LETTER
Burkholderia pseudomallei is the causative agent of melioidosis, a serious disease endemic in southeast Asia. Accurate identification of B. pseudomallei is important, since treatment of melioidosis requires prolonged antibiotics to prevent relapse (9). Although B. pseudomallei differs greatly from other Burkholderia species in pathogenicity and epidemiology, identification of B. pseudomallei is often difficult, as phenotypic tests and even 16S rRNA gene sequencing may not offer adequate discrimination from related species, such as B. thailandensis and B. cepacia complex (BCC) (3–6, 8).
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has recently emerged as a revolutionary technique for rapid bacterial identification at a low cost. Although MALDI-TOF MS has been shown to be useful for the identification of various nonfermenting Gram-negative bacilli, including some Burkholderia species (1), its application in identifying B. pseudomallei has not been explored.
Using 76 Burkholderia strains, we evaluated the performance of MALDI-TOF MS for the identification of B. pseudomallei (Table 1). All isolates were phenotypically identified by using the API 20NE system (bioMérieux) supplemented with biochemical methods. The identities of B. pseudomallei and B. thailandensis isolates were confirmed by groEL gene sequencing, and that of BCC by recA gene sequencing (6). All isolates were grown on horse blood agar at 37°C for 18 to 24 h and analyzed by the direct transfer method (2) (except that two BCC strains were analyzed by ethanol-formic acid extraction due to suboptimal results) in biosafety level II cabinets. Samples were processed in a MALDI-TOF MS spectrometer (Bruker Daltonik) with 1 μl of matrix solution (Bruker α-cyano). Spectra were obtained with an accelerating voltage of 20 kV in linear mode and analyzed within an m/z range of 2,000 to 20,000 Da. Spectra were analyzed with MALDI Biotyper 3.0 and Reference Library version 3.1.2.0 (Bruker Daltonik), which contained 41 Burkholderia main spectra (MSPs) comprising 26 species. Since B. pseudomallei is not represented in the Bruker database, 21 B. pseudomallei strains and, later, one B. thailandensis strain were added as reference strains.
Table 1.
Identification of Burkholderia species by MALDI-TOF MS
| Species | No. of strains of species | Origin or reference; strain(s) (no. of strains from origin) | No. of isolates identified usinga: |
|
|---|---|---|---|---|
| B. pseudomallei reference strains | B. pseudomallei and B. thailandensis reference strains | |||
| B. cepacia complex | 20 | Clinical isolates (n = 17) | 20 | 20 |
| BCCM/LMG collection; LMG1222, LMG14191, LMG20980 (n = 3) | ||||
| B. gladioli | 1 | BCCM/LMG collection; LMG2216 | 1 | 1 |
| B. pseudomallei | 52 | Clinical isolates (n = 15) | 52 | 52 |
| Veterinary isolates (n = 25) | ||||
| Environmental isolates (n = 12) | ||||
| B. thailandensis | 3 | 7 | 0 (misidentified as B. pseudomallei) | 3 |
Isolates were identified by MALDI-TOF MS using extended database with B. pseudomallei reference strains or with B. pseudomallei and B. thailandensis reference strains in this study.
The spectra obtained with B. pseudomallei compared to those obtained with other species are shown in Fig. 1. Using the Bruker database extended with B. pseudomallei reference strains, all isolates except for B. thailandensis were correctly identified (the score of the top match was ≥2.0, and the score of the second match was lower by ≥10%) (Table 1). Notably, the three B. thailandensis isolates were misidentified as B. pseudomallei. Further extension of the database with one additional B. thailandensis reference strain enabled the correct identification of two other B. thailandensis isolates. The misidentification of B. thailandensis by using the Bruker database is probably due to the inclusion of only one MSP from the species, which fails to cover intraspecies variability.
Fig 1.
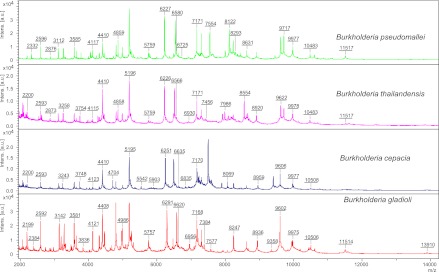
MALDI-TOF MS spectra of B. pseudomallei compared to those of other species. Intens. [a.u.], intensity in arbitrary units.
MALDI-TOF MS is potentially useful for accurate routine identification of B. pseudomallei and B. thailandensis. However, this requires optimization of the database by adding reference MSPs for B. pseudomallei and expanding the number of MSPs for B. thailandensis. While the present BCC isolates were correctly identified as BCC, species identification may require additional reference strains (1). Expansion of commercial databases with pathogens endemic in different localities is important to improve the usefulness of MALDI-TOF MS.
ACKNOWLEDGMENTS
We thank Wai-Sing Chan, Ting-Yin Wong, and Yu-Ting Tong for technical assistance.
This work is supported by the HKSAR Research Fund for the Control of Infectious Diseases (Commissioned Study) of the Health, Welfare and Food Bureau.
Footnotes
Published ahead of print 20 June 2012
Contributor Information
Susanna K. P. Lau, State Key Laboratory of Emerging Infectious Diseases The University of Hong Kong Hong Kong, China
Bone S. F. Tang, Department of Pathology Hong Kong Sanatorium Hospital Hong Kong, China
Shirly O. T. Curreem, Department of Microbiology The University of Hong Kong Hong Kong, China
Tsz-Ming Chan, Department of Pathology Hong Kong Sanatorium Hospital Hong Kong, China.
Paolo Martelli, Ocean Park Corporation Hong Kong, China.
Cindy W. S. Tse, Department of Pathology Kwong Wah Hospital Hong Kong, China
Alan K. L. Wu, Department of Pathology Pamela Youde Nethersole Eastern Hospital Hong Kong, China
Patrick C. Y. Woo, Research Centre of Infection and Immunology The University of Hong Kong Hong Kong, China
REFERENCES
- 1. Degand N, et al. 2008. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 46:3361–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eigner U, et al. 2009. Performance of a matrix-assisted laser desorption ionization-time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin. Lab. 55:289–296 [PubMed] [Google Scholar]
- 3. Godoy D, et al. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 41:2068–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ho CC, et al. 2011. Novel pan-genomic analysis approach in target selection for multiplex PCR identification and detection of Burkholderia pseudomallei, Burkholderia thailandensis, and Burkholderia cepacia complex species: a proof-of-concept study. J. Clin. Microbiol. 49:814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kiratisin P, Santanirand P, Chantratita N, Kaewdaeng S. 2007. Accuracy of commercial systems for identification of Burkholderia pseudomallei versus Burkholderia cepacia. Diagn. Microbiol. Infect. Dis. 59:277–281 [DOI] [PubMed] [Google Scholar]
- 6. Mahenthiralingam E, et al. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith MD, Angus BJ, Wuthiekanun V, White NJ. 1997. Arabinose assimilation defines a nonvirulent biotype of Burkholderia pseudomallei. Infect. Immun. 65:4319–4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spilker T, et al. 2009. Expanded multilocus sequence typing for Burkholderia species. J. Clin. Microbiol. 47:2607–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Woo PC, et al. 2003. Seronegative bacteremic melioidosis caused by Burkholderia pseudomallei with ambiguous biochemical profile: clinical importance of accurate identification by 16S rRNA gene and groEL gene sequencing. J. Clin. Microbiol. 41:3973–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]


