Abstract
Despite South Africa being one of the high-burden multidrug-resistant tuberculosis (MDR-TB) countries, information regarding the population structure of drug-resistant Mycobacterium tuberculosis strains is limited from many regions of South Africa. This study investigated the population structure and transmission patterns of drug-resistant M. tuberculosis isolates in a high-burden setting of South Africa as well as the possible association of genotypes with drug resistance and demographic characteristics. A total of 336 consecutive MDR-TB isolates from four provinces of South Africa were genotyped using spoligotyping and mycobacterial interspersed repetitive-unit–variable number tandem repeat (MIRU-VNTR) typing. Drug susceptibility testing for ofloxacin, kanamycin, and capreomycin was performed using the agar proportion method. The results showed that 4.8% of MDR-TB isolates were resistant to ofloxacin, 2.7% were resistant to kanamycin, and 4.5% were resistant to capreomycin, while 7.1% were extensively drug resistant (XDR), and the remaining 83.6% were susceptible to all of the second-line drugs tested. Spoligotyping grouped 90.8% of the isolates into 25 clusters, while 9.2% isolates were unclustered. Ninety-one percent of the 336 isolates were assigned to 21 previously described shared types, with the Beijing family being the predominant genotype in the North-West and Limpopo Provinces, while the EAI1_SOM family was the predominant genotype in the Gauteng and Mpumalanga Provinces. No association was found between genotypes and specific drug resistance patterns or demographic information. The high level of diversity and the geographical distribution of the drug-resistant M. tuberculosis isolates in this study suggest that the transmission of TB in the study settings is not caused by the clonal spread of a specific M. tuberculosis strain.
INTRODUCTION
Tuberculosis (TB) remains a major public health challenge worldwide. South Africa is ranked no. 3 of the world's 22 high-burden TB countries responsible for 80% of the world's TB burden with the second-highest prevalence of TB per capita in the world, at 998 cases per 100,000 of the population (33). The TB problem in South Africa is compounded by the high prevalence of human immunodeficiency virus (HIV) and the emergence and spread of drug-resistant Mycobacterium tuberculosis strains, especially, multidrug-resistant (MDR-TB) (resistant to at least isoniazid [INH] and rifampin [RIF]) and extensively drug-resistant (XDR-TB) strains (MDR-TB with additional resistance to any fluoroquinolone [FLQ] and to at least one of the three injectable second-line drugs, kanamycin [KAN], amikacin [AMK], and/or capreomycin [CAP]) (4, 32). In a nationwide survey in South Africa in 2008, 20.2% of all notified TB cases showed resistance to INH, and nearly half of these (9.6% of all cases) were MDR-TB (31). Since the outbreak of XDR-TB in Tugela Ferry in KwaZulu-Natal, XDR-TB has been identified in all provinces of South Africa (32). According to the data from the National Health Laboratory Service (NHLS) in 2010, 6.3% of the diagnosed MDR-TB cases are XDR-TB (18).
The design of strategies for the management of MDR- and XDR-TB depends on an understanding of the population structure, prevalence, and spread of drug-resistant strains that drives the epidemic. Genotyping is an important tool for understanding the origin and transmission patterns of drug-resistant strains. The IS6110-restriction fragment length polymorphism (RFLP) typing is the reference technique for genotyping of M. tuberculosis strains because of its high discriminatory power (27, 29). However, the method is laborious, requires large amounts (2 μg) of DNA, and has poor discriminatory power when applied to M. tuberculosis isolates with a low IS6110 copy number (15, 27). A number of PCR-based methods, such as spoligotyping (12) and mycobacterial interspersed repetitive-unit–variable number tandem repeat (MIRU-VNTR) typing (23), have been developed to compensate for the limitations of IS6110-RFLP typing. Spoligotyping is based on the amplification and detection of the presence or absence of nonrepetitive sequences called “spacers” found between direct repeat elements of the M. tuberculosis genome (12). This assay is simple, rapid, and highly reproducible, and results are expressed in a simple digital pattern, readily named, and entered into a database (12, 15). The MIRU-VNTR typing is based on amplification of multiple loci (either 12, 15, or 24), using primers specific to each repeat locus, and on the determination of the sizes of the amplicons, which reflects the number of the targeted MIRU-VNTR copies (15, 23, 24, 25). The MIRU-VNTR profiles are presented as multidigit numerical codes, and each digit represents the copy number in a locus (23, 24, 25). The discriminatory power of either spoligotyping or MIRU-VNTR typing alone is lower than that of IS6110-RFLP typing (15). However, if MIRU-VNTR typing is combined with spoligotyping, the discriminatory power approximates that of IS6110-RFLP typing (1, 8, 16).
Despite the high prevalence of MDR and XDR-TB in South Africa, nationwide data regarding the circulating drug-resistant strains are limited. Most of the earlier studies were from the Western Cape Province and were thus not representative of the epidemiology of drug-resistant strains in South Africa. Studies have shown that the global TB epidemiology is caused by different genotypes (3, 28, 30), and these genotypes occur at frequencies that differ between districts, cities, countries, and continents (3, 9, 29). In South Africa, the population structure of drug-resistant strains differs among the provinces. However, insufficient data are available from most of the provinces of South Africa, especially provinces in the northern region. Therefore, studies on the characterization of drug-resistant strains are needed in order to make accurate assessments regarding the population structure of drug-resistant strains, to estimate the transmission dynamics, and to determine prevalent families of circulating drug-resistant M. tuberculosis strains.
In this study, drug-resistant M. tuberculosis isolates from four provinces of South Africa (Gauteng, Limpopo, Mpumalanga, and North-West) were characterized using spoligotyping and MIRU-VNTR typing (12 loci) in order to understand the population structure and transmission patterns. Furthermore, the study explored the possible association of genotypes (cluster) with demographic information (age and sex) and drug resistance patterns.
MATERIALS AND METHODS
Study population.
In this study, a convenience sample was used, including 336 consecutive M. tuberculosis isolates identified as MDR-TB isolates from June 2007 to January 2008 at the NHLS tertiary laboratory, University of Limpopo. The NHLS laboratory is a high-throughput routine diagnostic laboratory that receives specimens for culture and drug susceptibility testing (DST) from the attached Dr. George Mukhari Hospital and the surrounding clinics and hospitals in the referring provinces of Gauteng, Limpopo, Mpumalanga, and North-West. The numbers of isolates from each province were as follows: 69.9% (235/336) of isolates were from Mpumalanga, 14.9% (50/336) of isolates were from Gauteng, 9.2% (31/336) of isolates were from North-West, and 5.9% (20/336) of isolates were from Limpopo. The isolates were collected from 22 hospitals and 53 clinics in Mpumalanga, 2 hospitals and 11 clinics in Gauteng, 5 hospitals and 16 clinics in North-West, and 2 hospitals and 9 clinics in Limpopo. During the study period, the confirmed numbers of MDR and XDR-TB cases were 275 and 3, respectively, for Mpumalanga, 532 and 20, respectively, for Gauteng, 186 and 2, respectively, for North-West, and 53 and 2, respectively, for Limpopo (13).
Demographic information, including sex, age, and clinic/hospital of specimen origin, were collected for all of the patients. Approval for the study protocol was obtained from the Ethics Committee of the Faculty of Health Sciences, University of Pretoria and University of Limpopo.
Culture and drug susceptibility testing.
Isolation of the recovered mycobacterial isolates from the clinical specimens was performed using the Bactec mycobacterial growth indicator tube 960 (MGIT 960) system (Becton, Dickinson, Sparks, MD) as described by the manufacturer. The isolates were identified as M. tuberculosis using Ziehl-Neelsen staining and confirmed with the Accuprobe method (Gen-Probe, Inc., San Diego, CA). Following identification, all of the M. tuberculosis isolates were tested for susceptibility to the first-line anti-TB drugs INH, RIF, ethambutol (EMB), and streptomycin (STR) with the MGIT AST SIRE kit (Becton, Dickinson, Sparks, MD).
Drug susceptibility testing using the agar proportion method.
Second-line anti-TB drug susceptibility testing was performed for KAN, OFX, and CAP using the agar proportion method in 7H11 medium containing 5 μg/ml, 2 μg/ml, and 10 μg/ml, respectively. The DST was done according to the Clinical and Laboratory Standards Institute (CLSI) procedures and recommended critical concentrations (17). Based on the DST profile, the M. tuberculosis isolates in this study were categorized into MDR-TB (susceptible to CAP, KAN, and OFX), pre-XDR-TB (resistant to one of the second-line drugs CAP, KAN, and OFX), and XDR-TB (resistant to OFX and CAP and/or KAN).
Genotyping.
Mycobacterial genomic DNA was extracted from 100 μl of 7H9 broth culture using the Amplicor respiratory specimen preparation kit (Roche Diagnostics, Germany) as described by the manufacturer. Spoligotyping was performed using a commercial kit (Isogen Life Science B.V., Utrecht, The Netherlands) as described previously (12).
The MIRU-VNTR typing was performed using 12 MIRU-VNTR loci (2, 4, 10, 16, 20, 23, 24, 26, 27, 31, 39, and 40). Each locus was amplified individually for all 336 M. tuberculosis isolates according to the protocol described by Le Flèche et al. (14). To amplify the different loci, locus-specific primers were used as described by Supply et al. (25). The amplicon size was determined by visual comparison with a 50-bp molecular marker (Fermentas, Vilnius, Lithuania). The number of alleles corresponding to the amplicon sizes was assigned using an allele-calling table described in the protocol of Supply et al. (26). The results from each of the 12 loci were combined to create the 12-digit allelic profiles.
Data analysis.
The spoligotyping results were entered in an Excel sheet as a binary code representing either a positive or negative hybridization result. Spoligotypes in binary format were entered in the SITVIT2 database (Pasteur Institute of Guadeloupe; http://www.pasteur-guadeloupe.fr/tb/bd_myco.html) which is an updated version of the previously released SpolDB4 database. The MIRU-VNTR pattern was analyzed using the MIRU-VNTRplus database (www.miru-vntrplus.org/). The results were entered into the database as numerical codes corresponding to the number of alleles at each locus. Dendrograms were constructed for spoligotyping, MIRU-VNTR, and the combination of both methodologies. The categorical coefficient was used to calculate the distance matrix, and the dendrograms were constructed using the unweighted-pair group method with arithmetic averages (UPGMA) algorithm. The evaluation of the discriminative power of each typing method alone as well as in combination was done using the Hunter-Gaston index (HGI), D = 1 − 1/N(N − 1) ∑nj = 1aj, where D is the index of discriminatory power, aj is the number of strains in the population that are indistinguishable from the jth strain, and N is the number of strains in the population. Allelic diversity of each locus was classified as “highly discriminant” (HGI, >0.6), “moderately discriminant” (HGI, >0.3), and “poorly discriminant” (HGI, <0.3) (19). A cluster was defined as two or more M. tuberculosis isolates from different patients with identical patterns. The clustering rate was defined as (nc − c)/n, where n is the total number of cases in the sample, c is the number of genotypes represented by at least two cases, and nc is the total number of cases in clusters of two or more patients (26).
The chi-square test or Fisher's exact test was performed to determine statistical association between genotypes and age, gender, and drug-resistant patterns. P values of <0.05 were considered significant.
RESULTS
A total of 336 MDR-TB isolates collected between June 2007 and January 2008 from four provinces of South Africa, Gauteng, Limpopo, Mpumalanga, and North-West, were genotyped by spoligotyping and MIRU-VNTR typing. The study population included 51% (171/336) males and 44% (146/336) females, while the genders of 5% (19/336) of the patients were not available. The patients' median age was 34.4 years (standard deviation [SD], 10.6 years), ranging from 6 to 69 years. Susceptibility testing results for second-line anti-TB drugs showed that 4.8% (16/336) were resistant to OFX, 2.7% (9/336) were resistant to KAN, and 4.5% (15/336) were resistant to CAP, 7.1% (24/336) were XDR-TB isolates, and the remaining 83.6% (281/336) were susceptible to all of the second-line anti-TB drugs tested.
Spoligotyping results for the 336 MDR-TB isolates produced 56 distinct patterns, including 25 clustered patterns and 31 unclustered (unique) patterns. The number of M. tuberculosis isolates per cluster ranged from 2 to 69 isolates. Comparison of spoligotyping results with the SpolDB4 database showed that 90.8% (305/336) of isolates belonged to 21 previously described shared types (STs), while 9.2% (31/336) were not found in the SpolDB4 database and were considered orphans. The clustering rate of spoligotyping was 86.3% (Table 1).
Table 1.
Discriminatory power for M. tuberculosis of spoligotyping and MIRU-VNTR typing alone and in combination
| Methodology | No. of: |
Clustering rate (%) | HGIa | |||
|---|---|---|---|---|---|---|
| Distinct patterns | Clusters | Clustered isolates | Unique isolates | |||
| Spoligotyping | 56 | 25 | 305 | 31 | 86.3 | 0.9029 |
| MIRU-VNTR | 324 | 11 | 23 | 313 | 3.6 | 0.9977 |
| Spoligotyping + MIRU-VNTR | 327 | 8 | 17 | 319 | 2.7 | 0.9998 |
HGI, Hunter-Gaston index.
Based on the spoligotypes, six major distinct families of TB were identified, including Beijing, East African Indian (EAI), Haarlem (H), Latin American and Mediterranean (LAM), T family, and S family, as well as Manu 1 and Manu 2 families. The Beijing family was the predominant genotype detected in the North-West and Limpopo Provinces, while the EAI1_SOM family was the predominant genotype in the Gauteng and Mpumalanga Provinces. Stratification of spoligotyping data by drug resistance is shown in Table 2. There was no significant association (>0.05) between the genotypes and age, gender, or specific drug susceptibility patterns. The distribution of genotypes in MDR, pre-XDR, and XDR isolates is shown in Table 2.
Table 2.
Distribution of the different M. tuberculosis families and drug susceptibility patterns in four provinces of South Africa
| Province | DST result | No. of Isolates | Spoligotype (n) |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beijing | EAI1_SOM | T1 | S | LAM3 | LAM4 | LAM9 | LAM11_ZWE | LAM10_CAM | LAM and S/convergent | X1 | X2 | X3 | H1 | H3 | T2-S | T2-T3 | T3 | T3_ETH | Manu1 | Manu2 | Unknown | |||
| Mpumalanga | MDR | 191 | 37 | 49 | 31 | 24 | 6 | 3 | 11 | 7 | 3 | 5 | 1 | 10 | 2 | 1 | 9 | 1 | 3 | 1 | 21 | |||
| Pre-XDR | 23 | 3 | 5 | 4 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 3 | ||||||||||||
| XDR | 21 | 6 | 5 | 3 | 2 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||
| Gauteng | MDR | 45 | 6 | 9 | 7 | 4 | 3 | 5 | 3 | 1 | 1 | 1 | 2 | 3 | ||||||||||
| Pre-XDR | 5 | 1 | 3 | 1 | ||||||||||||||||||||
| XDR | - | |||||||||||||||||||||||
| North-West | MDR | 27 | 10 | 2 | 4 | 3 | 2 | 2 | 1 | 1 | 2 | |||||||||||||
| Pre-XDR | 3 | 2 | 1 | |||||||||||||||||||||
| XDR | 1 | 1 | ||||||||||||||||||||||
| Limpopo | MDR | 17 | 6 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | |||||||||||||
| Pre-XDR | 1 | 1 | ||||||||||||||||||||||
| XDR | 2 | 1 | 1 | |||||||||||||||||||||
A high diversity of MIRU-VNTR patterns was obtained among the M. tuberculosis isolates. A total of 324 distinct patterns were obtained from the 336 MDR-TB isolates, which included 11 clustered patterns and 313 unclustered patterns. One cluster was comprised of three M. tuberculosis isolates, while the remaining 10 clusters were comprised of only two M. tuberculosis isolates each. The clustering rate of the MIRU-VNTR typing was 3.6% (Table 1). Analysis of the allelic diversity of the 12 MIRU-VNTR loci revealed that MIRU 26 was the most discriminatory locus, with nine alleles, followed by the MIRU loci 31, 40, and 16. The MIRU loci 4, 10, 23, 24, 27, and 39 were moderately discriminative. Among the 12 MIRU loci, MIRU 2 was less polymorphic, with only three alleles, while MIRU 20 showed only a single copy in all 336 M. tuberculosis isolates analyzed (Table 3).
Table 3.
Allelic polymorphism of the 12 MIRU-VNTR loci
| Allele locus | No. of strains with MIRU copy no. of: |
HGIa | Conclusion | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |||
| 2 | 17 | 307 | 12 | 0.162 | Poorly discriminatory | |||||||
| 4 | 227 | 35 | 1 | 74 | 0.534 | Moderately discriminatory | ||||||
| 40 | 61 | 12 | 152 | 89 | 13 | 10 | 0.684 | Highly discriminatory | ||||
| 10 | 1 | 1 | 9 | 154 | 150 | 18 | 1 | 1 | 1 | 0.589 | Moderately discriminatory | |
| 16 | 44 | 112 | 160 | 16 | 4 | 0.644 | Highly discriminatory | |||||
| 20 | 337 | 0 | Poorly discriminatory | |||||||||
| 23 | 2 | 172 | 158 | 4 | 0.519 | Moderately discriminatory | ||||||
| 24 | 177 | 123 | 23 | 7 | 7 | 1 | 2 | 0.585 | Moderately discriminatory | |||
| 26 | 7 | 71 | 25 | 17 | 132 | 21 | 52 | 9 | 3 | 0.766 | Highly discriminatory | |
| 27 | 4 | 19 | 189 | 120 | 4 | 0.542 | Moderately discriminatory | |||||
| 31 | 16 | 158 | 91 | 43 | 22 | 4 | 2 | 0.685 | Highly discriminatory | |||
| 39 | 2 | 78 | 196 | 55 | 5 | 0.5805 | Moderately discriminatory | |||||
HGI, Hunter-Gaston index.
The combination of spoligotyping and the MIRU-VNTR typing results provided the highest discriminatory power, with 327 distinct patterns, which included 8 cluster patterns and 319 unclustered patterns. One cluster was comprised of three M. tuberculosis isolates, and the remaining seven had only two M. tuberculosis isolates each (Table 1). The dendrogram for the combined typing by spoligotyping and MIRU-VNTR typing is shown in Fig. 1.
Fig 1.
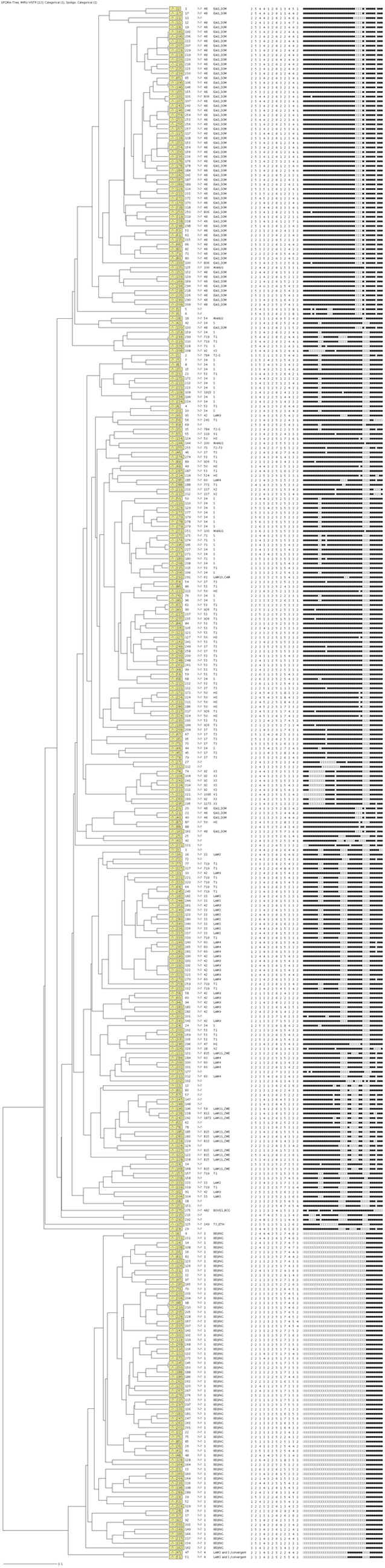
Combined dendrogram of 336 drug-resistant isolates analyzed using spoligotyping and mycobacterial interspersed repetitive-unit–variable number tandem repeat (MIRU-VNTR) typing.
The 336 isolates were from 119 hospitals/clinics in four provinces. The number of isolates received varied between the hospitals/clinics, where 65/119 (28 hospitals and 37 clinics) provided 2 to 27 and 55/119 (7 hospitals and 48 clinics) provided only 1 each. In order to determine possible transmission between the clustered isolates, the clustering and geographical origins of the isolates were compared for those hospitals/clinics that contributed two or more isolates. A possible epidemiological link was found in only eight clusters, each with two to six isolates, implying possible transmission. Six Beijing strains were isolated from patients attending the same clinic (Witbank Santa Center), while four strains were from patients attending in Themba Hospital in Mpumalanga. In Gauteng, four patients from the same hospital (Ga-Rankuwa) had isolates with the Beijing spoligotype. Another possible case of transmission was noted in which spoligotype families, including EAI1_SOM, S, H3, LAM_ZWE, LAM3, LAM4, and LAM9 were isolated from two or more patients attending the same hospital/clinic in Mpumalanga, Gauteng, and Limpopo Provinces. With MIRU-VNTR typing, 23 isolates were clustered into 11 clusters, but all of the clustered M. tuberculosis isolates were from different geographical settings. Using the combined typing results of spoligotyping and MIRU-VNTR typing, 17 M. tuberculosis isolates were clustered into 8 clusters; however, only two isolates were from the same clinic, while the remaining isolates were from different geographical settings.
DISCUSSION
Despite the high incidences of TB and drug resistance in South Africa, knowledge regarding the population structure of drug-resistant M. tuberculosis strains still remains limited in many regions of South Africa. This study is the first to include MDR-TB, pre-XDR, and XDR-TB isolates to determine the population structure of drug-resistant strains in the northern region of South Africa (Gauteng, Limpopo, Mpumalanga, and North-West Provinces). In this study, the prevalence of XDR-TB was 7.1% (24/336). The reported percentage of XDR-TB patients among the MDR-TB patients varies between countries, ranging between 3% and 19% (5, 6). A significant proportion of MDR-TB isolates (9.5%) in this study were resistant to a single marker for XDR-TB (pre-XDR), which raises concern that these pre-XDR-TB strains subsequently will become XDR-TB strains. Appropriate management of patients with pre-XDR is, therefore, important to minimize subsequent development into XDR-TB strains.
The Beijing family was the predominant genotype in the North-West and Limpopo Provinces in this study. In contrast to this study, Stavrum et al. (21) found the LAM and X genotypes were the predominant genotypes in the North-West and Limpopo Provinces, respectively, from strains isolated during 2001 to 2002. However, in the Western Cape region, the Beijing genotype is highly prevalent, where it represents 36.5% of the drug-resistant cases (11). The Beijing genotype is one of the most successful families and was initially found in China, but these strains have disseminated around the world (2, 3, 10).
It was interesting to find EAI1_SOM (ST 48) as the predominant genotype in Gauteng and Mpumalanga Provinces and as the second predominant strain in Limpopo and North-West Provinces. In contrast, Stavrum et al. (21) found T1 and LAM to be the most predominant genotypes in Gauteng and Mpumalanga Provinces, respectively, from strains isolated during 2001 to 2002. This EAI1_SOM family was first isolated in Somalia (3), and according to the SITVIT 2 database, it has been reported in eastern, northern, and southern Africa as well as Europe, Asia, and the Middle East. In a study by Chihot et al. (7), the frequency of EAI1_SOM was higher in Gauteng than in the Western Cape, Eastern Cape, and KwaZulu-Natal Provinces. In this study, it should be noted that the isolates from Gauteng Province included only one hospital and 12 clinics. Therefore, the isolates from this study could not be representative for the whole province.
Other significant families identified in this study included the T (mainly T1), LAM, and S families, which were among the major genotypes, while the X, H, and Manu families were among the minor families. The LAM family of strains in this study was represented by the LAM_ZWE, LAM3, LAM3 and S/convergent, and LAM9 genotypes. The LAM3 member of the LAM family, which is also known as F11, has been shown to be as successful as the Beijing genotype in contributing to the TB problem in the Western Cape Province of South Africa (22).
In spite of the predominance of the Beijing and EAI1_SOM families in this study, none of the associations with drug resistance were statistically significant. The Beijing family is generally considered to be associated with drug resistance; however, this is not true for all geographical settings (10). In the Western Cape Province alone, the Beijing family represents as many as 25% of the MDR-TB isolates (11). In this study, the EAI1_SOM family was predominant among MDR-TB isolates in Mpumalanga and Gauteng Provinces, while Beijing was predominant in the North-West and Limpopo Provinces.
The MIRU-VNTR genotyping results showed a high diversity among the M. tuberculosis isolates in this study. Genotyping with spoligotyping alone was the least discriminatory method (HDI) compared to MIRU-VNTR typing (HDI) and combined typing (HDI) with spoligotyping and MIRU-VNTR (HDI). The highest allelic diversity was observed for MIRU loci 26, 31, 40, and 16. It has been reported that loci 10, 16, 23, 26, and 40 were introduced as the loci with the most allele polymorphisms and loci 4, 20, 24, and 27 were the most poorly discriminated loci (20). However, in this study, MIRU loci 10 and 23 were moderately discriminative.
Even though the assessment of transmission pattern in this study may be difficult due to the relatively short period of sampling (6 months), it is still acceptable to interpret the transmission pattern. The comparison of the clustering and geographical origins of each of the isolates showed that the majority of the clustered M. tuberculosis isolates in this study were unique to their geographical setting. Generally, clustering indicates ongoing or recent transmission, while unique patterns indicate reactivation events (24). In this study, the lack of geographical links between most of the clustered M. tuberculosis isolates by spoligotyping suggests that active transmission of MDR-and XDR-TB strains may be limited in this region. Furthermore, when these isolates were typed by MIRU-VNTR and the combination of spoligotyping and MIRU-VNTR typing, the clustered isolates were differentiated and the numbers of epidemiological links among the M. tuberculosis isolates were significantly reduced (Table 1). The development of resistance in these settings could be through acquisition. Nevertheless, population-based studies are needed to confirm these findings. In contrast to this finding, clonal transmission of Beijing family (cluster R220) was reported as a major driver of the drug-resistant TB epidemic in Western Cape Province (11).
The limitations of the study include the short period of sampling, lack of information regarding the TB treatment history of the patients, and the fact that the study did not include M. tuberculosis pan-susceptible or mono-resistant isolates for comparison of the population structures in these groups.
Conclusions.
The present study provided new and relevant information regarding the population structure of MDR and XDR-TB strains in four provinces of South Africa (Gauteng, Limpopo, Mpumalanga, and North-West). The study demonstrated that the drug-resistant epidemic in this region is caused by a wide diversity of genotypes, with predominance of the Beijing and EAI1_SOM families. The high genetic diversity among the drug-resistant M. tuberculosis isolates indicated that the MDR and XDR-TB epidemic in this region is not caused by the clonal spread of a specific M. tuberculosis strain. Comparison of genotypes with the geographical origin of each isolate showed less of an epidemiological link among the isolates, suggesting a low level of active transmission during the period examined. Nevertheless, population-based studies over longer periods are needed to fully understand the epidemiology and spread of TB in this region. XDR-TB and pre-XDR-TB cases comprised a substantial fraction of the MDR-TB isolates investigated in this study, indicating the need for interventions to improve surveillance as well as rapid drug susceptibility testing. Based on the data obtained from this study, the following recommendations were made for the national TB control programs: greater vigilance, provision of rapid diagnostic assays, proper management of anti-TB drugs, and reduction of the occurrence of acquisition by providing support to patients to maximize adherence to prescribed regimens.
ACKNOWLEDGMENTS
We thank the staff members of NHLS/University of Limpopo (Medunsa Campus) and NHLS/University of Pretoria at the Tshwane Academic Division for assistance during the study.
The project was supported by a grant from the NHLS.
Footnotes
Published ahead of print 30 May 2012
REFERENCES
- 1. Barlow RE, et al. 2001. Comparison of variable number tandem repeat and IS6110-restriction fragment length polymorphism analyses for discrimination of high- and low-copy-number IS6110 Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 39:2453–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45–52 [DOI] [PubMed] [Google Scholar]
- 3. Brudey K, et al. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the Fourth International Spoligotyping Database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23 doi:10.1186/1471-2180-6-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention 3 November 2006, posting date Notice to readers: revised definition of extensively drug-resistant tuberculosis. MMWR Morb. Mortal. Wkly. Rep. 55:1176 www.cdc.gov/mmwr/preview/mmwrhtml/mm5543a4.htm [Google Scholar]
- 5. Centers for Disease Control and Prevention 2006. Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs—worldwide, 2000–2004. MMWR Morb. Mortal. Wkly. Rep. 55:301–305 [PubMed] [Google Scholar]
- 6. Centers for Disease Control and Prevention 2007. Extensively drug-resistant tuberculosis—United States, 1993–2006. MMWR Morb. Mortal. Wkly. Rep. 56:250–253 [PubMed] [Google Scholar]
- 7. Chihota VN, et al. 2012. Population structure of multi- and extensively drug-resistant Mycobacterium tuberculosis strains in South Africa. J. Clin. Microbiol. 50:995–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cowan LS, Mosher L, Diem L, Massey JP, Crawford JT. 2002. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol. 40:1592–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Filliol I, et al. 2002. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg. Infect. Dis. 8:1347–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glynn JR, Whiteley J, Bifani PJ, Kremer K, Van Soolingen D. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson R, et al. 2010. Drug-resistant tuberculosis epidemic in the Western Cape driven by a virulent Beijing genotype strain. Int. J. Tuberc. Lung. Dis. 14:119–121 [PubMed] [Google Scholar]
- 12. Kamerbeek J, et al. 1997. Simultaneous detection and strain differentiation of M. tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koornhof H, Ihekweazu C, Erasmus L, Coetzee G. 2011. Update on Corporate Data Warehouse-derived MDR- and XDR-TB statistics for eight provinces in South Africa, January 2007 to 30th June 2011. Commun. Dis. Surveill. Bull. 9(3):68–72 www.nicd.ac.za/assets/files/Bulletin%20August%202011(3).pdf [Google Scholar]
- 14. Le Flèche P, Fabre M, Denoeud F, Koeck JL, Vergnaud G. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2:37 doi:10.1186/1471-2180-2-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mathema B, Kurepina NE, Bifani PJ, Kreiswirth BN. 2006. Molecular epidemiology of tuberculosis: current insights. Clin. Microbiol. Rev. 19:658–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mazars E, et al. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. U. S. A. 98:1901–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. National Committee for Clinical Laboratory Standards 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes, vol. 23 Approved standard M24-A. National Committee for Clinical Laboratory Standards, Wayne, PA: [PubMed] [Google Scholar]
- 18. National Health Laboratory Services 2010. National Institute for Communicable Diseases annual report 2009. http://www.nicd.ac.za/assets/files/Annual_report_2009.pdf
- 19. Small PM, et al. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703–1709 [DOI] [PubMed] [Google Scholar]
- 20. Sola C, et al. 2003. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect. Genet. Evol. 3:125–133 [DOI] [PubMed] [Google Scholar]
- 21. Stavrum R, et al. 2009. High diversity of Mycobacterium tuberculosis genotypes in South Africa and preponderance of mixed infections among ST53 isolates. J. Clin. Microbiol. 47:1848–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Streicher EM, et al. 2004. Genotypic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 42:891–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Supply P, Magdalena J, Himpens S, Locht C. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991–1003 [DOI] [PubMed] [Google Scholar]
- 24. Supply P, et al. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762–771 [DOI] [PubMed] [Google Scholar]
- 25. Supply P, et al. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Supply P, et al. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Embden JD, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Soolingen D, et al. 1999. Molecular epidemiology of tuberculosis in the Netherlands: a nationwide study from 1993 through 1997. J. Infect. Dis. 180:726–736 [DOI] [PubMed] [Google Scholar]
- 29. Van Soolingen D. 2001. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Intern. Med. 249:1–26 [DOI] [PubMed] [Google Scholar]
- 30. Warren R, et al. 1999. DNA fingerprinting and molecular epidemiology of tuberculosis: use and interpretation in an epidemic setting. Electrophoresis 20:1807–1812 [DOI] [PubMed] [Google Scholar]
- 31. World Health Organization 2008. Global tuberculosis control: surveillance, planning, financing. World Health Organization, Geneva, Switzerland: http://www.who.int/tb/publications/global_report/en/ [Google Scholar]
- 32. World Health Organization 2010. Multidrug and extensively drug-resistant TB (M/XDR-TB) 2010 global report on surveillance and response. World Health Organization, Geneva, Switzerland: http://whqlibdoc.who.int/publications/2010/9789241599191_eng.pdf [Google Scholar]
- 33. World Health Organization 2010. Global tuberculosis control: surveillance, planning, financing. World Health Organization, Geneva, Switzerland: http://www.who.int/tb/publications/global_report/en/ [Google Scholar]


