| TR131 | CGACAAAGCCTGGAAGGAC | AGGCATCCAGATCCACTGAT | 30 |
| TR137 | ACAAGGTGGTGGTGCAGTC | GGGGAGGTCAAAGAAAGAGG | 33 |
| TR139 | ATCTCGAGCAGACCAGCATC | GTCAACTGGATCCGGAGAAA | 32 |
| TR149 | CTTCGGTCATCAAACAGCTTC | AGGGTGACCTGTGCGATATG | 33 |
| TR150 | ACGTGGCATCTCGATTGG | TCCCACGAGACCATCAGAAT | 30 |
| TR155 | CAACGTGGAATCTCAATACGC | CCCTTGAACAATTCGAGGAA | 31 |
| TR163 | AGGGCAAGGTTGTCGACTC | GCGAAGTCCTCGGCACTC | 30 |
| TR167 | CGGTCGTCACGATTACCAG | GAATAGAGCGTCGTGGTGGA | 33 |
| TR172 | CGTGTAGTCGCTTTGTGCTC | ACTAACCATCCCCCACGAC | 30 |
| TR179 | CCGAACGGTATAGGAGGTCA | TTCGTCATCAACGTGGTCAT | 33 |
| TR200 | ACATGACACGAACCCTCTGG | GCTATCTGGTGAGCGATGGT | 27 |
Period size refers to the size (no. of bp) of one copy of the tandem repeat in each MaVA locus.
For the PCR, DNA was extracted by boiling 200 μl of bacterial suspension at 100°C for 15 min, followed by a quick centrifugation to obtain the supernatant for use as the template DNA. Each locus of the TR was amplified using PCR master mix (Promega) with 20 pmol of the respective primer. The amplification assays were carried out with 35 cycles of denaturation at 94°C for 45 s, annealing at 61°C for 45 s, and extension at 72°C for 45 s. PCR products were analyzed by capillary electrophoresis using QIAxcel capillary electrophoresis system and QIAxcel DNA screening kit (Qiagen, Germany). The results were analyzed by BioCalculator software (Qiagen, Germany). The discriminatory power of the assay was evaluated by calculating the Hunter-Gaston discriminatory index (HGDI) (10). VNTR locus stability was studied by subculturing M. abscessus ATCC 19977 and 3 randomly selected clinical isolates for up to 60 passages in Middlebrook 7H9 broth and typing after every 5 passages. Assay reproducibility was assessed by repeating the typing of M. abscessus ATCC 19977. Primer specificity was tested against a few major Mycobacterium species (M. avium ATCC 25291, M. bovis BCG ATCC 35737, M. chelonae ATCC 19235, M. fortuitum ATCC 6841, M. tuberculosis H37Rv ATCC 27294, M. intracellulare ATCC 13950, M. kansasii ATCC 12478, M. scrofulaceum ATCC 19981, and M. smegmatis ATCC 607).
To further evaluate the MaVA, the same set of 38 isolates was typed by the multilocus sequence assay (MLSA) for Mycobacterium abscessus as described by Macheras et al. (13). The housekeeping genes amplified were argH, cya, glpK, gnd, murC, pta, and purH. The pgm gene used by Macheras et al. was omitted because it was not included in the Institute Pasteur Mycobacterium abscessus multilocus sequence typing (MLST) database (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Myco-abscessus.html) that we used to identify sequence types (STs) for our isolates. Two other small modifications had to be made to obtain positive amplification: (i) to amplify the cya gene, an in-house primer, cyaF (5′-GTGAAGCGGGCTAAAAAG-3′) was designed to replace the original ACF forward primer, and (ii) the annealing temperature was changed to 63°C for the amplification of argH. PCR products were purified and sent to a commercial laboratory for DNA sequencing by Applied Biosystems 3730xl DNA analyzer. The sequences were analyzed by the MLST module from CLC Genomics Workbench 5.0 (CLC bio, Denmark).
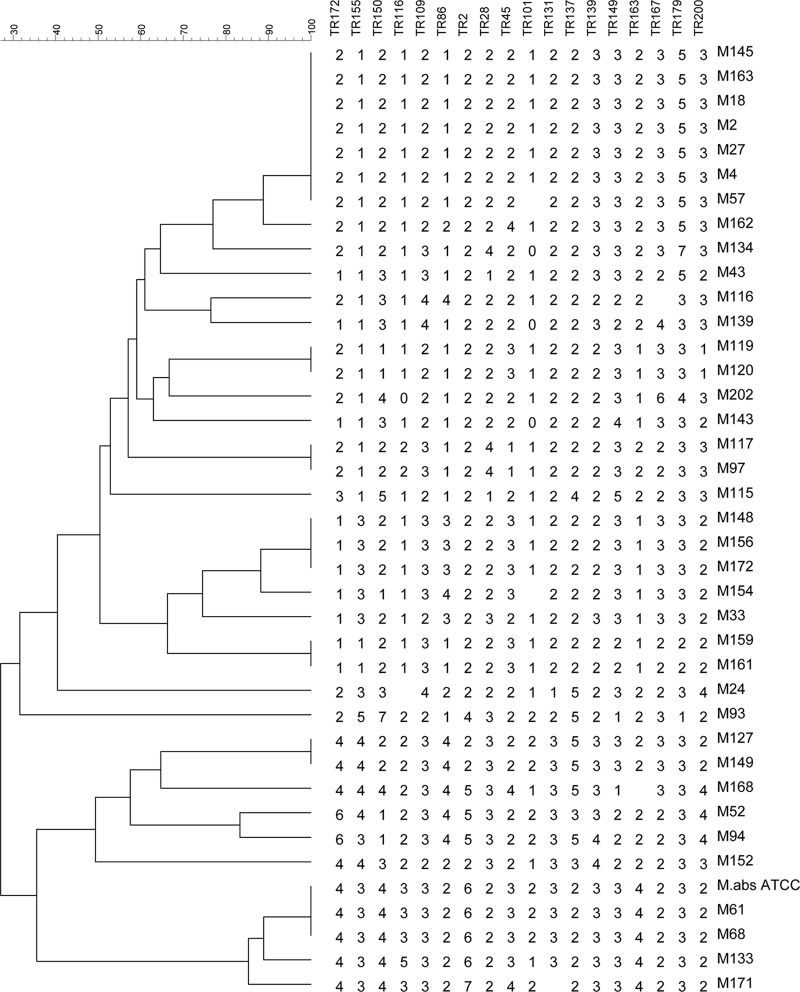
The MaVA results showed 100% typeability, 100% locus stability, and reproducibility of VNTR patterns. In 3 patients who had 2 isolates each, the paired isolates gave identical profiles. For the determination of discriminatory power, only one of each pair of isolates was included, leaving 35 unique isolates for the calculation of HGDI. The overall HGDI for all 18 TRs combined was 0.9563 (ranging from 0.3916 to 0.7109 for each locus), but the same discrimination index was attained with the use of only 6 loci (TR45, TR109, TR116, TR150, TR155, and TR172) (Table 2). The VNTR patterns of all 38 isolates and a reference strain were used to construct a dendrogram using Bionumerics v6.5 (Applied Math) and the unweighted-pair group method using average linkages (UPGMA). The dendrogram identified 25 genotypes and 7 clusters, of which 3 were made up of paired isolates from the same patients (Fig. 1). The 4 clusters of unique strains had 2 to 7 strains each. Two strains, M61 and M68, had a pattern identical to that of the reference Mycobacterium abscessus ATCC 19977.
Table 2.
Combined HGDI for 35 independent M. abscessus isolates
| Locus | No. of isolates with MaVA copy no.: |
HGDIa | Combined HGDIb |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | TR150 to TR155 | TR150 to TR109 | TR150 to TR116 | TR150 to TR45 | TR150 to TR167 | All loci | ||
| TR150 | 4 | 17 | 6 | 6 | 1 | 1 | 0.7109 | 0.8824 | 0.9328 | 0.9412 | 0.9563 | 0.9563 | 0.9563 | ||
| TR172 | 9 | 16 | 1 | 7 | 2 | 0.7008 | |||||||||
| TR155 | 19 | 11 | 4 | 1 | 0.6101 | ||||||||||
| TR109 | 15 | 17 | 3 | 0.5899 | |||||||||||
| TR116 | 1 | 21 | 8 | 3 | 1 | 0.5704 | |||||||||
| TR45 | 2 | 21 | 9 | 3 | 0.5798 | ||||||||||
| TR86 | 18 | 7 | 4 | 6 | 0.6723 | ||||||||||
| TR200 | 1 | 14 | 16 | 4 | 0.6353 | ||||||||||
| TR139 | 14 | 19 | 2 | 0.558 | |||||||||||
| TR167 | 13 | 19 | 1 | 1 | 0.5561 | ||||||||||
| TR179 | 1 | 1 | 22 | 1 | 9 | 1 | 0.5513 | ||||||||
| TR163 | 9 | 21 | 4 | 0.5508 | |||||||||||
| TR28 | 2 | 23 | 7 | 3 | 0.5328 | ||||||||||
| TR101 | 3 | 23 | 7 | 0.4754 | |||||||||||
| TR149 | 2 | 6 | 25 | 1 | 1 | 0.4689 | |||||||||
| TR131 | 1 | 25 | 8 | 0.4153 | |||||||||||
| TR2 | 27 | 1 | 3 | 3 | 1 | 0.4 | |||||||||
| TR137 | 27 | 2 | 1 | 5 | 0.3916 | ||||||||||
HGDI, Hunter-Gaston discriminatory index for each MaVA locus.
The shaded portions show the combined HGDIs for TR150 to TR155, TR150 to TR109, TR150 to TR116, TR150 to TR45, and TR150 to TR167, showing increasing discriminatory power from 0.8824 for 3 loci combined to a maximum of 0.9563 for 6 loci combined (matching the overall HGDI of 0.9563 for all 18 TRs combined, as shown).
Fig 1.
Dendrogram of 38 Mycobacterium abscessus strains based on MaVA results, created by using the Bionumerics v6.5 (Applied Math) UPGMA method. TR, tandem repeat. Strains M119 and M120, M159 and M161, and M127 and M149 were paired isolates from 3 patients. Strains M145 to M161 were identified with Mycobacterium massiliense, strain M24 with Mycobacterium bolletii, and strains M93 to M171 with Mycobacterium abscessus in the phylogenetic tree based on multilocus sequence data (Fig. 2).
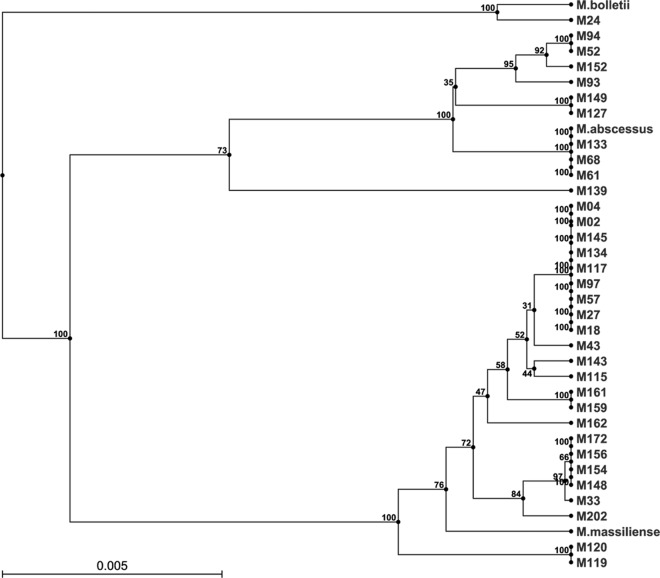
MLSA was done for 34 isolates as 4 isolates could not be retrieved after storage. Seventeen different allelic profiles or sequencetypes (STs) were identified from the data deposited in the Institute Pasteur Mycobacterium abscessus MLST database. All paired isolates from the same patients gave identical STs. With the exclusion of these paired STs, four clusters were identified with 2 to 9 strains in each cluster (data not shown). The discriminatory power appeared to be inferior to that of MaVA as the four clusters could each be further differentiated into different VNTR types. On the other hand, all VNTR clusters had only one ST each and the unique STs were also unique VNTR types. Hence, the MaVA performed better than the MLSA in the strain differentiation of Mycobacterium abscessus. A dendrogram based on the sequences of the 7 housekeeping genes and constructed using the CLC Genomics Workbench 5.0 UPGMA algorithm showed 23 strains clustering with the reference strain of Mycobacterium massiliense, 10 strains with Mycobacterium abscessus, and only one strain with Mycobacterium bolletii (Fig. 2). With one exception (M93), the MaVA phylogenetic grouping in Fig. 1 corresponded exactly to the subspecies grouping identified in the MLS-based phylogenetic tree (Fig. 2). Hence, besides strain differentiation, the MaVA is also potentially useful for the identification of M. abscessus subspecies.
Fig 2.
Dendrogram of 34 Mycobacterium abscessus strains based on multilocus sequencing with 7 housekeeping genes, constructed using the CLC Genomics Workbench 5.0 UPGMA algorithm. The reference strains used are Mycobacterium abscessus ATCC 19977, Mycobacterium massiliense type strain CIP108297, and Mycobacterium bolletii type strain CIP108541.
Molecular typing is a powerful public health tool for the study of disease epidemiology and transmission dynamics. In the hospital setting, it is invaluable for the investigation of outbreaks or pseudo-outbreaks and can provide assistance to clinical management as in the distinction of relapses from reinfection. To date, no entirely satisfactory molecular typing technique has been established for Mycobacterium abscessus. The VNTR assay we developed can potentially fill this void as it appears to be robust, has good discriminatory power, and requires only standard PCR and gel electrophoresis facilities, which have become affordable and available in many diagnostic laboratories. With a 5-h procedure time and a reagent cost of about $6 U.S. dollars for a 6-locus assay, it should be a cost-effective addition to the armamentarium in a routine microbiology laboratory with a molecular diagnostic service.
ACKNOWLEDGMENTS
This study was supported by a research grant (UM.C/625/1/HIR/16) from the University of Malaya, Malaysia.
All authors contributed to study planning, execution, and manuscript writing.
Footnotes
Published ahead of print 3 July 2012
REFERENCES
- 1. Adékambi T, et al. 2004. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J. Clin. Microbiol. 42:5493–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adékambi T, Berger P, Raoult D, Drancourt M. 2006. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56:133–143 [DOI] [PubMed] [Google Scholar]
- 3. Appelgren P, et al. 2008. Late-onset posttraumatic skin and soft-tissue infections caused by rapid-growing mycobacteria in tsunami survivors. Clin. Infect. Dis. 47:e11–e16 [DOI] [PubMed] [Google Scholar]
- 4. Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown-Elliott BA, Wallace RJ. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15:716–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cardoso AM, et al. 2008. Emergence of nosocomial Mycobacterium massiliense infection in Goiás, Brazil. Microbes Infect. 10:1552–1557 [DOI] [PubMed] [Google Scholar]
- 7. Choi G-E, et al. 2011. Efficient differentiation of Mycobacterium abscessus complex isolates to the species level by a novel PCR-based variable-number tandem-repeat assay. J. Clin. Microbiol. 49:1107–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Esther CR, Jr, Henry MM, Molina PL, Leigh MW. 2005. Nontuberculous mycobacterial infection in young children with cystic fibrosis. Pediatr. Pulmonol. 40:39–44 [DOI] [PubMed] [Google Scholar]
- 9. Fisher EJ, Gloster HM. 2005. Infection with Mycobacterium abscessus after Mohs micrographic surgery in an immunocompetent patient. Dermatol. Surg. 31:790–794 [DOI] [PubMed] [Google Scholar]
- 10. Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Griffith DE, et al. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care. Med. 175:367–416 [DOI] [PubMed] [Google Scholar]
- 12. Leão SC, Tortoli E, Euzéby JP, Garcia MJ. 2011. Proposal that Mycobacterium massiliense and Mycobacterium bolletii be united and reclassified as Mycobacterium abscessus subsp. bolletii comb. nov., designation of Mycobacterium abscessus subsp. abscessus subsp. nov. and emended description of Mycobacterium abscessus. Int. J. Syst. Evol. Microbiol. 61:2311–2313 [DOI] [PubMed] [Google Scholar]
- 13. Macheras E, et al. 2011. Multilocus sequence analysis and rpoB sequencing of Mycobacterium abscessus (sensu lato) strains. J. Clin. Microbiol. 49:491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mazars E, et al. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. U. S. A. 98:1901–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 16. Telenti A, et al. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175–178 [DOI] [PMC free article] [PubMed] [Google Scholar]