Abstract
Shiga toxin-producing Escherichia coli (STEC) is one of the most important groups of food-borne pathogens, and STEC strains belonging to the serotype O103:H2 can cause diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome in humans. STEC O103:non-H2 strains are also sometimes isolated from human patients, but their genetic characteristics and role in significant human enteric disease are not yet understood. Here, we investigated 17 STEC O103:non-H2 strains, including O103:H11, O103:H25, O103:HUT (UT [untypeable]), and O103:H− (nonmotile) isolated in Japan, and their characteristics were compared to those of STEC O103:H2 and other serotype STEC strains. Sequence analyses of fliC and eae genes revealed that strains possessed any of the following combinations: fliC-H2/eae-epsilon, fliC-H11/eae-beta1, and fliC-H25/eae-theta, where fliC-H2, -H11, and -H25 indicate fliC genes encoding H2, H11, and H25 flagella antigens, respectively, and eae-epsilon, -beta1, and -theta indicate eae genes encoding epsilon, beta1, and theta subclass intimins, respectively. Phylogenetic analysis based on the sequences of seven housekeeping genes demonstrated that the O103:H11/[fliC-H11] and O103:H25/[fliC-H25] strains formed two distinct groups, different from that of the O103:H2/[fliC-H2] strains. Interestingly, a group consisting of O103:H11 strains was closely related to STEC O26:H11, which is recognized as a most important non-O157 serotype, suggesting that the STEC O103:H11 and STEC O26:H11 clones evolved from a common ancestor. The multiplex PCR system for the rapid typing of STEC O103 strains described in the present study may aid clinical and epidemiological studies of the STEC O103:H2, O103:H11, and O103:H25 groups. In addition, our data provide further insights into the high variability of STEC stains with emerging new serotypes.
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) is one of the most important groups of food-borne pathogens worldwide because it can cause gastroenteritis that may be complicated by hemorrhagic colitis or hemolytic-uremic syndrome (HUS) (21). STEC O157:H7 is the main serotype responsible for outbreaks and sporadic cases of hemorrhagic colitis and HUS, but non-O157 serogroups (such as O26, O103, O111, and O145) can also be associated with severe illness in humans (16, 32).
Serotype O103:H2 is one of the most frequently isolated non-O157 STEC. It was first identified as a causative agent of HUS in 1992 (19), and since then both outbreaks and sporadic cases of diarrhea and HUS caused by STEC O103:H2 have been reported worldwide (2, 7, 14, 20, 34). STEC O103 strains expressing H antigens other than H2 are sometimes isolated from human patients. Sporadic cases of human infections with O103:H11 in Japan (37) and Canada (38) have been described previously, and it was recently shown that O103:H25 was responsible for outbreaks of HUS in Norway (35). Thus, STEC O103:non-H2 serotype strains have also become a threat to public health.
Our previous studies (12, 13) demonstrated that E. coli strains with the same O serogroup but different H types sometimes belong to different evolutionary lineages. Furthermore, most STEC strains possess various combinations of virulence genes and exhibit allelic variations of some genes, such as the stx gene on lambda-like prophages and eae (encoding the adhesin intimin) on the locus of enterocyte effacement (LEE) element, which may affect the pathogenicity of strains. Because O103:H2 is a major serotype of STEC, the prevalence and genotypic characteristics of these strains have been investigated in detail; however, little is known about the characteristics of STEC O103:non-H2 strains.
The aim of the present study was to compare STEC O103:non-H2 strains isolated from Japanese patients infected with STEC O103:H2 and other serotype STEC strains to identify their genetic characteristics and to explore their phylogenetic relationships to determine whether pathogenic non-H2 strains share similar molecular characteristics with other, better-characterized O103 strains.
MATERIALS AND METHODS
Bacterial strains.
The relevant characteristics of the 22 STEC O103 strains, including five O103:H2 strains used in the present study, are listed in Table 1. The strains were isolated from patients with gastrointestinal disease (including diarrhea and hemorrhagic colitis) from 2007 to 2011 in various prefectures of Japan. O serogroups of each strain were determined by agglutination tests with the anti-O103 serum (Denka Seiken Co., Ltd., Tokyo, Japan) according to the manufacturer's instructions. H types were determined using a set of anti-H sera purchased from Statens Serum Institut (Statens Serum Institut, Copenhagen, Denmark). Three STEC strains, O145:H− (092372), O121:H19 (071942), and O165:H− (071324), obtained from Osaka Prefectural Institute of Public Health and three different kinds of E. coli serotype strains, O128:H2 (100923), O130:H11 (102608), and O156:H25 (110085), obtained from Fukuoka Institute of Health and Environmental Sciences were used as controls for the phylogenetic analysis and the multiplex PCR assay described below, respectively. In addition, the sequences of five whole-genome-sequenced STEC strains were used: O157:H7 Sakai (accession number BA000007) (10), O26:H11 11368 (AP010953), O103:H2 12009 (AP010958) and O111:H− 11128 (AP010960) (24), and O104:H4 TY-2482 (AFVR01000000) (33).
Table 1.
Characteristics of STEC O103 serogroup strains
| Strain | Source (prefecture in Japan) | Yr | Sporadic/outbreaka | Clinical status or symptom(s)b | H serotypingc | fliC genotyped | stx gene | stx1 integration site | Intimin type | LEE integration site | ehx gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 072676 | Miyazaki | 2007 | Outbreak (5) | Di, Fe, BD | H11 | [H11] | stx1 | NDe | Beta1 | pheUf | + |
| 081163 | Yamaguchi | 2008 | Sporadic | Di, AP, Fe, BD | H11 | [H11] | stx1 | ND | Beta1 | pheU | + |
| 081319 | Kanagawa | 2008 | No data | Di, BD | H11 | [H11] | stx1 | ND | Beta1 | pheU | + |
| 082111 | Miyagi | 2008 | Sporadic | Di, AP | H11 | [H11] | stx1 | torS/Tg | Beta1 | pheU | + |
| 100207 | Miyazaki | 2010 | No data | Di, AP, Fe | H11 | [H11] | stx1 | torS/T | Beta1 | pheU | – |
| 100952 | Fukuoka | 2010 | Outbreak (2) | Di, AP, BD | H11 | [H11] | stx1 | ND | Beta1 | pheU | + |
| 102394 | Gifu | 2010 | Outbreak (2) | Di | H11 | [H11] | stx1 | torS/T | Beta1 | pheU | + |
| 101624 | Saitama | 2010 | Sporadic | Di, AP | HUT | [H11] | stx1 | torS/T | Beta1 | pheU | - |
| 110780 | Miyagi | 2011 | Sporadic | Di, AP, Fe | HUT | [H11] | stx1 | torS/T | Beta1 | pheU | + |
| 071049 | Osaka | 2007 | Sporadic | Di, AP | H- | [H11] | stx1 | ND | Beta1 | pheU | – |
| 080056 | Nagasaki | 2008 | Outbreak (3) | AP | H- | [H11] | stx1 | sbcB | Beta1 | pheU | – |
| 090688 | Yamaguchi | 2009 | Sporadic | Di, Fe | H25 | [H25] | stx1 | ND | Theta | ND | + |
| 070373 | Miyagi | 2007 | Sporadic | Di, AP, BD | HUT | [H25] | stx1 | ND | Theta | ND | + |
| 080984 | Yamagata | 2008 | Outbreak (2) | Di, BD | HUT | [H25] | stx1 | ND | Theta | ND | + |
| 082332 | Mie | 2008 | Sporadic | Di, AP | HUT | [H25] | stx1 | ND | Theta | ND | + |
| 080455 | Nara | 2008 | Sporadic | Di, AP, Fe, BD | HUT | [H25] | stx1 | ND | Theta | ND | + |
| 082589 | Yamagata | 2008 | Sporadic | Di, AP, Vo, BD | H2 | [H2] | stx1 | torS/T | Epsilon | ND | + |
| 092412 | Nagano | 2009 | Sporadic | AP, BD | H2 | [H2] | stx1 | torS/T | Epsilon | ND | + |
| 071556 | Fukuoka | 2007 | Outbreak (2) | Di, AP, BD | H2 | [H2] | stx1 | torS/T | Epsilon | ND | + |
| 111471 | Kagoshima | 2011 | Sporadic | BD | H2 | [H2] | stx1 | torS/T | Epsilon | ND | + |
| 111336 | Miyagi | 2011 | Sporadic | Di, AP | H2 | [H2] | stx1 | torS/T | Epsilon | ND | + |
| 111155 | Kagoshima | 2011 | Outbreak (4) | BD | HUT | [H2] | stx1 | torS/T | Epsilon | ND | + |
The numbers in parentheses indicate the numbers of confirmed patients (including asymptomatic carrier) in each outbreak.
Di, diarrhea; AP, abdominal pain; Fe, fever; Vo, vomiting; BD, bloody diarrhea.
HUT, untypeable; H−, nonmotile.
Types listed in square brackets were determined by sequence comparison of the fliC gene.
ND, not determined.
tRNA gene.
torS/T, torS-torT intergenic region.
PCR analysis of virulence markers.
The following 13 pathotype-associated genes were detected by PCR: stx1 and stx2 (4), ehxA (encoding enterohemolysin) (27) and eae (26), associated with enterohemorrhagic E. coli (EHEC) and/or enteropathogenic E. coli (EPEC); bfpA (encoding bundle-forming pilus) (9), associated with typical EPEC; elt (encoding heat-labile enterotoxin) and est (heat-stable enterotoxin) (39), associated with enterotoxigenic E. coli; astA (encoding heat-stable enterotoxin EAST1) (44) and aggR (encoding transcriptional activator of aggregative adherence fimbriae I expression) (6), associated with enteroaggregative E. coli (EAEC); ipaH (encoding invasive plasmid antigen H) (36), associated with enteroinvasive E. coli; cdtV (encoding cytolethal distending toxin [CDT] V, a member of the CDT family, associated with tissue damage [3]) (5); subAB (encoding subtilase cytotoxin) (22); and saa (encoding STEC autoagglutinating adhesin) (28). All PCRs were performed according to the protocols described previously.
Sequencing of fliC, eae, and seven housekeeping genes.
The H type was genetically determined by sequence comparison of the fliC gene. The entire coding region of fliC was amplified and sequenced using the primers F-FLIC-out (5′-TTAAATCCAGACCTGACCCGA-3′) and R-FLIC-out (5′-CCACAGCGAGTGTTTATCCAT-3′), and an additional primer F-FLIC1 (8) was used for internal sequencing of fliC. The entire coding region of eae was amplified and sequenced using two primer pairs: cesT-F9 and eae-R3 for N-terminal protein, and eae-F1 and escD-R1 for C-terminal protein (11). The internal regions of the seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) were PCR amplified and sequenced using the primers and protocol specified on the E. coli multilocus sequence typing (MLST) website (http://mlst.ucc.ie/mlst/dbs/ecoli).
MLSA.
The concatenated sequences (3,423 bp) of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) from O103 strains were used for multilocus sequence analysis (MLSA). In addition, the sequences of five whole-genome-sequenced STEC strains (O157:H7, O26:H11, O103:H2, O111:H−, and O104:H4) and three well-characterized STEC serotype strains (O121:H19, O165:H−, and O145:H−) were included in the analysis. E. coli reference strains, the ECOR collection, were also used for MLSA. Multiple alignments of sequences were constructed by using the CLUSTAL W program (41) in the MEGA4 software (40), and then neighbor-joining trees were generated by using the Tamura-Nei model. A bootstrap test with 1,000 replicates was used to estimate the confidence of the branching patterns of the tree. Sequences of the ECOR collection for MLSA and the sequence type (ST) of the STEC O103 strains were obtained from the E. coli MLST database (http://mlst.ucc.ie/mlst/dbs/ecoli).
Determination of Stx1 phage and LEE integration sites.
Thus far, seven genomic loci (torS-torT intergenic region, wrbA, yehV, prfC, sbcB, argW-tRNA, and ssrA-tmRNA) have been identified as integration sites of stx1-containing bacteriophages (Stx1 phages) (23). To determine integration sites for Stx1 phages on the chromosome, a universal PCR primer (Pstx1A-F, 5′-AAACCGCCCTTCCTCTGGAT-3′) targeted to the stx1A gene on the prophage and seven primers (Pstx1_tosRS-R, 5′-TTCAGGCTTTGTGCGGTGAG-3′; Pstx1_wrbA-R, 5′-CTCTCTGTTAACGGCGCTGGAT-3′; Pstx1_yehV-R, 5′-TGCCAGCGTGACAGAAGTTG-3′; Pstx1_prfC-R, 5′-ATCGGCATCATCACCAACGG-3′; Pstx1_sbcB-R, 5′-GCGGAACATCAATCAACGCCA-3′; Pstx1_argW-R, 5′-TCAACTTCTGGTTGGTCTCGC-3′; and Pstx1_ssrA-R, 5′-TCCTACCCGTACCCGCAAGTT-3′) targeted to the outside of each prophage region were designed on the basis of the genome sequences of the STEC strains O157:H7 Sakai, O26:H11 11368, O103:H2 12009, and O111:H− 11128. In addition, integration sites (pheV, pheU, and selC tRNA gene loci) of LEE elements were screened by primers described elsewhere (25). Long-range PCR screenings were performed by using TaKaRa LA Taq polymerase (TaKaRa Bio, Inc., Ohtsu, Japan).
Sequence analysis of the O103-antigen biosynthesis gene cluster and its flanking region.
The O103-antigen biosynthesis gene cluster and its flanking regions were amplified using a PCR primer pair, O55re-1F and O55re-1R (12). Each PCR product was sequenced by the shotgun method. Sequences were aligned using Sequencher software (v4.9; Gene Code Corp., Michigan), and sequence comparisons were performed by using in silico molecular cloning software (In Silico Biology, Yokohama, Japan).
Multiplex PCR assay.
The primers used for multiplex PCR and the lengths of the amplicons are listed in Table 2. A universal primer designed on the basis of the N-terminal sequences of fliC and specific primers designed on the basis of the highly diversified sequences (middle part) of each fliC gene were used. In addition, primers targeting the wzy (O103) gene were also used for control amplification. Multiplex PCR was performed with a 15-μl reaction mixture containing 10 ng of genomic DNA, 1× Kapa Taq buffer, each deoxynucleoside triphosphate at 0.3 mM, 2.5 mM MgCl2, 0.25 μM fliC_univ_F primer, 0.25 μM fliC_H2_R primer, 0.25 μM fliC_H25_R primer, 0.38 μM fliC_H11_R primer, 0.5 μM O103_wzy_F primer, 0.5 μM O103_wzy_R primer, and 0.4 U of Kapa Taq DNA polymerase (Kapa Biosystems, Woburn, MA). The thermocycling condition was 25 cycles of 94°C for 20 s, 57°C for 20 s, and 72°C for 30 s. The PCR products (2 μl) were electrophoresed in 1.5% in agarose gels in 0.5× TBE (25 mM Tris borate, 0.5 mM EDTA) and photographed under UV light after the gel was stained with ethidium bromide.
Table 2.
Primers used for multiplex PCR
| Target | Primer | Sequence (5′–3′) | Observed amplicon size (bp) |
|---|---|---|---|
| Universal forward primer for fliC | fliC_univ_F | ATGGCACAAGTCATTAATAC | |
| fliC (H11) | fliC_H11_R | TATTCTTAGCCGCTGCTGC | 755 |
| fliC (H2) | fliC_H2_R | TATCCTGATCAGAAGCCAGCA | 417 |
| fliC (H25) | fliC_H25_R | TGCGGGATAGATGTGATAGCA | 559 |
| wzy (O103) | O103_wzy_F | CTCTTGCTGCTATGAGCTTTG | 297 |
| O103_wzy_R | GCGGGGTCTTGTCATTTAAT |
Nucleotide sequence accession number.
The sequences of the two O103-antigen gene clusters from O103:H25 and O103:H11, and of the adk, fumC, gyrB, icd, mdh, purA, and recA genes were deposited in GenBank/EMBL/DDBJ database under accession numbers AB704860, AB704861, and AB704965 to AB705139, respectively.
RESULTS
Characterization of O103:non-H2 strains.
Seventeen STEC O103:non-H2 strains isolated from patients in Japan were investigated (Table 1). Six strains were isolated from disease outbreaks, nine were from sporadic cases, and two were from cases for which no information was available. Seven strains were classified as H11 type and one as H25 type by using agglutination assays. Seven additional strains were classified as HUT, because their H types could not be determined due to no or low agglutination, or because aggregation was observed for multiple anti-H antisera. The remaining two strains showed no motility.
fliC analysis.
The sequence analysis of fliC from all O103 strains examined showed that the amino acid sequences (487 amino acids [aa]) of two HUT (101624 and 110780) and two H− (071049 and 080056) strains were identical to those of H11-expressing O103 strains. The sequences (443 aa) of four HUT strains (070373, 080984, 082332, and 080455) were identical to that of H25-expressing O103 strain 090688, except for one amino acid difference in 080984. In addition, the sequence (494 aa) of OUT strain 111155 was identical to that of H2-expressing O103 strains and that of the fully sequenced O103:H2 strain. These results indicated that all of the control and experimental O103 strains were one of the following three H types: H2/[fliC-H2], H11/[fliC-H11], or H25/[fliC-H25] (Table 1). By comparison, the sequence identities of FliC between H2 and H11, between H11 and H25 (090688), and between H25 and H2 were 55.4, 50.4, and 49.4%, respectively.
PCR screening of virulence-related genes.
PCR-based screening for E. coli virulence-related genes showed that all O103 strains possessed stx1 and eae and that 18 of the strains examined carried ehx (Table 1). The remaining 10 genes (stx2, bfpA, elt, est, astA, aggR, ipaH, cdtV, subAB, and saa) included in the screen were absent from all strains examined.
eae typing.
The results of sequence analysis of eae from all O103 strains are shown in Table 1. The amino acid sequences of the H2/[H2] and H11/[H11] strains (948 and 939 aa, respectively) were identical to those of O103:H2 strain 12009 and O26:H11 strain 11368, respectively, indicating that H2/[H2] and H11/[H11] strains possess the eae genes encoding epsilon and beta1 subclass intimins (eae-epsilon and eae-beta1, respectively). In addition, the sequences (935 aa) of H25/[H25] strains were identical to that of O111:H− strain 11128, indicating that H25/[H25] strains possess eae-theta.
Integration site of Stx1 phages and LEE elements.
Long-range PCR screening targeting seven alternative integration sites of Stx1 phages was performed. All six H2/[H2] and five H11/[H11] strains were found to contain the Stx1 phage in the torS-torT intergenic region, and one H11/[H11] strain contained it in the sbcB locus (Table 1). The integration site in the other strains was not determined by these methods (Table 1). PCR screening analysis for three alternative integration sites of LEE showed that all H11/[H11] strains possess LEE elements in the pheU locus. The integration site of LEE in H2/[H2] and H25/[H25] was not determined (Table 1).
Phylogenetic relationship of O103 strains.
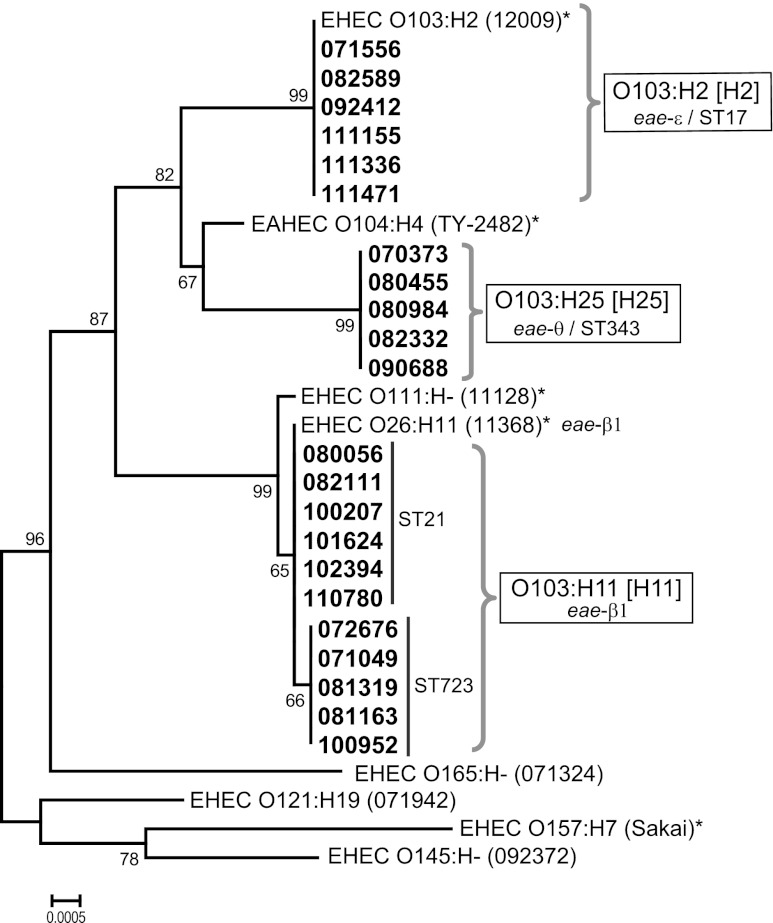
We analyzed the phylogenetic relationships among O103 strains and well-known strains from the STEC serotype collection. As shown in Fig. 1, the O103:H11/[H11] and O103:H25/[H25] strains formed two distinct groups, different from that of O103:H2/[H2] strains. The O103:H11/[H11] strains formed two groups with one nucleotide difference and were closely related to STEC O26:H11, while the O103:H25/[H25] strains were associated with Shiga toxin-producing EAEC O104:H4. The O103:H2/[H2] strains belonged to ST17 and the O103:H25/[H25] strains to ST343 (Fig. 1). One group of O103:H11/[H11] strains belonged to ST21, which was associated with O26:H11, and the other belonged to ST723 (Fig. 1). Compared to the sequences of the ECOR collection strains, three O103 groups belonged to the B1 phylogroup (data not shown). Pulsed-field gel electrophoresis pattern analysis revealed diverse populations of STEC O103:H11 and O103:H25 strains. For the O103:H11/[H11] classification, however, two strains (071049 and 101624) and three strains (082111, 100207, and 110780) differed by fewer than four bands within each of these two groups, indicating that they were genetically closely related (data not shown).
Fig 1.
Phylogenetic relationships of O103-serogroup strains among eight well-characterized STEC serotypes strains. The phylogenetic tree was constructed on the basis of the concatenated sequences of seven housekeeping genes by using the neighbor-joining algorithm. Bootstrap analysis was performed with 1,000 replicates.
Sequences of the O103-antigen biosynthesis gene cluster.
To gain more information about the genetic similarity of the O103-antigen encoding region among the three lineage groups, the sequences of the O103-antigen gene cluster of a representative strain from each lineage (072676 for O103:H11/[H11] and 080984 for O103:H25/[H25]) were determined and compared to that of STEC O103:H2 12009. The gene organization of the O103-antigen gene cluster was identical among the three strains, and their sequences were highly conserved except for three genes (ugd, rmlB, and galF) in the O103:[H25] strains (Fig. 2). In addition, the sequences of the O-antigen gene cluster and its flanking regions of O103:H11 were compared to those of O26:H11, which is closely related to O103:H11. As shown in Fig. 2, in addition to the flanking genes, three upstream genes (wzz, ugd, and gnd) and two downstream genes (rmlB and rmlF) in the O-antigen gene cluster were conserved between the O103 and O26 strains (94.1 to 99.7% identity).
Fig 2.
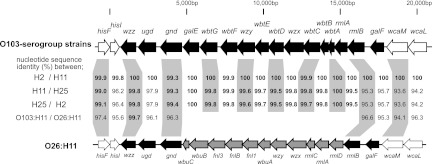
Comparison of O103-antigen biosynthesis gene clusters and their flanking regions. The genetic organization of the O103-antigen gene cluster and its flanking regions from O103 serotype strains is shown at top and that from STEC O26:H11 11368 (AP010953) is shown at the bottom. Genes associated with O-antigen biosynthesis are indicated by black arrows, and flanking genes are indicated by white arrows. O26-specific genes are indicated by gray arrows. Nucleotide sequence identities (%) between O103:H2 and O103:H11, between O103:H11 and O103:H25, and between O103:H25 and O103:H2 are shown in the middle. In addition, sequence identities between O103:H11 and O26:H11 are also shown.
Multiplex PCR.
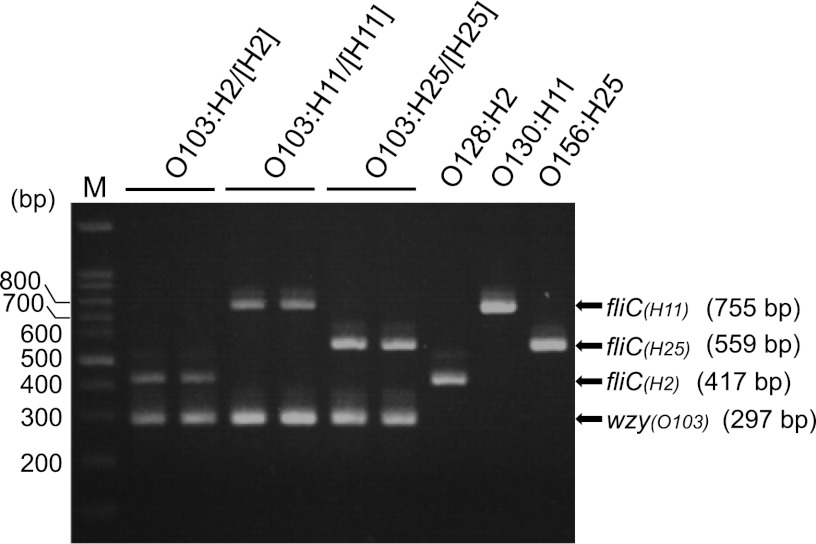
We developed a multiplex PCR system for classifying the pathogenic O103 strains that were confirmed to possess the stx and/or eae gene(s). Because fliC alleles encoding each of the H2, H11, and H25 antigens were lineage-specific among the STEC O103 strains (Fig. 1), this multiplex PCR method targeting fliC provided a rapid way to classify STEC O103 strains into three clonal groups. On the basis of the sequences of the O103-antigen gene clusters obtained in the present study, primers targeting the wzy (O103) gene were also designed for control amplification. The validity of the multiplex PCR system was confirmed using 22 STEC O103 control strains and three different H-antigen serotype control strains (O128:H2, O130:H11, and O156:H25). All PCR products matched the predicted sizes of the fliC (H2) (417 bp), fliC (H11) (755 bp), fliC (H25) (559 bp), and wzy (O103) (297 bp) genes, and the expected band patterns (Fig. 3).
Fig 3.
Multiplex PCR products of STEC O103 strains representing three groups. The strains used were 082589 and 111155 for O103:H2/[H2], 072676 and 071049 for O103:H11/[H11], 090688 and 082332 for O103:H25/[H25] and three non-O103 strains expressing either H2, H11, or H25 antigen. M, 100-bp DNA ladder markers.
DISCUSSION
Although STEC O103:H2, O26:H11 and O111:H− strains belong to the E. coli B1 phylogroup and are closely related, especially O26:H11 and O111:H-, genomic analyses support the hypothesis that independent acquisition of Stx phages, LEE elements and many other virulence-related genes has driven the emergence of each STEC (24).
In the present study, 17 STEC O103-serogroup strains were classified into three distinct clonal groups coincident with variations in their fliC and eae genes (Fig, 1). A key finding was that strains belonging to the O103:H11/[H11] group were closely related to STEC O26:H11, suggesting that the STEC O103:H11 and STEC O26:H11 clones evolved from a common ancestor with one or more exchange(s) of the region encoding O-antigen biosynthesis. It is known that EHEC O157:H7 emerged from an O55:H7-like EPEC ancestor by specific events including acquisition of the O157-antigen biosynthesis gene cluster by horizontal gene transfer (43), and a previous genome-wide sequence comparison showed that a large region of up to 130 kb including the O-antigen gene cluster was replaced by the result of recombination events (17). From the present sequence comparison of the O-antigen gene cluster and its flanking region between STEC O103:H11 and STEC O26:H11, a level of sequence conservation comparable to that of housekeeping genes (representing the backbone of the chromosome and nearly 100% conserved on the basis of the sequences of genes for MLSA) was not observed in the neighboring genes except for wzz (99.7%), suggesting that replacement of the region containing the O-antigen gene cluster occurred across a larger region.
Beutin et al. (1) demonstrated considerable diversity among STEC/EPEC O103 strains, which was investigated by MLST and eae typing. O103:H2 strains were predominantly positive for eae-epsilon, whereas an O103:H11 strain, whose MLST profile was different from those of the O103:H2 strains, was positive for eae-beta1. Ogura et al. (23) demonstrated that LEE elements are generally found at specific loci within the clonal groups and, among all six STEC O26:H11/H− strains tested, LEE elements with eae-beta1 were located at the pheU-tRNA locus. The O103:H11/[H11] strains tested also carried LEE elements with eae-beta1 at the pheU locus (Table 1), suggesting that, after acquiring a LEE element with eae-beta1 in the pheU locus, a LEE-positive common ancestor divided into the two clonal groups of STEC O26:H11 and O103:H11. On the other hand, the presence and location of Stx phages are known to be unsteady even within a clonal group. Stx1 phages in O157:H7 strains have been found in at least three different loci: sbcB, yehV, and argW (23). It is known that STEC O26:H11 strains carried the Stx1 phage at the wrbA locus (23); in contrast, five of the O103:H11/[H11] strains studied here carried the Stx1 phage in the torS-torT intergenic region, which was previously found to be an integration site in STEC O103:H2 (23), and one O103:H11/[H11] strain carried the Stx1 phage at the sbcB locus, which was found to be an integration site in O157:H7 (23). The remaining 10 strains characterized here had unknown integration sites. These results suggested that the Stx1 phage has integrated into different sites of the genome even among closely related strains, and it is not clear when the lineages associated with STEC O26:H11 and O103:H11 acquired the Stx1 phage(s).
A few cases of infection associated with STEC O103:H25 have been reported (30, 31, 42), and most isolates were found to be Stx1-producing strains. In 2006, however, an outbreak caused by Stx2-producing O103:H25 strains in Norway was reported (35). Among the 17 cases, 10 were children who developed HUS. The sequences of seven housekeeping genes for MLSA from Stx2-producing O103:H25 NVH-734 (GenBank accession no. AGSG01000000) (15) were identical to those of the Stx1-producing O103:H25 strains that we investigated, indicating that they belonged to the same clonal group (data not shown).
Although serotypes O103:H11 and O103:H25 are rare causes of EHEC disease, these serotypes used here were obtained from patients with diarrhea and hemorrhagic colitis. Because these O103 strains were the only bacteria known to cause these conditions, it is likely that the isolated strains caused these conditions. Thus, these serotype strains could be a threat to human health, and caution should be exercised around them. The clinical isolates characterized here were geographically and temporally dispersed, suggesting that these pathogens are widespread throughout Japan. Precise O/H serotyping of STEC strains isolated from human and food sources is required for validation. In many cases, the O-serogroup classification of STEC strains provides enough information to presume its clonal relatedness to well-known O-serogroup strains. Our STEC O103 clinical isolates, however, belonged to three distinct clonal groups. Despite the fact that these strains had diverse genetic backgrounds, they all carried the EHEC marker genes stx1, eae, and/or ehx. Although the H type can be a useful phenotypic marker for classifying strains, we could not determine the H type of some O103 isolates, because of unclear agglutination or lack of bacterial motility. As many researchers have shown before (8, 18, 29), sequence variation in the fliC gene could be a proxy for these agglutination tests. In the present study, on the basis of sequence variation in fliC genes, we developed a multiplex PCR method for such classification of STEC O103 strains. The PCR-based methodologies described in the present study may be utilized to aid clinical and epidemiological studies of the STEC O103 serogroup strains.
In conclusion, we demonstrated that STEC O103 from patients formed three distinct groups, and the group comprising O103:H11 strains was closely related to STEC O26:H11. These findings suggest that the STEC O103:H11 and O26:H11 clones evolved from a common ancestor and provide further insights into the high variability of STEC strains with emerging new serotypes.
ACKNOWLEDGMENTS
We thank Atsuko Akiyoshi and Hitomi Satou for technical assistance and the Sendai City Institute of Public Health and Miyagino Public Health Department for contributing bacterial strains.
This study was supported by Grants-in-Aid for Young Scientists from the Japan Society for the Promotion of Science (A.I. and S.I.); a Health Labor Sciences Research Grant from the Ministry of Health, Labor, and Welfare, Japan; a Scientific Research Grant on Priority Areas from the University of Miyazaki; and the Program to Disseminate Tenure Tracking System from the Japanese Ministry of Education, Culture, Sports, Science, and Technology.
Members of the EHEC Study Group include the following: Kazumi Horikawa, Yoshiki Etoh, and Shachiko Ichihara, Fukuoka Institute of Health and Environmental Sciences; Junji Seto, Yamagata Prefectural Institute of Public Health; Shuji Yoshino, Miyazaki Prefectural Institute for Public Health and Environment; Yutaka Shiraki, Gifu Prefectural Research Institute for Health and Environmental Sciences; Kiyoshi Tominaga, Yamaguchi Prefectural Institute of Public Health and Environment; Takashi Hatakeyama, Miyagi Prefectural Institute of Public Health and Environmental; Madoka Hamada, Kagoshima Prefectural Institute for Environmental Research and Public Health; Hiromi Nakamura, Osaka City Institute of Public Health and Environmental Sciences; Sumie Kohori, Saitama City Institute of Health Science and Research; Yuji Migita, Nagasaki Prefectural Institute for Environmental Research and Public Health; Hitomi Kasahara, Nagano Environmental Conservation Research Institute; Misao Hashida, Nara Prefectural Institute for Hygiene and Environment; Yuki Nagai, Mie Prefecture Health and Environment Research Institute; members of the Sagamihara City Laboratory of Public Health; and Kazuko Seto, Osaka Prefectural Institute of Public Health.
Footnotes
Published ahead of print 20 June 2012
REFERENCES
- 1. Beutin L, Kaulfuss S, Herold S, Oswald E, Schmidt H. 2005. Genetic analysis of enteropathogenic and enterohemorrhagic Escherichia coli serogroup O103 strains by molecular typing of virulence and housekeeping genes and pulsed-field gel electrophoresis. J. Clin. Microbiol. 43:1552–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beutin L, Krause G, Zimmermann S, Kaulfuss S, Gleier K. 2004. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 42:1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bielaszewska M, Sinha B, Kuczius T, Karch H. 2005. Cytolethal distending toxin from Shiga toxin-producing Escherichia coli O157 causes irreversible G2/M arrest, inhibition of proliferation, and death of human endothelial cells. Infect. Immun. 73:552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cebula TA, Payne WL, Feng P. 1995. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J. Clin. Microbiol. 33:248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cergole-Novella MC, et al. 2007. Distribution of virulence profiles related to new toxins and putative adhesins in Shiga toxin-producing Escherichia coli isolated from diverse sources in Brazil. FEMS Microbiol. Lett. 274:329–334 [DOI] [PubMed] [Google Scholar]
- 6. Czeczulin JR, Whittam TS, Henderson IR, Navarro-Garcia F, Nataro JP. 1999. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 67:2692–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eklund M, Scheutz F, Siitonen A. 2001. Clinical isolates of non-O157 Shiga toxin-producing Escherichia coli: serotypes, virulence characteristics, and molecular profiles of strains of the same serotype. J. Clin. Microbiol. 39:2829–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fields PI, et al. 1997. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for identification of E. coli O157:H7 and O157:NM. J. Clin. Microbiol. 35:1066–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gunzburg ST, Tornieporth NG, Riley LW. 1995. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J. Clin. Microbiol. 33:1375–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hayashi T, et al. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11–22 [DOI] [PubMed] [Google Scholar]
- 11. Hyma KE, et al. 2005. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J. Bacteriol. 187:619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iguchi A, Ooka T, Ogura Y, Asadulghani Nakayama K, Frankel G, Hayashi T. 2008. Genomic comparison of the O-antigen biosynthesis gene clusters of Escherichia coli O55 strains belonging to three distinct lineages. Microbiology 154:559–570 [DOI] [PubMed] [Google Scholar]
- 13. Iguchi A, et al. 2011. Wide distribution of O157-antigen biosynthesis gene clusters in Escherichia coli. PLoS One 6:e23250 doi:10.1371/journal.pone.0023250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kappeli U, Hachler H, Giezendanner N, Beutin L, Stephan R. 2011. Human infections with non-O157 Shiga toxin-producing Escherichia coli, Switzerland, 2000–2009. Emerg. Infect. Dis. 17:180–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. L'Abee-Lund TM, et al. 2012. The highly virulent 2006 Norwegian EHEC O103:H25 outbreak strain is related to the 2011 German O104:H4 outbreak strain. PLoS One 7:e31413 doi:10.1371/journal.pone.0031413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lathrop S, Edge K, Bareta J. 2009. Shiga toxin-producing Escherichia coli, New Mexico, USA, 2004–2007. Emerg. Infect. Dis. 15:1289–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leopold SR, et al. 2009. A precise reconstruction of the emergence and constrained radiations of Escherichia coli O157 portrayed by backbone concatenomic analysis. Proc. Natl. Acad. Sci. U. S. A. 106:8713–8718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Madic J, et al. 2010. Simplex and multiplex real-time PCR assays for the detection of flagellar (H-antigen) fliC alleles and intimin (eae) variants associated with enterohaemorrhagic Escherichia coli (EHEC) serotypes O26:H11, O103:H2, O111:H8, O145:H28, and O157:H7. J. Appl. Microbiol. 109:1696–1705 [DOI] [PubMed] [Google Scholar]
- 19. Mariani-Kurkdjian P, et al. 1993. Identification of a clone of Escherichia coli O103:H2 as a potential agent of hemolytic-uremic syndrome in France. J. Clin. Microbiol. 31:296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muraoka R, et al. 2007. An enterohemorrhagic Escherichia coli O103 outbreak at a nursery school in Miyazaki Prefecture, Japan. Jpn. J. Infect. Dis. 60:410–411 [PubMed] [Google Scholar]
- 21. Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Newton HJ, et al. 2009. Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement. Emerg. Infect. Dis. 15:372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ogura Y, et al. 2007. Extensive genomic diversity and selective conservation of virulence-determinants in enterohemorrhagic Escherichia coli strains of O157 and non-O157 serotypes. Genome Biol. 8:R138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ogura Y, et al. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:17939–17944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ooka T, et al. 2012. Clinical Significance of Escherichia albertii. Emerg. Infect. Dis. 18:488–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oswald E, et al. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paton AW, Paton JC. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paton AW, Paton JC. 2002. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J. Clin. Microbiol. 40:271–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramos-Moreno AC, Cabilio-Guth BE, Baquerizo-Martinez M. 2006. Can. the fliC PCR-restriction fragment length polymorphism technique replace classic serotyping methods for characterizing the H antigen of enterotoxigenic Escherichia coli strains? J. Clin. Microbiol. 44:1453–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramotar K, Henderson E, Szumski R, Louie TJ. 1995. Impact of free verotoxin testing on epidemiology of diarrhea caused by verotoxin-producing Escherichia coli. J. Clin. Microbiol. 33:1114–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rivas M, et al. 2006. Characterization and epidemiologic subtyping of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome and diarrhea cases in Argentina. Foodborne Pathog. Dis. 3:88–96 [DOI] [PubMed] [Google Scholar]
- 32. Rivas M, et al. 2008. Risk factors for sporadic Shiga toxin-producing Escherichia coli infections in children, Argentina. Emerg. Infect. Dis. 14:763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rohde H, et al. 2011. Open-source genomic analysis of Shiga-toxin-producing Escherichia coli O104:H4. N. Engl. J. Med. 365:718–724 [DOI] [PubMed] [Google Scholar]
- 34. Saito S, et al. 1998. A familial outbreak of verotoxin-producing Escherichia coli O103:H2 infection in which a calf was the suspected infectious source. Kansenshogaku Zasshi. 72:707–713 (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 35. Schimmer B, et al. 2008. Outbreak of haemolytic uraemic syndrome in Norway caused by stx2-positive Escherichia coli O103:H25 traced to cured mutton sausages. BMC Infect. Dis. 8:41 doi:10.1186/1471-2334-8-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sethabutr O, et al. 2000. Detection of PCR products of the ipaH gene from Shigella and enteroinvasive Escherichia coli by enzyme-linked immunosorbent assay. Diagn. Microbiol. Infect. Dis. 37:11–16 [DOI] [PubMed] [Google Scholar]
- 37. Seto K, Taguchi M, Kobayashi K, Kozaki S. 2007. Biochemical and molecular characterization of minor serogroups of Shiga toxin-producing Escherichia coli isolated from humans in Osaka prefecture. J. Vet. Med. Sci. 69:1215–1222 [DOI] [PubMed] [Google Scholar]
- 38. Spika JS, Michel P, Milley D, Wilson J, Waters J. 1998. Shiga toxin-producing Escherichia coli infections in Canada, p 23–29 In Kaper JB, O'Brien AD. (ed), Escherichia coli O157:H7 and other Shiga-producing E. coli strains. ASM Press, Washington, DC [Google Scholar]
- 39. Stacy-Phipps S, Mecca JJ, Weiss JB. 1995. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J. Clin. Microbiol. 33:1054–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 41. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thompson LH, Giercke S, Beaudoin C, Woodward D, Wylie JL. 2005. Enhanced surveillance of non-O157 verotoxin-producing Escherichia coli in human stool samples from Manitoba. Can. J. Infect. Dis. Med. Microbiol. 16:329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wick LM, Qi W, Lacher DW, Whittam TS. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 187:1783–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamamoto T, Echeverria P. 1996. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect. Immun. 64:1441–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]