Abstract
The increase in the capacity and reduction in cost of whole-genome sequencing methods present the imminent prospect of such data being used routinely in real time for investigations of bacterial disease outbreaks. For this to be realized, however, it is necessary that generic, portable, and robust analysis frameworks be available, which can be readily interpreted and used in real time by microbiologists, clinicians, and public health epidemiologists. We have achieved this with a set of analysis tools integrated into the PubMLST.org website, which can in principle be used for the analysis of any pathogen. The approach is demonstrated with genomic data from isolates obtained during a well-characterized meningococcal disease outbreak at the University of Southampton, United Kingdom, that occurred in 1997. Whole-genome sequence data were collected, de novo assembled, and deposited into the PubMLST Neisseria BIGSdb database, which automatically annotated the sequences. This enabled the immediate and backwards-compatible classification of the isolates with a number of schemes, including the following: conventional, extended, and ribosomal multilocus sequence typing (MLST, eMLST, and rMLST); antigen gene sequence typing (AGST); analysis based on genes conferring antibiotic susceptibility. The isolates were also compared to a reference isolate belonging to the same clonal complex (ST-11) at 1,975 loci. Visualization of the data with the NeighborNet algorithm, implemented in SplitsTree 4 within the PubMLST website, permitted complete resolution of the outbreak and related isolates, demonstrating that multiple closely related but distinct strains were simultaneously present in asymptomatic carriage and disease, with two causing disease and one responsible for the outbreak itself.
INTRODUCTION
Nucleotide sequence-based typing methods, such as multilocus sequence typing (MLST) (34) and antigen gene sequencing typing (AGST) (40), have demonstrated (i) that DNA sequences provide robust means of characterizing bacteria and (ii) the value of widely accepted portable nomenclatures, based on curated reference systems (25). Developments in parallel sequencing technologies, leading to reduced costs and increased capacity, have resulted in whole-genome sequencing (WGS) approaches increasingly being employed in the analysis of bacterial pathogens (35). The challenge is to combine whole-genome sequencing and sequence typing methods to provide clinical microbiologists and public health practitioners tools that they can easily use and data that they can readily interpret (30) without complex and computer-intensive analysis techniques.
The classification of disease isolates has five main clinical purposes: (i) diagnosis, that is, identification of the infectious agent; (ii) detection of transmission between individuals; (iii) outbreak detection, that is, establishing the spread of particular strains locally or regionally; (iv) longer-term and evolutionary studies to identify the emergence of particularly pathogenic or virulent variants; (v) assessment of vaccine preventability. Each of these has a different public health role and requires different levels of typing resolution. Whole-genome sequence data potentially provide the ultimate level of genetic characterization, but for such data to be exploited effectively and efficiently it is necessary to analyze them in a hierarchical manner, extracting that information necessary to resolve the clinical question being addressed.
Genome sequencing has been used for epidemiological typing in other organisms but has usually involved the identification of single-nucleotide polymorphisms (SNPs) by mapping short reads against a reference to generate phylogenetic trees (8, 17, 28, 36). This requires a suitable reference and is only applicable to closely related isolates. For clinical and public health use, however, it is also important to be able to readily extract relevant information from the genome that can place the isolate in context with existing typing methods and inform control measures, even when isolates are distantly related. The Bacterial Isolate Genome Sequence Database (BIGSdb) platform (26) facilitates sequence extraction and indexing of loci within the bacterial genome sequence data by using an MLST-like approach, which is based on alleles at multiple genetic loci. Within BIGSdb, any number of loci can be defined, and these can be grouped in multiple ways for analysis as coherent sets. This allows, for example, successive typing schemes to be employed, which provide incrementally increasing levels of resolution by the inclusion of an expanding number of loci. The appropriate level of discrimination can then be chosen on the basis of the question being asked.
In order to provide a straightforward and rapid means of exploiting whole-genome sequence data in this hierarchical fashion, we have combined the ability of the BIGSdb software (26) to identify, index, and extract genetic variation data rapidly and effectively with the NeighborNet clustering algorithm (6) and the SplitsTree visualization tool (22) to provide a highly scalable method of establishing the relationships among clinical isolates from whole-genome sequence data, at a wide range of genetic resolutions, rapidly and with minimal computational requirements. The approach is demonstrated by a reanalysis of a well-characterized meningococcal disease outbreak at the University of Southampton, United Kingdom, that occurred in October 1997 (13, 15). This analysis confirmed the conclusions reached by MLST, AGST, and epidemiological analysis, and also provided additional information on the variation of isolates in individual transmission events and the relationship of the isolates to the global variation of the disease-causing clone.
MATERIALS AND METHODS
Bacterial isolates.
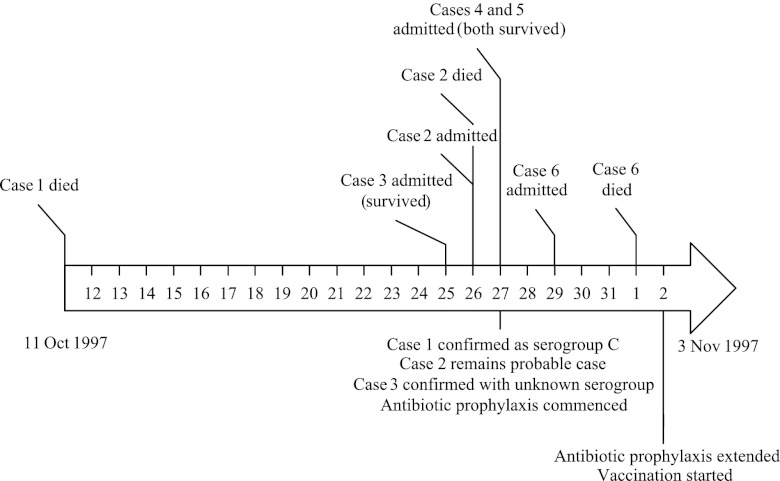
A total of 6 cases of meningococcal disease occurred in the University over a 3-week period (Fig. 1, cases 1 to 6): meningococcal isolates were available for three of these (cases 1, 3, and 6). Case 1 was treated as a single case, whereas the other cases occurred 2 weeks later. Two isolates from remote cases (remote cases 1 and 2) were also investigated, as they were epidemiologically suspected to be part of the outbreak. None of the patients was known to any other, although cases 2, 4, and 5 all attended the same nightclub on the same night and became ill 3 to 4 days later (15). All of the disease isolates were serogroup C, and one case without an isolate was demonstrated by PCR assay (4) to have a group C meningococcal infection. Serological tests did not discriminate the disease isolates, with case 1 characterized as C:2a:NST and cases 3 and 6 as C:NT:P1.5. Expression of serotype and sero-subtype can be intermittent, resulting in a nontypeable (NT) result, and these strains were initially assumed to be the same (15). A carriage isolate was obtained from a direct contact of case 1 (carrier 1). During outbreak investigation and control at the time, 587 students were examined for meningococcal carriage. A total of 147 meningococci were isolated (25%), with only 6 of these serogroup C (4.1% of meningococci; carriage rate, 1.02%), and bacterial cultures were available for four of these (carriers 2 to 5) (15). All 10 isolates were retrieved from storage at −80°C in Mueller-Hinton broth plus glycerol and were inoculated onto Columbia horse blood agar (Oxoid) and incubated for 24 h at 37°C in a 5% CO2 atmosphere. Genomic DNA was also prepared from FAM18 (3) and a serogroup C meningococcal isolate used in the original molecular investigation of the outbreak (13).
Fig 1.
Timeline of the outbreak.
Whole-genome sequencing, data assembly, and upload to the PubMLST database.
Genomic DNA was prepared from cell suspensions of the plate cultures with the Wizard Genomic DNA purification kit (Promega). Standard Illumina multiplex libraries were generated with 1 μg of genomic DNA sheared to between 200 and 300 bp using a Covaris E210 acoustic shearing device. DNA fragments were end-repaired, and a 3′ nontemplated adenosine residue was ligated to the Illumina multiplexing adaptor oligonucleotide for sequencing. Up to 12 libraries were pooled and run together in an equimolar ratio for sequencing per flow cell lane on the Illumina Genome Analyser II platform, and 76-bp paired-end reads were generated. Genome sequence data were assembled using Velvet, version 1.2.01 (49), with optimal parameters determined by the VelvetOptimiser script. The resultant assemblies were uploaded to the Neisseria PubMLST database (http://pubmlst.org/neisseria/), which runs the BIGSdb platform (26) (Neisseria PubMLST ID 662-667 and 669-672; a direct link is available through http://pubmlst.org/neisseria/seqbin/xxx, where xxx is the ID number) to facilitate analysis.
Data analysis and visualization.
The PubMLST BIGSdb software includes an autotagger functionality, which scans deposited sequences against defined loci. This process runs in the background and automatically updates isolate records with allele numbers and marks regions on the assembled contiguous sequences (contigs) present in the database for any genes with sequences that exactly match alleles defined in the PubMLST sequence definition database. At the time of analysis (March 2012), approximately 1,200 Neisseria loci had been defined, including most of the core genome, MLST loci, the PorA and FetA variable regions, and capsular region. Autotagging identified known alleles at these loci, enabling the strain types to be immediately extracted. Manual scanning and curation were performed to identify any new alleles within these isolates. These were submitted to PubMLST for assignment, and the genome data were subsequently rescanned to complete allelic assignment.
Genome comparison.
The BIGSdb Genome Comparator tool, implemented within the PubMLST website, was employed to compare the WGS data obtained for the isolates. This tool can use either loci defined within the database or an annotated reference genome as the comparator for analysis (26). When a reference genome is employed, the coding sequences within the annotation are extracted and compared against the assembled contigs for the isolate genomes under comparison with BLAST. Unique allele sequences at each locus are designated with an integer starting at 1 (representing identity to the reference sequence). One of the outputs is a color-coded whole-genome MLST (wgMLST) profile, which facilitates comparison among isolates. This output is also broken down into variable loci: those that are identical in all isolates, those that are missing in all, and those where a sequence is incomplete due to the locus being situated at the end of an assembly contig. Alignments of variable loci can be generated so that the underlying sequence differences can be assessed. The isolates were compared at: (i) the MLST loci; (ii) 20 eMLST loci (9); (ii) the 53 rMLST loci (24); and (iv) all loci defined in the FAM18 genome annotation (3). A distance matrix based on the number of variable alleles was generated automatically, and the isolates were resolved into networks with the NeighborNet algorithm (6) either “live” within the PubMLST website or by using a stand-alone instance of SplitsTree4 (22). A step-by-step guide to replicate this analysis on the PubMLST.org site is provided in the supplemental material. For detailed analysis of sequence differences among isolates of a single strain type, repetitive sequences due to multiple copies of insertion element transposases and pseudogenes were removed when using the FAM18 annotation as the source of comparator coding sequences, as these are currently poorly assembled from these data. Paralogous loci that matched multiple alleles were also removed from the analysis.
RESULTS
Strain types.
Since the initial analysis of this disease outbreak (13), there have been some changes in the nomenclature of PorA typing, and FetA (FrpB) typing has been introduced for the meningococcus (25). Notwithstanding changes in nomenclature, the strain types obtained from each of the isolates by automated annotation of the whole-genome sequence data were identical to those generated by serological methods and conventional MLST and antigen sequence typing (13). There were a total of five distinct strain types identified using the current and older nomenclatures, with two from cases of disease: strain 1, from case 1 and carrier 1; strain 2, from cases 3 and 6 and remote cases 1 and 2; strain 3, from carrier 2; strain 4, from carrier 3; and strain 5, from carriers 4 and 5 (Table 1). In the original report of this outbreak (13), strain 3 (from an asymptomatic carrier) was incorrectly reported as ST-11; both this analysis and conventional MLST identified this as ST-67, a single-locus variant of ST-11 at the fumC locus. This error was a typographical mistake in the earlier paper, a likely consequence of the fumC locus being added to the Neisseria MLST scheme while the original analysis was being completed (21, 34) and which was not noticed until the current analysis was undertaken.
Table 1.
Properties of ST-11 complex meningococci isolated during the outbreak, compared with two reference isolates
| Strain | Isolate description | Serological type | Strain type from genome sequencea | Allele designationb |
|||||
|---|---|---|---|---|---|---|---|---|---|
| ′porB | ′tbpB | fHbp | ′penA | ′rpoB | folP/dhpS (NEIS1609) | ||||
| Reference 1 | FAM18 (USA 1983) | C:P1.5,2:F1-30:ST-11 (cc11) | 2-2 | 1 | 22 | 1 | 9 | 1 | |
| Reference 2 | UK 1993 | C:2a:P1.5 | C:P1.5-1,10-4:F3-6:ST-11 (cc11) | 2-2 | 90 | 92 | 3 | 4 | 14 |
| Strain 1 | Case 1 | C:2a:NT | C:P1.5c,2:F3-6:ST-11 (cc11) | 2-36 | 90 | 110 | 3 | 4 | 14 |
| Carrier 1 | C:2a:NT | C:P1.5c,2:F3-6:ST-11 (cc11) | 2-36 | 90 | 669 | 3 | 4 | 14 | |
| Strain 2 (outbreak strain) | Case 3 | C:NT:P1.5 | C:P1.5-1,10-4:F3-6:ST-50 (cc11) | 2-37 | 90 | 92 | 3 | 4 | 14 |
| Case 6 | C:NT:P1.5 | C:P1.5-1,10-4:F3-6:ST-50 (cc11) | 2-37 | 90 | 92 | 3 | 4 | 14 | |
| Remote case 1 | C:NT:P1.5 | C:P1.5-1,10-4:F3-6:ST-50 (cc11) | 2-37 | 90 | 92 | 3 | 4 | 14 | |
| Remote case 2 | C:NT:P1.5 | C:P1.5-1,10-4:F3-6:ST-50 (cc11) | 2-37 | 90 | 92 | 3 | 4 | 14 | |
| Strain 3 | Carrier 2 | C:NT:P1.5,2 | C:P1.5,2:F1-1:ST-67 (cc11) | 2-39 | 1 | 22 | 1 | 9 | 1 |
| Strain 4 | Carrier 3 | C:2a:NT | C:P1.Δ,Δ:F1-30:ST-52 (cc11) | 2-2 | 1 | 22 | 1 | 9 | 1 |
| Strain 5 | Carrier 4 | C:2a:P1.15 | C:P1.19-3,15:F5-5:ST-51 (cc11) | 2-38 | 1 | 22 | 57 | 9 | 1 |
| Carrier 5 | C:2a:P1.15 | C:P1.19-3,15:F5-5:ST-51 (cc11) | 2-38 | 1 | 22 | 57 | 9 | 1 | |
As defined by Jolley et al. in 2007 (25).
Prefixing of name with a prime symbol indicates that the locus is defined as a fragment of the complete gene coding sequence.
Insertion element IS1301 interrupts PorA VR1. Designations are defined by the peptide sequence of variable loops (40) and were only made for sequences that were predicted to be expressed.
In addition to loci used currently for strain type nomenclature, allelic data on the following genes or gene fragments (indicated with a leading prime mark) were extracted: ′porB, encoding part of the serotyping antigen; ′tbpB, encoding part of the transferrin binding protein B (18); fHbp, encoding an investigational vaccine antigen (5); ′penA, encoding part of penicillin binding protein 2, which is responsible for penicillin susceptibility (45); ′rpoB, encoding part of RNA polymerase, which is responsible for rifampin susceptibility (42); and folP (dhpS), encoding dihydropterate synthase, which is responsible for sulfonamide susceptibility (14, 38). These data were consistent with the strain types and indicated that all the isolates were likely susceptible to penicillin and rifampin. Strains 1 and 2 and reference strain 2 were predicted to be sulfonamide sensitive and the remaining strains were predicted to be resistant.
The presence of the IS1301 insertion element, reported to be found in the ET-15 variant of the ST-11 clonal complex but not in other members of this complex (10), was determined by querying the genome sequences against the IS1301 sequence, using BLAST within the PubMLST.org website. IS1301 was detected in strain 1 and strain 2 isolates as well as in the UK1993 reference isolate. It was not found in FAM18 or any of the other isolates. Presence of the element disrupted contig assembly due to repeat sequences, so its location and frequency in the genome could not be determined precisely. In strain 1, IS1301 interrupted the PorA variable region 1, splitting the gene onto two assembly contigs.
Relationships of strains revealed by different comparisons.
With the seven-locus MLST comparison, the case 1 and carrier 1 isolates (strain 1; ST-11) were indistinguishable from FAM18 and the United Kingdom reference isolate. The four outbreak isolates were single-locus variants of ST-11 (strain 2; ST-50), as were two of the carrier isolates, carrier 2 (strain 3; ST-67) and carrier 3 (strain 4; ST-52). Carriers 4 and 5 were double-locus variants of ST-11 (strain 5; ST-51). The overall relationship was a star phylogeny centered on ST-11 (Fig. 2A). Increasing the number of housekeeping alleles to 20 using eMLST (9) provided more resolution, particularly in separating the case 1 and carrier 1 (strain 1) isolates from the two reference isolates. There was also some evidence for the other carrier isolates being more closely related to each other and to FAM18 than to the remaining isolates (Fig. 2B). A comparison at the 53 rMLST loci (2, 24) grouped the United Kingdom reference isolate with the strain 2 outbreak isolates, related to but distinct from the strain 1 isolates. These were distinct from the carrier isolates and FAM18 (Fig. 2C). The final comparison, based on the 944 loci that varied in at least one isolate, out of a total of 1,975 coding sequences annotated in the FAM18 genome, confirmed that there were two distinct groups of disease isolates present and that there were three groups of very closely related isolates, corresponding to the strains identified by MLST combined with antigen gene sequencing: case 1 and carrier 1 (strain 1), cases 3 and 6 and remote cases 1 and 2 (strain 2), and carrier 4 and 5 (strain 5) (Fig. 2D).
Fig 2.
NeighborNet graphs generated using the Genome Comparator tool of BIGSDB with seven-locus MLST data (A), 20-locus extended MLST data (B), rMLST data (C), or the 944 variable loci (D) identified when the FAM18 annotation was compared against all isolates.
Whole-genome variation within strain types.
Only minor differences (8 to 26 loci) were confirmed among isolates designated belonging to the same strain (Fig. 3; see also Tables S1 to S3 in the supplemental material). As the many repetitive regions in the meningococcal chromosome were not reliably assembled from short read data alone, potentially leading to incorrect assignments for small coding regions in repetitive elements, these were excluded from this analysis. These incorrect assignments did not affect global analyses, since the proportion of potentially misassigned loci was small relative to the total number of coding sequences. Genome Comparator initially identified 38 loci that appeared to vary between the two isolates representing strain 1 based on the coding sequences annotated in the FAM18 genome. The majority of these were repetitive sequences from putative insertion element transposases or hypothetical protein pseudogenes. After removing these from the comparison, eight differences were observed in coding sequences that were a mixture of mutations, allelic replacement, and insertions/deletions (see Table S1). Four of these differences were in adjacent genes (NMC0347 to NMC0350), including fHbp, which encodes the vaccine component factor H binding protein, with a higher number of nucleotide changes observed compared to those in single genes, indicative of a single recombination event. Differences in the Opa1800 coding sequence between the case isolate and the carrier isolate which was epidemiologically presumed to have been acquired from it were a single synonymous mutation near the 3′ end and loss of a single pentanucleotide (CTCTT) repeat in the signal peptide-encoding region, which is known to affect phase variation (20, 43).
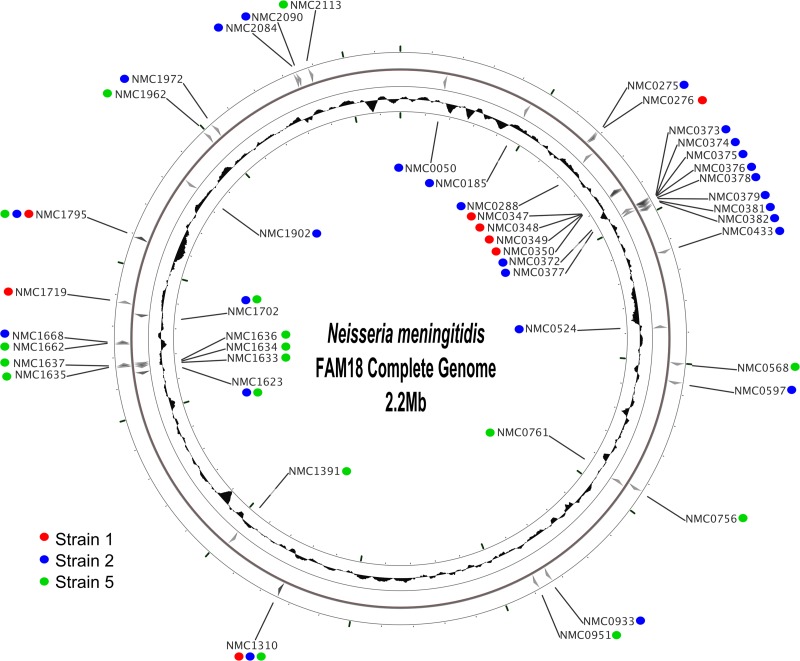
Fig 3.
Location of nucleotide changes among isolates corresponding to each of strains 1, 2, and 5, mapped onto the FAM18 genome. The red circles represent the loci that varied within isolates designated strain 1 (8 loci varied, none of which were single base changes); the blue circles represent loci that varied within strain 2 (27 loci varied, 10 of which were single base changes); the green circles represent the loci that varied within strain 5 (17 loci varied, 7 of which were single base changes). Full details of the locus changes can be found in Tables S1 to S3 of the supplemental material. The figure was generated using CGView (44).
Among the four isolates from the later local and remote cases, representing strain 2, Genome Comparator initially identified 77 variable loci (with between 32 and 56 variable loci between pairs of isolates). After filtering loci for repetitive and paralogous sequences, there were 36 allelic differences (see Table S2 in the supplemental material). Different alleles were seen in a run of 10 adjacent genes (NMC0372 to NMC0382) in a single isolate (case 6), while these were identical in the other three isolates (NMC0380 was removed from the analysis due to missing data). This run occurred over a region of approximately 13 kbp and, as with the run of adjacent loci in strain 1, each locus in this run had many more nucleotide changes than seen in other loci. It was likely that these changes resulted from a single recombination event, although the size was larger than the previously estimated average recombination tract length of 1.1 kbp for meningococci (27). In the original description of the molecular typing of this outbreak (13), the pulsed-field gel electrophoresis (PFGE) patterns for cases 3 and 6 were slightly different, which was interpreted as resulting from a chromosomal rearrangement. We analyzed the alleles we found to be different between these two isolates for recognition sites for the three restriction enzymes used in the PFGE: NheI, SfiI, and SpeI. There was only one predicted recognition site for any of the PFGE restriction enzymes in the variable alleles of these isolates. This was for SpeI in NMC1310 (NEIS1310), which was present in the isolates for both case 3 and case 6. This suggests that the original explanation of spontaneous rearrangement remains likely.
A total of 39 variable loci were identified between the two isolates designated strain 5. Once repetitive and paralogous sequences were removed, 17 allelic differences were confirmed (see Table S3 in the supplemental material). Differences were seen within five adjacent genes (NMC1633 to -1637) and, as in the other strains, the number of nucleotide differences within these genes was higher than for nonadjacent variable loci.
DISCUSSION
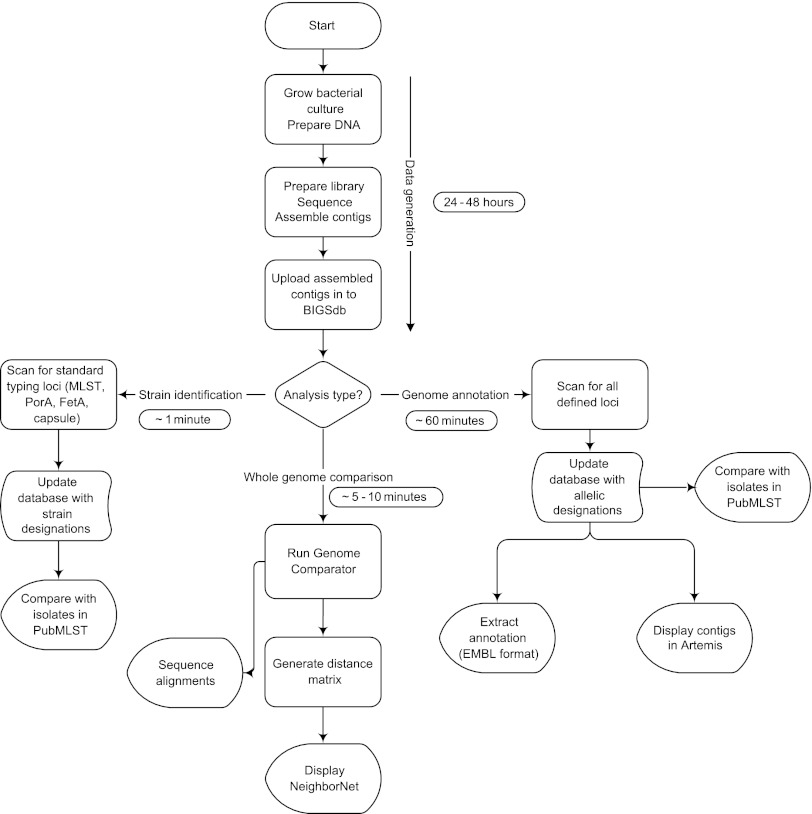
Here we have presented a practical approach to the exploitation of bacterial genomic data in a clinical setting by using computationally nonintensive, open access software tools. These are simple to use and produce easily understood “plain language” reports and graphical representations of the data. De novo assembly of high-coverage short-read data permits samples to be examined efficiently, without the need to map individual nucleotide changes to a reference genome, and the hierarchical locus-by-locus analysis approach enables the rapid interpretation of the resultant data to resolve a range of clinical and epidemiological questions. Specifically, the calculation of distance matrices from genome-wide allelic comparisons is fast and robust for the effects of horizontal genetic exchange. With larger numbers of loci, this provides very high resolution, and NeighborNet (6) is an effective visualization tool that represents the relationships of isolates from such matrices accurately and quickly, being generated in real time on the PubMLST website. The approach is both generalizable and highly scalable, permitting the analysis of large numbers of isolates. With current technology the whole process can be completed in less than 48 h from the receipt of a clinical specimen, with a large proportion of this time devoted to culturing of the isolate. Illumina sequencing followed by de novo assembly has been shown to be accurate for bacterial genomes (12, 19, 37, 41). The bioinformatics analysis following de novo assembly takes only a few minutes to an hour, depending on the level of comparison required (Fig. 4).
Fig 4.
The BIGSdb process workflow. Following sequencing, rapid analysis for strain identification and genome comparison can be performed in minutes.
For clinical purposes, it is necessary to identify the disease-causing bacterium and its likely properties, and from whole-genome sequence data this can be readily achieved by analysis against reference sequences. Identification of bacterial species is conventionally achieved by analysis of the 16S rRNA loci (7), which can be extracted from whole-genome sequence data, but where whole-genome sequence data are available, rMLST, which indexes variation at the ribosomal protein subunit genes, provides greater resolution (24). Where suitable curated collections of reference alleles exist, it is possible to extract simultaneously additional information, such as sequence type, likely antimicrobial resistance, serotype, and virulence, providing both backward compatibility and the nomenclature that is essential for reporting results (48). The Neissera MLST website used for the present analysis has a wide range of such curated schemes (25, 45), and similar schemes exist for numerous other bacteria of medical importance (24, 31). In the example presented here, data from only a limited number of loci expressed with widely accepted typing nomenclature (Table 1) (25) provided the information necessary to manage these cases of disease (13, 15).
While the seven-locus MLST combined with AGST was sufficient to resolve the outbreak (13), important elements of the epidemiology of the ST-11 complex were not resolved at this level of resolution. As had been shown previously (13), a total of five distinct strains belonging to the ST-11 complex were present in the university town in a period of just 19 days. Carriage rates of these strains were very low (15), as was typical for serogroup C members of the ST-11 complex in the United Kingdom and other countries in the period just before the introduction of the serogroup C meningococcal vaccine in 1999, which has eliminated such outbreaks (23, 32, 33). The rMLST and wgMLST analyses resolved these related strains into two distinct groups: one group related to the FAM18 isolate, and the other, including the outbreak strain, more closely related to the isolate obtained in the United Kingdom in 1993 (Fig. 2C and D). Comparison with these reference strains indicated that all five strains were likely to have been introduced independently at around the same time into the University transmission system, with two likely transmissions outside the University (remote cases 1 and 2). This observation demonstrates the complexity of transmission of the meningococcus and the importance of high-resolution typing in resolving the epidemiology of this organism. In the case of this outbreak, molecular typing was of public health significance, as it demonstrated that the outbreak strain was distinct from a case of disease that occurred 2 weeks earlier, and that both these strains associated with invasive disease were distinct from three other ST-11 complex strains circulating in the student population at the time. The conclusions of the outbreak investigation would have been very different if lower-resolution techniques, such as serological-based typing and PFGE alone, were used, potentially affecting both outbreak management and public health policy (15).
The very-high-resolution information available from whole-genome data enabled the investigation of short-chain transmission by comparing closely related isolates using Genome Comparator, with the annotation of the FAM18 genome sequence as a reference. The relatively large number of genetic changes seen within closely related isolate pairs, which included point mutations, indels, and horizontal genetic exchange, seems unlikely to have been accumulated during the course of the outbreak but was more likely to be due to the variation in the population of organisms that were circulating in carriage at that time. As this was present in two of the three strains examined, this suggests that infection of individuals with a number of closely related but distinct variants may be common with the meningococcus. While this seems the most likely explanation, it is still possible that these changes occurred during the outbreak, although investigations of short transmission chains in other species have reported fewer nucleotide changes between isolates (11, 16, 28). Further data about the clock rate and whether mutation rates vary in different clonal complexes would be needed to categorically rule this out, and analysis of further outbreaks, especially within close-knit or isolated communities, would be of value here. It was also of note that two genes, a restriction-modification protein (NMC1310 [NEIS1310]) and a hypothetical protein (NMC1795 [NEIS1795]), were variable in all of the strains examined, but it is unclear if this had any functional significance.
During the late 1990s, elevated levels of serogroup C meningococcal disease were experienced by a number of countries. This was a consequence of the spread of a variant of the ST-11 clonal complex with a propensity to attack young adults and to cause localized outbreaks (1, 29, 46). These outbreaks often occurred among first-year students in university halls of residence, or among new recruits at military establishments, likely due to the introduction of susceptible hosts. Although MLST characterized this variant as the central genotype of the ST-11 complex, ST-11, it was distinct from other members of the clonal complex based on multilocus enzyme electrophoresis (MLEE), by virtue of a mutation in the fumC gene that was outside the region included in the fragment sequenced for MLST, and was therefore referred to by its electrophoretic type (ET), ET-15 (47). ET-15 isolates can also be differentiated from other members of the complex by the presence of the IS1301 insertion sequence (10). In this investigation, reference strain 2 (United Kingdom, 1993), strain 1, and strain 2 (the outbreak) possessed both the fumC mutation and IS1301, and they were therefore designated ET-15 and were predicted to be sensitive to sulfadiazine (39). These strains formed a separate clade by rMLST and wgMLST, demonstrating that the ET-15 strains are indeed distinct from other ST-11 complex isolates. It is noteworthy that only these strains were involved in disease and that they were not recovered from carriage in this outbreak, other than from the direct contact of case 1.
The establishment of MLST as the primary typing method for Neisseria and many other species has been achieved only by the presence and maintenance of authoritative Web-accessible databases. Curation of these databases requires teams of individuals with the resources to assess and define new alleles or antigen types. While much of this allele definition work may be automatable, the presence of authoritative nomenclature servers overseen by a responsible body is essential for genomic analysis based on a gene-by-gene approach. Without a defined and universally agreed nomenclature, analysis has to be continually repeated with new pairwise comparisons required every time new data are added to an analysis. The PubMLST Neisseria database is now indexing this allelic diversity, and all new genomes deposited within it will be scanned automatically for known variants. Periodic scanning of the database by curators will identify new allelic variants, which will then be defined and available for automated scanning.
In conclusion, the BIGSdb platform and the whole-genome MLST approach facilitated the resolution of a disease outbreak of a genetically diverse pathogen by using high-throughput sequencing, and the Web-based database tools rapidly extracted clinically and epidemiologically relevant information in a scalable manner. It was possible to compare the data from the isolates under investigation with any subset of loci that has been deposited in the reference database, for example, the rMLST and eMLST schemes, and to use the annotation of a reference genome to get a level of resolution from the whole genome, wgMLST. The recently described rMLST scheme (24), which uses the 53 ribosomal protein genes, resolved the outbreak with higher resolution than standard MLST and had the advantage that the loci are universally present. In a clinical setting, with genomic data available, rMLST can be applied to any bacterial species, including currently undescribed bacterial species, first at the level of species and then to the level of high-resolution strain typing, all with a single set of loci.
Supplementary Material
ACKNOWLEDGMENTS
M.C.J.M. is a Wellcome Trust Senior Research Fellow supported by Wellcome Trust grant 087622/Z/08/Z. Sequencing performed at the Wellcome Trust Sanger Institute was supported by Wellcome Trust grant 098051.
Footnotes
Published ahead of print 11 July 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Ashton FE, et al. 1991. Emergence of a virulent clone of Neisseria meningitidis serotype 2a that is associated with meningococcal group C disease in Canada. J. Clin. Microbiol. 29:2489–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bennett JS, et al. 2012. A genomic approach to bacterial taxonomy: an examination and proposed reclassification of species within the genus Neisseria. Microbiology 158:1570–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bentley SD, et al. 2007. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genet. 3:e23 doi:10.1371/journal.pgen.003023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borrow R, et al. 1997. Non-culture diagnosis and serogroup determination of meningococcal B and C infection by a sialyltransferase (siaD) PCR ELISA. Epidemiol. Infect. 118:111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brehony C, Wilson DJ, Maiden MC. 2009. Variation of the factor H-binding protein of Neisseria meningitidis. Microbiology 155:4155–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bryant D, Moulton V. 2004. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21:255–265 [DOI] [PubMed] [Google Scholar]
- 7. Clarridge JE. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Croucher NJ, et al. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Didelot X, Urwin R, Maiden MC, Falush D. 2009. Genealogical typing of Neisseria meningitidis. Microbiology 155:3176–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Elias J, Vogel U. 2007. IS1301 fingerprint analysis of Neisseria meningitidis strains belonging to the ET-15 clone. J. Clin. Microbiol. 45:159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eyre DW, et al. 2012. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2:3001124 doi:10.1136/bmjopen-2012-001124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Farrer RA, Kemen E, Jones JD, Studholme DJ. 2009. De novo assembly of the Pseudomonas syringae pv. syringae B728a genome using Illumina/Solexa short sequence reads. FEMS Microbiol. Lett. 291:103–111 [DOI] [PubMed] [Google Scholar]
- 13. Feavers IM, et al. 1999. Multilocus sequence typing and antigen gene sequencing in the investigation of a meningococcal disease outbreak. J. Clin. Microbiol. 37:3883–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fiebelkorn KR, Crawford SA, Jorgensen JH. 2005. Mutations in folP associated with elevated sulfonamide MICs for Neisseria meningitidis clinical isolates from five continents. Antimicrob. Agents Chemother. 49:536–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gilmore A, Jones G, Barker M, Soltanpoor N, Stuart JM. 1999. Meningococcal disease at the University of Southampton: outbreak investigation. Epidemiol. Infect. 123:185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grad YH, et al. 2012. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc. Natl. Acad. Sci. U. S. A. 109:3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harris SR, et al. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harrison OB, Maiden MC, Rokbi B. 2008. Distribution of transferrin binding protein B gene (tbpB) variants among Neisseria species. BMC Microbiol. 8:66 doi:10.1186/1471-2180-8-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hillier LW, et al. 2008. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods 5:183–188 [DOI] [PubMed] [Google Scholar]
- 20. Hobbs MM, et al. 1998. Recombinational reassortment among opa genes from ET-37 complex Neisseria meningitidis isolates of diverse geographical origins. Microbiology 144:157–166 [DOI] [PubMed] [Google Scholar]
- 21. Holmes EC, Urwin R, Maiden MCJ. 1999. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol. Biol. Evol. 16:741–749 [DOI] [PubMed] [Google Scholar]
- 22. Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 23. Ibarz-Pavon AB, et al. 2011. Changes in serogroup and genotype prevalence among carried meningococci in the United Kingdom during vaccine implementation. J. Infect. Dis. 204:1046–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jolley KA, et al. 2012. Ribosomal multi-locus sequence typing: universal characterisation of bacteria from domain to strain. Microbiology 158:1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jolley KA, Brehony C, Maiden MC. 2007. Molecular typing of meningococci: recommendations for target choice and nomenclature. FEMS Microbiol. Rev. 31:89–96 [DOI] [PubMed] [Google Scholar]
- 26. Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595 doi:10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jolley KA, Wilson DJ, Kriz P, McVean G, Maiden MC. 2005. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol. Biol. Evol. 22:562–569 [DOI] [PubMed] [Google Scholar]
- 28. Köser CU, et al. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 366:2267–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kremastinou J, et al. 1999. Recent emergence of serogroup C meningococcal disease in Greece. FEMS Immunol. Med. Microbiol. 23:49–55 [DOI] [PubMed] [Google Scholar]
- 30. Larsen MV, et al. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50:1355–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maiden MC. 2006. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60:561–588 [DOI] [PubMed] [Google Scholar]
- 32. Maiden MC, et al. 2008. Impact of meningococcal serogroup C conjugate vaccines on carriage and herd immunity. J. Infect. Dis. 197:737–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maiden MC, Stuart JM, UK Meningococcal Carriage Group 2002. Carriage of serogroup C meningococci 1 year after meningococcal C conjugate polysaccharide vaccination. Lancet 359:1829–1831 [DOI] [PubMed] [Google Scholar]
- 34. Maiden MCJ, et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Medini D, et al. 2008. Microbiology in the post-genomic era. Nat. Rev. Microbiol. 6:419–430 [DOI] [PubMed] [Google Scholar]
- 36. Morelli G, et al. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet. 42:1140–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nishito Y, et al. 2010. Whole genome assembly of a natto production strain Bacillus subtilis natto from very short read data. BMC Genomics 11:243 doi:10.1186/1471-2164-11-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qvarnstrom Y, Swedberg G. 2000. Additive effects of a two-amino-acid insertion and a single-amino-acid substitution in dihydropteroate synthase for the development of sulphonamide-resistant Neisseria meningitidis. Microbiology 146:1151–1156 [DOI] [PubMed] [Google Scholar]
- 39. Ringuette L, Lorange M, Ryan A, Ashton F. 1995. Meningococcal infections in the province of Quebec, Canada, during the period 1991 to 1992. J. Clin. Microbiol. 33:53–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Russell JE, Jolley KA, Feavers IM, Maiden MC, Suker J. 2004. PorA variable regions of Neisseria meningitidis. Emerg. Infect. Dis. 10:674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salzberg SL, et al. 2012. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res. 22:557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Skoczynska A, Ruckly C, Hong E, Taha MK. 2009. Molecular characterization of resistance to rifampicin in clinical isolates of Neisseria meningitidis. Clin. Microbiol. Infect. 15:1178–1181 [DOI] [PubMed] [Google Scholar]
- 43. Stern A, Meyer TF. 1987. Common mechanism controlling phase and antigenic variation in pathogenic Neisseriae. Mol. Microbiol. 1:5–12 [DOI] [PubMed] [Google Scholar]
- 44. Stothard P, Wishart DS. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21:537–539 [DOI] [PubMed] [Google Scholar]
- 45. Taha MK, et al. 2007. Target gene sequencing to characterize the penicillin G susceptibility of Neisseria meningitidis. Antimicrob. Agents Chemother. 51:2784–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tribe DE, et al. 2002. Increase in meningococcal disease associated with the emergence of a novel ST-11 variant of serogroup C Neisseria meningitidis in Victoria, Australia, 1999–2000. Epidemiol. Infect. 128:7–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vogel U, Claus H, Frosch M, Caugant DA. 2000. Molecular basis for distinction of the ET-15 clone within the ET-37 complex of Neisseria meningitidis. J. Clin. Microbiol. 38:941–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vogel U, et al. 2012. Ion torrent personal genome machine sequencing for genomic typing of Neisseria meningitidis for rapid determination of multiple layers of typing information. J. Clin. Microbiol. 50:1889–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zerbino D. 2010. Using the Velvet de novo assembler for short-read sequencing technologies. Curr. Protoc. Bioinformatics 11:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.