Abstract
Single-nucleotide polymorphism (SNP) arrays have become a popular technology for disease-association studies, but they also have potential for studying the genetic differentiation of human populations. Application of the Affymetrix GeneChip Human Mapping 500K Array Set to a population of 102 individuals representing the major ethnic groups in the United States (African, Asian, European, and Hispanic) revealed patterns of gene diversity and genetic distance that reflected population history. We analyzed allelic frequencies at 388, 654 autosomal SNP sites that showed some variation in our study population and 10% or less missing values. In spite of the small size (23-31 individuals) of each subpopulation, there were no fixed differences at any site between any two subpopulations. As expected from the African origin of modern humans, greater gene diversity was seen in Africans than in either Asians or Europeans, and the genetic distance between Asians and European populations was significantly lower than that between either of these two populations and Africans. Principal components analysis applied to a correlation matrix among individuals was able to separate completely the major continental groups of humans (Africans, Asians, and Europeans), while Hispanics overlapped all three of these groups. Genes containing two or more markers with extraordinarily high genetic distance between subpopulations were identified as candidate genes for health differences between subpopulations. The results show that, even with modest sample sizes, genome-wide SNP genotyping technologies have great promise for capturing signatures of gene frequency difference between human subpopulations, with applications in areas as diverse as forensics and the study of ethnic health disparities.
Recent years have seen an increasing interest in health disparities among human subpopulations [1-3] and the possibility that genetic differences among human populations may play some role in health differences [4-6]. Substantial genetic evidence supports the hypothesis that modern humans originated in Africa [7-9]. Populations whose ancestors emigrated out of Africa to Europe and Asia came to differ from the African population in allelic frequencies as a result both of population bottlenecks in emigration and subsequent genetic drift. As a result, major geographic subdivisions of the human population are known to differ from one another in allelic frequencies at numerous loci, from allozymes [10] to Alu insertion polymorphisms [11] to single-nucleotide polymorphisms [12]. However, relatively few fixed differences occur between major geographic subdivisions of humans; most known differences involve allele frequency differences, not the complete replacement of one allele by another. Moreover, if studied on a fine geographic scale, much of the gene frequency difference between continents appears to be clinal [13].
The diseases for which studies have shown differences of incidence between human subpopulations are typically complex diseases whose etiology is thought to involve both environmental and, possibly, genetic factors [14-19]. The genetic contribution to complex diseases is thought to be polygenic and to involve small contributions from a number of relatively common allelic variants, in sharp contrast to Mendelian disease genes subject to a mutation-selection balance [5, 20]. Such common allelic variants might plausibly be among those that differ in allelic frequency between major human subpopulations because of either genetic drift or adaptation to local environmental conditions. Furthermore, described differences between human populations with respect to complex disease phenotypes typically involve frequency differences, as would be expected if gene frequency differences play a role.
Array technologies have become an important tool in assaying human genetic variation and in genetic disease-association studies. Here we apply the Affymetrix GeneChip Human Mapping 500K Array Set to the SNP500 Cancer population, a population of 102 individuals representing major U.S. ethnic groups [21]. The 500,000 SNPs assayed by this system were chosen to provide markers evenly distributed along the chromosomes and having relatively high gene diversity (heterozygosity) in the human population on the basis of gene frequency data from the HapMap project [22]. We first analyzed the results for the expected signatures of human population history. These included gene frequency differences at these SNP sites between populations in our sample representing the major historic subdivisions of humans (African, European, and Asian), as well as evidence of admixture, particularly in the U.S. Hispanic populations. In looking for these signatures, we test both whether the SNP500 population shows evidence of population history and whether the 500K Array Set can be used to discover aspects of human population history. Next we searched for evidence of protein-coding genes containing more markers with elevated inter-population gene frequency differences than expected by chance, on the expectation that such genes may contribute to phenotypic differences between populations, possibly including complex disease phenotypes.
RESULTS
Gene Diversity and Genetic Distance
We computed overall mean and median gene diversity in the SNP500 Cancer population for 388, 654 autosomal SNP loci assayed by the Affymetrix GeneChip Human Mapping 500K Array Set. The mean (0.281) and median (0.290) gene diversity in our study population were quite high (Table 1), reflecting the manufacturer’s choice of high-diversity markers. Nonetheless, there were significant differences among subpopulations with respect to gene diversity, supported by both parametric and nonparametric tests (Table 1). The African subpopulation showed the highest mean and median values of gene diversity, both of which differed significantly from those of other subpopulations (Table 1). The Hispanic subpopulation interestingly showed significantly greater mean and median gene diversity than either European or Asian subpopulations, but significantly less than the African subpopulation (Table 1).
Table 1.
Gene diversity at 388, 654 autosomal SNP sites.
| Population | Mean ± S.E. | Median (range) |
|---|---|---|
| African | 0.27804 ± 0.00026 | 0.28733 (0.00000-0.50000) |
| European | 0.25281 ± 0.00029 1 | 0.26977 4 (0.00000-0.50000) |
| Asian | 0.24535 ± 0.00029 1,2 | 0.28733 4,5 (0.00000-0.50000) |
| Hispanic | 0.26874 ± 0.00027 1,2,3 | 0.28733 4,5,6 (0.00000-0.50000) |
| All | 0.28112 ± 0.00025 | 0.29066 (0.00976-0.50000) |
Significantly different from African (2-tailed P < 0.001; paired t-test)
Significantly different from European (2-tailed P < 0.001; paired t-test)
Significantly different from Asian (2-tailed P < 0.001; paired t-test)
Significantly different from African (2-tailed P < 0.001; Wilcoxon signed-rank test)
Significantly different from European (2-tailed P < 0.001; Wilcoxon signed rank test)
Significantly different from Asian (2-tailed P < 0.001; Wilcoxon signed rank test)
The mean and median values of the genetic distance between African and Asian subpopulations were significantly greater than those between African and European (Table 2). This difference may reflect European admixture in the case of African Americans. The Hispanic subpopulation showed the lowest mean and median genetic distance with the European subpopulation, significantly lower than those between Hispanic and African or Hispanic and Asian (Table 2). On the other hand, the Hispanic subpopulation showed significantly lower mean and median genetic distance to the African subpopulation than did the European subpopulation (Table 2). Likewise, the Hispanic population showed significantly lower mean and median genetic distance to the Asian subpopulation than did the European subpopulation (Table 2).
Table 2.
Genetic distance between populations at 388, 654 autosomal SNP sites.
| Comparison | Mean ± S.E. | Median (range) |
|---|---|---|
| African-European | 0.03718 ± 0.00008 | 0.02014 (0.00000-0.68377) |
| African-Asian | 0.04223 ± 0.00009 1 | 0.02198 5 (0.00000-0.68377) |
| European-Asian | 0.02404 ± 0.00006 1,2 | 0.01019 5,6 (0.00000-0.51099) |
| African-Hispanic | 0.02559 ± 0.00005 1 | 0.01250 5 (0.00000-0.54437) |
| European-Hispanic | 0.00978 ± 0.00004 3 | 0.00431 7 (0.00000-0.20600) |
| Asian-Hispanic | 0.01878 ± 0.00004 4 | 0.01047 6 (0.00976-0.46839) |
Significantly different from African-European distance (2-tailed P < 0.001; paired t-test)
Significantly different from African-Asian distance (2-tailed P < 0.001; paired t-test)
Significantly different from African-Hispanic distance and from Asian-Hispanic distance (2-tailed P < 0.001 in each case; paired t-tests).
Significantly different from European-Asian distance (2-tailed P < 0.001; paired t-test)
Significantly different from African-European distance (2-tailed P < 0.001; Wilcoxon signed rank test)
Significantly different from African-Asian distance (2-tailed P < 0.001; Wilcoxon signed rank test)
Significantly different from African-Hispanic distance and from Asian-Hispanic distance (2-tailed P < 0.001 in each case; Wilcoxon signed rank tests).
Significantly different from European-Asian distance (2-tailed P < 0.001; Wilcoxon signed rank test)
In spite of gene frequency differences between populations, none of the 388, 654 sites showed a fixed difference between any two subpopulations; that is, there was no case in which one allele was fixed in one subpopulation and another allele was fixed in another subpopulation. When gene diversities and genetic distances were calculated only for the SNP sites with no missing values (N = 133,826), the patterns were similar to those seen in the larger data set (not shown).
Genetic Correlations Among Individuals
We analyzed genetic correlations among individuals in order to determine to what extent gene frequency information at these SNP sites could be used to separate major continental groups of humans. There were 150, 557 SNPs with no missing values for the African, Asian, and European subpopulations. When pairwise correlations were calculated among the 79 individuals in these three subpopulations, the mean of 1017 within-subpopulations correlations (0.652 ± 0.001 S.E.) differed significantly from the mean of 2064 between-population correlations (0.570 ± 0.001; P < 0.001; randomization test).
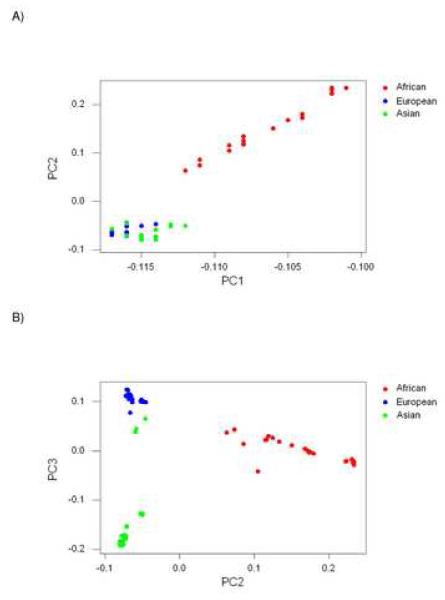
We extracted principal components from the 79 × 79 matrix of genetic correlation coefficients between individuals in the African, Asian, and European subpopulations. The first principal component (PC1) accounted for 60.3% of the variance in the matrix. Loadings on PC1 were relatively high in members of the African subpopulation in comparison to Asian and European subpopulations, although Africans were not completely separated from the other subpopulations along this axis (Figure 1A). Thus PC1 seemed to reflect mainly patterns of gene frequency originating in Africa. PC2, which accounted for 4.0% of the variance, clearly contrasted Africans, on the one hand, and Asians and Europeans, on the other hand (Figure 1A). PC3, which accounted for 2.6% of the variance, contrasted Asians with Europeans (Figure 1B).
Figure 1.
Plots of loadings on principal components (PCs) extracted from matrix of genetic correlations at 150, 557 SNP sites in 79 individuals (excluding Hispanics): (A) PC2 vs. PC1; (B) PC3 vs. PC2.
When loadings on PC2 and PC3 were plotted on the same set of axes, the three major continental subpopulations (Africans, Asians, Europeans) were completely separated from each other in two-dimensional space (Figure 1B). No other principal component beyond PC1, PC2, and PC3 accounted for more than 1% of the variance in the data. The majority of Asians showed strongly negative loadings on PC3, strongly contrasting with both Europeans and Africans on this axis (Figure 1B). These individuals (N = 21) all had reported ancestry in East Asia. By contrast, three Asians clustered much closer to Europeans on PC3 (Figure 1B). The latter three had reported ancestry in the Indian subcontinent.
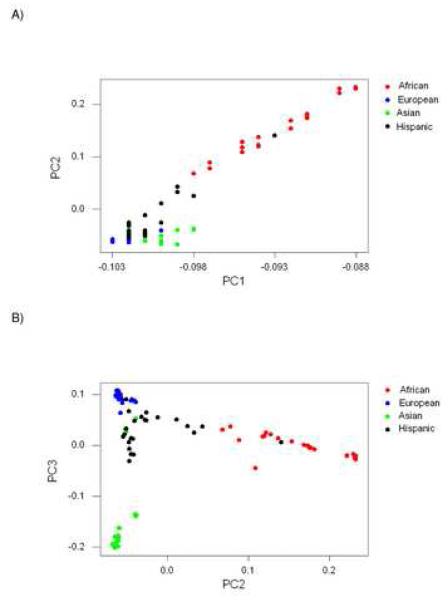
When the Hispanic subpopulation was included, there were 133, 826 sites with no missing values. Correlations among individual genotypic scores at these sites were estimated, and principal components were extracted from the 102 × 102 correlation matrix. PC1, accounting for 61.5% of the variance, again had high loadings on Africans, although one Hispanic individual fell in the midst of the Africans along this axis (Figure 2A). Other Hispanics grouped with Europeans and Asians along PC1 (Figure 2A). PC2, accounting for 3.2% of the variance, separated Africans from the other subpopulations, except for one Hispanic individual (Figure 2A). PC3, accounting for 2.0% of the variance, separated one group of Asians from other Asians, Europeans, and most Hispanics (Figure 2B). No other principal component accounted for as much as 1% of the variance.
Figure 2.
Plots of loadings on principal components (PCs) extracted from matrix of genetic correlations at 133, 826 SNP sites in 102 individuals (including Hispanics): (A) PC2 vs. PC1; (B) PC3 vs. PC2.
Thus, when Hispanics were included in the analyses, the first three principal components took a similar form to those obtained when Hispanics were excluded. The loadings on non-Hispanics of PC1, PC2, and PC3 were very similar in the data set including Hispanics (Figure 2) to the corresponding values for the data set excluding Hispanics (Figure 2). As a result, the three other subpopulations were still completely separated in two-dimensional space by PC2 and PC3 even when Hispanics were included (Figure 2B). However, Hispanics themselves could not be separated from the other subpopulations by principal component analysis.
Genes with High Divergence
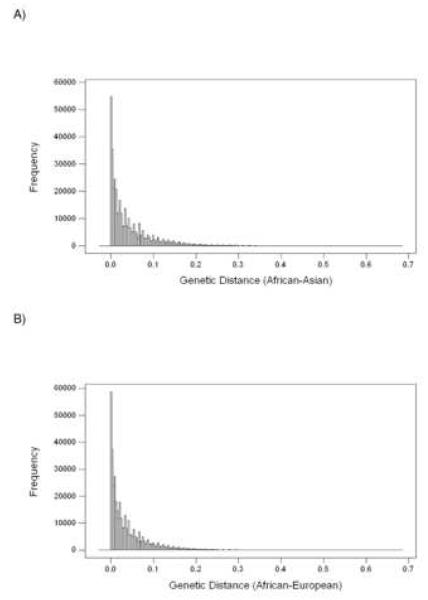
Genetic distances at the 388, 654 SNP sites for both the African-Asian and African-European comparisons showed strongly positively skewed distributions (Figure 3). Skewness was 2.31 in the case of the African-Asian distances and 2.41 in the case of African-European distances. These high positive skewness values indicate a distribution with a long right tail. We chose genes having two or more SNPs with genetic distance > 0.37 between African and Asian populations as candidates for major allelic frequency differences between these two populations (Table 3). Randomization tests applied to each chromosome indicated that the probability of two or more such SNPs occurring in a single gene by chance was less than 5 × 10−5. Likewise, we chose genes having two or more SNPs with genetic distance > 0.34 between African and European populations as candidates for major allelic frequency differences between these two populations (Table 4). Randomization tests applied to each chromosome indicated that the probability of two or more SNPs with genetic distances greater than these cut-off values occurring in a single gene by chance was less than 5 × 10−5 in every case.
Figure 3.
Histograms illustrating frequency distributions of genetic distance at 388, 654 SNP sites between (A) African and Asian; and (B) African and European subpopulations.
Table 3.
Genes with two or more SNPs showing significantly (P < 5 × 10−5) high African-Asian genetic distance.
| Chromosome | Locus (protein function) | SNPs with high genetic distance 1 |
Known nonsynonymous SNPs 1 |
|---|---|---|---|
| 1 |
LYST (lysosomal trafficking regulator) |
rs7522053 (I3); rs7529320 (I3); rs9970096 (I14); rs57533575 (I28); rs6667717 (I39) |
rs6665568 (E20); rs7541091 (E23); rs2753327 (E35) |
| 2 | SEC15L2 (sec15-like 2) | rs11686713 (I3); rs6546753 (I20) |
rs2048173 (E1) |
| 2 |
GLI2 (GLI-Kruppel family member) |
rs895552 (I2); rs895553 (I2); rs2871874 (I2); rs11122834 (I2); rs1466044 (I2) rs1466042 (I2); rs1107445 (I2) |
rs13427953 (E5); rs12618388 (E8); rs3099537 (E8) |
| 3 |
FHIT (fragile histidine triad) |
rs750793 (I8); rs294451 (I8) |
-- |
| 4 |
SLC30A9 (solute carrier family 30, zinc transporter, member 9) |
rs2581441 (I2); rs10433709 (I8); rs804191 (I11); rs11051 (E18, 3′UTR) |
rs1047626 (E2); rs1801962 (E12) |
| 4 |
CCDC4 (coiled-coil domain containing 4) |
rs6447132 (I3); rs57664565 (I3) |
-- |
| 4 |
SPATA5 (spermatogenesis associated 5) |
rs307029 (I14); rs307032 (I14) |
rs28716389 (E16) |
| 4 |
PALLD (paladin, cytoskeletal associated protein) |
rs4616716 (I3); rs436442 (I16) |
rs7655494 (E2); rs7671781 (E2); rs54293759 (E12) |
| 5 | CDH9 (cadherin 9, type 2) | rs6871019 (I2); rs10805793 (I2) |
rs2288466 (E2); rs2288467 (E2) |
| 6 |
BAI3 (brain-specific angiogenesis inhibitor 3) |
rs4706854 (I16); rs7759645 (I16) |
rs2183071 (E28) |
| 8 |
XKR6 (Kell blood group complex subunit-related family, member 6) |
rs11773990 (I1); rs10108618 (I1) |
rs13255844 (E1) |
| 9 |
IARS (isoleucine tRNA synthase) |
rs115757 (I19); rs471478 (I30); rs10761149 (I31) |
rs5556155 (E27); rs11547887 (E27); rs17855985 (E27); rs556155 (E32) |
| 12 |
POLR3B (DNA-directed RNA polymerase III, polypeptide B) |
rs11112952 (I6); rs2246353 (I12); rs12305703 (I12) |
rs17038460 (E18) |
| 12 |
MPHOSPH9 (M-phase phosphoprotein 9) |
rs1716168 (I12); rs1716167 (I13) |
rs1260318 (E18) |
| 20 |
HM13 (minor histocompatibility 13; signal peptide peptidase) |
rs6058022 (I1); rs11906851 (I11); rs6120719 (I11) |
rs17855400 (E7); rs1044419 (E8) |
| 21 |
BACE2 (beta-site APP-cleaving enzyme 2) |
rs766850 (I1); rs6517656 (I1); rs12329755 (I1) |
-- |
SNPs are designated by dbSNP I.D. Abbreviations: I = intron; E = exon. Boldface indicates nonsynonymous SNPs showing frequency differences between the two populations in HapMap frequency data.
Table 4.
Genes with two or more SNPs showing significantly high (P < 5 × 10−5) African-European genetic distance.
| Chromosome | Locus (protein function) | SNPs with high genetic distance 1 |
Known nonsynonymous SNPs 1 |
|---|---|---|---|
| 1 |
HIVEP3 (HIV-1 enhancer binding protein 3) |
rs2077354 (I1); rs4526604 (I1); rs710235 (I1); rs4284254 (I1); |
rs17363472 (E4); rs2810566 (E4); rs2181280 (E4) rs2146315 (E4); rs2291344 (E8); rs2483689 (E8); rs9439043 (E9) rs11809423 (E9) |
| 1 |
GLIS1 (GLIS family zinc finger protein 1) |
rs12062528 (I1); rs12120383 (I2) |
rs4307514 (E3) |
| 2 |
MAP4K3 (mitogen-activated protein kinase kinase kinase kinase 3) |
rs6721049 (I1); rs7596222 (I1) |
rs13005084 (E1) |
| 2 |
GLI2 (GLI-Kruppel family member) |
rs895552 (I2); rs895553 (I2); rs2871874 (I2); rs11122834 (I2); rs1466044 (I2) rs1107445 (I2); rs2311803 (I3) |
rs13427953 (E5); rs12618388 (E8); rs3099537 (E8) |
| 3 |
ULK4 (unk-51-like kinase 4) |
rs9864051 (I19); rs9814300 (I8) |
rs1052501 (E9); rs3774372 (E10); rs17063572 (E11); rs4973986 (E12); rs17215589 (E13); rs6769117 (E29); rs12488691 (E29) |
| 4 |
ARHGAP10 (rho GTPase activating protein 10) |
rs6535556 (I1); rs6822971 (I1); rs11729081(I1); rs17614025 (I10); rs3990920 (I1); rs1194409 (I13) |
rs2276832 (E21) |
| 4 |
LRBA (LPS-responsive vesicle trafficking, BEACH and anchor containing) |
rs11099780 (I2); rs2407548 (I2); rs1201202 (I5); rs7690977 (I22); rs57665654 (I34); rs1993109 (I35) |
rs1782360 (E23); rs13151295 (E24); rs13151294 (E24); rs17027133 (E30); rs3749574 (E55); rs2290846 (E57) |
| 5 | SEPT8 (septin 8) | rs402959 (I1); rs30527 (I1) |
-- |
| 7 |
ICA1 (islet cell autoantigen 1) |
rs6977682 (I3); rs1861032 (I4) |
rs17847180(E7) |
| 8 |
SLC20A2 (solute carrier family 20, phosphate transporter, member 2) |
rs7832529 (I5); rs3888124 (I10) |
-- |
| 8 |
ST18 (suppressor of tumorigenicity 18, carcinoma, zinc finger protein) |
rs12674582 (I2); rs7459810 (I2); rs7388668 (I2) |
rs2303460 (E28) |
| 8 |
XKR4 (Kell blood group complex subunit-related family, member 4) |
rs7844897 (I1); rs16921658 (I1); rs13439780 (I2) |
-- |
| 9 |
ZNF782(zinc finger protein 782) |
rs10124033 (I3); rs7859940 (I5); rs12236125 (I5) |
rs4645656 (E6); rs7870376 (E6); |
| 9 |
TNFSF8 (tumor necrosis factor ligand superfamily, member 8) |
rs3181360 (I1); rs3181362 (I3) |
-- |
| 11 |
GRM5 (glutamate receptor, metabotropic 5) |
rs10741500 (I2); rs4488199 (I3); rs992259 (I3) |
-- |
| 14 | GPHN (gephyrin) | rs7156737 (I1); rs8007677 (I2); rs8022328 (I3); rs723432 (I5); rs8013401 (I8); rs10142059 (I8); rs8003929 (I10); rs10138850 (I16); rs2281676 (I20) |
-- |
| 15 |
APBA2 (amyloid beta A4 precursor protein-binding family A, member 2) |
rs4424881 (I1); rs6495913 (I2) |
rs8040932 (E3); rs1046394 (E11); rs4581676 (E11) |
| 15 | BRUNOL6 (bruno-like 6) | rs2959925 (I2); rs1291497(I3); rs4777498(E13, 3′UTR) |
rs17855555(E1); rs17852852 (E12) |
| 16 |
ARHGAP17 (Rho GTPase activating protein 17) |
rs8045868 (I10); rs8045281 (I10) |
rs28365822 (E19) |
| 17 |
PCTP (phosphatidylcholine transfer protein) |
rs2960070(I1); rs2960067 (I2) |
rs12941739 (E1) |
| 20 | CDH26 (cadherin-like 26) | rs187751(I5); rs168982 (I5); |
rs6128739 (E6); rs6128740 (E6) |
| 21 |
USP25 (ubiquitin specific peptidase 25) |
rs2823504 (I17); rs2823507 (I18) |
rs2279797 (E18) |
SNPs are designated by dbSNP I.D. Abbreviations: I = intron; E = exon. Boldface indicates nonsynonymous SNPs showing frequency differences between the two populations in HapMap frequency data.
Almost all of the SNPs showing high inter-population divergence in these genes were in located in introns (Tables 3 and Table 4), reflecting the choice of SNPs in the Affymetrix GeneChip Human Mapping 500K Array Set. We searched dbSNP for reported nonsynonymous SNPs in the exons of the same genes (Tables 3 and Tables4), since nonsynonymous SNPs are most likely to have phenotypic effects. Certain of these nonsynonymous SNPs showed inter-population frequency differences, paralleling those of the high-divergence SNPs (Tables 3 and Tables 4). However, no population frequency data were available for the majority of nonsynonymous SNPs in these genes.
DISCUSSION
Application of the Affymetrix GeneChip Human Mapping 500K Array Set to a population of 102 individuals representing the major ethnic groups in the United States (African, Asian, European, and Hispanic) revealed patterns of gene diversity and genetic distance that reflected population history. We analyzed allelic frequencies at 388, 654 autosomal SNP sites that showed some variation in our study population and 10% or less missing values. In spite of the small size (23-31 individuals) of each subpopulation, there were no fixed differences at any site between any two subpopulations. Subpopulations of African, Asian, and European origin showed gene frequency signatures consistent with the history of the human continental populations. As expected from the African origin of modern humans [7-9], greater gene diversity was seen in Africans than in either Asians or Europeans, and the genetic distance between Asians and European populations was significantly lower than that between either of these two populations and Africans.
Principal components were extracted from the correlation matrix among individuals belonging to African, Asian, and European subpopulations at 150, 557 SNP sites with no missing values in any individual. The first principal component, accounting for about 60% of the variance, appeared to reflect the African origin of all three subpopulations. On the other hand, the second and third principal components set up contrasts between the subpopulations. Although together accounting for 6.6% of the variance, the second and third principal components perfectly separated these three subpopulations. Thus, in spite of the absence of fixed differences, gene frequency data alone in samples of modest size provided sufficient information to separate the major continental groups of humans. This was of interest in that the SNPs chosen for the Affymetrix 500K Array Set were not chosen for their utility in distinguishing among human subpopulations.
Because of complex population histories, human subpopulations are not discrete categories from a genetic point of view [9, 13, 23]. While separating the major continental groups, the principal component analysis also captured aspects of population history indicative of gradation between continents. In the case of Asians, individuals with ancestry in East Asia were much more strongly separated from Europeans than those with ancestry in the Indian subcontinent (Figure 1B). The latter clustered much closer to Europeans than did other Asians, as is expected given the history of gene flow across the Eurasian landmass [13] and consistent with results reported for microsatellite data [24].
When Hispanics were added to the analysis, they were not separable from the other subpopulations and in fact overlapped all three. While Hispanics showed significantly lower genetic distances from Europeans than from Africans or Asians, Hispanics were significantly closer to both Africans and Asians than were Europeans. These results are consistent of a predominant ancestry from Europe in the Hispanic population, but with substantial African and Asian (Native American) contributions, as has been hypothesized previously on the basis of both genetic and historical data [25]. Thus, there was a clear signature in the data of the admixed nature of the Hispanic subpopulation and the contribution of all three major continental subpopulations. This finding is of interest in that it suggests that a similar approach might be used to test for admixture in other populations whose history is not as well documented as that of U.S. Hispanics.
We identified certain protein-coding loci including two or more SNPs with high African-Asian genetic distance and certain loci including two or more SNPs with high African-European genetic distance. These loci can be considered candidates for playing a role in phenotypic differences between subpopulations, possibly including differences with respect to complex diseases phenotypes. Both of these lists of candidate genes included many with known or probable involvement in disease or disease related processes, including certain autoimmune diseases and cancers. For example, genes with high African-Asian frequency differences included a number of genes known or believed to be involved in disease-related processes. Some of these genes are associated with Mendelian diseases. The SNPs studied here are clearly not involved in Mendelian disease, nor is their evidence that the populations studied differ with respect to the known diseases associated with these loci. Nonetheless, the fact that mutation at a locus can cause Mendelian disease is evidence of the functional importance of the locus.
The genes with high African-Asian frequency differences included the following (disease-related functions and references in parentheses): LYST (Chediak-Higashi syndrome [26]); GLI2 (brain and pituitary developmental anomalies [27]); FHIT (tumor suppression [28]; BAI3 (glioblastoma [29]); IARS (autoimmune polymyositis [30]); HM13 (signal peptide cleaving for HLA-E presentation [31]); BACE (Alzheimer’s disease [32]). In addition to GLI2, genes with high African-European frequency differences included the following: HIVPE3 (HIV-1 infection [33]); GLIS1 (psoriasis [34]); ICA1 (Type I diabetes [35]); TNFSF8 (Hodgkin’s lymphoma [36]); APBA2 (Alzheimer’s disease [37]).
The LYST gene, which showed high genetic distances between Africans and Asians, plays an important role in vesicle trafficking and thus in the immune system. A recessive mutation at this causes Chediak Higashi syndrome, a severe immune deficiency accompanied by hypopigmentation [26]. Mutations at the orthologous mouse gene can cause not only a similar immune disorder but also coat color changes [38]. LYST was included in a recent survey of genes potentially affecting human skin pigmentation genes and was reported to show a signature of extended haplotype homozygosity in a Yoruban population [39]. Interestingly, the LRBA gene, which showed genetic distances between Africans and Europeans, encodes a protein that shares sequence homology (including BEACH domains) with the product of the LYST gene and likewise having a role in vesicle trafficking [40-41].
In recent years there has been a tendency to examine the human genome for evidence of positive selection, including directional selection leading to differences between subpopulations in allelic frequency. On the other hand, several widely used methods of testing for positive selection have been criticized for being overly non-conservative statistically and based on flawed statistical and biological reasoning [42-43]. The rarity of fixed differences between major human groups is evidence that any differences in directional selection between these groups must be either quite recent or relatively weak.
Whatever the truth about the prevalence of positive selection in humans, there is evidence that purifying selection, which acts to eliminate deleterious mutations, is by far more prevalent is human coding regions than is positive selection [12, 44-47]. A SNP site subject to ongoing purifying selection will tend to show reduced genetic distance between subpopulations [45]. Thus, whether allelic frequency differences between subpopulations are due to selection or to genetic drift, it can be concluded that purifying selection is generally relaxed in the case of SNPs that show differences between subpopulations [48].
Moreover, from the point of view of identifying the genetic basis of health differences between human subpopulations, it may not be particularly important whether drift or selection is responsible for a given difference. In either case, the relaxation of purifying selection on a given SNP site implies that the allelic differences at this site are not likely to be deleterious. This in turn implies that major health differences between human subpopulations are likely to involve not deleterious genes per se but rather deleterious gene-by-environment interactions. Several current hypotheses to account for ethnic differences with respect to complex disease incidence are consistent with this view. For example, the high level of type II diabetes in Native Americans is believed to involve gene-by-environment interaction [49].
In conclusion, the present study shows that genotyping a large number of SNPs with array technology provides substantial information relevant to population history. Even with relatively small samples, it is possible to assign individuals to the major geographic subpopulations of humans (African, Asian, and European) on the basis of allelic frequency patterns at highly polymorphic SNP sites. The latter finding is not without interest for forensic applications of this technology. In addition, the array technology can be used to identify genes with unusually high frequency differences among subpopulations, some of which may contributed to phenotypic differences between racial and ethnic groups, including health differences. The substantially more extensive samples expected to be available in the near future will further enhance the usefulness of array technology in addressing a variety of basic and applied questions in human population genetics.
MATERIALS AND METHODS
Study Population and Genotyping
Genotype data were obtained for each of 102 unique anonymized individuals of diverse geographic origin (the SNP500 Cancer population [21]). Each individual was assigned to one of four subpopulations, on the basis of self-described ethic group affiliation. The four subpopulations were chosen to represent four major U.S. ethnic groups: 24 African/African-American (here designated “African”); 31 Caucasian (here designated “European); 24 Asian; and 23 Hispanic [21]. This panel of anonymized samples was obtained from Coriell Cell Repositories (Coriell Institute for Medical Research, Camden NJ, USA) has been widely used as a resource for SNP and assay information [21]. None of the individuals are related, and previous analyses of SNP data showed differentiation among African, European, and Asian subpopulations consistent with known human population history [12].
Genotype data were obtained using the Affymetrix GeneChip Human Mapping 500K Array Set (http://www.affymetrix.com/products/arrays/specific/500k.affx), which uses two different arrays to type, respectively ~262,000 and ~238,000 SNPs. Of the SNPs assayed, 63.4% are neither in introns nor in mRNA of predicted genes; 35.1% are in introns; 0.8% are in exons; 0.6% are in 3′UTRs; and 0.1% are in 5′UTRs. The software supplied by the manufacturer was used to determine SNP calls. For purpose of analyses, we excluded SNPs on the X chromosome (N = 4826); all SNPs for which less than 94 (90%) of the 102 individuals had missing data (i.e., unresolved genotype calls; N = 96, 915); and all SNPs for which no diversity was observed in the study population (N = 4463). The resulting data set included 388, 654 autosomal SNPs.
For analyses comparing individuals, we also used data sets including only SNPS for which there were no missing data for any individual. There were 133,826 such sites when the whole study population was used, and 150, 557 such sites when the Hispanic group was excluded.
Statistical Methods
At each SNP site (locus), the gene diversity (“heterozygosity”) was estimated by 1 - ∑ xi2 where xi is the frequency of the ith allelic variant at the site (ref. 50, p. 177). For a given bi-allelic SNP locus, the genetic distance (allelic frequency divergence) between each pair of subpopulations was computed using the formula
where x1 and y1 are the frequencies of the first allele in each of the two subpopulations, respectively, and x2 and y2 are the frequencies of the second allele in each of the two subpopulations, respectively. The average of d over all loci is the average genetic distance (DA) (Ref. 50, p. 216). Skewness was used as a measure of the symmetry of distribution of d [51]; skewness is zero in the case of a perfectly symmetrical distribution and positive in the case of a distribution with an extended right “tail.”
Using the restricted data sets with no missing values, we computed pairwise genetic correlation coefficients between individuals by assigning genotypic values (+1 for the BB homozygote, 0 for the heterozygote, and −1 for the AA homogygote, where A and B arbitrarily designate the two alleles at the locus). In order to test the hypothesis that genetic correlation coefficients within subpopulations differed from those between populations, we used a randomization test because the correlation coefficients are not independent of one another. In this test, we created 1000 pseudo-data sets by sampling (with replacement) from the observed correlation coefficients. Each pseudo-data set included the same number of within-subpopulation correlations and between-population correlations as did the real data, and randomly sampled values were randomly assigned to each of the two categories. Principal component analysis [52] was applied to the correlation matrix in order to identify principal orthologous axes in the matrix of correlation coefficients among individuals.
In order to identify genes that showed highly divergent allelic frequencies between populations, we identified genes having two or more SNPs in our data set with significantly higher minimum d than expected for two SNPs chosen at random from SNPs in our data set. We applied a Monte Carlo procedure (randomization test) to identify the significance level for minimum d for a random pair of genes. For each chromosome, we sampled at random (with replacement) 200,000 pairs of SNPs; and we computed minimum d for each pair. Because chromosomes differed with respect to both the number of SNPs and the distribution of d, we applied the Monte Carlo procedure separately to each chromosome; but then, for a conservative test, we used for each chromosome a cut-off based on the upper tail of minimum d values observed in randomizations for any of the chromosomes. In the case of d between African and European populations, a minimum of d = 0.34 for two or SNPs in a gene provided a two-tailed significance level of 5 × 10−5 or better for any one of the chromosomes. Likewise, in the case of d between African and Asian populations, a minimum of d = 0.37 for two or SNPs in a gene provided a two-tailed significance level of 5 × 10−5 or better for any one of the chromosomes. Each of these cut-off values corresponded to approximately the top 0.1% of d values between the two populations compared across all chromosomes. All statistical analyses were conducted using the Minitab statistical package, release 13 (http://www.minitab.com/).
Acknowledgments
This project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. NO1-CO-12400. Partial support was provided by N.I.H. grant GM43940 to A.L.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lillie-Blanton M, Parsons PE, Gayle H, Dievler A. Racial differences in health: not just black and white, but shades of gray. Annu. Rev. Public Health. 1996;17:411–448. doi: 10.1146/annurev.pu.17.050196.002211. [DOI] [PubMed] [Google Scholar]
- 2.Whaley AL. Ethnicity/race, ethics, and epidemiology. J. Natl. Med. Assoc. 2006;95:736–742. [PMC free article] [PubMed] [Google Scholar]
- 3.Shavers VL. Racism and health inequity among Americans. J. Natl. Med. Assoc. 2006;98:386–396. [PMC free article] [PubMed] [Google Scholar]
- 4.Garte S. The racial genetics paradox in biomedical research and public health. Public Health Reports. 2006;117:421–425. doi: 10.1016/S0033-3549(04)50181-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nature Reviews Genetics. 2002;3:611–621. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- 6.Bamshad M. Genetic influences on health: does race matter? JAMA. 2005;294:937–946. doi: 10.1001/jama.294.8.937. [DOI] [PubMed] [Google Scholar]
- 7.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC. African populations and the evolution of human mitochondrial DNA. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 8.Nei M, Takezaki N. The root of the phylogenetic tree of human populations. Mol. Biol. Evol. 1996;13:170–177. doi: 10.1093/oxfordjournals.molbev.a025553. [DOI] [PubMed] [Google Scholar]
- 9.Kidd KK, Pakstis AJ, Speed WC, Kidd JR. Understanding human DNA sequence variation. J. Hered. 2004;95:406–420. doi: 10.1093/jhered/esh060. [DOI] [PubMed] [Google Scholar]
- 10.Nei M, Roychoudhury AK. Evolutionary relationships of human populations on a global scale. Mol. Biol. Evol. 1993;10:927–943. doi: 10.1093/oxfordjournals.molbev.a040059. [DOI] [PubMed] [Google Scholar]
- 11.Jorde LB, Wooding SP. Genetic variation, classification and ‘race.’. Nature Genetics. 2004;36:528–533. doi: 10.1038/ng1435. [DOI] [PubMed] [Google Scholar]
- 12.Hughes AL, Packer B, Welch R, Bergen AW, Chanock SJ, Yeager M. Effects of natural selection on inter-population divergence at polymorphic sites in human protein-coding loci. Genetics. 2005;170:1181–1187. doi: 10.1534/genetics.104.037077. M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serre D, Pääbo S. Evidence for gradients of human genetic diversity within and among continents. Genome Research. 2004;14:1679–1685. doi: 10.1101/gr.2529604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barnes KC. Genetic epidemiology of health disparities in allergy and clinical immunology. J. Allergy Clin. Immunol. 2006;117:243–254. doi: 10.1016/j.jaci.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 15.Chen H, Hernandez W, Shriver MD, Ahaghotu CA, Kittles RA. ICAM gene cluster SNPs and prostate cancer risk in African Americans. Hum. Genet. 2006;120:69–76. doi: 10.1007/s00439-006-0184-3. [DOI] [PubMed] [Google Scholar]
- 16.Cross RK, Jung C, Wasan S, Joshi G, Sawyer R, Roghmannn MC. Racial differences in disease phenotypes in patients with Crohn’s disease. Inflamm. Bowel Dis. 2006;12:192–198. doi: 10.1097/01.MIB.0000217767.98389.20. [DOI] [PubMed] [Google Scholar]
- 17.Dominick KL, Baker TA. Racial and ethnic differences in osteoarthritis: prevalence, outcomes, and medical care. Ethn. Dis. 2004;14:558–566. [PubMed] [Google Scholar]
- 18.Marshall MC., Jr. Diabetes in African Americans. Postgrad. Med. J. 2005;81:734–740. doi: 10.1136/pgmj.2004.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stansbury JP, Jia H, Williams LS, Vogel WB, Duncan PW. Ethnic disparities in stroke: epidemiology, acute care, and postacute outcomes. Stroke. 2005;36:374–386. doi: 10.1161/01.STR.0000153065.39325.fd. [DOI] [PubMed] [Google Scholar]
- 20.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 21.Packer B, Yeager M, Staats B, Welch R, Crenshaw A, Kiley M, Eckert A, Beerman M, Miller E, Bergen A, Rothman N, Strausberg R, Chanock SJ. SNP500Cancer: a public resource for sequence validation and assay development for genetic variation in candidate genes. Nucleic Acids Res. 2004;32:D528–D532. doi: 10.1093/nar/gkh005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bamshad M, Wooding S, Salisbury BA, Stephens JC. Deconstructing the relationship between genetics and race. Nature Reviews Genetics. 2004;5:598–609. doi: 10.1038/nrg1401. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 25.Hanis CL, Hewett-Emmett D, Bertin TK, Schull WJ. Origins of U.S. Hispanics. Implications for diabetes. Diabetes Care. 1991;14:618–627. doi: 10.2337/diacare.14.7.618. [DOI] [PubMed] [Google Scholar]
- 26.Barbosa MD, Nguyen QA, Tchernev VT, Ashley JA, Detter JC, Blaydes SM, Brandt SJ, Chotai D, Hodgman C, Solari RC, Lovett M, Kingsmore S. Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature. 1996;282:262–265. doi: 10.1038/382262a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roessler E, Du Y-Z, Mullot JL, Casas E, Allen WP, Gillessen-Kaesbach G, Roeder ER, Ming JE, Ruiz I, Altaba A, Muenke M. Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features. Proc. Natl. Acad. Sci. USA. 2003;100:13424–13429. doi: 10.1073/pnas.2235734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pekarsky Y, Garrison PN, Palamarchuk A, Zanesi N, Aqeilan RI, Huebner K, Barnes LD, Croce CM. Fhit is a physiological target of the protein kinase Src. Proc. Natl. Acad. Sci. USA. 2004;191:3775–3779. doi: 10.1073/pnas.0400481101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiratsuchi T, Nishimori H, Ichise H, Nakamura Y, Tokino T. Cloning and characterization of BAI2 and BAI3 novel genes homologous to brain-specific angiogenesis inhibitor 1 (BAI1) Cytogenet. Cell Genet. 1997;79:103–108. doi: 10.1159/000134693. [DOI] [PubMed] [Google Scholar]
- 30.Targoff IN. Autoantibodies in polymyositis. Rheum. Dis. Clin. N. Am. 1992;18:455–482. [PubMed] [Google Scholar]
- 31.Weihofen A, Binns K, Lemburg MK, Ashmann K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 32.Bennett BD, Babu-Khan S, Loeloff R, Louis J-C, Curran E, Citron M, Vassar R. Expression analysis of BACE2 in brain and peripheral tissues. J. Biol. Chem. 2000;275:20647–20651. doi: 10.1074/jbc.M002688200. [DOI] [PubMed] [Google Scholar]
- 33.Hicar MD, Liu Y, Allen CE, Wu L-C. Structure of the human zinc finger protein HIVEP3: molecular cloning, expression, exon-intron structure, and comparison with paralogous genes HIVEP1 and HIVEP2. Genomics. 2001;71:89–100. doi: 10.1006/geno.2000.6425. [DOI] [PubMed] [Google Scholar]
- 34.Nakanishi N, Kim Y-S, Nakajima T, Jetten AM. Regulatory role for Krüppel-like zinc-finger protein Gli-similar 1 (Glis1) in PMA-treated and psoriatic epidermis. J. Invest. Dermatol. 2006;126:49–60. doi: 10.1038/sj.jid.5700018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pietropaolo M, Castaño L, Babu S, Buelow R, Kuo Y-L, Martin S, Martin A, Powers AC, Prochazka M, Naggert J, Leiter EH, Eisenbarth GS. Islet cell autoantigen 69 kD (ICA69): molecular cloning and characterization of a novel diabetes-associated autoantigen. J. Clin. Invest. 1993;92:359–371. doi: 10.1172/JCI116574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith CA, Gruss HJ, Davis T, Anderson D, Farrah T, Baker E, Sutherland GR, Brannan CI, Copeland NG, Jenkins NA, Grabstein KH, Gliniak B, McAlister IB, Fanslow W, Alderson M, Falk B, Gimpel S, Gillis S, Din WS, Goodwin RG, Armitage RJ. CD30 antigen, a marker for Hodgkin’s lymphoma, is a receptor whose ligand defines an emerging family of cytokines with homology to TNF. Cell. 1993;73:1349–1360. doi: 10.1016/0092-8674(93)90361-s. [DOI] [PubMed] [Google Scholar]
- 37.McLoughlin DD, Miller CC. The intracellular cytoplasmic domain of the Alzheimer’s disease amyloid precursor protein interacts with phosphotyrosine-binding domain proteins in the yeast two-hybrid system. FEBS Lett. 1996;18:197–200. doi: 10.1016/s0014-5793(96)01128-3. [DOI] [PubMed] [Google Scholar]
- 38.Runkel F, Büssow H, Seburn KL, Cox GA, Ward DM, Kaplan J, Franz T. Grey, a novel mutation in the murine Lyst gene, causes the beige phenotype by skipping of exon 25. Mammalian Genome. 2006;17:203–210. doi: 10.1007/s00335-005-0015-1. [DOI] [PubMed] [Google Scholar]
- 39.Izagirre N, García I, Junquera C, de la Rúa C, Alonso S. A scan for signatures of positive selection in candidate loci for skin pigmentation in humans. Mol. Biol. Evol. 2006;23:1697–1706. doi: 10.1093/molbev/msl030. [DOI] [PubMed] [Google Scholar]
- 40.Wang J-W, Howson J, Haller E, Kerr WG. Identification of a novel lipopolysaccharide-inducible gene with key features of both a kinase anchor proteins and chs1/beige proteins. J. Immunol. 2001;166:4586–4595. doi: 10.4049/jimmunol.166.7.4586. [DOI] [PubMed] [Google Scholar]
- 41.Gebauer D, Li J, Jogl G, Shen Y, Myszka DG, Tong L. Crystal structure of the PH-BEACH domains of human LRBA/BGL. Biochemistry. 2001;43:14873–14880. doi: 10.1021/bi049498y. [DOI] [PubMed] [Google Scholar]
- 42.Hughes AL, Friedman R, Glenn NL. The future of data analysis in evolutionary genomics. Current Genomics. 2006;7:227–234. [Google Scholar]
- 43.Sabeti PC, Schaffner SK, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 44.Freudenberg-Hua Y, Freudenberg J, Kluck N, Cichon S, Propping P, Nöthen MM. Single nucleotide variation analysis in 65 candidate genes for CNS disorders in a representative sample of the European population. Genome Res. 2003;13:2271–2276. doi: 10.1101/gr.1299703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hughes AL, Packer B, Welch R, Bergen AW, Chanock SJ, Yeager M. Widespread purifying selection at polymorphic sites in human protein-coding loci. Proc. Natl. Acad. Sci. USA. 2003;100:15754–15757. doi: 10.1073/pnas.2536718100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sunyaev S, Ramensky V, Koch I, Lathe WS, III, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Human Mol. Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Z, Fu Y-X, Hewett-Emmett D, Boerwinkle E. Investigating single nucleotide polymorphism (SNP) density in the human genome and its implications for molecular evolution. Gene. 2003;312:207–213. doi: 10.1016/s0378-1119(03)00670-x. [DOI] [PubMed] [Google Scholar]
- 48.Hughes AL, Packer B, Welch R, Chanock SJ, Yeager M. High level of functional polymorphism indicates a unique role of natural selection at human immune system loci. Immunogenetics. 2005;57:821–827. doi: 10.1007/s00251-005-0052-7. [DOI] [PubMed] [Google Scholar]
- 49.Mulligan CJ, Hunley K, Cole S, Long JC. Population genetics, history, and health patterns in Native Americans. Annu. Rev. Genomics Hum. Genet. 2004;5:295–315. doi: 10.1146/annurev.genom.5.061903.175920. [DOI] [PubMed] [Google Scholar]
- 50.Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York: 1987. [Google Scholar]
- 51.Dudewicz EJ, Mishra SN. Modern Mathematical Statistics. Wiley; New York: 1988. [Google Scholar]
- 52.Johnson R, Wichern D. Applied Multivariate Statistical Methods. 3rd Edition. Prentice Hall; Englewood Cliffs, NJ: 1992. R. [Google Scholar]