Abstract
Predictive modeling suggests that actual carbapenem MIC results are more predictive of clinical patient outcomes than categorical classification of the MIC as susceptible, intermediate, or resistant. Some have speculated that current CLSI guidelines' suggested thresholds are too high and that clinical success is more likely if the MIC value is ≤1 mg/liter for certain organisms. Patients treated with carbapenems and with positive blood cultures for Pseudomonas aeruginosa, Acinetobacter baumannii, or extended-spectrum beta-lactamase (ESBL)-producing Gram-negative bacteria were considered for evaluation in this clinical retrospective cohort study. Relevant patient demographics and microbiologic variables were collected, including carbapenem MIC. The primary objective was to define a risk-adjusted all-cause hospital mortality breakpoint for carbapenem MICs. Secondarily, we sought to determine if a similar breakpoint existed for indirect outcomes (e.g., time to mortality and length of stay [LOS] postinfection for survivors). Seventy-one patients met the criteria for study inclusion. Overall, 52 patients survived, and 19 died. Classification and regression tree (CART) analysis determined a split of organism MIC between 2 and 4 mg/liter and predicted differences in mortality (16.1% versus 76.9%; P < 0.01). Logistic regression controlling for confounders identified each imipenem MIC doubling dilution as increasing the probability of death 2-fold (adjusted odds ratio [aOR] 2.0; 95% confidence interval [CI], 1.3 to 3.2). Secondary outcomes were similar between groups. This study revealed that patients with organisms that had a MIC of ≥4 mg/liter had worse outcomes than patients whose isolates had a MIC of ≤2 mg/liter, even after adjustment for confounding variables. We recommend additional clinical studies to better understand the susceptibility breakpoint for carbapenems.
INTRODUCTION
It has become well known that inactive antimicrobial therapy and delay of active antimicrobial therapy are associated with less than optimal treatment outcomes (20, 21, 25, 27). Recently, several authors have demonstrated that actual MIC results may be more predictive of clinical patient outcomes than categorical classification of MICs as susceptible, intermediate, or resistant; such is the case for fluoroquinolones (13), expanded-spectrum cephalosporins (3), and extended-spectrum penicillins (34). We postulated that a similar relationship might exist within the classification susceptible for carbapenems, with the highest MIC values most predictive of worse outcomes. This expectation is buttressed by mathematical models that predict current dosing strategies are not likely to obtain optimal outcomes for carbapenem MICs of >1 mg/liter for Enterobacteriaceae (5). Mathematical models such as these have been used to justify the alterations of the carbapenem susceptibility breakpoint to ≤1 mg/liter for Enterobacteriaceae (15).
Carbapenems are often used as front-line drugs for Gram-negative bacterial infections due to progressive resistance among other extended-spectrum beta-lactams (28). Unfortunately, carbapenem resistance as documented in large surveillance studies has been increasing, most notably among Pseudomonas aeruginosa, Acinetobacter baumannii, and extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae (30). Genetically, carbapenem resistance is complex, multifactorial, and dependent on the individual organism-carbapenem pair. For instance, A. baumannii is most commonly resistant to carbapenems as a result of carbapenem-hydrolyzing oxacillinase (OXA) enzymes (16), whereas resistance to carbapenems in P. aeruginosa is most frequently due to metallo-beta-lactamases, drug efflux pumps, and deletion of membrane porin channels (31). Enterobacteriaceae in the United States are most frequently carbapenem resistant due to the spread of Klebsiella pneumoniae-producing carbapenemase (KPC) enzymes (2). To simplify the complexities of genetic resistance, organism resistance to individual agents is commonly standardized through phenotypic MIC procedures (8). Such classification provides an objective endpoint for standardization that may be interpreted as an interval variable and allows a common metric for assessment of different organism genera and species to the same antibiotic. Current research suggests that the MIC is more important than the genetic resistance mechanism; that is, MICs vis-à-vis pharmacokinetic and pharmacodynamic targets do not differ on the basis of the genetic resistance mechanism produced. Similar pharmacokinetic/pharmacodynamic targets exist for Klebsiella pneumoniae regardless of carbapenemase production (7, 12, 15) and for Enterobacteriaceae regardless of extended-spectrum beta-lactamase production (1, 4, 11). While it is known that MICs may not entirely classify all organism-carbapenem pairs, such data are critical for the treating clinician hoping to complete real-time assessments for patients and act appropriately with antimicrobial therapy.
Current Clinical and Laboratory Standards Institute (CLSI) guidelines label strains of A. baumannii with carbapenem MIC values of ≤4 mg/liter as susceptible (10), while the P. aeruginosa susceptibility breakpoint was recently lowered to ≤2 mg/liter in the most recent guidelines. This was due to speculation that a threshold of 4 mg/liter for resistance may in fact be too high and that clinical failure is more likely if the MIC value is ≥2 mg/liter (5). The primary goal of this study was to determine if a clinical mortality breakpoint existed for carbapenems in patients treated with carbapenems for Gram-negative bloodstream infections. Secondarily, we sought to determine if a similar breakpoint existed for indirect outcomes, such as time to mortality and length of stay (LOS) postinfection for survivors.
MATERIALS AND METHODS
This retrospective, cohort study was conducted at Northwestern Memorial Hospital (NMH) located in Chicago, IL. Patients treated with imipenem or meropenem and with positive blood cultures for Pseudomonas aeruginosa, Acinetobacter baumannii, and ESBL-producing Gram-negative bacteria, as identified by the NMH Microbiology Laboratory, from 1 January 2005 through 7 August 2008 were considered for inclusion. Each patient was only included once, using the first positive culture result within the hospital admission. Patients were included if they were ≥18 years of age and received ≥48 h of treatment with a carbapenem. Patients were excluded if they were <18 years of age, if the sensitivity tests for the infecting organism to imipenem or meropenem were not available, or if they received either imipenem or meropenem for less than 48 h. This study was reviewed and approved by Institutional Review Boards at Northwestern University and Midwestern University.
Organism identification and susceptibility classification were completed with the Vitek 2 system (bioMérieux, Balmes-les-Grottes, France) and by manual biochemical identification when necessary. Antimicrobial susceptibility testing was performed on all isolates by the Vitek 2 system as approved by the United States Food and Drug Administration to determine MICs. Organism susceptibility was interpreted according to CLSI guidelines (8). In cases of simultaneous Gram-negative bacteremia, the most resistant organism was used for analyses.
Patient demographics and data variables were collected by a trained reviewer using inpatient electronic medical records, pharmacy databases, and clinical microbiology databases. Patients were assessed for demographics (age, gender, and race), location of hospitalization (intensive care unit [ICU] versus non-ICU), serum creatinine on the day of culture, hepatic dysfunction, solid organ or hematological transplant, diabetes, immunosuppression, composite comorbidity score, number of previous hospitalizations within 12 months of culture, days of hospital stay postinfection, presence of central venous access, presence of polymicrobial bacteremia (excluding coagulase-negative Staphlyococcus), infecting organism, source of infection, organism imipenem MIC, carbapenem infusion scheme (30 min or 3 h), and in-hospital mortality from any cause. Patients at Northwestern Memorial Hospital minimally receive guideline-concordant or more aggressive doses of antibiotics (18) with appropriate renal function adjustments. This is ensured by utilization of standardized, prepopulated, computerized order sets and prospective clinical pharmacist participation in the care of each patient. Any deviations from this procedure that may be represented in this study sample were likely random. For imipenem, dosing was according to weight and renal function for severe, life-threatening infections (i.e., a target of 4 g/day unless reduced according to weight and/or renal function) (26). Meropenem was dosed at a minimum of 3 g per day divided every 8 h in patients with normal renal function. Patients with creatinine clearance between 16 and 30 ml/min minimally received 1 g every 12 h, and patients with creatinine clearance less than 15 ml/min received 500 mg daily. Meropenem infusion schemes were either as a 3-h extended infusion (patients receiving meropenem after 1 January 2008) or as a 30-min infusion (patients receiving meropenem prior to 31 December 2007).
Definitions.
ICU-onset infection was defined as the patient being located in the ICU when the culture returned positive for a Gram-negative bacteremia. Concurrent renal dysfunction was defined as acute or chronic renal dysfunction. Acute renal dysfunction was a rise of serum creatinine of 0.5 mg/dl or 50% from patient baseline to immediately before the infection. Chronic renal dysfunction was defined as a physician diagnosis of chronic kidney dysfunction (CKD) (as indicated in the patient's medical history). Hepatic dysfunction was defined as any liver enzymes greater than three times the upper normal limit at the time of culture. Immunosuppression was defined as steroid use, chemotherapy use, or documented presence of HIV/AIDS or immunosuppressant agents. Comorbidity was quantified with the Deyo modification of the Charlson comorbidity index and was calculated for each patient using ICD-9 codes assigned at hospital admission (6, 14). The source of the bloodstream infection was considered from the attending physician's written diagnosis. Receipt of other active therapy was defined as use of amikacin, tobramycin, gentamicin, cefepime, ceftazidime, piperacillin-tazobactam, colistin, tigecycline, ciprofloxacin, levofloxacin, or moxifloxacin given in conjunction with either meropenem or imipenem. Sensitivities were interpreted according to the CLSI guidelines in place at the time of isolate identification (8).
Statistical analysis.
The primary outcome analyzed was all-cause hospital morality based on organism MIC. Secondary analyses included time to mortality in days and length of stay (LOS) in days postculture for surviving patients. An exploratory analysis was also performed that separated organisms into the CLSI specific categorizations of: Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii. Data analysis, except where otherwise specified, was performed using Intercooled Stata version 11.1 (College Station, TX). Descriptive statistical analyses were performed on all study variables. To determine breakpoints, a binary recursive partitioning methodology, classification and regression tree (CART) modeling, was utilized to attempt to define a split between interval variables and outcomes utilizing SPSS v.17 and the Decision Trees add-on v.17 (SPSS, Inc., Chicago, IL). Sensitivity and specificity were calculated for each possible breakpoint.
Inferential statistics were performed as follows for bivariate analyses. Chi-square and Fisher's exact analyses were performed where appropriate for nominal data; Student's t tests or Wilcoxon rank sum tests were conducted for interval data. Initial odds ratios (ORs) with 95% confidence intervals (CIs) were calculated from bivariate logistic regression. Multivariate models were created to assess the dependent primary outcome of mortality and control for all relevant confounders listed in Table 1 as a forward stepwise procedure by adding variables with a plausible relationship to the dependent outcomes and significant at the level P < 0.2 in bivariate analyses (i.e., baseline differences between MIC stratifications or differences between those with hospital mortality) to the logistic regression equation. Models were assessed by adding each variable iteratively to the multivariate model according to significance. Model selection was completed by assessing twice the difference in the log likelihood ratios between the models and comparing the product against a chi-square distribution with the appropriate degrees of freedom (17, 17a). Probabilities of outcomes adjusting for confounders were calculated from adjusted odds ratios (aORs). The selected breakpoint was then assessed in bivariate and multivariate models to determine predictability of mortality. Models were repeated replacing the identified breakpoints with log base 2 (log2) MIC as an independent variable to provide results for MIC in its commonly reported form, doubling dilution. These finalized models were assessed to potentially allow for further inclusion of organism type (i.e., A. baumannii, P. aeruginosa, or Enterobacteriaceae class) using the same model building criteria described above. All tests were two-tailed, and alpha was fixed at 0.05 for final interpretations. To determine the final model fit, the Hosmer and Lemeshow goodness-of-fit test was employed after grouping data into a maximum of 10 quantiles (17, 17a). A jackknife procedure was used to systematically eliminate one observation (i.e., n − 1) from the multivariate model until each observation had been dropped once. Models were then pooled to allow the estimation of the variance of the coefficient.
Table 1.
Baseline demographics stratified by imipenem MIC and mortality
| Parameter | Demographic result by: |
|||
|---|---|---|---|---|
| Imipenem MIC |
Patient mortality |
|||
| ≤2 mg/liter (n = 58) | ≥4 mg/liter (n = 13) | Survived (n = 52) | Died (n = 19) | |
| General clinical | ||||
| Age, mean, yr (SD) | 54.7 (17.7) | 56.3 (13.4) | 54.3 (18.1) | 55.9 (12.7) |
| Female, n (%) | 17 (30.4) | 5 (38.5) | 16 (30.7) | 6 (31.6) |
| Wt, mean, kg (SD) | 82.9 (28.4) | 82.7 (20.6) | 80.2 (22.1) | 88 (37.5) |
| Charlson score, median (IQRe) | 2 (1–3.5) | 3 (2–5) | 2 (0.5–4) | 2 (1–4) |
| Any transplant, n (%) | 11 (19.7) | 4 (30.1) | 10 (19.2) | 5 (26.3) |
| Hepatic dysfunction, n (%) | 13 (23.2) | 3 (23.1) | 8 (15.4) | 8 (42.1)a |
| Concurrent renal dysfunction, n (%) | 21 (37.5) | 7 (53.9) | 20 (38.5) | 9 (47.4) |
| Diabetes, n (%) | 14 (25) | 2 (15.4) | 13 (25) | 3 (15.8) |
| Malignancy, n (%) | 23 (41.1) | 8 (61.6) | 21 (40.4) | 10 (52.6) |
| ANCf ≤ 500, n (%) | 8 (14.3) | 4 (30.8) | 8 (15.4) | 4 (21.1) |
| Immunosuppression, n (%) | 32 (57.1) | 7 (53.9) | 28 (53.4) | 11 (57.9) |
| Serum creatinine on day of culture, mean, mg/dl (SD) | 2.1 (2) | 1.6 (1.2) | 2.1(2) | 1.6 (1.1) |
| Bloodstream infection factors | ||||
| Unidentified source, n (%) | 20 (35.7) | 2 (15.4) | 19 (36.6) | 3 (15.8) |
| Respiratory source, n (%) | 6 (10.7) | 8 (61.5)b | 6 (11.5) | 9 (47.4)c |
| Urinary source, n (%) | 14 (25) | 3 (23.1) | 16 (30.1) | 2 (10.5) |
| Polymicrobial bacteremia, n (%) | 11 (19.6) | 4 (30.8) | 13 (25) | 2 (10.5) |
| Central line, n (%) | 49 (87.5) | 13 (100) | 44 (84.6) | 19 (100) |
| In ICU at onset of infection, n (%) | 21 (37.5) | 10 (76.9)c | 16 (30.8) | 15 (79)b |
| Hospitalization(s) in last 12 mo, mean (SD) | 2.5 (2.6) | 1.9 (3.3) | 2.5 (2.7) | 2.1 (2.8) |
| Extended infusion, n (%) | 36 (64.3) | 6 (46.2) | 30 (57.7) | 12 (63.2) |
| Meropenem, n (%) | 37 (63.8) | 9 (69.2) | 31 (59.6) | 15 (78.9) |
| Other active therapy,d n (%) | 18 (32.1) | 6 (46.2) | 17 (32.7) | 7 (36.8) |
| MIC of ≥4 mg/liter, n (%) | 3 (6) | 10 (52.6) | ||
| Organism species, n (%) | ||||
| Enterobacteriaceae (n = 46) | 43 (76.8) | 1 (7.7)b | 39 (75.0) | 7 (36.8)c |
| Pseudomonas aeruginosa (n = 19) | 12 (21.4) | 7 (53.9)a | 10 (19.2) | 9 (47.4)a |
| Acinetobacter baumannii (n = 6) | 1 (1.7) | 5 (38.5)b | 3 (5.8) | 3 (15.8) |
P ≤ 0.05.
P ≤ 0.001.
P ≤ 0.01.
Any noncarbapenem reported as susceptible or intermediate to infecting organism.
IQR, interquartile range.
ANC, absolute neutrophil count.
RESULTS
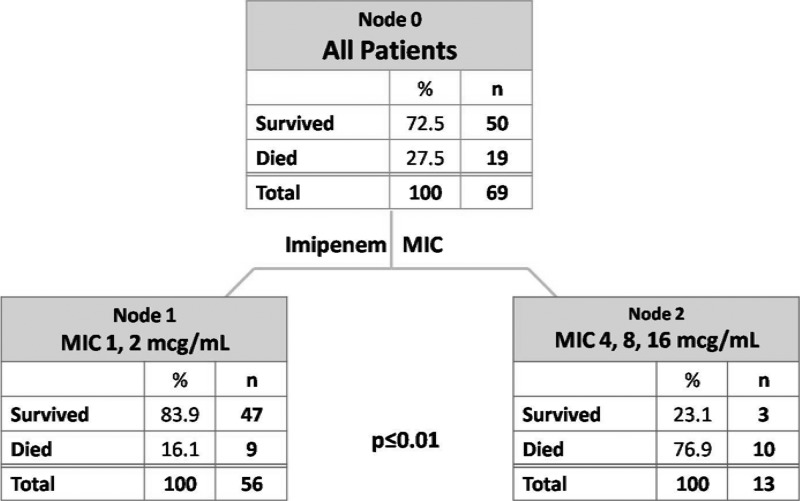
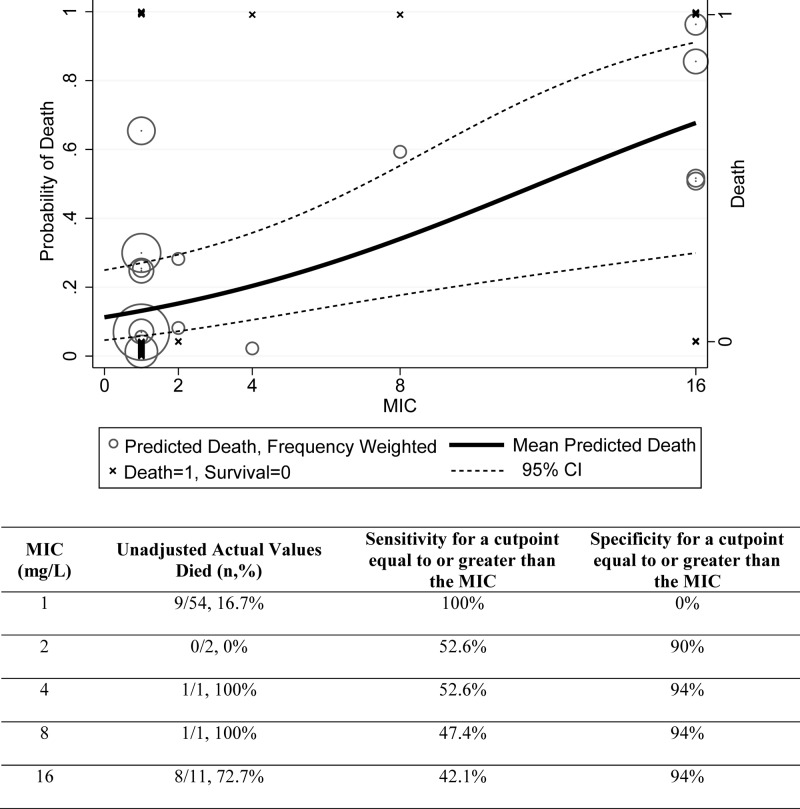
Seventy-one patients met the criteria for study inclusion during the study period. An optimal split of mortality was determined by CART analysis and was found between imipenem MICs of ≤2 mg/liter and ≥4 mg/liter (16.1% versus 76.9%; P < 0.01) (Fig. 1). Sensitivity and specificity were maximized with this categorical break of a MIC of ≥4 mg/liter of 52.6% and 94%, respectively (Fig. 2). This classification scheme correctly predicted 82.6% of cases. Patients were accordingly stratified to MICs of ≤2 mg/liter and ≥4 mg/liter with baseline demographics and clinical characteristics presented in Table 1. There were 58 patients in the group with the imipenem MIC of ≤2 mg/liter and 13 patients in the group with an imipenem MIC of ≥4 mg/liter. Patients in the higher-category MIC group tended to be more ill, with notable differences existing in the number of patients having a respiratory source of infection (P < 0.001) and residing in the ICU at the onset of infection (P = 0.014). Differences existed in baseline organism MICs and mortality as a function of organism classification.
Fig 1.
Classification and regression tree model stratifying mortality by MIC.
Fig 2.
Adjusted probability of death and actual mortality rates according to imipenem MIC. As shown, the probability of death was adjusted for the factors hepatic dysfunction, urinary source of infection, and ICU residence at the onset of infection.
Baseline demographics and clinical characteristics of patients stratified by mortality are listed in Table 1. Overall, there were 52 patients who survived and 19 who died. In addition to a higher incidence of hepatic dysfunction (P = 0.02), patients who died also had higher incidence of a respiratory source as the infection source (P = 0.002) and were in the ICU at the onset of bacteremia (P < 0.001). At the bivariate level, one of the most profound differences was the MIC category, where 76.9% of those with a MIC of ≥4 mg/liter died versus 16.1% who died with a MIC of ≤2 mg/liter (P < 0.01). Receipt of other active therapy and type of carbapenem infusion scheme (i.e., prolonged versus intermittent infusion) were not associated with differences in mortality.
The stepwise logistic regression model constructed with a MIC of ≥4 mg/liter as a categorical variable identified the following as predictors of mortality: MIC of ≥4 mg/liter (OR, 27.9; 95% CI, 3.8 to 202.9; P = 0.001), hepatic dysfunction (OR, 5.9; 95% CI, 1.1 to 30.2; P = 0.034), and ICU at onset of infection (OR, 5.8; 95% CI, 1.3 to 26.3; P = 0.024) (Table 2). The logistic regression, constructed with imipenem log2 (to represent doubling dilutions), identified each imipenem MIC doubling dilution as increasing the probability of death 2-fold (aOR, 2.0; 95% CI, 1.3 to 3.2; P = 0.003) (Fig. 2). Being in the ICU at onset of infection (OR, 5.9; 95% CI, 1.3 to 25.6; P = 0.018) was also a significant predictor of mortality when analyzed with this model. Other variables that were significantly different between groups when evaluated using bivariate analysis were included in all multivariate models but did not remain significant. Jackknife procedures employed for the final logistic regression equations demonstrated agreement with the result for the cutpoint of ≥4 mg/liter with an aOR of 27.9 (95% CI, 1.4 to 546.1) and an increasing aOR of 2.0 for each doubling dilution increase (95% CI, 1.1 to 3.7). The Hosmer and Lemeshow goodness-of-fit test did not detect a lack of model fit for the imipenem log2 multivariate model (P = 0.94) or the CART-derived breakpoint multivariate model (P = 0.55) (17, 17a). Although this statistic cannot prove good model fit, categorical agreement for mortality at each MIC value was high (Fig. 2). Finally, inclusion of organism category to the above finalized multivariate models did not change any interpretations; organism class was not a predictive variable (P = 0.69 for log2 MIC analysis and P = 0.96 for MIC of ≥4 mg/liter analysis).
Table 2.
Multivariate analysis of predictors for mortality
| Parameter | Result of multivariate analysis with predictor: |
|||||
|---|---|---|---|---|---|---|
| Imipenem MIC of ≥4 mg/liter |
Log2 imipenem MIC |
|||||
| OR for mortality | 95% CI | P value | OR for mortality | 95% CI | P value | |
| Hepatic dysfunction | 5.87 | 1.14–30.2 | 0.034 | 4.66 | 0.99–21.9 | 0.052 |
| Patient in ICU at onset of infection | 5.75 | 1.26–26.3 | 0.024 | 5.87 | 1.35–25.58 | 0.018 |
| Urinary source | 0.14 | 0.017–1.10 | 0.062 | 0.18 | 0.024–1.28 | 0.086 |
| MIC of ≥4 mg/liter | 27.9 | 3.82–202.9 | 0.001 | |||
| Log2 imipenem MIC | 2.03 | 1.28–3.23 | 0.003 | |||
Regarding secondary outcomes at the bivariate level, time to mortality (P = 0.98) and LOS postinfection for survivors (3.3-day increase for each doubling dilution; P = 0.2) differed numerically but not with statistical significance. In both subset analyses, power was constrained as only 19 and 50 patients, respectively, were included. Multivariate models attempting to control for confounders were no more instructive.
DISCUSSION
To our knowledge, the results presented herein are the first clinical data demonstrating increasingly poor outcomes with incrementally increasing MICs for Gram-negative organisms with MICs classified (and previously classified) as susceptible by CLSI carbapenem breakpoints. We have described a risk-adjusted increased probability of death from Gram-negative bloodstream infections for organisms with increasing MICs even within the susceptible range. Binary recursive partitioning demonstrated an optimal breakpoint of a MIC of ≥4 mg/liter for mortality prediction. Similarly, a trend toward increased LOS for survivors was seen with a MIC of ≥4 mg/liter, even though power was more constrained in this analysis. These clinical outcome data support mathematical suggestions that a susceptibility breakpoint of a MIC of ≤4 mg/liter for imipenem and meropenem may be too high to meet established pharmacokinetic/pharmacodynamic targets for Gram-negative bacilli when using current FDA-approved doses (5, 15). It appears that obtaining optimal outcomes for patients with elevated MICs may be unachievable even with prolonged infusion schemes (22, 23, 32). We noted no difference between patients receiving intermittent infusion or prolonged infusion over 3 h, though we had limited power to assess its effect.
In June 2009, a CLSI working group evaluating the current breakpoints for Enterobacteriaceae recommended lowering the susceptibility breakpoint for carbapenems (excluding ertapenem) from ≤4 mg/liter to ≤1 mg/liter (Table 3) (15). These recommendations were constructed on the basis of (i) clinical data, (ii) MIC distributions and cutoffs, and (iii) pharmacokinetic/pharmacodynamic data derived from animal exposure-response studies and Monte Carlo simulations (5, 15, 22, 23, 32). Further discussion ensued regarding resetting the breakpoints for the carbapenems with activity against nonfermenting Gram-negative organisms to the same level, as has been recently done for Enterobacteriaceae, and the change for the P. aeruginosa breakpoint to >2 mg/liter being resistant was made in 2012 (15). Multiple mathematical simulations of various dosing schemes of both imipenem and meropenem indicate that a susceptibility breakpoint of ≥4 mg/liter for imipenem and meropenem against P. aeruginosa and A. baumannii may be too high (22, 23, 32).
Table 3.
Carbapenem susceptibility breakpoints by agency
| Agencya | Susceptibility breakpoint (mg/liter) forb: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
Enterobacteriaceae |
P. aeruginosa |
A. baumannii |
|||||||
| Dor | Imi | Mer | Dor | Imi | Mer | Dor | Imi | Mer | |
| CLSI | ≤1 | ≤1 | ≤1 | ≤2 | ≤2 | ≤2 | ≤4 | ≤4 | |
| FDA | ≤0.5 | ≤4 | ≤4 | ≤2 | ≤4 | ≤4 | ≤1 | ≤4 | ≤4 |
| EUCAST | ≤1 | ≤2 | ≤2 | ≤1 | ≤4 | ≤2 | ≤1 | ≤2 | ≤2 |
CLSI, Clinical and Laboratory Standards Institute; FDA, Food and Drug Administration; EUCAST, European Committee on Antimicrobial Susceptibility Testing.
Dor, doripenem; Imi, imipenem; Mer, meropenem.
The complexity of the current recommendations of breakpoints across different regulatory bodies can be found in Table 3. Universally lowering the breakpoints for carbapenems would aid in reducing the confusion surrounding the guidelines and improve an epidemiological split between organisms with genetic underpinnings for reduced susceptibility phenotypes from those that represent wild-type organisms that would require de novo resistance mutations. Some strains of Klebsiella pneumoniae have been known to test as susceptible on sensitivity analysis when the old breakpoints are used and then behave as resistant when exposed to carbapenems (35). This was further corroborated when the Centers for Disease Control and Prevention (CDC) reported in January 2009 that there were strains of Enterobacteriaceae that were considered susceptible (i.e., MIC of ≤4 mg/liter) according to laboratory sensitivity tests but contained carbapenemases, allowing them to behave as resistant organisms (29). This evidence also supports the suggestion to lower the MIC value of carbapenems.
Clinical studies have shown concordance with antimicrobial, pharmacokinetic/pharmacodynamic mathematical modeling and have described poor clinical outcomes with increasing MICs, even within the classification of susceptible. Tam et al. reported a retrospective cohort study of patients with P. aeruginosa bloodstream infections that had reduced susceptibilities to piperacillin-tazobactam, defined as 32 mg/liter or 64 mg/liter (CLSI susceptibility breakpoint of ≤64 mg/liter) (34). Patients treated with piperacillin-tazobactam were compared to patients receiving other active therapy, using 30-day mortality as the primary outcome. There was a significant difference in mortality rates between the piperacillin-tazobactam group and the control group (85.7% versus 22.2%; P = 0.004), and time to mortality was also shorter in the piperacillin-tazobactam group (P < 0.001). When controlling for the APACHE II score in the multivariate analysis, empirical treatment with piperacillin-tazobactam was an independent predictor of mortality (OR, 220.5; 95% CI, 3.8 to 12,707.4; P = 0.009). Notably, the piperacillin-tazobactam susceptibility breakpoints for P. aeruginosa were lowered to ≤16 mg/liter by the CLSI in 2012 (10).
DeFife et al. evaluated Gram-negative bacteremia treated with levofloxacin in a retrospective cohort study (n = 312) and stratified patients based on levofloxacin MIC (13). Patients were classified as having a low MIC (≤0.25 mg/liter), intermediate MIC (0.5 mg/liter), or high MIC (1 or 2 mg/liter), all of which are considered susceptible according to the CLSI susceptibility breakpoint of 2 mg/liter for levofloxacin. Patients in the high-MIC group had a longer time to clearance of bacteremia (mean of 2.1 days versus 1.0 day; P < 0.001) and longer LOS postculture (mean of 16.4 days versus 7.3 days; P = 0.02).
Bhat et al. evaluated 204 episodes of Gram-negative bacteremia treated with FDA-approved doses of cefepime (1 or 2 g every 12 h) (3). A CART analysis identified that a cefepime MIC of ≥8 mg/liter predicted increased 28-day mortality (58.4% versus 21.4%; P = 0.001). Multivariate analysis including cefepime MICs of 8 mg/liter and ≥16 mg/liter as independent variables revealed both were predictive of 28-day mortality (aOR of 9.1, 95% CI of 2.2 to 37.5, and P = 0.002 and aOR of 7.5, 95% CI of 1.9 to 29.2, and P = 0.004, respectively). The authors suggested that when utilizing cefepime at 1 or 2 g every 12 h, a MIC of ≥8 mg/liter should not be considered susceptible. However, there were few patients with pathogens that had a MIC of 8, and many of them received doses much lower (e.g., ≤2 g/day) than are commonly used in contemporary clinical practice. The CLSI has recently revised the susceptibility breakpoints for Enterobacteriaceae to many cephalosporins, but did not change the susceptibility breakpoints of ≤8 mg/liter for cefepime as mathematical models predict pharmacodynamic success when doses of ≥3 g/day are used (9).
While our results are compelling, limitations to these data do exist. First, this was a constrained-enrollment, retrospective study at a single institution, and only associations can be suggested. Despite having a limited number of patients, especially in the middle MIC classifications, we noted a high categorical agreement with our breakpoint of 2 mg/liter (Fig. 2). In an effort to minimize the relative constraint of subject numbers in any categorical MIC, we utilized a jackknife procedure to omit observations iteratively and systematically to assess if any singular result drove outcomes. Results using the jackknife procedure were isomorphic. Patients with isolates showing a MIC of ≥4 mg/liter were much more likely to die even after controlling for confounders. It should be noted, however, that if the single patient with a MIC of 4 mg/liter survived instead of died, the same analyses performed would suggest a breakpoint of 8 mg/liter. Such uncertainty underscores the need for more robust clinical outcome data, especially in the MIC range of 2 to 8 mg/liter. Second, automated testing using the standard Vitek 2 card [Vitek 2 AST-GN 24 (22229) card] was a clinical microbiology constraint and necessitated surrogate MICs for 20 cultures. In all cases in which surrogate MICs were used, isolates were tested against imipenem, and patients were treated with meropenem. Universally, isolates were found at either the lowest imipenem MIC value (i.e., ≤1 mg/liter; n = 16) or the highest MIC value (i.e., ≥16 mg/liter; n = 4). Of those testing at the highest MIC value, all were A. baumannii (n = 4), where imipenem MICs are generally equal to or lower than meropenem MICs (16). Thus, imipenem did not likely overcall resistance for meropenem treatment. In the remaining cases (n = 12), all were Enterobacteriaceae, where carbapenem discordance is not known to exist (16, 19, 24, 33). Third, our subgroup analysis on organism category was unproductive, likely due to a lack of power. Fourth, while we recognize that automated susceptibility testing such as Vitek 2 is subject to limitations, these methods are approved by the FDA for categorical susceptibility interpretation and are utilized widely in clinical practice. Thus, we believe our results apply to Vitek 2 MICs and further study is warranted by other methodologies, such as broth or agar dilution. Fifth, since doripenem and ertapenem were not evaluated, it is not clear that results can be extrapolated to those patient populations. Finally, due to constraints in our sample size, models were not improved when the variable “other active therapy” was added to our models. Thus, we cannot comment on the utility of such a practice. Given our results and the noted limitations above, we offer the following recommendation. Until a larger study is performed with (i) MICs in the range of 2 mg/liter to 8 mg/liter, (ii) better representation from genera of organisms, and (iii) MIC results tested by broth or agar dilution, we believe that it is prudent to consider Gram-negative organisms with an imipenem MIC of ≥4 mg/liter as potentially clinically resistant.
We have shown that there is empirical clinical evidence for the consideration of lowering the breakpoints of carbapenems for Gram-negative organisms. Minimally, MIC data should be considered before utilizing any carbapenem therapy where resistance is presumed. This study revealed that patients with organisms that had a MIC of ≥4 mg/liter had increased mortality compared to patients whose isolates had lower MICs. We support recommendations to empirically lower the breakpoints for carbapenems for Gram-negative organisms.
ACKNOWLEDGMENTS
We acknowledge Emily Steadman for assistance with data collection and database management.
This work was supported in part by a Chicago College of Pharmacy Summer Research grant, Midwestern University, Downers Grove, IL. Midwestern University had no role in the writing of this article or the decision to submit the manuscript for publication.
All authors declare they have had no relevant conflicts of interest related to this article within the previous 2 years. No author has been paid to write this article by a pharmaceutical company. The corresponding author has had full access to all data and has had final responsibility for the decision to submit the article for publication.
Footnotes
Published ahead of print 9 July 2012
REFERENCES
- 1. Ambrose PG, Bhavnani SM, Jones RN, Craig WA, Dudley MN. 2004. Use of pharmacokinetic-pharmacodynamic and Monte Carlo simulation as decision support for the re-evaluation of NCCLS cephem susceptibility breakpoints for Enterobacteriaceae, abstr A-138. Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother [Google Scholar]
- 2. Arnold RS, et al. 2011. Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South. Med. J. 104:40–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhat SV, et al. 2007. Failure of current cefepime breakpoints to predict clinical outcomes of bacteremia caused by gram-negative organisms. Antimicrob. Agents Chemother. 51:4390–4395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhavnani SM, Ambrose PG, Craig WA, Dudley MN, Jones RN. 2006. Outcomes evaluation of patients with ESBL- and non-ESBL-producing Escherichia coli and Klebsiella species as defined by CLSI reference methods: report from the SENTRY Antimicrobial Surveillance Program. Diagn. Microbiol. Infect. Dis. 54:231–236 [DOI] [PubMed] [Google Scholar]
- 5. Bhavnani SM, et al. 2010. Pharmacokinetic-pharmacodynamic basis for CLSI carbapenem susceptibility breakpoint changes, abstr A-1382. Abstr. 50th Intersci. Conf. Antimicrob. Agents Chemother [Google Scholar]
- 6. Charlson ME, Pompei P, Ales KL, MacKenzie CR. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J. Chronic Dis. 40:373–383 [DOI] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute 2009. Subcommittee on Antimicrobial Susceptibility Testing, January 11–13, summary minutes. http://www.clsi.org/Content/NavigationMenu/Committees/Microbiology/AST/ArchiveofPreviousEvents/Summary_MinutesASTJan2009.pdf Accessed on 14 July 2011
- 8. Clinical and Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing. M100-S119. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing. M100-S120. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing. M100-S122. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11. Craig WA, Bhavnani SM, Ambrose PG, Dudley MN, Jones RN. 2005. Evaluation of clinical outcomes among patients with ESBL producing Enterobacteriaceae treated with cephalosporin monotherapy, abstr K-1291. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother [Google Scholar]
- 12. Craig WA, et al. 2008. Impact of KPCs on the in vivo activity of three carbapenems in the neutropenic mouse-thigh infection model, abstr A-029. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother [Google Scholar]
- 13. Defife R, Scheetz MH, Feinglass JM, Postelnick MJ, Scarsi KK. 2009. Effect of differences in MIC values on clinical outcomes in patients with bloodstream infections caused by gram-negative organisms treated with levofloxacin. Antimicrob. Agents Chemother. 53:1074–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deyo RA, Cherkin DC, Ciol MA. 1992. Adapting a clinical comorbidity index for use with ICD-9-CM administrative databases. J. Clin. Epidemiol. 45:613–619 [DOI] [PubMed] [Google Scholar]
- 15. Dudley MN, et al. 25 January 2010, posting date CLSI Working Group. http://www.clsi.org/Content/NavigationMenu/Committees/Microbiology/AST/January2010ASTMeetingPresentations/EnterobacteriaceaeWGCarbapenemBreakpointDiscussionSlides.pdf Accessed on 28 June 2011
- 16. Esterly JS, Qi C, Malczynski M, Scheetz MH. 2010. Predictability of doripenem susceptibility in Acinetobacter baumannii isolates based on other carbapenem susceptibilities and bla OXA gene status. Pharmacotherapy 30:354–360 [DOI] [PubMed] [Google Scholar]
- 17. Hosmer DW, Lemeshow S. 2000. Applied logistic regression, 2nd ed, p 31–46 Wiley & Sons, Hoboken, NJ [Google Scholar]
- 17a. Hosmer DW, Lemeshow S. 2000. Applied logistic regression, 2nd ed, p 91–142 Wiley & Sons, Hoboken, NJ [Google Scholar]
- 18. Infectious Diseases Society of America 2011. Standards, practice guidelines, and statements developed and/or endorsed by IDSA. http://www.idsociety.org/Content.aspx?id=9088 Accessed 5 July 2011
- 19. Jones RN, Sader HS, Fritsche TR, Rhomberg PR. 2006. Carbapenem susceptibility discords among Acinetobacter isolates. Clin. Infect. Dis. 42:158. [DOI] [PubMed] [Google Scholar]
- 20. Kang CI, et al. 2005. Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob. Agents Chemother. 49:760–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kollef KE, et al. 2008. Predictors of 30-day mortality and hospital costs in patients with ventilator-associated pneumonia attributed to potentially antibiotic-resistant gram-negative bacteria. Chest 134:281–287 [DOI] [PubMed] [Google Scholar]
- 22. Krueger WA, et al. 2005. Evaluation by Monte Carlo simulation of the pharmacokinetics of two doses of meropenem administered intermittently or as a continuous infusion in healthy volunteers. Antimicrob. Agents Chemother. 49:1881–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee LS, et al. 2010. Comparison of 30-min and 3-h infusion regimens for imipenem/cilastatin and for meropenem evaluated by Monte Carlo simulation. Diagn. Microbiol. Infect. Dis. 68:251–258 [DOI] [PubMed] [Google Scholar]
- 24. Lesho E, Wortmann G, Moran K, Craft D. 2005. Fatal Acinetobacter baumannii infection with discordant carbapenem susceptibility. Clin. Infect. Dis. 41:758–759 [DOI] [PubMed] [Google Scholar]
- 25. Lodise TP, Jr, et al. 2007. Predictors of 30-day mortality among patients with Pseudomonas aeruginosa bloodstream infections: impact of delayed appropriate antibiotic selection. Antimicrob. Agents Chemother. 51:3510–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Merck 2010. Primaxin IV package insert. Merck & Co., Whitehouse Station, NJ [Google Scholar]
- 27. Micek ST, et al. 2005. Pseudomonas aeruginosa bloodstream infection: importance of appropriate initial antimicrobial treatment. Antimicrob. Agents Chemother. 49:1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Michalopoulos A, Falagas ME. 2010. Treatment of Acinetobacter infections. Expert Opin. Pharmacother. 11:779–788 [DOI] [PubMed] [Google Scholar]
- 29. Patel JB. 27 September 2007, posting date Carbapenem resistance in Enterobacteriaceae. Centers for Disease Control and Prevention, Atlanta, GA: http://www.state.nj.us/health/phel/documents/teleconf/carbapenem_powerpoint.ppt Accessed on 14 July 2011 [Google Scholar]
- 30. Rhomberg PR, Jones RN. 2009. Summary trends for the Meropenem Yearly Susceptibility Test Information Collection Program: a 10-year experience in the United States (1999–2008). Diagn. Microbiol. Infect. Dis. 65:414–426 [DOI] [PubMed] [Google Scholar]
- 31. Rodriguez-Martinez JM, Poirel L, Nordmann P. 2009. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:4783–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sakka SG, et al. 2007. Population pharmacokinetics and pharmacodynamics of continuous versus short-term infusion of imipenem-cilastatin in critically ill patients in a randomized, controlled trial. Antimicrob. Agents Chemother. 51:3304–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scheetz MH, Esterly JS, Malczynski M, Postelnick M, Qi C. 2009. Impact of dissimilar susceptibility breakpoints for doripenem on susceptibility and carbapenem discordance for Pseudomonas aeruginosa. Diagn. Microbiol. Infect. Dis. 64:465–467 [DOI] [PubMed] [Google Scholar]
- 34. Tam VH, et al. 2008. Outcomes of bacteremia due to Pseudomonas aeruginosa with reduced susceptibility to piperacillin-tazobactam: implications on the appropriateness of the resistance breakpoint. Clin. Infect. Dis. 46:862–867 [DOI] [PubMed] [Google Scholar]
- 35. Weisenberg SA, Morgan DJ, Espinal-Witter R, Larone DH. 2009. Clinical outcomes of patients with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae after treatment with imipenem or meropenem. Diagn. Microbiol. Infect. Dis. 64:233–235 [DOI] [PMC free article] [PubMed] [Google Scholar]