Abstract
Nucleic acid amplification methods such as the PCR have had a major impact on the diagnosis of viral infections, often achieving greater sensitivities and shorter turnaround times than conventional assays and an ability to detect viruses refractory to conventional isolation methods. Their effectiveness is, however, significantly influenced by assay target sequence variability due to natural diversity and rapid sequence changes in viruses that prevent effective binding of primers and probes. This was investigated for a diverse range of enteroviruses (EVs; species A to D), human rhinoviruses (HRVs; species A to C), and human parechovirus (HPeV) in a multicenter assay evaluation using a series of full-length prequantified RNA transcripts. RNA concentrations were quantified by absorption (NanoDrop) and fluorescence methods (RiboGreen) prior to dilution in buffer supplemented with RNase inhibitors and carrier RNA. RNA transcripts were extremely stable, showing minimal degradation after prolonged storage at temperatures between ambient and −20°C and after multiple freeze-thaw cycles. Transcript dilutions distributed to six referral laboratories were screened by real-time reverse transcriptase PCR assays using different primers and probes. All of the laboratories reported high assay sensitivities for EV and HPeV transcripts approaching single copies and similar amplification kinetics for all four EV species. HRV detection sensitivities were more variable, often with substantially impaired detection of HRV species C. This could be accounted for in part by the placement of primers and probes to genetically variable target regions. Transcripts developed in this study provide reagents for the ongoing development of effective diagnostics that accommodate increasing knowledge of genetic heterogeneity of diagnostic targets.
INTRODUCTION
Infections with picornaviruses, human rhinoviruses (HRVs), enteroviruses (EVs), and human parechoviruses (HPeVs) are frequent in human populations worldwide. Diseases associated with these viruses range from the common cold and exacerbation of asthma and bronchitis to severe infections of the central nervous system (CNS) and myocardium. EVs and HRVs are classified as members of the Enterovirus genus (27, 46), a diverse group of human, monkey, and other mammalian viruses, while HPeV is a member of the Parechovirus genus, along with the rodent Ljungan virus (13, 19, 49). The 93 human EVs fall into four genetically distinct species, human EV species A (EV-A) to EV-D (20, 46). Species B variants (echoviruses, Coxsackie B viruses, and Coxsackie A virus 9 [CAV-9]) are the mostly frequently identified viral causes of CNS-associated infections in Western countries and, along with species A serotype EV71, in Southeast Asia. Human rhinoviruses fall into three species, HRV-A to -C, containing 75, 25, and >60 types, respectively (45, 46). The disease associations of different HRV species and types are similar, although with increasing evidence of greater disease severity reported for species C (reviewed in reference 29). The common HPeV variants found in Europe are types 1, 3, 4, and 6, with a recently described association between type 3 infections and severe neonatal infections leading to sepsis (5, 6, 14).
Screening, surveillance, and investigation of the disease associations of different EV, HRV, and HPeV types require assays that are effective for the range of genetic variants found in diagnostic samples. Assays that target the highly conserved 5′ untranslated regions (UTRs) of these viruses (typically by real-time PCR) (2, 9, 18, 37, 51) therefore need to accommodate naturally occurring sequence variability in this region to ensure equal sensitivity. Similarly, virus species and type identification through amplification and sequencing of coding regions such as VP1 in the case of EVs and HPeV (14, 35, 36) and VP4 for HRVs (31, 42) require often quite degenerate primer sequences to allow effective amplification of these more divergent regions of the genome.
In the current study, we have addressed one of the major problems with evaluating the performance of screening and virus typing assays for EVs and PeVs through the creation of a set of standardized RNA transcript controls. Each is quantified in absolute numbers of RNA copies, and together they represent the wide range of naturally occurring EV, HRV, and HPeV variability. While RNA transcripts have been widely used as individual controls for the diagnostic screening of several human viruses, including noroviruses, flaviviruses, EVs, and rhinoviruses (11, 28, 44, 47, 48), this study extends their use to create larger panels representing the full genetic diversity of EV-A to -D, rhinoviruses A and B, recombinant (Ca) and nonrecombinant (Cc) variants of HRV-C (17), and HPeV type 1 (HPeV-1). Comparative evaluation of the sensitivity and amplification dynamics of a wide range of currently implemented real-time PCRs for these viruses, along with several different typing methods, provides considerable insights into the performance of these assays and how they may be potentially improved. For example, their use has revealed frequent potential sensitivity problems with HRV-C detection.
MATERIALS AND METHODS
Human EV, rhinovirus, and HPeV-1 transcripts.
Full-length cDNA clones of CAV-16 (accession number U05876), echovirus 7 (E7) and E30 (AF465516 and GB-27 [unpublished]), CAV-21 (D00538), and EV70 (D00820) representing EV-A to -D (A, CAV-16; B, E7, E30; C, CAV-21; D, EV70) were kindly provided by D. J. Evans, University of Warwick. Rhinovirus species A (HRV-A1b; D00239) and B (HRV-B14; X01087) and the HPeV-1 clone (Harris isolate; FM242866) were provided by G. Stanway, University of Essex. For recombinant HRV-C (HRV-Ca) and nonrecombinant (HRV-Cc) variants, 5′ UTR-VP4-partial VP2 clones were assembled from amplified sequences from the variants R4636/07 (HRV-Cpat19; Cc) and R3092/06 (HRV-C40; Ca) (45).
Plasmids were linearized at the 3′ end and purified by phenol-chloroform extraction and ethanol precipitation. Sense orientation RNA transcripts were generated by T7 RNA polymerase using a MEGAscript in vitro RNA transcription kit (Ambion UK) according to the manufacturer's protocol. Transcribed RNA was DNase treated prior to precipitation with lithium chloride. Newly transcribed RNA was analyzed for integrity on a denaturing RNA-agarose gel with 2.2 M formaldehyde.
RNA quantification.
RNA transcript concentrations were quantified by using two methods, a NanoDrop ND-1000 quantifying optical density at 260 nm and the Quant-iT RiboGreen RNA quantification system, according to manufacturer's protocol (Invitrogen UK). The RNA concentrations determined by the two assays correlated closely (data not shown). RNA concentrations were converted to numbers of genome copies by assuming a mean molecular mass of each base of 330 g/mol. RNA was diluted in RNA storage solution (1 mM sodium citrate, 0.1 mM EDTA, pH 6.0; Ambion UK) containing 0.05 μg/ml herring sperm carrier RNA and 0.1 U/ml RNasin (New England BioLabs UK). Dilutions of RNA were aliquoted and stored at −20°C prior to testing distribution and distribution to referral laboratories. For long-term storage, RNA in storage solution was archived in aliquots at −80°C.
Transcript amplification by real-time PCR.
Dilution series of transcripts (105 to 10−2 copies/μl) were amplified singly or in replicate using routine real-time reverse transcriptase PCR (RT-PCR) assays designed for diagnostic testing for EV, HRV, and HPeV by five laboratories performing real-time PCR detection of EVs, HPeV, and HRV. These were the Specialist Virology Laboratory, Edinburgh Royal Infirmary, Edinburgh, United Kingdom (3); the Regional Virology Laboratory, Gartnavel Hospital, Glasgow, United Kingdom; the Regional Virology Laboratory, Royal Victoria Hospital, Belfast, United Kingdom; the Health Protection Agency (HPA) laboratory, Bristol, United Kingdom; the Department of. Medical Microbiology, Academic Medical Centre, Amsterdam, The Netherlands (EV and HPeV only); and the Department of Virology, University of Turku, Turku, Finland (EV and HRV only). Assays used different primers, probes, and amplification conditions.
Specialist Virology Laboratory, Edinburgh, United Kingdom.
Real-time PCR assays for EVs and HPeVs were performed as previously described (13). HRV screening was performed under the same reaction conditions and with the primers and probes previously described (43).
Regional Virology Laboratory, Gartnavel Hospital, Glasgow, United Kingdom.
Real-time PCR assays for EVs and HPeVs were performed as previously described (3). HRV screening was done with the primers and probes previously described (7).
Regional Virology Laboratory, Belfast, United Kingdom.
Single-step TaqMan RT-PCR assays targeting the 5′ UTR were used to detect EV and HRV. Assays used the Superscript III Platinum One-Step Quantitative RT-PCR System (Invitrogen, Carlsbad, CA) in 10-μl reaction mixture volumes comprising 0.2 μl Superscript III RT Taq mix, 5 μl ×2 Reaction Mix (containing 0.4 mM each deoxynucleoside triphosphate [dNTP]), 3.5 mM MgSO4, 0.4 μM each primer, 0.2 μM probe, and nuclease-free water to a volume of 8 μl. Two microliters of transcript was added as the template, giving a final reaction volume of 10 μl. Real-time RT-PCR was performed in 96 white-well plates using the Roche 480 LightCycler II (Roche, Mannheim, Germany). Cycling conditions were as follows: 50°C for 15 min, 95°C for 5 min, and 45 cycles of 95°C for 10 s and 60°C for 60 s. The primer and probe sequences used for EV and HRV detection were as follows: EV 1A, TCC TCC GGC CCC TGA ATG; EV 1B, GAA ACA CGG ACA CCC AAA GTA; EV 1P, 6-carboxyfluorescein (FAM)-CGGTTCCGCYRCAGA-MGBNFQ; HRV 1A, AGC CTG CGT GGC TGC CTG; HRV 1A2, CCT GCG TGG CGG CCA RC; HRV 1B, CCC AAA GTA GTY GGT CCC RTC C; HRV 1P, FAM-TCC TCC GGC YCC TGA ATG-MGBNFQ.
Department of Medical Microbiology, Amsterdam, The Netherlands.
RNA was directly transcribed by rHex cDNA reaction (40 μl) (5), and 5 μl of transcribed cDNA was amplified by real-time PCR for EV and HPeV as previously described (4).
Department of Virology, University of Turku, Turku, Finland.
HRV and EV transcripts were detected in RT-PCR assays with universal primers from the 5′ UTR (38). RNA transcripts were reverse transcribed with Moloney murine leukemia virus (MMLV) RNase H transcriptase (Promega) in a reaction mixture containing 40 U of the RT enzyme, 4 U of RNasin RNase inhibitor, 500 nM dNTP, 1.2 μM ENRI4− primer (GAAACACGGACACCCAAAGTA), RT buffer, and 5 μl of RNA transcript in a total volume of 20 μl. cDNA synthesis was carried out at 42°C for 1 h. Amplifications with SYBR green detection were performed in 25-μl reaction mixtures with Maxima SYBR master mix (Fermentas), 600 nM ENRI3+ (CGGCCCCTGAATGCGGCTAA) and ENRI4− primers, and 5 μl of cDNA using a Rotor Gene 6000 instrument (Corbett Research). The amplification program included the following steps: 15 min at 95°C; 45 cycles of 15 s at 94°C, 30 s at 65 to 56°C (touchdown, 1°C/cycle for the first 10 cycles), and 40 s at 72°C; and melting at 72 to 95°C at increments of 1°C/5 s. Amplifications with proprietary FAM (HRV)- or Cy5 (EV)-labeled probes were performed analogously with Maxima Probe master mix (Fermentas) without melting curve generation.
HPA, Bristol, United Kingdom.
Reverse transcription was performed in 25-μl volumes using 100 U MMLV RT (Promega) and 0.5 mg/ml random hexamers for 30 min at 37°C, followed by 10 min at 95°C. Real-time PCR was performed in 20-μl reaction volumes consisting of ABI Fast Universal Master Mix (Applied Biosystems) with 5 μl cDNA and primers and probes as described below. A two-temperature thermal cycling protocol (95°C denaturation and 60°C annealing/extension) was used. The primers and probe used for rhinovirus detection were as follows: HuRV-MG1F forward primer, GACARGGTGTGAAGAGCC (300 nM); HuRV-MG1R reverse primer, CAAAGTAGTYGGTCCCATCC (300 nM); HuRV-MG2F forward primer, GACATGGTGTGAAGACYC (300 nM); HuRV-MGP TaqMan probe, (JOE/BHQ) TCCTCCGGCCCCTGAATGYGGCTAA (100 nM). The primers and probe used for EV detection were as follows: EV-F forward primer, CCCCTGAATGCGGCTAATC (300 nM); EV-R reverse primer, ATTGTCACCATAAGCAGCCA (300 nM); EV68aR reverse primer, GTCACCATTAGCAGTCATAAAAGTAA (300 nM); EV-P TaqMan probe, (FAM/BHQ) CGGAACCGACTACTTTGGGTGTCCGT (100 nM). The primers and probes used for PeV detection were as follows: PeV-CCF forward primer, CACTAGTTGTAAGGCCCACGAA (300 nM); PeV-CCR reverse primer, GGCCCCAGATCAGATCCA (300 nM); PeV-CCP TaqMan probe, (Cy5/BHQ) CAGTGTCTCTTGTTACCTGCGGGTACCTTCT (100 nM) (10).
Results from different referral laboratories were normalized to take into account differences in RNA (and, where relevant, cDNA) volumes in different assays.
Other PCR assays.
EV, HRV, and HPeV sequences were amplified by a range of virus typing and reference tests used in the Specialist Virology Laboratory, University of Edinburgh. EV and HRV transcripts were assayed in six replicates by nested PCRs using the primer pairs from the 5′ UTR (50) and the VP4/partial VP2 region (50). VP1 regions were amplified for EV-A, -B, and -D as previously described (16, 26). Species C sequences were amplified by newly designed primers from VP2 (outer sense, position 1172 [5′ base numbered in the poliovirus Leon type 3 isolate, accession number K01392], TCN MRR GGR TGG TGG TGG AA; inner sense, position 1223, TTY GGN CAR AAY ATG TAY TAY CAY TA; outer antisense, position 1731, CCR TTR AAY TCR CWR CAC ATN GG; inner antisense, position 1629, CCC CAR TTR TTR TGY TTN RCC AT). HPeV sequences were amplified in the 5′ UTR (15) and the VP3-VP1 junction (14).
RNA stability.
EV-B (E30) and -A (CAV-16) transcripts were investigated for stability at different temperatures. A 100-μl volume of a 104-copy/μl dilution of each was incubated in storage solution for up to 30 days at ambient temperature, 4°C, and 37°C; an aliquot of each was also freeze-thawed three times. To determine the possible contribution of residual contaminating template DNA to RT-PCR results, control reaction mixtures without a reverse transcription step were prepared in parallel; all control reaction mixtures were negative at the highest transcript concentration tested (60,000 RNA copies/reaction mixture).
Nucleotide sequence accession numbers.
Composite sequences of R4636/07 (HRVCpat19; Cc) and R3092/06 (HRV-C40; Ca) have been deposited in GenBank and assigned accession numbers JX276744 and JX276745, respectively.
RESULTS
Amplification of transcripts by real-time RT-PCR.
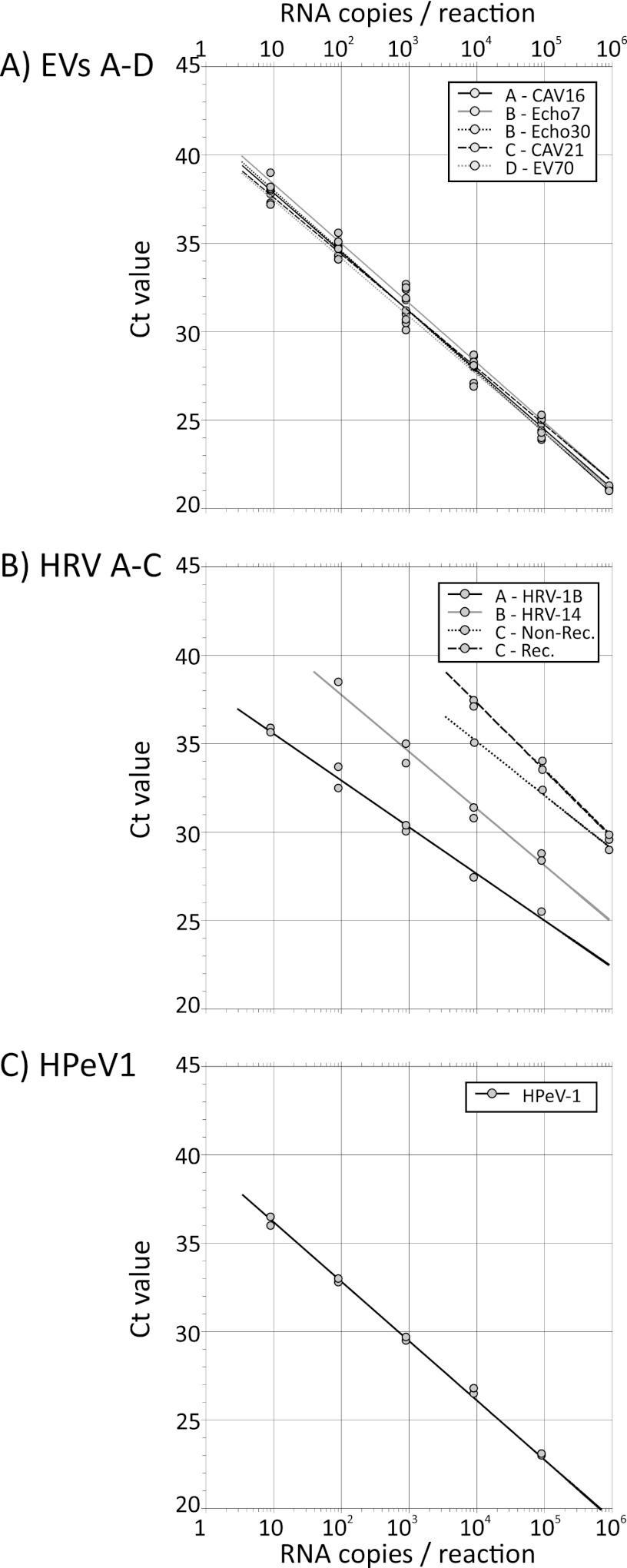
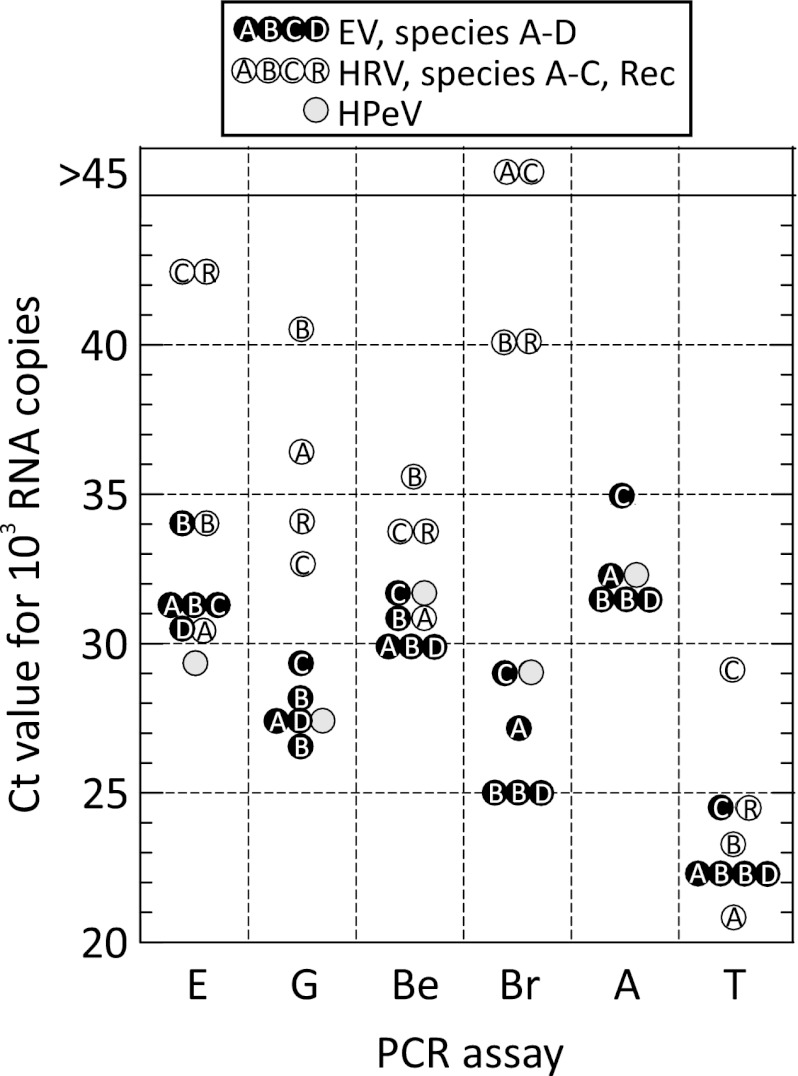
Dilution series of the RNA transcripts ranging from 105 to 10−2 copies/μl were assayed by a previously described EV/HPeV multiplexed PCR method (3) (Fig. 1). The five EV transcripts showed highly reproducible amplification dynamics by real-time PCR with a linear relationship between the log10-transformed RNA input copy number (x axis) and the cycle threshold (CT) value. Amplification efficiency was close to 100% (data not shown). For each transcript, the assay endpoint sensitivity lay between 0.9 and 9 RNA copies (Fig. 2A), apart from negative results for one of two of the replicate assays of E30 and EV70. Assay of the EV transcripts by other laboratories yielded similarly consistent results (Fig. 3), although there was marked variability in amplification rates between the laboratories (CT value of a nominal 1,000 RNA copies), ranging from around 32 (Edinburgh and Amsterdam) to 23 (Turku). There was a similarly wide range of endpoint sensitivities (0.4 to 20 RNA copies) between the laboratories (Fig. 2A), although sensitivity was generally consistent among the five EV transcripts tested by each laboratory. Two-log reduced sensitivity for the species C transcript was observed on testing by the Bristol and Turku laboratories and additionally for species D by the latter laboratory.
Fig 1.
Replicate testing of EV (A), HRV (B), and HPeV (C) transcripts using the multiplexed PCR from the Specialist Virology Laboratory, Edinburgh Royal Infirmary. The nonrecombinant and recombinant HRV-C sequences correspond to HRV-Cpat19 (Cc) and HRV-C40 (Ca), respectively.
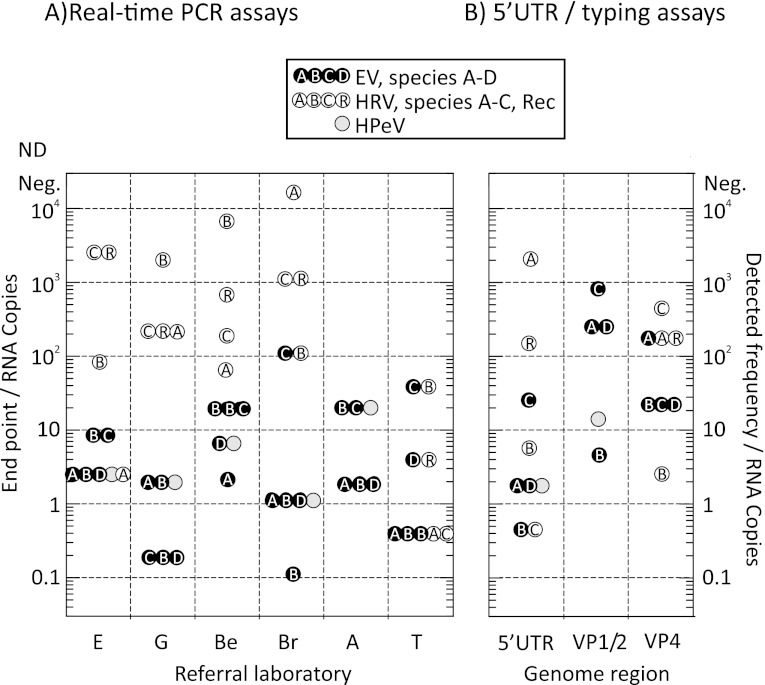
Fig 2.
Endpoint sensitivities for EV, HRV, and HPeV transcript sequences of 5′ UTR-based real-time assays from referral laboratories (A) and 5′ UTR and coding region nested PCR assays (the latter used for typing) (B). For real-time PCR, mean values are shown where testing was carried out in replicate; for typing assays, endpoints were calculated by 6-fold replicate testing in 10-fold dilution steps. Laboratory abbreviations: E, Specialist Virology Laboratory, Edinburgh Royal Infirmary, Edinburgh, United Kingdom; G, Regional Virology Laboratory, Gartnavel Hospital, Glasgow, United Kingdom; Be, Regional Virology Laboratory, Royal Victoria Hospital, Belfast, United Kingdom; Br, HPA, Bristol, United Kingdom; A, Department of. Medical Microbiology, Academic Medical Centre, Amsterdam The Netherlands; T, Department of Virology, University of Turku, Turku, Finland.
Fig 3.
Amplification rate (extrapolated or interpolated CT values for 1,000 input RNA copies) with mean values shown where testing was carried out in replicate for each real-time assay for EV, HRV, and HPeV transcripts. Laboratory abbreviations are as in Fig. 2.
Much more variable amplification dynamics were observed with HRV transcripts, with assays from many laboratories showing reduced sensitivity for species B and C transcripts. For example, amplification of the HRV-A transcript in the multiplexed assay in Edinburgh showed amplification dynamics similar to those of EV-A to -D, while amplification of HRV-B and -C was much slower (Fig. 1B and 3A) and showed reduced assay sensitivity (Fig. 2A). Comparison of testing for HRV transcripts from each laboratory assay showed a consistent trend toward lower amplification rates and endpoint sensitivities for HRV than for EV and HPeV transcripts (Fig. 2A and 3), particularly for species C. In marked contrast, all of the assays detected HPeV RNA sequences with rates and sensitivities similar to those for EVs.
The observed variability of assay sensitivities and amplification efficiencies could be accounted for, at least in part, by the existence of sequence mismatches of primer and probe sequences and the target sequences (Fig. 4). The primers used for EV detection were targeted in conserved regions between positions 416 and 606 within the 5′ UTR. With the exception of the sense primers used in Edinburgh and Glasgow (11), none of the primers or probes mismatched transcript target sequences. However, the one or two mismatches between the sense primer and EV evidently had little effect on assay performance; similar detection efficiencies were observed for transcripts from all four species (Fig. 2A and 3).
Fig 4.
Primer matches to EV and HRV transcript sequences. Mismatches are highlighted with shaded boxes. Sequence alignments are numbered on the basis of the CAV-16 sequence (accession no. U05876).
Primers and probes used for HRV detection were, in general, similarly conserved in the target regions among HRV variants (Fig. 4B). As for EV, few, if any, mismatches were observed with any of the real-time PCR methods, although many assays showed reduced sensitivity for species C rhinoviruses. In the Edinburgh real-time PCR, the poor sensitivity for species C might plausibly have arisen through the two mismatches toward the 3′ end of the antisense primer. All of the HPeV primers and probes showed perfect matches to the HPeV1 transcript (data not shown).
Finally, although the transcripts used in the assay represent a substantial proportion of the naturally occurring sequence diversity of EVs, HRVs, and HPeV, additional variability is shown among the full currently described data set of known types within each species. To illustrate this, strict and 95% consensus sequences were constructed from alignments of each of the described types within EVs and HRVs (Fig. 4). Primers and probes showed many potential mismatches with one or more described variants within EVs and HRVs at numerous positions. For example, both sense and antisense primers fir HRV used by Edinburgh and Glasgow targeted regions in the 5′ UTR that were quite variable, with many positions showing less than 95% conservation among the broader set of HRV types (Fig. 4B). Better conservation was observed in EVs (Fig. 4A) and HPeV-1 to -6 (data not shown).
Sensitivity and specificity of primers used for EV and rhinovirus typing.
RNA transcripts were amplified by nested PCR assays for capsid-coding regions (VP4/partial VP2) of EVs and HRVs, VP1 (EV-A, -B, and -D), partial VP2 (EV-C), and partial VP3/VP1 (HPeV), which are used for (sero)type identification (Fig. 2B; see Table S1 in the supplemental material). Dilutions ranging from 105 to 10−2 copies/μl were assayed in 6-fold replicates. Frequencies of positives at each dilution were used to calculate detected RNA concentrations that were compared with input copy numbers to determine assay sensitivity. Amplification of EV, HRV, and HPeV RNA transcripts by nested PCR in the 5′ UTR showed relatively high sensitivity and close concordance between the input RNA copy numbers and the RNA copy numbers detected (Fig. 2B). The sensitivities of the amplification methods for the coding regions VP4/VP2 (EVs and HRVs), VP1 (EV-A, -B, and -D), VP2 (EV-C), and VP3/VP1 (HPeV) ranged from equal sensitivity to an approximately 100-fold reduction. While these values may vary between serotypes within a species (and could not be assessed with the current panel of transcripts), these findings provide evidence that typing assay sensitivity is broadly comparable between types.
RNA stability.
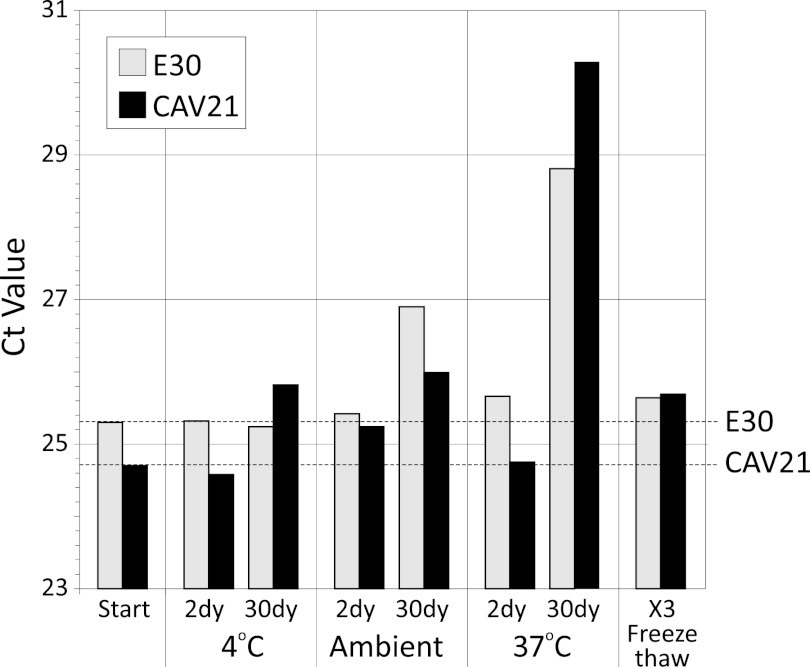
Dilution of transcript RNA in a low-pH, RNase-free solution containing carrier RNA was designed to enhance their longer-term stability and thus the reproducibility of assay evaluation in referral laboratories. To investigate how stable the RNA preparations actually were, two representative EV transcripts (E30, CAV-21) were subjected to a range of temperatures and freeze-thaw cycles and their RNA content was assessed by real-time PCR (Fig. 5). No or minimal changes in CT values (reflecting residual RNA concentrations) were observed on freezing-thawing or incubation for up to 30 days at 4°C, while increases of approximately 2- and 8-fold were observed on incubation at room temperature and 37°C for 30 days (corresponding to approximately 4-fold and 250-fold reductions of amplifiable RNA sequences).
Fig 5.
Stability of E30 and CAV-21 RNA transcripts determined by real-time PCR assay (CT values, y axis) after incubation at different temperatures and for different durations (24 h and 30 days [dy]) or freezing and thawing three times (x axis).
DISCUSSION
RNA transcripts of EV-A to -D, HPeV-1, and rhinoviruses A, B, Ca, and Cc were developed for use as molecularly calibrated RNA standards for validation and comparison of the assay sensitivities of a range of real-time PCRs and typing protocols. This Clinical Virology Network-initiated study is a response to the growing need for independent external validation of molecular-analysis-based diagnostic assays that have become the standard method of virus detection (33, 40). For all three target groups of viruses, substantial sequence diversity in most parts of the genome complicates the development of effective assays capable of detection and identification of all species and types. For each, the 5′ UTR is the most conserved region and is almost invariably targeted by existing real-time PCRs for diagnostic screening. In the case of EVs and HPeVs, screening of cerebrospinal fluid (CSF) in cases of suspected viral meningitis and neonatal sepsis requires high assay sensitivity and specificity due to the low viral loads in this compartment (8, 21, 32, 34). Recent developments of highly sensitive multiplexed PCR assays therefore represent a significant advantage in the screening of patients with these clinical presentations. Indeed, the similarity of the sensitivities of each of the real-time assays evaluated for different EV species provides reassurance that the rarity of species C and D detection in CSF samples cannot be directly attributed to assay insensitivity.
Rhinovirus screening presents a different set of difficulties. Despite its genetic diversity, PCR primers and probes for the 5′ UTR can also accommodate most of the sequence variability between species. However, these target the same conserved regions found in EVs and lead to substantial cross-amplification of EVs by HRV primers and vice versa (12). As demonstrated by the highly variable results from real-time screening of transcript dilutions, there were also substantial differences in assay sensitivity for different HRV species, most markedly for recombinant and nonrecombinant species C variants that were frequently undetected or showed ≥2-log reductions in sensitivity compared to species A and B (Fig. 2). These findings are consistent with previous studies documenting the difficulty in species C detection by PCR. Combined with its inability to be grown in cell culture, this may account for its relatively late discovery in 2006 to 2007 (1, 22–25, 30, 39). The demonstration of largely ineffective detection of species C will lead to improved assay design and modified primer/probe sequences. These can be reevaluated with the transcripts as part of its validation process; this process is under way in Edinburgh.
The same transcripts were used to evaluate the sensitivity of EV, HRV, and HPeV typing assays based on the amplification and sequencing of coding region sequences. This is necessary not only because sequence variability in the 5′ UTR is so restricted as to preclude (sero)type identification, but the occurrence of recombination between the 5′ UTR and the rest of the genome (41) and the consequent absence of species-specific 5′ UTR sequences prevent reliable species identification. A variety of typing assays have been developed, in the case of EV and PeVs, in the VP1 region (5, 14, 35, 36), where sequences have been shown to be highly predictive of EV and HPeV types. Similarly, VP4 sequences provide a reliable indication of HRV types, a more conserved region of the capsid gene that can be amplified with a common set of primers (31, 42, 50).
Variable amplification efficiencies with nested primers from the different capsid-encoding regions were observed (Fig. 2B), observations that derive from primer mismatches with target sequences in the transcripts that cannot be fully accommodated through the use of degenerate bases. However, sensitivity differences from screening (5′ UTR-based) assays and from calculated input RNA copy numbers were rarely greater than 100-fold (and usually much lower). In the specific cases of EV-B and HPeV, almost equivalent sensitivities compared to screening PCRs may underlie the previously described high frequency of successfully typed CSF samples despite their generally low viral loads (13, 14, 26).
The use of an RNA dilution buffer specifically designed to prevent RNA degradation through the incorporation of RNase inhibitors and a low concentration of carrier RNA proved highly effective at maintaining high stability despite a variety of mistreatments (Fig. 5). The minimal effect of freezing-thawing and its stability at both 4°C and ambient temperature suggest that dilution series of the RNA transcript standards can be relatively easily distributed without the need for frozen shipment. Combined with their lack of infectivity and extremely low cost of production and distribution, these transcripts can be readily supplied to diagnostic laboratories as a contribution to their quality assurance mechanisms and ongoing assay evaluations.
In summary, this study has successfully developed and evaluated a series of RNA transcripts that capture much of the diversity of EV and HRV and provide the means for laboratories to readily evaluate their screening and typing assays in absolute (RNA copy number) terms. These reagents could contribute substantially to comparisons of assay sensitivities between laboratories, troubleshooting, and ongoing assay development as our understanding of picornavirus diversity increases.
Supplementary Material
ACKNOWLEDGMENT
We thank Tiina Ylinen, Department of Virology, University of Turku, Turku, Finland, for technical help.
Footnotes
Published ahead of print 27 June 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Arden KE, McErlean P, Nissen MD, Sloots TP, Mackay IM. 2006. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J. Med. Virol. 78:1232–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arola A, Santti J, Ruuskanen O, Halonen P, HyypiÄ T. 1996. Identification of enteroviruses in clinical specimens by competitive PCR followed by genetic typing using sequence analysis. J. Clin. Microbiol. 34:313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bennett S, et al. 2011. Rapid simultaneous detection of enterovirus and parechovirus RNAs in clinical samples by one-step real-time reverse transcription-PCR assay. J. Clin. Microbiol. 49:2620–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benschop K, Molenkamp R, van der Ham A, Wolthers K, Beld M. 2008. Rapid detection of human parechoviruses in clinical samples by real-time PCR. J. Clin. Virol. 41:69–74 [DOI] [PubMed] [Google Scholar]
- 5. Benschop KS, et al. 2006. Human parechovirus infections in Dutch children and the association between serotype and disease severity. Clin. Infect. Dis. 42:204–210 [DOI] [PubMed] [Google Scholar]
- 6. Boivin G, Abed Y, Boucher FD. 2005. Human parechovirus 3 and neonatal infections. Emerg. Infect. Dis. 11:103–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brittain-Long R, et al. 2008. Multiplex real-time PCR for detection of respiratory tract infections. J. Clin. Virol. 41:53–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Casas I, et al. 2001. Molecular characterization of human enteroviruses in clinical samples: comparison between VP2, VP1, and RNA polymerase regions using RT nested PCR assays and direct sequencing of products. J. Med. Virol. 65:138–148 [PubMed] [Google Scholar]
- 9. Chapman NM, Tracy S, Gauntt CJ, Fortmueller U. 1990. Molecular detection and identification of enteroviruses using enzymatic amplification and nucleic acid hybridization. J. Clin. Microbiol. 28:843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corless CE, et al. 2002. Development and evaluation of a ‘real-time’ RT-PCR for the detection of enterovirus and parechovirus RNA in CSF and throat swab samples. J. Med. Virol. 67:555–562 [DOI] [PubMed] [Google Scholar]
- 11. Dierssen U, Rehren F, Henke-Gendo C, Harste G, Heim A. 2008. Rapid routine detection of enterovirus RNA in cerebrospinal fluid by a one-step real-time RT-PCR assay. J. Clin. Virol. 42:58–64 [DOI] [PubMed] [Google Scholar]
- 12. Harvala H, et al. 2012. High detection frequency and viral loads of human rhinovirus species A to C in fecal samples; diagnostic and clinical implications. J. Med. Virol. 84:536–542 [DOI] [PubMed] [Google Scholar]
- 13. Harvala H, et al. 2011. Comparison of human parechovirus and enterovirus detection frequencies in cerebrospinal fluid samples collected over a 5-year period in Edinburgh—HPeV type 3 identified as the most common picornavirus type. J. Med. Virol. 83:889–896 [DOI] [PubMed] [Google Scholar]
- 14. Harvala H, et al. 2009. Aetiological role of human parechovirus type 3 in neonatal sepsis identified by direct typing assay on cerebrospinal fluid. J. Infect. Dis. 199:1753–1760 [DOI] [PubMed] [Google Scholar]
- 15. Harvala H, et al. 2008. Epidemiology and clinical associations of human parechovirus respiratory infections. J. Clin. Microbiol. 46:3446–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harvala H, et al. 2011. Detection and genetic characterization of enteroviruses circulating among wild populations of chimpanzees in Cameroon: relationship with human and simian enteroviruses. J. Virol. 85:4480–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang T, et al. 2009. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One 4:e6355 doi:10.1371/journal.pone.0006355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hyypiä T, Auvinen P, Maaronen M. 1989. Polymerase chain reaction for human picornaviruses. J. Gen. Virol. 70:3261–3268 [DOI] [PubMed] [Google Scholar]
- 19. Hyypiä T, et al. 1992. A distinct picornavirus group identified by sequence analysis. Proc. Natl. Acad. Sci. U. S. A. 89:8847–8851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hyypiä T, Hovi T, Knowles NJ, Stanway G. 1997. Classification of enteroviruses based on molecular and biological properties. J. Gen. Virol. 78:1–11 [DOI] [PubMed] [Google Scholar]
- 21. Iturriza-Gómara M, Megson B, Gray J. 2006. Molecular detection and characterization of human enteroviruses directly from clinical samples using RT-PCR and DNA sequencing. J. Med. Virol. 78:243–253 [DOI] [PubMed] [Google Scholar]
- 22. Kistler A, et al. 2007. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J. Infect. Dis. 196:817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lamson D, et al. 2006. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004-2005. J. Infect. Dis. 194:1398–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lau SK, et al. 2007. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J. Clin. Microbiol. 45:3655–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee WM, et al. 2007. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS One 2:e966 doi:10.1371/journal.pone.0000966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leitch EC, et al. 2009. Direct identification of human enterovirus serotypes in cerebrospinal fluid by amplification and sequencing of the VP1 region. J. Clin. Virol. 44:119–124 [DOI] [PubMed] [Google Scholar]
- 27. Lindberg AM, Johansson S. 2002. Phylogenetic analysis of Ljungan virus and A-2 plaque virus, new members of the Picornaviridae. Virus Res. 85:61–70 [DOI] [PubMed] [Google Scholar]
- 28. Lu X, et al. 2008. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J. Clin. Microbiol. 46:533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mackay IM. 2008. Human rhinoviruses: the cold wars resume. J. Clin. Virol. 42:297–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McErlean P, et al. 2007. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J. Clin. Virol. 39:67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McIntyre CJ, McWilliam-Leitch EC, Savolainen-Kopra C, Hovi T, Simmonds P. 2010. Analysis of genetic diversity and sites of recombination in human rhinovirus species C. J. Virol. 84:10297–10310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mirand A, et al. 2008. Prospective identification of enteroviruses involved in meningitis in 2006 through direct genotyping in cerebrospinal fluid. J. Clin. Microbiol. 46:87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Niesters HG. 2002. Clinical virology in real time. J. Clin. Virol. 25(Suppl 3):S3–S12 [DOI] [PubMed] [Google Scholar]
- 34. Nix WA, Oberste MS, Pallansch MA. 2006. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J. Clin. Microbiol. 44:2698–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Norder H, Bjerregaard L, Magnius LO. 2001. Homotypic echoviruses share aminoterminal VP1 sequence homology applicable for typing. J. Med. Virol. 63:35–44 [PubMed] [Google Scholar]
- 36. Oberste MS, et al. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olive DM, et al. 1990. Detection and differentiation of picornaviruses in clinical samples following genomic amplification. J. Gen. Virol. 71(Pt 9):2141–2147 [DOI] [PubMed] [Google Scholar]
- 38. Peltola V, et al. 2008. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J. Infect. Dis. 197:382–389 [DOI] [PubMed] [Google Scholar]
- 39. Renwick N, et al. 2007. A recently identified rhinovirus genotype is associated with severe respiratory-tract infection in children in Germany. J. Infect. Dis. 196:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saldanha J. 2001. Validation and standardisation of nucleic acid amplification technology (NAT) assays for the detection of viral contamination of blood and blood products. J. Clin. Virol. 20:7–13 [DOI] [PubMed] [Google Scholar]
- 41. Santti J, Hyypiä T, Kinnunen L, Salminen M. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Savolainen C, Blomqvist S, Mulders MN, Hovi T. 2002. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 83:333–340 [DOI] [PubMed] [Google Scholar]
- 43. Scheltinga SA, Templeton KE, Beersma MF, Claas EC. 2005. Diagnosis of human metapneumovirus and rhinovirus in patients with respiratory tract infections by an internally controlled multiplex real-time RNA PCR. J. Clin. Virol. 33:306–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwaiger M, Cassinotti P. 2003. Development of a quantitative real-time RT-PCR assay with internal control for the laboratory detection of tick borne encephalitis virus (TBEV) RNA. J. Clin. Virol. 27:136–145 [DOI] [PubMed] [Google Scholar]
- 45. Simmonds P, et al. 2010. Proposals for the classification of human rhinovirus species C into genotypically-assigned types. J. Gen. Virol. 91:2409–2419 [DOI] [PubMed] [Google Scholar]
- 46. Stanway G, et al. 2005. Family Picornaviridae, p 757–778 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses Elsevier/Academic Press, London, United Kingdom [Google Scholar]
- 47. Tan EL, Chow VT, Quak SH, Yeo WC, Poh CL. 2008. Development of multiplex real-time hybridization probe reverse transcriptase polymerase chain reaction for specific detection and differentiation of Enterovirus 71 and Coxsackievirus A16. Diagn. Microbiol. Infect. Dis. 61:294–301 [DOI] [PubMed] [Google Scholar]
- 48. Trujillo AA, et al. 2006. Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J. Clin. Microbiol. 44:1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wigand R, Sabin AB. 1961. Properties of ECHO types 22, 23 and 24 viruses. Arch. Gesamte Virusforsch. 11:224–247 [DOI] [PubMed] [Google Scholar]
- 50. Wisdom A, McWilliam Leitch C, Gaunt E, Harvala H, Simmonds P. 2009. Screening respiratory samples for human rhinoviruses (HRV) and enteroviruses: comprehensive VP4/2-typing reveals high incidence and genetic diversity of HRV species C. J. Clin. Microbiol. 47:3958–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zoll GJ, et al. 1992. General primer-mediated polymerase chain reaction for detection of enteroviruses: application for diagnostic routine and persistent infections. J. Clin. Microbiol. 30:160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.