Abstract
We report a role for Borrelia microti as a cause of relapsing fever in Iran supported by robust epidemiological evidence. The molecular identity of this spirochete and its relation with other relapsing fever borreliae have, until now, been poorly delineated. We analyzed an isolate of B. microti, obtained from Ornithodoros erraticus ticks, by sequencing four loci (16S rRNA, flaB, glpQ, intragenic spacer [IGS]) and comparing these sequences with those of other relapsing fever borreliae. Phylogenetic analysis using concatenated sequences of 16S rRNA, flaB, and glpQ grouped B. microti alongside three members of the African group, B. duttonii, B. recurrentis, and B. crocidurae, which are distinct from B. persica, the most prevalent established cause of tick-borne relapsing fever in Iran. The similarity values for 10 concatenated sequences totaling 2,437 nucleotides ranged from 92.11% to 99.84%, with the highest homologies being between B. duttonii and B. microti and between B. duttonii and B. recurrentis. Furthermore, the more discriminatory IGS sequence analysis corroborated the close similarity (97.76% to 99.56%) between B. microti and B. duttonii. These findings raise the possibility that both species may indeed be the same and further dispel the one-species, one-vector theory that has been the basis for classification of relapsing fever Borrelia for the last 100 years.
INTRODUCTION
Relapsing fever (RF), as the name implies, is characterized by recurrent febrile episodes. The causative agents of the disease, several Borrelia species, are transmitted by soft ticks of the genus Ornithodoros. The only exception is the spirochete Borrelia recurrentis, the etiological agent of epidemic relapsing fever that is transmitted by the human body louse (Pediculus humanus humanus) (4). Tick-borne relapsing fever (TBRF) is a disease endemic to Iran, with 1,415 confirmed cases from the entire country occurring from 1997 to 2006 (16). Borrelia persica accounts for most of these cases in locations where its argasid tick vector, Ornithodoros tholozani, is commonly encountered. Other Borrelia species, including Borrelia microti, Borrelia latyschewii, and Borrelia baltazardii, have also been described in Iran (14, 19). Unlike some TBRF Borrelia, Iranian species are not named after their specific argasid tick vectors; B. persica is transmitted by O. tholozani, B. microti is transmitted by O. erraticus, and B. latyschewii is transmitted by O. tartakowskyi. No argasid tick vector has yet been described for B. baltazardii (14). There are no recent reports on the occurrence of B. latyschewii and B. baltazardii relapsing fevers in Iran; in contrast, the epidemiological evidence for B. microti causing human infections is strong (12, 16). The spirochete was described in 1947 following isolation from Microtus spp. in the Kazeroun area, southern Iran. Later, following reports of sporadic human cases from the same area, the spirochetes were isolated from O. erraticus ticks collected from rodent burrows (12). O. erraticus tick infection rates may reach 50%, and the occurrence of TBRF in areas without O. tholozani ticks suggests a potential role for B. microti as the second TBRF causative agent in Iran (16). Relapsing fever cases in Hormozgan Province, south Iran, which comprise 3.7% of total RF cases reported, are mostly from localities in which O. erraticus ticks predominate (16). The distribution of O. erraticus ticks in Iran covers a vast area from the central to the south of country; these ticks are not inclined to enter domestic dwellings and are commonly found in burrows of wild rodents, including Tatera spp., Meriones spp., and Microtus spp. (12, 13).
Previously, the nomenclature of Borrelia species was based on the cospecies concept, taking into account the association of the Borrelia spp. with a specific tick, its geographical location, and virulence tests using laboratory animals (1). Virulence testing successfully differentiated most Iranian TBRF-causing borreliae; however, some isolates remained ambiguous (1, 13). Molecular analysis of B. persica and its taxonomic position with other Borrelia species has recently been assessed (2, 11, 21); however, there are few data on other Iranian Borrelia species. An initial study designed to provide species-specific PCR for detection of B. persica and B. microti using glpQ and 16S rRNA revealed the divergence between these two Iranian species (18).
Here, we present a more comprehensive report on the molecular characterization of B. microti, a potential agent of TBRF in Iran, using four loci, and demonstrate its close relationship to B. duttonii and B. recurrentis, two well-characterized RF causative agents in Africa.
MATERIALS AND METHODS
An isolate of Borrelia was originated from O. erraticus ticks that were collected from rodent burrows by scraping the walls of tunnels and sieving the contents. Tick collections were made in the Hesark region, south of Karaj City in Alborz Province, Iran, during 2000 (M. Assmar, personal communication). The isolate was maintained via continual passage in outbred white laboratory mice aged 8 to 10 weeks for about 9 successive years. Virulence tests confirmed that this isolate was distinct from B. persica. Unlike B. persica, which causes a high spirochetemia in adult guinea pigs (2, 13), in the present study, inoculation of 200 μl of infected blood samples with spirochetes at concentrations ranging from 250 to 2,750 spirochetes per microliter caused occult infections (not detectable in fresh blood by dark-field microscopy or by examination of Giemsa-stained slides) in adult guinea pigs, with concentrations reaching as high as 2.1 × 107 spirochetes per ml in adult mice 2 to 4 days postinoculation. These infections were followed by 2 to 3 relapses.
Between 200 and 300 μl of blood was collected from adult mice 3 to 4 days following inoculation, when spirochete concentrations reached 1.2 × 107 to 2.1 × 107 per ml of blood. DNA extraction from blood samples was performed using a Miniprep DNA extraction kit (Qiagen, Germany) according to the manufacturer's instructions, and partial sequences of the 16S rRNA, flagellin (flaB), and glycerophosphodiester phosphodiesterase (glpQ) genes as well as an intragenic spacer (IGS) region were amplified using the primers and conditions detailed by others (2, 7, 11, 21). Final reaction volumes of 25 μl contained 20 pmol of each primer, 2 mM MgCl2, 10 mM Tris-HCl, 50 mM KCl, 200 μM deoxynucleoside triphosphates, 1 U of Taq (Roche, Mannheim, Germany), and 3 μl of DNA. A negative control containing all reagents except DNA was included in all amplifications. PCR products were resolved on 1% agarose gels in 1× TAE (Tris-acetate-EDTA) buffer, and amplicons of the expected size were purified using a gel band purification kit (Pharmacia, Piscataway, NJ) according to the manufacturer's recommendations and later sequenced in both directions using the same primers at concentrations of 10 pmol at the SeqLAb laboratory in Germany. The 16S rRNA (1,052-bp), glpQ (667-bp), and flaB (718-bp) sequences of our isolate and those of nine other relapsing fever Borrelia species from GenBank (Table 1) were concatenated using the BioEdit sequence alignment program (version 7.1.3.0) (10) and aligned by using Clustal X software (27). The distances between concatenated sequences were calculated, and phylogenetic trees were constructed by using the Jukes-Cantor option of the neighbor-joining method in a complete deletion procedure using MEGA 4 software (26). The robustness of the topologies was estimated through 1,000 bootstrap replications. The IGS region (526 bp) of B. microti obtained in this study and the IGS regions from three Old World Borrelia species, including B. duttonii, B. recurrentis, and B. crocidurae (Table 2), were compared, and distances and a phylogenetic tree were obtained as described above.
Table 1.
Details of 16S rRNA, glpQ, and flaB gene sequences used in this study
| Species | Gene | GenBank accession no. | Reference |
|---|---|---|---|
| B. duttonii | 16S rRNA | AF107364 | 6 |
| glpQ | DQ346787 | 8 | |
| flaB | DQ346833 | 8 | |
| B. microti | 16S rRNA | JF803950 | This study |
| glpQ | JF825473 | This study | |
| flaB | JF825472 | This study | |
| B. recurrentis | 16S rRNA | AF107367 | 6 |
| glpQ | DQ346777 | 8 | |
| flaB | DQ346814 | 8 | |
| B. crocidurae | 16S rRNA | GU350713 | 28 |
| glpQ | GU357579 | 28 | |
| flaB | GU357619 | 28 | |
| B. persica | 16S rRNA | U42297 | 21 |
| glpQ | HM161661 | 22 | |
| flaB | HM194740 | 22 | |
| B. hispanica | 16S rRNA | GU350708 | 28 |
| glpQ | GU357574 | 28 | |
| flaB | GU357614 | 28 | |
| B. hermsii | 16S rRNA | EU203150 | 24 |
| glpQ | EU194845 | 24 | |
| flaB | EU194843 | 24 | |
| B. coriaceae | 16S rRNA | U42286 | 21 |
| glpQ | AF247158 | 20 | |
| flaB | D82864 | 9 | |
| B. parkeri | 16S rRNA | AY934598 | 23 |
| glpQ | AY934635 | 23 | |
| flab | AY934623 | 23 | |
| B. turicatae | 16S rRNA | AY934610 | 23 |
| glpQ | AY934641 | 23 | |
| flaB | AY934629 | 23 |
Table 2.
Details of IGS sequences used in this study
| Species | Strain designation | GenBank accession no. | Reference |
|---|---|---|---|
| B. duttonii | TzBd6 | GQ401250 | 7 |
| B. duttonii | TzBd19 | GQ401251 | 7 |
| B. recurrentis | A11 | DQ000278 | 25 |
| B. recurrentis | Br12 | GQ401266 | 7 |
| B. recurrentis | SJC13 | GQ401252 | 7 |
| B. duttonii | Bd4 | GQ401270 | 7 |
| B. duttonii | MA/15 | DQ000280 | 25 |
| B. duttonii | MA/18 | DQ000281 | 25 |
| B. recurrentis | A1 | DQ000277 | 25 |
| B. recurrentis | SJC4 | GQ401246 | 7 |
| B. recurrentis | Br5 | GQ401247 | 7 |
| B. recurrentis | Br7 | GQ401278 | 7 |
| B. duttonii | Ly | DQ000279 | 25 |
| B. duttonii | TzBd4 | GQ401257 | 7 |
| B .duttonii | Bd6 | GQ401268 | 7 |
| B. duttonii | Bd10 | GQ401261 | 7 |
| B. duttonii | TzBd14 | GQ401262 | 7 |
| B. microti | IR-1 | JQ436580 | This study |
| B.duttonii | 1120K3 | GU350721 | 28 |
| B. duttonii | CR2A | GU350722 | 28 |
| B. duttonii | WM | DQ000282 | 25 |
| B. duttonii | Tzbd1 | GQ401245 | 7 |
| B. duttonii | Bd9 | GQ401244 | 7 |
| B. duttonii | Bd11 | GQ401243 | 7 |
| B. crocidurae | Achema | GU350723 | 28 |
Nucleotide sequence accession numbers.
The sequence data for the 16S rRNA, glpQ, flaB, and IGS loci were submitted to GenBank with accession numbers JF803950, JF825472, JF825473, and JQ436580, respectively.
RESULTS
A total of 2,437 nucleotides were concatenated over three loci for 10 relapsing fever borreliae, revealing sequence homologies of from 92.11% to 99.84% (Table 3). B. microti showed the greatest homology (99.84%) with B. duttonii. The glpQ sequences of these two species were 100% identical over the entire 667 bp determined. Comparison with other published glpQ nucleotide sequences showed 99.85% homology with B. recurrentis, 97.72% with B. crocidurae, 96.15% with B. hispanica, 87.82% with B. persica, 85.49% with B. hermsii, 84.39% with B. turicatae, and 84.20% with B. parkeri and B. coriaceae. Over 718-bp flaB gene sequences, B. microti differed at three nucleotides (99.56% homology) from both B. duttonii and B. recurrentis; with this sequence, the highest homology (100%) was between B. duttonii and B. recurrentis. Our flaB sequence exhibited 100% identity with another sequence from Iran previously deposited in GenBank (accession no. JF70895; unpublished data). Sequencing of the 16S rRNA over 1,052 bp gave a poor resolution for separating these Borrelia species, with similarity values ranging from 98.17% to 100% and with only a single nucleotide difference separating B. microti from B. duttonii (99.9% homology). It is noteworthy that B. microti shared the same nucleotide difference with all of the eight other species assessed and showed 100% identity with another B. microti 16S rRNA sequence previously submitted from Iran (GenBank accession no. JF681792; unpublished data). High DNA similarities were also observed between B. microti and B. recurrentis at three loci (16S rRNA, 99.62%; glpQ, 99.85%; and flaB, 99.56%). All three genes, the 16S rRNA, flaB, and glpQ genes, clustered B. microti with the Old World group (B. duttonii, B. recurrentis, B. crocidurae, and B. hispanica) yet separate from B. persica; thus, we show only the phylogenetic tree based on concatenated sequences for three loci in B. microti and nine other relapsing fever agents (Fig. 1). The IGS region of B. microti displayed 97.76% to 99.56% identity with B. duttonii, 98.01% identity with B. recurrentis, and 94.58% identity with B. crocidurae. The greatest IGS homologies were seen with B. duttonii strain CR2A, giving 99.56% identity; strain 1120k, giving 99.34% identity; and strains MW, Tzbd1, Bd9, and Bd11, demonstrating 99.12% identity. Strain CR2A was obtained from O. erraticus ticks in West Africa, and strain 1120K was obtained from O. moubata ticks in Congo. The phylogenetic position of B. microti based on its IGS sequence is shown in the supplemental material.
Table 3.
Levels of similarity for three concatenated loci (16S rRNA, flaB, and glpQ) totaling 2,437 bp
| Species | % similarity |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| B. microti | B. duttonii | B. recurrentis | B. crocidurae | B. hispanica | B. persica | B. hermsii | B. turicatae | B. parkeri | B. coriaceae | |
| B. microti | 100 | |||||||||
| B. duttonii | 99.84 | 100 | ||||||||
| B. recurrentis | 99.67 | 99.84 | 100 | |||||||
| B. crocidurae | 98.97 | 99.05 | 98.97 | 100 | ||||||
| B. hispanica | 98.21 | 98.21 | 98.13 | 98.17 | 100 | |||||
| B. persica | 93.46 | 93.51 | 93.42 | 93.60 | 94.11 | 100 | ||||
| B. hermsii | 92.67 | 92.67 | 92.58 | 92.67 | 92.66 | 92.31 | 100 | |||
| B. turicatae | 92.62 | 92.71 | 92.62 | 92.80 | 92.84 | 92.86 | 95.56 | 100 | ||
| B. parkeri | 92.58 | 92.67 | 92.58 | 92.67 | 92.84 | 92.77 | 95.34 | 99.05 | 100 | |
| B. coriaceae | 92.20 | 92.20 | 92.11 | 92.11 | 92.51 | 92.21 | 93.96 | 95.33 | 95.20 | 100 |
Fig 1.
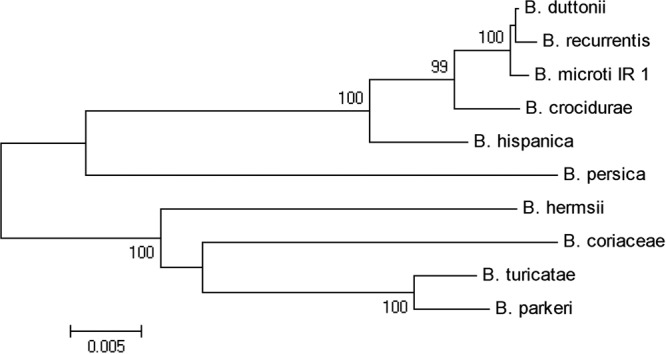
Phylogenetic tree based on concatenation of 16S rRNA, glpQ, and flaB DNA sequences. The scale bar corresponds to a distance of 0.005. The accession numbers of gene sequences used for construction of the tree are shown in Table 1.
DISCUSSION
The aim of this study was to establish the taxonomy of B. microti. Preliminary molecular analysis of B. microti based on partial sequences of the glpQ and 16S rRNA genes (GenBank accession numbers EU914142 and EU914144, respectively) suggested a close association of this species with the African TBRF group rather than B. persica (18). The four targets of B. microti sequenced herein all corroborated homology between our isolate and African species, particularly B. duttonii and B. recurrentis. Concatenated coding genes can be used to draw phylogenetic inference, while IGS, being noncoding, provides a more discriminatory typing tool. Our findings revealed a high degree of homology between B. microti and isolates of B. duttonii and B. recurrentis, while typing of IGS proved to be more discriminatory than typing of other loci but could show overlap between subgroups of B. duttonii and B. recurrentis (7, 25, 28). The greatest nucleotide homology between B. microti and B. duttonii over the loci analyzed (Fig. 1; see Fig. S1 in the supplemental material) raises the possibility that B. microti and B. duttonii may belong to a single species. As full genomic sequences are now becoming available, it might be better to consider this cluster of strains to be variants within a complex rather than individual species (15). This would embrace both the within-species variation disclosed by IGS typing and the between-species diversity reported herein.
Transmission of B. duttonii is associated with living in traditional mud-built dwellings that are commonly infested with O. moubata ticks, while exposure to B. microti, in common with B. crocidurae, appears to correlate with sleeping in the open and proximity to tick-infested burrows; these conditions are more likely to be provided in southern Iran, owing to the long summers and very mild winters. Intriguingly, humans are the only known vertebrate hosts for B. duttonii (and its derived louse-borne variant, B. recurrentis). This was recently challenged through the finding of B. duttonii DNA in blood samples of chickens and swine living close to humans, suggesting potential alternative reservoirs for infection (17); however, the competence of transmission from these hosts remains to be substantiated. In contrast, isolation of B. microti from Microtus spp. suggests that it may be maintained by enzootic cycles that involve rodents and other small mammals (12, 13) and that thus resemble the life cycle of B. crocidurae (3, 5, 29). Interestingly, the isolate of B. duttonii, which showed the greatest homology with our B. microti isolate, was derived from O. erraticus ticks in West Africa (strain CR2A), initially being classified as B. crocidurae rather than B. duttonii (28). It is tempting to speculate whether such as yet undisclosed links hold the key to the evolutionary origins of these isolates.
Differential pathogenicity has historically been used to differentiate species of TBRF. B. duttonii is known to cause severe infection and significant perinatal mortality in East Africa (5), while infection with B. microti presents as a milder form of the disease in Iran (13). Again, this might parallel the milder TBRF seen to be associated with B. crocidurae infection in West Africa (5). Borrelia duttonii isolates Bd9, Bd11, and WM were from spirochetemic febrile patients, and strain TzBd1was from an asymptomatic blood donor in Tanzania (7, 25). Whether the difference in virulence is due to adaptation to different tick vectors is open to question; this can be assessed following cross-vector exchanges of spirochetes derived from O. erraticus and O. moubata. Molecular analysis of isolates of B. microti from various geographical regions together with in situ analysis from either clinical or vector samples will help to provide more accurate insights into the evolutionary origins of B. microti.
Supplementary Material
ACKNOWLEDGMENTS
This work was part of a research project on relapsing fever funded by the Pasteur Institute of Iran (grant no. 500).
We thank M. Assmar for providing the information regarding collection and identification of the ticks and N. Piazak for his technical assistance.
Footnotes
Published ahead of print 20 June 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Assous MV, Wilamowski A. 2009. Relapsing fever borreliosis in Eurasia—forgotten, but certainly not gone! Clin. Microbiol. Infect. 15:407–414 [DOI] [PubMed] [Google Scholar]
- 2. Assous MV, Wilamowski A, Bercovier H, Marva E. 2006. Molecular characterization of tickborne relapsing fever Borrelia, Israel. Emerg. Infect. Dis. 12:1740–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bouattour A, et al. 2010. Borrelia crocidurae infection of Ornithodoros erraticus (Lucas, 1849) ticks in Tunisia. Vector Borne Zoonotic Dis. 10:825–830 [DOI] [PubMed] [Google Scholar]
- 4. Burgdorfer W, York N. 1976. The epidemiology of the relapsing fevers, p 191–200 In Johnson ERC. (ed), Biology of parasitic spirochetes. Academic Press, New York, NY [Google Scholar]
- 5. Cutler SJ, Abdissa A, Trape JF. 2009. New concepts for the old challenge of African relapsing fever borreliosis. Clin. Microbiol. Infect. 15:400–406 [DOI] [PubMed] [Google Scholar]
- 6. Cutler SJ, et al. 1999. Successful in vitro cultivation of Borrelia duttonii and its comparison with Borrelia recurrentis. Int. J. Syst. Bacteriol. 49(Pt 4):1793–1799 [DOI] [PubMed] [Google Scholar]
- 7. Cutler SJ, Bonilla EM, Singh RJ. 2010. Population structure of East African relapsing fever Borrelia spp. Emerg. Infect. Dis. 16:1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cutler SJ, Scott JC, Wright DJM. 2008. Phylogenic relationship between Borrelia recurrentis and Borrelia duttonii. Int. J. Med. Microbiol. 298(Suppl 1):193–20217765656 [Google Scholar]
- 9. Fukunaga M, Okada K, Nakao M, Konishi T, Sato Y. 1996. Phylogenetic analysis of Borrelia species based on flagellin gene sequences and its application for molecular typing of Lyme disease borreliae. Int. J. Syst. Bacteriol. 46:898–905 [DOI] [PubMed] [Google Scholar]
- 10. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 41:95–98 [Google Scholar]
- 11. Halperin T, et al. 2006. Detection of relapsing fever in human blood samples from Israel using PCR targeting the glycerophosphodiester phosphodiesterase (GlpQ) gene. Acta Trop. 98:189–195 [DOI] [PubMed] [Google Scholar]
- 12. Janbakhsh B, Ardelan A. 1977. The nature of sporadic cases of relapsing fever in Kazeroun area, southern Iran. Bull. Soc. Pathol. Exot. Filiales 70:587–589 [PubMed] [Google Scholar]
- 13. Karimi U. 1981. Relapsing fever and its epidemiology. Pasteur Institute of Iran, Tehran, Iran: (In Farsi.) [Google Scholar]
- 14. Karimi Y, Hovind-Hougen K, Birch-Andersen A, Asmar M. 1979. Borrelia persica and B. baltazardi sp. nov.: experimental pathogenicity for some animals and comparison of the ultrastructure. Ann. Microbiol. 130B:157–168 [PubMed] [Google Scholar]
- 15. Lescot M, et al. 2008. The genome of Borrelia recurrentis, the agent of deadly louse-borne relapsing fever, is a degraded subset of tick-borne Borrelia duttonii. PLoS Genet. 4:e1000185 doi:10.1371/journal.pgen.1000185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Masoumi Asl H, et al. 2009. The epidemiology of tick-borne relapsing fever in Iran during 1997-2006. Travel Med. Infect. Dis. 7:160–164 [DOI] [PubMed] [Google Scholar]
- 17. McCall PJ, et al. 2007. Does tick-borne relapsing fever have an animal reservoir in East Africa? Vector Borne Zoonotic Dis. 7:659–666 [DOI] [PubMed] [Google Scholar]
- 18. Oshaghi MA, et al. 2011. Discrimination of relapsing fever Borrelia persica and Borrelia microtti by diagnostic species-specific primers and polymerase chain reaction-restriction fragment length polymorphism. Vector Borne Zoonotic Dis. 11:201–207 [DOI] [PubMed] [Google Scholar]
- 19. Piazak N, Seyyed Rashti MA, Assmar M. 2000. Distribution of Ornithodoros tartakowskyi and its infection rate with Borrelia latyschevii in Serakhs area, Khorasan Province. Iranian J. Public Health 1–4:103–108 (In Farsi.) [Google Scholar]
- 20. Porcella SF, et al. 2000. Serodiagnosis of louse-Borne relapsing fever with glycerophosphodiester phosphodiesterase (GlpQ) from Borrelia recurrentis. J. Clin. Microbiol. 38:3561–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ras NM, et al. 1996. Phylogenesis of relapsing fever Borrelia spp. Int. J. Syst. Bacteriol. 46:859–865 [DOI] [PubMed] [Google Scholar]
- 22. Safdie G, et al. 2010. Molecular characterization of Borrelia persica, the agent of tick borne relapsing fever in Israel and the Palestinian Authority. PLoS One 5:e14105 doi:10.1371/journal.pone.0014105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwan TG, et al. 2005. Phylogenetic analysis of the spirochetes Borrelia parkeri and Borrelia turicatae and the potential for tick-borne relapsing fever in Florida. J. Clin. Microbiol. 43:3851–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwan TG, Schrumpf ME, Hinnebusch BJ, Anderson DE, Jr, Konkel ME. 1996. GlpQ: an antigen for serological discrimination between relapsing fever and Lyme borreliosis. J. Clin. Microbiol. 34:2483–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Scott JC, Wright DJM, Cutler JC. 2005. Typing African relapsing fever spirochetes. Emerg. Infect. Dis. 11:1722–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 27. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Toledo A, et al. 2010. Phylogenetic analysis of a virulent Borrelia species isolated from patients with relapsing fever. J. Clin. Microbiol. 48:2484–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vial L, et al. 2006. Incidence of tick-borne relapsing fever in west Africa: longitudinal study. Lancet 368:37–43 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


