Abstract
The percentage of invasive penicillin-nonsusceptible pneumococci (PNSSP) isolated in Italy in the seven-valent pneumococcal conjugate vaccine (PCV7) era moderately increased in comparison to the pre-PCV7 era. Increase of nonvaccine serotypes was observed among PNSSP. The most frequent PNSSP clones were the same as those identified in the pre-PCV7 era, although they were present in different proportions. Clonal expansion, emergence of new clones, and acquisition of penicillin resistance by established clones contributed to the maintenance of penicillin resistance.
TEXT
Streptococcus pneumoniae is a leading cause of illness and death in children and adults worldwide (11, 18). Over 90 pneumococcal serotypes are known, many of which can cause invasive pneumococcal disease (IPD) (9). After the introduction of the heptavalent pneumococcal conjugate vaccine (PCV7), licensed in the United States in 2000, the rate of IPD due to vaccine serotypes (VS) decreased dramatically (23), while the rate of IPD due to serotypes not included in the vaccine (nonvaccine serotypes [NVS]) increased, a phenomenon called serotype replacement (10, 20). An indirect effect of PCV7 was the decrease of infections due to penicillin-nonsusceptible S. pneumoniae (PNSSP), which were most commonly associated with VS (14). However, due to serotype replacement, infections due to penicillin-resistant NVS increased (14).
PCV7 has been available in Italy since 2001; in 2008, PCV7 coverage was 55% on a national basis, with wide regional variations (12). A previous study, performed on pneumococcal isolates obtained during the years 2001 to 2003, showed that approximately 12% were penicillin resistant and that resistant strains belonged to several international clones (7). In this study, we describe the serotypes, antimicrobial resistance profiles, and molecular characterizations of PNSSP recovered after the implementation of PCV7, in comparison with the prevaccine era, in order to gain insight into the evolution of the pneumococcal population in Italy.
The nationwide Italian surveillance of invasive bacterial diseases (http://www.simi.iss.it/files/Report_MBI.pdf) includes the shipping (on a voluntary basis) of invasive pneumococcal isolates to the Istituto Superiore di Sanità for serotyping and further characterization. All isolates obtained in the years from 2006 to 2010 were examined in this study. Serotyping was performed by latex agglutination and the Quellung reaction (7), and antibiotic susceptibility to penicillin (PEN), ceftriaxone (CRO), erythromycin (ERY), clindamycin (CLI), tetracycline (TET), and chloramphenicol (CHL) was tested by Etest (AB Biodisk, Solna, Sweden). The results were interpreted following the breakpoints of the 2010 Clinical and Laboratory Standards Institute (CLSI) (5) for meningitis, considering as PNSSP the isolates showing MICs of ≥0.12 μg/μl. Results for PEN and CRO (parenteral) were evaluated also using the CLSI breakpoints for nonmeningitis cases. The definitions of low-level (MIC, 0.12 to 1 μg/μl) and high-level (MIC, >1 μg/μl) PEN resistance were used for epidemiological purposes and for comparison with previous studies. The presence of ERY resistance genes was examined in all PNSSP by PCR (16). Pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and pbp2b/pbp2x restriction analysis were used to define clonal groups, as previously described (6, 7). At least one isolate from each PFGE type containing <10 isolates and at least 3 isolates from each PFGE type containing ≥10 isolates were submitted to MLST. The sequence types (STs) obtained were considered related if they were identical, a single-locus variant (SLV), or a double-locus variant (DLV) of each other. In order to identify pneumococcal clones, STs were compared to those of the international clones of the Pneumococcal Molecular Epidemiology Network (PMEN; http://www.sph.emory.edu/PMEN/) and to those available at the MLST website (http://www.mlst.net) by using eBURST to define clonal complexes (CCs). Therefore, each clone was identified by the related PMEN clone, its ST, and the corresponding CC.
Overall, 778 invasive isolates were obtained in the years 2006 to 2010, representing approximately one-third of all the reported IPD cases, with 219 isolates (28.1%) obtained from children under 5 years old. Among these isolates, 218 (28.0%) belonged to VS and 560 (72.0%) to NVS. The most frequent serotypes, in ranked order, were 19A (13.7%), 1 (12.3%), 14 (10.7%), 3 (9.4%), and 7F (9.1%).
A total of 112 isolates (14.4%) were PNSSP (MIC, ≥0.12 μg/μl), and of these, 40 (35.7%) were isolated from children under 5 years old. Sixty-three out of 112 PNSSP (56.2%) were VS, and 49 (43.8%) belonged to NVS. The serotypes recovered are shown in Table 1. High-level resistance to PEN (MIC, >1 μg/ml) was found in 34 isolates and was mostly associated with serotypes 9V, 14, 19A, and 23F. Using the CLSI breakpoints for meningitis, 41 PNSSP (36.6%) were resistant or intermediate to CRO (MIC, ≥1 μg/μl). However, by considering the CLSI breakpoints for nonmeningitis cases, only 10 (8.9%) of the isolates under study were intermediate or resistant to PEN (MIC, ≥4 μg/μl), and only 14 (12.5%) were intermediate or resistant to CRO (MIC, ≥2 μg/μl). As for the other antibiotics, 84 isolates (75%) were resistant to ERY, 79 (70.5%) to CLI, 71 (63.4%) to TET, and 14 (12.5%) to CHL. Resistance to ERY was associated with the presence of erm(B) in 70 isolates, of mef(E) in 3 isolates, and of double gene erm(B) mef(E) in 6 isolates (Table 1). Multidrug resistance, that is, resistance to at least three different classes of antibiotics, was found in 81 (72.3%) PNSSP.
Table 1.
Penicillin-nonsusceptible pneumococci isolated in 2006 to 2010 in Italy
| VS or NVS | Serotype (no. of isolates) | No. of PFGE typesa | No. of Ery-resistant isolates | Ery resistance gene(s) (no. of isolates) | No. of MDR isolates | Most frequent antibiotype in MDR isolates |
|---|---|---|---|---|---|---|
| VS | 14 (26) | 2 | 16 | erm(B) (16) | 16 | PEN, CRO, ERY, CLIN, TET |
| 23F (12) | 3 | 10 | erm(B) (10) | 10 | PEN, CRO, ERY, CLIN, CHL | |
| 19F (11) | 4 | 9 | erm(B) (8); erm(B) and mef(E) (1) | 9 | PEN, ERY, CLIN, TET | |
| 9V (8) | 1 | 1 | mef(E) (1) | 1 | PEN, CRO, ERY, TET | |
| 6B (6) | 3 | 6 | erm(B) (6) | 6 | PEN, ERY, CLIN, TET | |
| NVS | 19A (20) | 4 | 17 | erm(B) (12); erm(B) and mef(E) (5) | 17 | PEN, ERY, CLIN, TET |
| 24F (11) | 1 | 11 | erm(B) (11) | 11 | PEN, ERY, CLIN | |
| 15A (8) | 1 | 8 | erm(B) (8) | 8 | PEN, ERY, CLIN, TET | |
| 6A (5) | 2 | 5 | erm(B) (2); mef(E) (3) | 2 | PEN, ERY, CLIN, TET | |
| 6C (1) | 1 | 0 | NTb | 0 | ||
| 7F (1) | 1 | 0 | NT | 0 | ||
| 11A (1) | 1 | 0 | NT | 0 | ||
| 15B (1) | 1 | 1 | erm(B) (1) | 1 | PEN, ERY, CLIN, TET | |
| 23B (1) | 1 | 0 | NT | 0 |
Indicates the number of PFGE types found within each serotype.
NT, not tested.
A good correspondence between PFGE and MLST results was found. The 8 most common clones, representing 104 out of 112 (92.9%) PNSSP, were all PMEN clones (Table 2). The most abundant clone was Spain9V-3/ST156/CC156, followed by Sweden15A-25/ST63/CC63 and Denmark14-32/ST230/CC230 clones. The 6 largest clones included multiple serotypes, with Colombia 23F-26/ST338/CC156 and Sweden15A-25/ST63/CC63 including 6 and 5 serotypes, respectively (Table 2). Clonal diversity within each serotype is shown in Table 2: among VS, serotypes 19F, 23F, and 6B showed the highest genetic diversity, and among NVS, serotype19A showed the highest genetic diversity. While in the pre-PCV7 era PNSSP of serotype 19A was only found associated with Sweden15A-25/ST63/CC63 (6), in this study 19A was associated with four clones (Table 2).
Table 2.
Genotypic characteristics of penicillin-nonsusceptible pneumococcal clinical isolates in Italy, 2006 to 2010
| PFGE type (no. of isolates) | ST(s) | CCa | PMEN cloneb | Serotype(s) (no. of isolates) | MIC range (μg/ml) |
pbp gene restriction profile(s)c |
|||
|---|---|---|---|---|---|---|---|---|---|
| PEN | CRO | ERY | pbp2b | pbp2x | |||||
| 1 (30) | 143, 557, 2916 | 156 | Spain9V-3/ST156 | 9V (8), 14 (22) | 0.25–2 | 0.5–4 | 0.12–≥256 | 6, 34, 37 | 2, 28, 39 |
| 2 (26) | 63, 2543, 3816 | 63 | Sweden15A-25/ST63 | 15A (8), 15B (1), 19A (8), 19F (6), 23F (3) | 0.12–1 | 0.06–1 | 0.12–≥256 | 10 | 8, 26, 27, 28, 29, 31, 36, 39 |
| 3 (16) | 230, 276, 2013 | 230 | Denmark14-32/ST230 | 19A (5), 24F (11) | 0.25–1 | 0.12–2 | 0.12–≥256 | 27 | 2, 8, 41 |
| 34 (9) | 81 | 81 | Spain23F-1/ST81 | 23F (7), 19F (2) | 1–4 | 0.5–2 | ≥256 | 6 | 2 |
| 59 (9) | 338, 1131, 2689, 7121 | 156 | Colombia23F-26/ST338 | 6C (1), 7F (1), 19A (2), 19F (2), 23B (1), 23F (2) | 0.12–0.5 | ≤0.03–0.12 | 0.12 | 19 | 11, 22, 28, 29 |
| 85 (6) | 320 | 320 | Taiwan19F-14/ST236 | 19A (5), 19F (1) | 2–4 | 1–2 | ≥256 | 36 | 28, 40 |
| 4 (4) | 315 | 315 | Poland6B-20/ST315 | 6B (4) | 0.12–0.25 | 0.06–0.12 | ≥256 | 12 | 22 |
| 63 (4) | 550 | 15 | England14-9/ST9 | 14 (4) | 0.5 | 0.25–0.5 | 0.12–≥256 | 19 | 26, negd |
| 50 (3) | 473 | 473 | None | 6A (3) | 0.25–0.5 | 0.06–0.12 | 0.25–8 | 5 | 38 |
| 6 (2) | 675e | Singleton | None | 6A (2) | 0.25 | 0.12 | ≥256 | 28 | 12 |
| 35 (1) | 90 | 156 | Spain6B-2/ST90 | 6B (1) | 1 | 1 | ≥256 | 22 | 12 |
| 64 (1) | 331 | Singleton | None | 11A (1) | 0.25 | 0.25 | 0.12 | 35 | 28 |
| 92 (1) | NDf | ND | ND | 6B (1) | 2 | 1 | 64 | 33 | 12 |
Clonal complex as defined using information from the MLST website (www.mlst.net).
A related PMEN clone was inferred by the ST associated with the PFGE type (STs were considered related to a PMEN reference clone if they were identical SLVs or DLVs.
See text for details.
No PCR product was amplified.
Defined on the basis of a previous assignation (7).
ND, not determined.
Overall, 13 and 15 different restriction profiles were detected for pbp2b and pbp2x genes, respectively, and 28 different composite pbp2b/pbp2x profiles were obtained (Table 2). pbp2b restriction profiles appeared to be specific for the different clones and conserved over time, strongly supporting the use of pbp2b restriction profiles as typing markers for PNSSP strains. On the contrary, half of the pbp2x restriction profiles were shared between different PNSSP clones, indicating a frequent horizontal exchange of this gene. The presence of the same pbp2x profile in two different clones could also be a marker of a capsular switch phenomenon. As reported by Brueggemann et al. (3), pbp2x can be involved in recombinational events that include the capsular locus between different pneumococcal clones.
When the results were compared with those obtained in the pre-PCV7 era (2001 to 2003), differences in the relative frequencies of PNSSP, VS, NVS, and clones were observed. PNSSP represented approximately 14% of the invasive pneumococci in the PCV7 era, a percentage slightly higher than that found in the period antecedent to PCV7 implementation, which was 10% to 12% (7). Among PNSSP, VS decreased from 78.9% to 56.2%, with a concurrent increase in NVS from 21.1% to 43.8%. The proportion of VS among PNSSP from 2006 to 2010 was 2-fold higher than that among the nonselected invasive strains from the same period, indicating that most of the antibiotic resistance was still associated with VS.
Within NVS, an increment in the proportion of 19A, 24F, 15A, and 6A was noted, while 35F, which was well represented in the prevaccine era among PNSSP (7), was not detected.
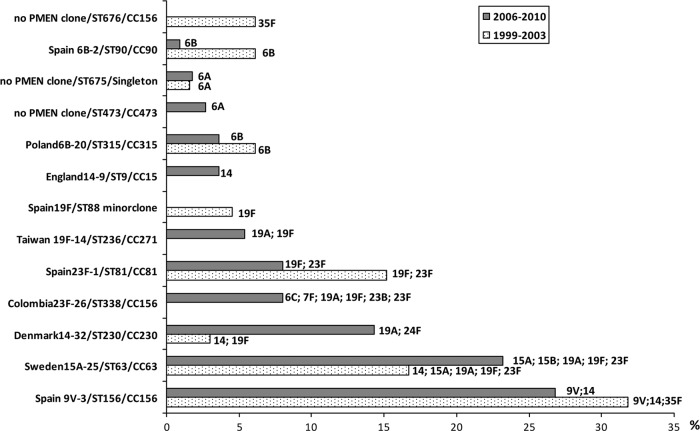
Seven clones, accounting for 76.8% of the PNSSP isolated in the PCV7 era, were recovered in both periods, although in different proportions (Fig. 1). Clonal expansion appears to represent the primary mechanism for serotype replacement and for the maintenance of antimicrobial resistance. Two clones, Sweden15A-25/ST63/CC63, expressing multiple VS and NVS serotypes, and Denmark14-32/ST230/CC230, expressing NVS 19A and 24F, expanded in the PCV7 era, ranking second and third, respectively. The increase of these two clones, associated with both carriage and disease, has been reported in several countries (1, 8, 17, 22). Denmark14-32/ST230/CC230 was detected in Italy as far back as 1997, in association with serotype 24F isolates (19).
Fig 1.
Bar chart showing changes in the relative frequencies of the major PNSSP clones causing IPD in Italy identified before (1999 to 2003) and after (2006 to 2010) the introduction of PCV7. The serotypes expressed by each clone are indicated.
We observed that some VS clones, present in Italy before the introduction of PCV7, appeared to be more refractory to extinction than others; for instance, Spain9V-3/ST156, which ranked first in both periods, showed a limited decrease, in comparison with Spain23F-1/ST81 and Spain6B-2/ST90. The presence of the same antibiotic-resistant clones in the periods before and after PCV7 implementation, and also of an evolution in their proportions leading to persistence of antimicrobial resistance, has already been documented in a colonization study from Portugal (22).
Other mechanisms that seem to have significantly contributed to the evolution of PNSSP in Italy are introduction of new clones and acquisition of penicillin resistance by established clones. For example, Taiwan19F-14/ST236/CC320 and Colombia23F-26/ST338/CC156 clones were not detected in the prevaccine era (10) and emerged in Italy only in 2008 and 2007, respectively. These two clones have different characteristics: Taiwan19F-14/ST236/CC320 comprises isolates with high-level penicillin resistance, intermediate or full resistance to ceftriaxone (MIC range, 1 to 2 μg/ml), and resistance to erythromycin (MIC, >256 μg/ml), while Colombia23F-26/ST338/CC156 comprises isolates with low-level penicillin resistance that are fully susceptible to the other antibiotics. Both these clones have expanded successfully in different areas of the globe. Taiwan19F-14/ST236/CC320 spread rapidly in Asian countries before PCV7 introduction (4, 21); in the United States it has been associated with the emergence of MDR serotype 19A isolates after PCV7 implementation (2). Colombia23F-26/ST338/CC156 has been increasingly found in the United States and Japan (13, 17). De novo acquisition of penicillin resistance has been detected in 2 PNSSP clones, new clone/ST473/CC473 and England14-9/ST9/CC15, both found in the pre-PCV7 era only in association with penicillin-susceptible isolates (6).
It has been suggested that PCV7 acts as a “serotype filter,” because its effect on the prevalence of a particular strain is predictable on the basis of the serotype of the strain, with little effect on the genetic background (15). Our findings about the high clonal diversity of serotype 19A PNSSP, the emergence of new PNSSP clones, and the disappearance of others seem to indicate that the vaccine exerts a significant effect also on the genetic background of pneumococci.
Although PCV7 had an important impact in preventing IPD, a second-generation vaccine containing 6 additional serotypes (PCV13) has been developed. PCV13 was introduced in Italy at the end of 2010, and according to our study it should cover 78.5% of infections due to PNSSP. Serotypes 15A and 24F represent the most abundant PNSSP serotypes not covered by the new vaccine. Continuous surveillance of the pneumococcal population following the introduction of PCV13 is essential to evaluate the impact of the new vaccine on the evolution of the pneumococcal population.
ACKNOWLEDGMENTS
We acknowledge the use of the pneumococcal MLST database, which is located at Imperial College, London, and is funded by the Wellcome Trust.
This study was supported in part by grants from the Italian Ministry of Health (CCM; Surveillance of Invasive Bacterial Diseases, 2009, and Laboratory Surveillance of Infections Caused by Bacterial Pathogens submitted to European Surveillance or by Bioterrorism Agents, 2010).
Footnotes
Published ahead of print 2 July 2012
REFERENCES
- 1. Aguiar SI, et al. 2010. Denmark14-230 clone as an increasing cause of pneumococcal infection in Portugal within a background of diverse serotype 19A lineages. J. Clin. Microbiol. 48:101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beall BW, et al. 2011. Shifting genetic structure of invasive serotype 19A pneumococci in the United States. J. Infect. Dis. 203:1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brueggemann AB, Pai R, Crook DW, Beall B. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 3:e168 doi:10.1371/journal.ppat.0030168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choi EH, et al. 2008. Streptococcus pneumoniae serotype 19A in children, South Korea. Emerg. Infect. Dis. 14:275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing, 20th informational supplement. M100-S20 CLSI, Wayne, PA [Google Scholar]
- 6. Gherardi G, et al. 2009. Population structure of invasive Streptococcus pneumoniae isolates in Italy prior to the implementation of the 7-valent conjugate vaccine (1999–2003). Eur. J. Clin. Microbiol. Infect. Dis. 28:99–103 [DOI] [PubMed] [Google Scholar]
- 7. Gherardi G, et al. 2007. Antibiotic-resistant invasive pneumococcal clones in Italy. J. Clin. Microbiol. 45:306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hanage WP, et al. 2007. Diversity and antibiotic resistance among nonvaccine serotypes of Streptococcus pneumoniae carriage isolates in the post-heptavalent conjugate vaccine era. J. Infect. Dis. 195:347–352 [DOI] [PubMed] [Google Scholar]
- 9. Hausdorff WP, Bryant J, Paradiso PR, Siber GR. 2000. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin. Infect. Dis. 30:100–121 [DOI] [PubMed] [Google Scholar]
- 10. Hsu HE, et al. 2009. Effect of pneumococcal conjugate vaccine on pneumococcal meningitis. N. Engl. J. Med. 360:244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang SS, et al. 2011. Healthcare utilization and cost of pneumococcal disease in the United States. Vaccine 29:3398–3412 [DOI] [PubMed] [Google Scholar]
- 12. ICONA Working Group 2009. ICONA 2008: national vaccination coverage survey among children and adolescents. Rapporti ISTISAN 09/29. Istituto Superiore di Sanitá, Rome, Italy: http://www.iss.it/binary/publ/cont/09_29_web.pdf. (In Italian.) [Google Scholar]
- 13. Imai S, et al. 2009. High prevalence of multidrug-resistant Pneumococcal Molecular Epidemiology Network clones among Streptococcus pneumoniae isolates from adult patients with community-acquired pneumonia in Japan. Clin. Microbiol. Infect. 15:1039–1045 [DOI] [PubMed] [Google Scholar]
- 14. Kyaw MH, et al. 2006. Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N. Engl. J. Med. 354:1455–1463 [DOI] [PubMed] [Google Scholar]
- 15. Lipsitch M, et al. 2007. Strain characteristics of Streptococcus pneumoniae carriage and invasive disease isolates during a cluster-randomized clinical trial of the 7-valent pneumococcal conjugate vaccine. J. Infect. Dis. 196:1221–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monaco M, Camilli R, D'Ambrosio F, Del Grosso M, Pantosti A. 2005. Evolution of erythromycin resistance in Streptococcus pneumoniae in Italy. J. Antimicrob. Chemother. 55:256–259 [DOI] [PubMed] [Google Scholar]
- 17. Moore MR, et al. 2008. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J. Infect. Dis. 197:1016–1027 [DOI] [PubMed] [Google Scholar]
- 18. O'Brien KL, et al. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902 [DOI] [PubMed] [Google Scholar]
- 19. Pantosti A, et al. 2002. A novel, multiple drug-resistant, serotype 24F strain of Streptococcus pneumoniae that caused meningitis in patients in Naples, Italy. Clin. Infect. Dis. 35:205–208 [DOI] [PubMed] [Google Scholar]
- 20. Rosen JB, et al. 2011. Geographic variation in invasive pneumococcal disease following pneumococcal conjugate vaccine introduction in the United States. Clin. Infect. Dis. 53:137–143 [DOI] [PubMed] [Google Scholar]
- 21. Shin J, Baek JY, Kim SH, Song JH, Ko KS. 2011. Predominance of ST320 among Streptococcus pneumoniae serotype 19A isolates from 10 Asian countries. J. Antimicrob. Chemother. 66:1001–1004 [DOI] [PubMed] [Google Scholar]
- 22. Simões AS, et al. 2011. Clonal evolution leading to maintenance of antibiotic resistance rates among colonizing pneumococci in the PCV7 era in Portugal. J. Clin. Microbiol. 49:2810–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whitney CG, et al. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737–1746 [DOI] [PubMed] [Google Scholar]