Abstract
Early stages of mucosal infection are potential targets for HIV-1 prevention. CD4 is the primary receptor in HIV-1 infection whereas DC-SIGN likely plays an important role in HIV-1 dissemination, particularly during sexual transmission. To test the hypothesis that an inhibitor simultaneously targeting both CD4 and DC-SIGN binding sites on gp120 may provide a potent anti-HIV strategy, we designed constructs by fusing the extracellular CD4 and DC-SIGN domains together with varied arrangements of the lengths of CD4, DC-SIGN and the linker. We expressed, purified and characterized a series of soluble CD4-linker–DC-SIGN (CLD) fusion proteins. Several CLDs, composed of a longer linker and an extra neck domain of DC-SIGN, had enhanced affinity for gp120 as evidenced by molecular-interaction analysis. Furthermore, such CLDs exhibited significantly enhanced neutralization activity against both laboratory-adapted and primary HIV-1 isolates. Moreover, CLDs efficiently inhibited HIV-1 infection in trans via a DC-SIGN-expressing cell line and primary human dendritic cells. This was further strengthened by the results from the human cervical explant model, showing that CLDs potently prevented both localized and disseminated infections. This is the first time that soluble DC-SIGN-based bifunctional proteins have demonstrated anti-HIV potency. Our study provides proof of the concept that targeting both CD4 and DC-SIGN binding sites on gp120 represents a novel antiviral strategy. Given that DC-SIGN binding to gp120 increases exposure of the CD4 binding site and that the soluble forms of CD4 and DC-SIGN occur in vivo, further improvement of CLDs may render them potentially useful in prophylaxis or therapeutics.
INTRODUCTION
The majority of HIV-1 infections are acquired by mucosal exposure, with sexual transmission as the leading mode of HIV-1 infection worldwide. The distribution of dendritic cells (DCs) in the cervicovagina and colorectum may allow them to be one of the first cell types to contact HIV-1 (21, 34, 57). DCs may capture HIV-1 by attachment receptors, in particular DC-SIGN, and transfer the virus to permissive CD4+ T cells, resulting in trans infection (12, 19, 37, 55). In addition, DC-SIGN binding to HIV-1 increases the local concentration of the virus on the DC surface and can enhance cis infection via the low levels of CD4 and CCR5 on DCs (38). Both HIV-1-captured and -infected DCs can efficiently release virus particles to CD4+ T cells at the points of cell contact termed virological synapses (41). Evidence from a colorectal explant study indicated that DC-SIGN accounts for 90% of HIV-1 binding on mucosal mononuclear cells (23). Our previous study using a cervicovaginal model demonstrated that simultaneous blockade of CD4 and mannose-binding C-type lectin receptors (MCLRs) including DC-SIGN is required to inhibit HIV-1 uptake and dissemination by migratory cells (28). Given their critical roles likely played in HIV-1 transmission, CD4 and DC-SIGN are important targets for the development of topical microbicides.
HIV-1 entry and transmission involve complex interactions between viral envelope glycoprotein (Env) and receptors on host cells. The binding of gp120 to CD4 is virtually universal among HIV-1 isolates. Soluble CD4 (sCD4), which acts as a receptor decoy to prevent the engagement of HIV-1 Env with cell-surface CD4, represents a promising competitive viral attachment inhibitor. However, despite its efficient neutralization activity against laboratory-adapted HIV-1 strains, sCD4 showed poor antiviral activity against primary HIV-1 isolates, and very large doses of sCD4 were required to achieve modest reductions of viral loads in vivo (33). This is likely due to the relatively lower Env-binding affinity of sCD4 per se compared with that of target cell-bound CD4 (52). Although PRO-542 (CD4-IgG2), a tetrameric fusion protein between CD4 and immunoglobulin G, is much more potent in vitro than the parental monomer, the translation of this improvement to clinical use remains uncertain (2, 51). The interaction of HIV-1 with DC-SIGN does not result in direct infection of DCs but instead enhances cis and/or trans infection. Several studies have shown that antagonists against DC-SIGN inhibit DC-SIGN-mediated HIV-1 transmission (7, 42, 46), whereas the antiviral activity of sDC-SIGN seems more complex (25, 40). Although sDC-SIGN decreases the capture of HIV-1 by DC-SIGN (39), sDC-SIGN binding to HIV-1 Env can also increase the exposure of the CD4 binding site on gp120, which in turn contributes to enhancement of infection (25), compromising the development of DC-SIGN as a single agent.
We hypothesized that an inhibitor against both CD4 and DC-SIGN binding sites on gp120 might represent a better anti-HIV strategy and that an sCD4–DC-SIGN fusion protein could have potent antiviral activity. As a fusion protein, the binding of sDC-SIGN to Env may not only enhance the engagement of sCD4 to gp120 but also block the DC-SIGN binding sites on gp120 to prevent HIV-1 transmission. In the current study, we designed, expressed, purified and characterized a series of soluble CD4-linker–DC-SIGN (CLD) fusion proteins. We assessed the protein oligomeric state and gp120 binding affinity of CLDs and tested their anti-HIV activity against several laboratory-adapted and primary isolates in cis infection of target cells. We further used a DC-SIGN-expressing cell line and primary dendritic cells to examine the anti-HIV potency in trans infection of target cells. The capability of inhibiting HIV-1 infection and dissemination was also evaluated in a human cervical explant model. Our findings demonstrate that several CLDs had significantly enhanced avidity to gp120 and much improved anti-HIV activity and could potently prevent both localized and disseminated infections of HIV-1. Our results support the concept that targeting both CD4 and DC-SIGN binding sites on gp120 represents a novel antiviral strategy and may have implications for the development of CD4/DC-SIGN-based therapeutic and prophylactic antiretrovirals.
MATERIALS AND METHODS
Plasmids, cell lines, viruses and proteins.
The env clones BaL, MWS2 and CH811 in pcDNA3.1 and viruses HIV-1BaL and HIV-1RF were described previously (29, 31, 32). The U87-CD4.CCR5 cell line and pNL4-3.Luc.R-E- were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIH. The 293T cell line was purchased from the American Type Culture Collection. Vector pET28a(+) was from Novagen. The Raji/DC-SIGN cell line and anti-DC-SIGN antibodies (MAbs) 507 and 526 were described previously (28, 34). Anti-CD4 MAb RPA-T4 was from BD Biosciences. Mannan was from Sigma-Aldrich. Protein CN54 gp140 was described previously (32).
Design and genetic engineering of expression constructs.
The CD4 and DC-SIGN DNA sequences were amplified from a human CD4 gene in pcDNA3.1 (31) and a human DC-SIGN gene in pcDNA3.1 (39), respectively. For a list of PCR primers used in this study see Table S1 in the supplemental material. Amplified DNA fragments were subsequently cloned into pET28a(+) after corresponding enzyme digestion. All recombinant DNA clones were confirmed by sequencing.
Protein expression and purification.
The E. coli strain Rosetta (Novagen) was used for protein expression as described previously (39). After induction with isopropyl-β-d-thiogalactoside for 4 h, the bacteria were collected and lysed by ultrasonic treatment. The insoluble fraction in the lysates was washed twice with phosphate-buffered saline (PBS) supplemented with 1 M guanidine hydrochloride (GuHCl) and resuspended in denaturation buffer (10 mM Tris, 500 mM NaCl, 6 M GuHCl, 5 mM dithiotreitol, pH 8.0). The denatured recombinant proteins were refolded for 24 h at 4°C in refolding buffer (10 mM Tris, 500 mM NaCl, 3 mM CaCl2, 10% glycerol, 3 mM glutathione [GSH], 0.3 mM glutathione disulfide [GSSG], pH 8.0) (14, 53). After refolding, the solutions were loaded onto a pre-equilibrated nickel-charged chelating Sepharose Fast Flow column (GE Healthcare). Proteins were purified according to the manufacturer's instructions.
SDS-PAGE and Western blotting.
Purified recombinant CLDs were resolved by 12% SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore) using the Microarray System (Bio-Rad). After being blocked in PBS plus 4% nonfat milk at 4°C overnight, the membrane was washed with TBS-T and subsequently incubated with anti-DC-SIGN MAb for 2 h at room temperature. After washes with TBS-T, the membrane was incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody for 2 h at room temperature. Luminometric detection of target proteins was achieved with SuperSignal West Dura Chemiluminescent Substrate (Pierce, Thermo Scientific) and visualized by a charge-coupled-device camera (Alpha Innotech).
Analytical ultracentrifugation.
Analytical ultracentrifugation was performed at 20°C using a Beckman Optima XL-I analytical ultracentrifuge equipped with an An-60Ti rotor. Purified recombinant proteins in dialysis buffer (10 mM Tris-HCl, pH 7.4, 200 mM NaCl, 5 mM CaCl2 and 0.1 mM glutathione) were loaded into 12-mm-path-length cells and centrifuged at 50,000 rpm for sCD4 and sDC-SIGN and 40,000 rpm for C35NDs60c. A total of 200 absorbance scans (280 nm) were recorded and data were analyzed with a C(s) distribution of the Lamm equation solutions calculated by the SEDFIT program (16, 47). Protein partial specific volume values, solution density and solution viscosity were calculated with SEDNTERP.
Binding kinetics analysis.
The buffer of tested protein was exchanged into phosphate-buffered saline (pH 7.4) with a desalt spin column (Thermo Scientific). CN54 gp140 was biotinylated by mixing with sulfo-NHS-LC biotinylation reagents in PBS for 30 min at room temperature according to the manufacturer's instructions (Pierce). The interactions between gp140 and CLDs were measured on a Forte-Bio Octet RED system (ForteBio) (1). This system monitors interference of light reflected from the surface of a sensor to measure the thickness of molecules bound to the sensor surface. The biotinylized gp140 (5 μg/ml) was immobilized on streptavidin biosensors. After reaching baseline, sensors were dipped into different concentrations of tested proteins for association and then moved into running buffer (3 mM CaCl2, 0.1% bovine serum albumin [BSA], and 0.05% Tween 20 in PBS, pH 7.4) for dissociation. A buffer-only reference was subtracted from all curves. Octet Molecular Interaction System software was used for data analysis.
Binding in the presence of antibodies against CD4 or/and DC-SIGN was also performed. C35NDs60c was immobilized on an anti-His biosensor. In the first association phase, sensors were incubated in running buffer (20 mM Tris, 150 mM NaCl, 3 mM CaCl2, 0.1% BSA, and 0.05% Tween 20, pH 8.0) supplemented with MAbs against CD4 (20 μg/ml RPA-T4) and/or DC-SIGN (507 + 526; 20 μg/ml each) as an association buffer for 1,800 s. Then the second association phase was performed for another 1,800 s in the presence or absence of 5 μg/ml CN54 gp140. Dissociation was subsequently performed in running buffer for 1,800 s.
Measurement of cytotoxicity.
Cytotoxicity of fusion proteins was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay as described previously (30). TZM-bl cells were seeded in 96-well plates overnight and subsequently cultured in medium containing serially diluted proteins for 3 days. Medium was then removed and 100 μl of MTT solution [medium containing 0.5 mg/ml 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] was added to each well and incubated for 4 h at 37°C. After removal of MTT solution, 100 μl of acid-isopropanol (0.04 N HCl in isopropanol) was added and the optical density was read with a Modulus Microplate Luminometer (Turner BioSystems).
Infection.
Stocks of pseudotyped reporter viruses were prepared by cotransfecting 293T cells with Env expression constructs and plasmid pNL4-3.Luc.R-E- as described previously (32). U87-CD4.CCR5 or TZM-bl cells (1 × 104 cells/well) were seeded in 96-well plates 1 day prior to infection. Env-pseudotyped viruses, HIV-1BaL or HIV-1RF, were incubated with serially diluted CLDs for 1 h at 37°C and then the mixtures containing the viruses and the proteins were added to the preseeded cells. The luciferase activity of the cell lysate was determined 48 h postinfection with a Modulus Microplate Luminometer (Turner BioSystems). The 50% and 90% inhibitory concentration values (IC50 and IC90) were calculated using Prism 4.1 (GraphPad).
Virus capture and transfer assay.
HIV-1 was preincubated with CLDs at 37°C for 1 h in round-bottom 96-well plates, while Raji/DC-SIGN cells (1.5 × 105 cells/well) were pretreated with 1 mg/ml mannan. Following incubation, Raji/DC-SIGN cells were added into CLD-pretreated HIV-1 while the same amount of HIV-1 was added to mannan-pretreated Raji/DC-SIGN cells. Following incubation at 37°C for another 2 h, unbound viruses were extensively washed with PBS. For the HIV-1 capture assay, washed Raji/DC-SIGN cells were lysed directly and p24 was measured as described previously (27, 28). For the virus transfer assay, washed Raji/DC-SIGN cells were then cocultured with U87-CD4.CCR5 cells. Forty-eight hours later, cells were lysed and luciferase activity was determined.
Human monocyte-derived DCs (MDDCs) were generated from a highly enriched population of CD14+ monocytes. Briefly, peripheral blood mononuclear cells (PBMCs) were isolated using a Ficoll-Hypaque density gradient followed by negative selection using the Monocyte Isolation Kit according to the manufacturer's protocol (Miltenyi Biotec). To obtain immature MDDCs (iMDDCs), monocytes were cultured in the presence of IL-4 (500 U/ml; R&D Systems) and GM-CSF (800 U/ml; R&D Systems) for 7 days (3, 28). iMDDCs were used for HIV-1 capture and transfer assay as described above.
Culture and infection of human cervical tissues.
Cervical tissues were obtained from women undergoing planned therapeutic hysterectomy in the absence of any cervical pathology at Hubei Hospital of Traditional Chinese Medicine or St George's Hospital London, with written consent obtained from all tissue donors according to the local research ethics committee. In brief, 3 mm × 3 mm explants were cultured in 200 μl of supplemented RPMI 1640 (28). Explants were preincubated in the presence or absence of C15D or antibody against CD4 or DC-SIGN for 1 h at 37°C. In dose-response experiments, explants were preincubated in the presence of serially diluted C35NDs60c, sCD4 or sDC-SIGN. After a 2-h exposure to HIV-1BaL at 37°C, cervical explants were extensively washed to remove unbound viruses and inhibitors and then cultured for 9 days at 37°C in fresh plates. For migratory cell experiments, after exposure to viruses, explants were cultured in the presence of 100 ng/ml recombinant human MIP-3β (R&D Systems) for 24 h at 37°C. Cells emigrating out of the explants were washed and cocultured with PM1 cells (0.5 × 105 cells/well in a 96-well plate) at 37°C. In all cases, supernatants were collected and stored at −80°C before subsequent measurement of p24 by enzyme-linked immunosorbent assay (ELISA) (28).
RESULTS
Design and construction of plasmids expressing recombinant CD4-linker–DC-SIGN fusion proteins.
The CD4 binding site lies on the gp120 neutralizing domain, while DC-SIGN recognizes mannose type glycans distributed around the outer domain (22, 45). Due to the distance between CD4 and DC-SIGN binding sites on gp120, an optimal linker is required to ensure that both components of the fusion protein can access their binding sites. Because the Gly4Ser repeat has been broadly used in the construction of fusion proteins, in the current study, we designed 3, 4, 5, and 7 Gly4Ser repeat linkers and linkers of 5 and 7 Gly4Ser repeats integrated with DC-SIGN neck domain (see Tables S1 and S2 in the supplemental material). Since CD4 D1D2 domains affect the binding activity to gp120, while gp120-binding residues are all located within the first N-terminal domain (D1) of CD4 (13), we designed CD4-linker–DC-SIGN fusion proteins containing CD4 D1D2 domains or the first 87 amino acids (aa) or 106 aa of the CD4 D1 domain. In addition, modifications were made in the C35D construct by truncating the CD4 moiety (mC35D and sC35D contained the first 108 aa and 87 aa of CD4 D1, respectively) or adding the DC-SIGN neck domain (C25ND and C35ND). A Ser-to-Cys mutation at amino acid 60, which can form a disulfide bond with gp120 (8), was introduced into C35ND, designated C35NDs60c (Fig. 1a).
Fig 1.
Schematic diagrams of CLD-expressing plasmids and biochemical characterization of purified fusion proteins. (a) Schematic representation of fusion proteins. Abbrevations: sCD4, the N-terminal 183 aa of CD4; mC, the N-terminal 106 aa of CD4; sC, the N-terminal 87 aa of CD4; N, DC-SIGN neck domain; sDC-SIGN, DC-SIGN carbohydrate-recognition domain (CRD); 15, 20, 25, 35, the number of amino acids; s60c, a Cys-to-Ser mutation at aa 60 on CD4. (b) Schematics of CLD-expressing plasmids. All CD4 and DC-SIGN moieties for CLDs were cloned into pET-28a(+) using restriction enzyme sites NdeI, BamHI and HindIII. Restriction enzyme sites and linker sequences were introduced into CD4 and DC-SIGN sequences by PCR. (c) SDS-PAGE and Western blot analysis of C35NDs60c. Purified C35NDs60c was resolved in 12% SDS-PAGE in reducing or nonreducing condition, followed by detection with Coomassie blue staining or Western blotting (reducing condition only). Lane 1, Western blotting; C35NDs60c was detected by MAb 507 against DC-SIGN, followed by an HRP-conjugated secondery antibody; lane 2 and lane 3, Coomassie blue staining of C35NDs60c in reducing and nonreducing conditions, respectively; lane 4, molecular marker in reducing condition. (d) Analytical ultracentrifugation in sedimentation velocity mode. CD4, peaks at 1.9 S and 3.8 S, corresponding to apparent molecular masses of 21.5 kDa and 57.4 kDa; DC-SIGN, peaks at 2.0 S, corresponding to an apparent molecular mass of 22.4 kDa; C35NDs60c, peaks at 6.3 S and 9.2 S, corresponding to apparent molecular masses of 214.8 kDa and 398.2 kDa. One of three independent experiments is shown.
We generated nine pET28a constructs encoding the following recombinant fusion proteins: C15D, C20D, C25D, C35D, mC35D, sC35D, C25ND, C35ND and C35NDs60c. Two pET28a plasmids expressing sCD4 and sDC-SIGN, respectively, were also constructed (see Table S1 and Table S2 in the supplemental material). The schematic diagrams of the constructs are illustrated in Fig. 1a and 1b.
Expression, refolding, purification and characterization of CLDs.
The fusion proteins produced in E. coli Rosetta existed mainly as inclusion bodies. Modulating factors that impact protein expression, such as host bacterial strain, IPTG concentration and inducing temperature, had little effect on the solubility of CLDs. In order to obtain soluble proteins, protein refolding was performed by slowly adding the denatured recombinant proteins into the refolding buffer. Final concentration was kept at 30 to 50 μg/ml to avoid protein aggregation and precipitation. All recombinant proteins in this study contained a His-tag at the N-terminus. The refolded proteins were purified with an Ni-resin column and target proteins were eluted with buffer containing 300 mM imidazole. Imidazole-free proteins were obtained by dialysis performed twice at 4°C.
An example of the purified fusion proteins, C35NDs60c, was analyzed by SDS-PAGE and Western blotting (Fig. 1c). Reducing SDS-PAGE (Fig. 1c, lane 2) and Western blot analysis revealed that C35NDs60c was correctly purified, with an expected band around 60 kDa. C35NDs60c migrated mainly as a single band in both nonreducing and reducing SDS-PAGE (Fig. 1c, lanes 2 and 3). CD4, a moiety of the fusion protein, has three oxidized isoforms, representing reduced protein (R) and the predominant disulfide-bonded CD4 isoforms (O1/O2) (8), while only O1 is in the functional state. C35NDs60c electrophoresed slightly faster in nonreducing SDS-PAGE than in reducing SDS-PAGE, indicating that it was in an O1 state containing correct disulfide bonds.
Oligomeric state of recombinant CLDs.
The function of recombinant fusion proteins can be impacted by their oligomeric state. Analytical ultracentrifugation (AUC) was conducted to evaluate the molecular mass of the proteins. The distributions C(s) of protein sedimentation coefficients are shown in Fig. 1d. The predicted monomer molecular masses of sDC-SIGN, sCD4 and C35NDs60c were 18.4 kDa, 22.1 kDa and 60.5 kDa, respectively. sDC-SIGN had a main peak at 2.0 S, corresponding to an apparent molecular mass of 22.4 kDa. sCD4 showed a major peak around 1.9 S, corresponding to an apparent molecular mass of 21.5 kDa. sCD4 also had a minor peak with a molecular mass of 57.4 kDa, but reducing SDS-PAGE confirmed that sCD4 had no band around 57.4 kDa (data not shown). The minor peak in the C(s) distribution of sCD4 might be dimers caused by incorrectly formed disulfide bonds. C35NDs60c demonstrated a major peak at 6.3 S, corresponding to an apparent molecular mass of 214.8 kDa and a minor peak at 9.2 S, corresponding to an apparent molecular mass of 398.2 kDa. In nonreducing SDS-PAGE, C35NDs60c showed mainly as monomers (60.5 kDa) (Fig. 1b, lane 3), although oligomers were detected, some of which likely formed in nonreducing conditions (8). The 214.8-kDa complex in the AUC indicated that C35NDs60c existed mainly as tetramers. The neck domain of DC-SIGN may have contributed to this tetramer formation (49). During the refolding procedure C35NDs60c may also have formed a higher oligomer caused by incorrectly formed disulfide bonds, which would explain the existence of the minor peak. Taken together, the AUC results showed that the D1D2 domain of CD4 and the carbohydrate-recognition domain of DC-SIGN existed mainly in monomer form while the majority of the C35NDs60c were tetramers.
CLDs demonstrate enhanced binding affinity to gp120.
We performed direct binding analysis on a Forte-Bio Octet RED system. A streptavidin biosensor immobilized with biotinylated CN54 gp140 was immersed in different concentrations of CLDs (all protein molar concentrations were calculated based on monomers). To characterize protein interactions, two protein-binding models: a Langmuir 1:1 model and a bivalent model, were applied (see Fig. S1 in the supplemental material). χ2 was the sum of squared deviations, where deviation was the difference between the actual data point and the fitted curve. Values close to zero indicated a good curve fit (5). As indicated by the calculated χ2 shown in Table 1, CLDs fitted preferably to the bivalent binding model, whereas the control proteins sCD4 and sDC-SIGN fitted with the Langmuir binding model. These results demonstrated that, unlike sCD4 and sDC-SIGN, CLDs interacted with gp140 in a bivalent manner, indicating that both CD4 and DC-SIGN moieties of CLDs could effectively reach their respective binding sites on gp140, either simultaneously or sequentially. Moreover, as shown in Table 1, the affinity of CLDs with a 35-aa linker to gp140 was obviously higher than that of sCD4 (KD = 1.21E−09 M) and sDC-SIGN (KD = 1.43E−08 M). The difference of affinity between CLDs with a 35-aa linker and C15D implied that the length of linkers impacted the interactions between CLDs and gp120. Linkers in appropriate length benefitted the binding of CLD moieties to gp120, while short linkers hindered such interactions. The linker with 35 aa seemed long enough to allow effective bindings of the fusion proteins to gp140. Other modifications on CLDs, such as adding the DC-SIGN neck domain and/or introducing the Cys-to-Ser mutation into CD4, also enhanced the affinity of CLDs to gp140. Compared with the off rate of CD4 (Kdis = 6.25E−05 S−1), CLDs with the 35-aa linker dissociated more slowly, indicating the formation of more stable complexes with gp140.
Table 1.
Kinetic parameters of gp140-CLD bindinga
| Parameterd | C35NDs60c | C35ND | C35D | C15D | sCD4b | sDC-SIGNc |
|---|---|---|---|---|---|---|
| KD1 (M) | 3.20E–10 | 1.66E–09 | 1.04E–09 | 2.62E–08 | 1.21E–09 | 1.43E–08 |
| KD2 (M) | 1.17E–09 | 2.11E–10 | 5.96E–10 | 1.95E–09 | ||
| Ka1 (1/ms) | 9.05E+04 | 1.41E+04 | 1.41E+04 | 1.09E+04 | 5.16E+04 | 1.57E+04 |
| Ka2 (1/ms) | 1.12E+04 | 8.28E+04 | 6.99E+04 | 1.10E+04 | ||
| Kdis1 (1/s) | 2.90E–05 | 2.34E–05 | 1.46E–05 | 2.86E–04 | 6.25E–05 | 2.24E–04 |
| Kdis2 (1/s) | 1.31E–05 | 1.75E–05 | 4.17E–05 | 2.14E–05 | ||
| χ2Langmuir | 12.52 | 5.39 | 5.51 | 4.08 | 0.49 | 0.05 |
| χ2bivalent | 0.38 | 0.10 | 0.59 | 0.91 | 0.90 | 0.52 |
Data are representative of three independent experiments.
N-terminal 183 aa of CD4.
Carbohydrate recognition domain of DC-SIGN.
KD, equilibrium (affinity) constant; Ka, association rate constant; Kdis, dissociation rate constant.
When the CD4 or DC-SIGN domain of C35NDs60c was blocked by antibodies, the binding of gp140 to the C35NDs60c-immobilized biosensor decreased, and a combination of anti-CD4 and anti-DC-SIGN antibodies had an additive effect in inhibiting C35NDs60c-gp140 interaction in the association phase (see Fig. S2 in the supplemental material). In the dissociation phase, the gp140-C35NDs60c complex appeared to dissociate more slowly than the antibody-C35NDs60c complex. The difference at the end of the dissociation phase might have been due to a competitive binding of gp140 to C35NDs60c by replacing anti-CD4 or/and anti-DC-SIGN antibodies (see Fig. S2 in the supplemental material). The binding results together indicate that gp140 can bind to both CD4 and DC-SIGN domains of C35NDs60c and that the avidity between C35NDs60c and gp140 is likely higher than that between C35NDs60c and the antibodies used in this study.
CLDs inhibit HIV-1 infection in cis.
Because the binding of gp120 to CD4 is virtually universal among HIV-1 isolates, regardless of viral tropism, we expected that CLDs would be active against both R5 and X4 strains. We initially tested the antiviral potency of C15D against HIV-1BaL (R5) and HIV-1RF (X4) in TZM-bl cells. The IC50s were 60.5 nM and 77.8 nM, respectively (Table 2), indicating that CLDs can neutralize HIV-1 regardless of viral tropism. Subsequent experiments were carried out by using BaL Env-pseudotyped HIV-1 to infect U87-CD4.CCR5 cells. All CLDs demonstrated anti-HIV activity (Table 2). Compared with sCD4 (IC50 = 25.3 nM), C35D showed enhanced neutralization activity (IC50 = 15.7 nM), while the CLDs with shorter linkers (C15D, C20D and C25D) had similar or lower antiviral activity. It seemed that linker length was a crucial factor affecting the anti-HIV activity of the CLDs, probably by interfering with CLD-gp120 interactions. The IC90 of the CLDs had a trend similar to that of IC50.
Table 2.
Anti-HIV activity of variant CLD formsa
| Parameter | sCD4b | C15D | C20D | C25D | C35D | C25ND | C35ND | C35NDs60c |
|---|---|---|---|---|---|---|---|---|
| HIV-1BaLc | ||||||||
| IC90 (nM) | 148.1 ± 10.4 | 335.7 ± 53.2 | NDf | ND | ND | ND | ND | ND |
| IC50 (nM) | 27.1 ± 3.6 | 60.5 ± 12.9 | ND | ND | ND | ND | ND | ND |
| HIV-1RFc | ||||||||
| IC90 (nM) | 199.8 ± 7.2 | 384.2 ± 44.6 | ND | ND | ND | ND | ND | ND |
| IC50 (nM) | 34.5 ± 2.2 | 77.8 ± 15.4 | ND | ND | ND | ND | ND | ND |
| BaLd | ||||||||
| IC90 (nM) | 153.9 ± 4.3 | 355.2 ± 30.4 | 308.2 ± 30.7 | 126.6 ± 25.3 | 79.6 ± 25.1 (1.9 ↓) | 30.7 ± 3.6 (5.0 ↓) | 26.7 ± 2.0 (5.8 ↓) | 28.1 ± 1.3 (5.5 ↓) |
| IC50 (nM) | 25.3 ± 1.8 | 66.9 ± 5.2 | 55.9 ± 7.3 | 26.4 ± 4.9 | 15.7 ± 4.7 (1.6 ↓) | 4.9 ± 1.0 (5.2 ↓) | 5.3 ± 0.5 (4.8 ↓) | 3.3 ± 0.3 (7.7 ↓) |
| MSW2d | ||||||||
| IC90 (nM) | >1,000 | ND | ND | ND | ND | 977.1 ± 77.6 | 990.7 ± 85.2 | 65.6 ± 13.5 (>15.2 ↓) |
| IC50 (nM) | >1,000 | ND | ND | ND | ND | 578.6 ± 53.1 (>1.7 ↓) | 667.3 ± 98.3 (>1.5 ↓) | 13.0 ± 3.7 (>76.9 ↓) |
| CH811d | ||||||||
| IC90 (nM) | >1,000 | ND | ND | ND | ND | 423.5 ± 47.6 (>2.4 ↓) | 354.7 ± 60.4 (>2.8 ↓) | 36.0 ± 3.5 (>27.8 ↓) |
| IC50 (nM) | 277.8 ± 69.1 | ND | ND | ND | ND | 56.7 ± 12.9 (4.9 ↓) | 42.8 ± 17.6 (6.5 ↓) | 4.7 ± 0.6 (59.1 ↓) |
| CC50 (nM)e | >1,600 | >1,600 | >1,600 | >1,600 | >1,600 | >1,600 | >1,600 | >1,600 |
Data are mean ± SD of at least three independent experiments (↓, fold decrease), with each condition performed in triplicate. Bold data imply that CLDs had significantly increased anti-HIV-1 activity. All protein molar concentrations were calculated based on monomer.
N-terminal 183 aa of CD4.
Replication-competent HIV-1.
Env-pseudotyped HIV-1.
CC50, 50% cytotoxicity concentration. Highest concentration tested.
ND, not determined.
CD4-truncated CLDs showed decreased neutralization activity. The IC50s of mC35D and sC35D were over 1,000 nM, at least 40 -fold higher than that of C35D. The DC-SIGN neck domain has seven complete and one incomplete 23-aa-long repeats, which form helical stretches to aggregate as tetramers. Adding the neck domain may also lengthen the linker. The IC50s of C25ND and C35ND were enhanced to 4.9 nM and 5.3 nM, respectively. The similar IC50s of the two fusion proteins imply that a 25-aa linker length is sufficient and the extra 10 aa had little effect on the neutralization activity. The IC50 of C35NDs60c (IC50 = 3.3 nM) was 1.6-fold lower than that of C35ND and 7.7-fold lower than that of sCD4.
As C25ND, C35ND and C35NDs60c exhibited better anti-HIV activity than CLDs with linkers lacking the DC-SIGN neck domain, neutralization potency against primary HIV-1 Envs was further investigated. Two primary HIV-1 Env clones were used, including MWS2, a clade C Env cloned from semen of a subject known to have infected women by vaginal intercourse, and CH811, a clade B Env isolated from a Chinese patient's blood sample. As shown in Table 2, C25ND and C35ND exhibited much weaker neutralization activity against MWS2 and CH811 than that of HIV-1BaL. Remarkably, C35NDs60c demonstrated much stronger bioactivity than C25ND and C35ND, possessing potent neutralization activity against the two primary HIV-1 Envs, with an IC50 of 13.0 nM and 4.7 nM, respectively. None of the CLDs was cytotoxic to the tested cells (Table 2).
Previous studies by others revealed that sCD4 or sDC-SIGN at suboptimal concentrations could enhance HIV-1 infection (25, 48, 54). We tested whether CLD had a similar effect. Our results indicated that C35NDs60c did not enhance HIV-1 infection at least in the tested concentration range (see Fig. S3a in the supplemental material).
CLDs inhibit HIV-1 capture and transfer via DC-SIGN-expressing cells and iMDDCs.
Raji/DC-SIGN cells were used to assess the anti-HIV capability of CLDs against virus capture and trans infection. As shown in Fig. 2a and 2b, sCD4 exhibited little effect in inhibiting HIV-1 capture by DC-SIGN but rendered the bound virus noninfectious to U87-CD4.CCR5 cells. Interestingly, all CLDs tested at the same concentration (1,000 nM) demonstrated similar trends to sDC-SIGN in interfering with virus capture by Raji/DC-SIGN cells, as well as similar capacity as sCD4 in inhibiting virus trans-infection from Raji/DC-SIGN cells to U87-CD4.CCR5 cells.
Fig 2.
CLDs inhibit HIV-1 capture and transfer by Raji/DC-SIGN cells and iMDDCs. BaL Env-pseudotyed HIV-1 was preincubated with or without inhibitor for 1 h at 37°C before the addition to Raji/DC-SIGN cells or iMDDCs. Cells pretreated with or without mannan were exposed to viruses as controls. After exposure to virues for 2 h at 37°C, cells were extensively washed and either lysed for capture assay or cocultured with U87-CD4.CCR5 cells for transfer assay. (a) HIV-1 captured by Raji/DC-SIGN cells. Medium alone was defined as 100% and its p24 concentration was 1.79 ng/ml. (b) RLU of HIV-1 trans infection from Raji/DC-SIGN cells to U87-CD4.CCR5 cells. Medium alone was defined as 100%. (c) HIV-1 captured by iMDDCs. Medium alone was defined as 100% and its p24 concentration was 2.87 ng/ml. (d) RLU of HIV-1 trans infection from iMDDCs to U87-CD4.CCR5 cells. Medium alone was defined as 100%. Data shown are mean ± SD of three independent experiments, with each condition performed in triplicate.
To confirm the anti-HIV activity of CLDs in trans-infection, similar experiments were conducted in iMDDCs. Similar to controls sDC-SIGN and mannan, all CLDs demonstrated less potency against virus capture than that observed in Raji/DC-SIGN cells (Fig. 2c). Differences between the DC-SIGN-expressing cell line and iMDDCs were likely due to differences in the receptor repertoire of iMDDCs, which also express other attachment receptors in addition to DC-SIGN. Nevertheless, CLDs still possessed better efficacy than sCD4 and sDC-SIGN to suppress virus uptake by iMDDCs (Fig. 2c). Among those, C35NDs60c inhibited more than 50% of HIV-1 capture. Despite the incomplete inhibition of HIV-1 capture, all CLDs showed enhanced potency against trans infection, almost completely blocking virus transfer from iMDDCs to U87-CD4.CCR5 cells (Fig. 2d). These results imply that HIV-1 may bind to iMDDCs via additional attachment receptors other than DC-SIGN, but CLDs can render the bound virus noninfectious.
Considering that saturated concentrations of proteins were used in the above study, we performed additional dose-response assays. In the neutralization assay, C35NDs60c was much more potent than a combination of sCD4 and sDC-SIGN (see Fig. S3a in the supplemental material). In viral capture and transfer assays, C35NDs60c also demonstrated better antiviral activities than sCD4 and sDC-SIGN in combination (see Fig. S3b and S3c).
CLDs inhibit both localized mucosal infection and dissemination pathways.
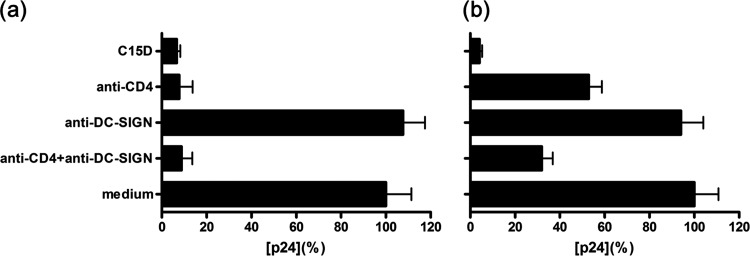
In the absence of a suitable animal model for HIV-1, ex vivo culture of human tissue explants has been generally accepted as an alternative to mimic in vivo physiological conditions. We conducted experiments to determine whether CLDs could inhibit HIV-1BaL infection and dissemination in cervical tissues. As a conceptual study, we initially used C15D to conduct our experiment. Concentrations of all proteins were used at 30 μg/ml (∼750 nM for C15D). As seen in Fig. 3a and 3b, anti-CD4 antibody blocked localized infection but was incapable of inhibiting virus dissemination in mucosal tissues. Although antibodies against DC-SIGN might decrease the capture of the virus by migrating cells within mucosa, receptors other than DC-SIGN could play a role in transmission. A combination of anti-CD4 and anti-DC-SIGN antibodies was required to block HIV-1BaL infection and dissemination in cervical tissues. Of note, C15D not only efficiently inhibited localized infection but also prevented disseminated infection by migratory cells, with better efficacy than the combination of anti-CD4 and anti-DC-SIGN antibodies. In addition, we performed dose-response experiments using sCD4, sDC-SIGN and an improved CLD (C35NDs60c) against HIV-1BaL infection in human cervical explants. While sDC-SIGN had little inhibitory effect against HIV-1 infection in the tested concentration range, C35NDs60c demonstrated significantly enhanced antiviral activity, with an IC50 one log lower than that of sCD4 (see Fig. S4 in the supplemental material). Our data together suggest that further improvement of CLDs may render them potentially useful in prophylaxis or therapeutics, in particular for microbicide development.
Fig 3.
CLD inhibits localized mucosal HIV-1 infection and dissemination. Human cervical explants were preincubated with or without inhibitors for 1 h at 37°C before exposure to HIV-1BaL for 2 h at 37°C. After incubation, explants were extensively washed and cultured in the presence of 100 ng/ml of MIP-3β for 48 h. Emigrating cells were collected, washed, and cocultured with PM1 cells. The explants were cultured in separate wells. Data are representative of three independent experiments, with each condition performed in triplicate. Data shown are p24 antigen (mean± SD) released from both (a) cultured explants and (b) PM1 cocultured migratory cells at day 9. p24 in the absence of inhibitor was defined as 100%, and p24 concentrations were 1.56 and 2.74 ng/ml for the cervical explants and PM1 cocultured migratory cells, respectively. Anti-CD4, RPA-T4. Anti-DC-SIGN, MAb 507 and 526.
DISCUSSION
Effective microbicides need to protect against all potential routes of HIV-1 transmission across mucosal surfaces. Based on our previous findings that HIV-1 uptake and dissemination by migratory dendritic cells (DCs) can occur through CD4 and mannose binding C-type lectin receptor DC-SIGN (28), in the current study, we designed, expressed, purified and characterized a series of bifunctional CD4-linker–DC-SIGN fusion proteins (CLDs). We demonstrate that several of the CLDs had enhanced gp120-binding affinity and much improved antiviral activity against HIV-1 infection and dissemination.
Soluble CD4 (sCD4) has poor antiviral activity against primary HIV-1 isolates (33). In order to improve the anti-HIV potency, several strategies have been employed to date to make fusion proteins, for instance, protein polymerization. Immunoglobulin G (IgG) is usually used as a frame protein for making fused oligomers. PRO-542, a tetramer CD4 wherein the Fv portions of both the heavy and light chains of human IgG2 have been replaced with the D1D2 domains of human CD4, binds to gp120 with high affinity (2). However, the increased size gained by fusing to the antibody may diminish the neutralization potency of the candidate molecule (10). The large size likely also makes IgG-anchored fusion protein less able to penetrate into tissues (35). When we designed and characterized a range of CLDs, we observed that the integration of the DC-SIGN neck domain into fusion proteins greatly increased tetramer formation and significantly enhanced the antiviral potency against both laboratory-adapted and primary isolates. This novel polymer strategy could be extended to design other fusion proteins aimed to enhance bioactivity.
Design of sCD4-based fusion proteins simultaneously targeting different binding sites on gp120 provides another alternative to improve anti-HIV potency. Most fusion proteins designed to date, including sCD4-17b, m35-sCD4 and CD4HC-(GS7)-IgGE51, have been focused on the CD4 binding site and CD4-binding-induced epitopes on gp120 (9, 36, 56). Unlike the molecules described above, CLDs, which bind to the CD4-binding site and glycans on gp120, were designed to interrupt the interactions between gp120 and the entry receptor CD4 as well as the attachment receptor DC-SIGN. Such a unique binding mode not only enhanced the engagement of sCD4 to gp120, but also inhibited the DC-SIGN binding sites on gp120. Although CD4 and DC-SIGN have little direct interaction, DC-SIGN binding to gp120 causes an allosterically induced exposure of the CD4 binding site and therefore facilitates a more stable binding of CD4 to gp120. Indeed, CLDs not only enhanced the neutralization activity, but also inhibited cis and trans infection of HIV-1 in both cellular and human cervical explant models. Given that there is limited research on sDC-SIGN-based antiretrovirals and that sDC-SIGN alone is unlikely to be used in practice, our study provides evidence that DC-SIGN fused to other proteins targeting gp120 may provide an important new strategy for enhancing anti-HIV potency. As DC-SIGN functions as an attachment receptor for a range of enveloped viruses such as dengue, hepatitis C, Ebola, and so on (17, 26), the method of fusing DC-SIGN with other functional moieties may be extended to prevent or treat these additional types of viruses.
Several CLDs, including C35D, C25ND, C35ND and C35NDs60c, demonstrated much improved neutralizing activity against HIV-1. In contrast, C15D, C20D and C25D had similar or decreased antiviral activity compared with that of sCD4. This implies that the fusion of proteins expressing two components binding to different sites on gp120 per se does not guarantee enhanced bioactivity. The amino acids involved in CD4 recognition distribute discontinuously around C2, C3 and V5 regions of gp120 (4, 24, 45), whereas DC-SIGN interacts with discontinuously distributed high-mannose oligosaccharides on gp120 (20). Short linkers likely hindered the two moieties of the fusion proteins from simultaneously interacting with gp120, while a linker with suitable length could render CLDs with higher gp120-binding avidity and better anti-HIV potency. To this end we did not observe significant differences in terms of anti-HIV activity when C25ND and C35ND were tested against both laboratory-adapted and primary isolates, including one cloned from semen of a subject known to have infected women by vaginal intercourse. This result suggests that a linker of 25 amino acids integrated with an extra DC-SIGN neck domain may be sufficient for simultaneous binding of the two moieties to gp120, at least for the isolates tested.
The signaling pathway activated by the binding of HIV-1 to DC-SIGN is thought to cause immunosuppressive responses and trigger HIV-1 transmission and replication (18). In addition to masking the CD4-binding site on gp120, CLDs interrupted gp120 glycan–DC-SIGN interaction and inhibited the uptake of the virus by host cells, avoiding potential triggering of the downstream signaling pathways. Several carbohydrate-binding agents, including cyanovirin-N and griffithsin, also target gp120 glycans and have potent antiviral activity (6). However, those proteins originated from bacteria or plants. Whether they can be used prophylactically or therapeutically requires further evaluation. Given that the soluble forms of CD4 and DC-SIGN occur normally in vivo and that the Gly4Ser repeat linker is poorly immunogenic (43, 44), further improvement of CLDs may render them potentially useful as prophylactic or therapeutic agents.
Beyond a novel antiretroviral proof-of-concept, CLDs designed in the current study may have additional potential. CD4 engagement to gp120 induces the exposure of immunogenic epitopes, including V3 and chemokine receptor binding sites. Immunization with the CD4-gp120 complex has been shown to enhance viremia control in nonhuman primates (11, 15). In those studies, immunogens were cross-linked CD4-gp120 complexes or recombinant fusion proteins of CD4 and gp120, in which the native epitopes on gp120 might have been affected. In contrast, CLDs, especially C35NDs60c, have a lower off rate than sCD4, facilitating the formation of a more stable CD4-gp120 complex, probably induced by DC-SIGN engagement. Given that DC-SIGN binding to HIV-1 gp120 increases exposure of the CD4 binding site (25), it will be interesting to determine in future studies whether CLD-bound gp120/gp140 complexes can be used as immunogenic components to elicit better neutralizing antibodies.
Despite the described favorable characteristics of CLDs, further improvements in antiviral activity are needed. As both CD4 and DC-SIGN components used to make CLDs do not contain glycans, we used a bacterial expression system to produce proteins. Due to their poor solubility, the target proteins were expressed mainly in the form of inclusion bodies. Even though purified target proteins showed high anti-HIV activity after refolding, a certain degree of incorrect formation of disulfide bonds or/and aggregation occurred, and this could affect their bioactivity. While careful refolding technology could decrease such wrong disulfide bonding and aggregation, this process would be difficult to scale up. Other expression systems, such as transgenic plants (50), may provide an alternative to produce proteins on a larger scale.
In conclusion, this is the first time that sDC-SIGN-based bifunctional proteins have demonstrated anti-HIV potency. The designed and expressed CLDs are novel bifunctional proteins with increased gp120 binding avidity. CLDs inhibit HIV-1 infection and dissemination in cell lines, primary dendritic cells and mucosal cervical tissues.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Ministry of Science and Technology of China (2010CB530100, 2012ZX10001006-02) (Q.H.), the National Natural Science Foundation of China (30872357) (Q.H.) (30770100) (T.D.), the Chinese Academy of Sciences (KSCX2-YW-R-144) (QH), and the Hotung Trust (Q.H., G.G., R.S.).
We thank Wei Jin for assistance in the preparation of dendritic cells and Wenjie Huang for assistance in protein lyophilization.
Footnotes
Published ahead of print 11 June 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Abdiche Y, Malashock D, Pinkerton A, Pons J. 2008. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 377:209–217 [DOI] [PubMed] [Google Scholar]
- 2. Allaway GP, et al. 1995. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res. Hum. Retroviruses 11:533–539 [DOI] [PubMed] [Google Scholar]
- 3. Arrighi JF, et al. 2004. DC-SIGN-mediated infectious synapse formation enhances X4 HIV-1 transmission from dendritic cells to T cells. J. Exp. Med. 200:1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ashkenazi A, et al. 1990. Mapping the CD4 binding site for human immunodeficiency virus by alanine-scanning mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 87:7150–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baleux F, et al. 2009. A synthetic CD4-heparan sulfate glycoconjugate inhibits CCR5 and CXCR4 HIV-1 attachment and entry. Nat. Chem. Biol. 5:743–748 [DOI] [PubMed] [Google Scholar]
- 6. Balzarini J. 2007. Targeting the glycans of glycoproteins: a novel paradigm for antiviral therapy. Nat. Rev. Microbiol. 5:583–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berzi A, et al. 2012. A glycomimetic compound inhibits DC-SIGN-mediated HIV infection in cellular and cervical explant models. AIDS 26:127–137 [DOI] [PubMed] [Google Scholar]
- 8. Cerutti N, et al. 2010. Stabilization of HIV-1 gp120-CD4 receptor complex through targeted interchain disulfide exchange. J. Biol. Chem. 285:25743–25752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen W, Xiao X, Wang Y, Zhu Z, Dimitrov DS. 2010. Bifunctional fusion proteins of the human engineered antibody domain m36 with human soluble CD4 are potent inhibitors of diverse HIV-1 isolates. Antiviral Res. 88:107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen W, Zhu Z, Feng Y, Dimitrov DS. 2008. Human domain antibodies to conserved sterically restricted regions on gp120 as exceptionally potent cross-reactive HIV-1 neutralizers. Proc. Natl. Acad. Sci. U. S. A. 105:17121–17126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DeVico A, et al. 2007. Antibodies to CD4-induced sites in HIV gp120 correlate with the control of SHIV challenge in macaques vaccinated with subunit immunogens. Proc. Natl. Acad. Sci. U. S. A. 104:17477–17482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Witte L, et al. 2007. Syndecan-3 is a dendritic cell-specific attachment receptor for HIV-1. Proc. Natl. Acad. Sci. U. S. A. 104:19464–19469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Esser U, et al. 2000. Molecular function of the CD4 D1 domain in coreceptor-mediated entry by HIV type 1. AIDS Res. Hum. Retroviruses 16:1845–1854 [DOI] [PubMed] [Google Scholar]
- 14. Ferrer M, et al. 1997. Construction and characterization of a radio-iodinatable mutant of recombinant human CD4. J. Immunol. Methods 210:215–225 [DOI] [PubMed] [Google Scholar]
- 15. Fouts T, et al. 2002. Crosslinked HIV-1 envelope-CD4 receptor complexes elicit broadly cross-reactive neutralizing antibodies in rhesus macaques. Proc. Natl. Acad. Sci. U. S. A. 99:11842–11847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garbett NC, Mekmaysy CS, Chaires JB. 2010. Sedimentation velocity ultracentrifugation analysis for hydrodynamic characterization of G-quadruplex structures. Methods Mol. Biol. 608:97–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geijtenbeek TB, den Dunnen J, Gringhuis SI. 2009. Pathogen recognition by DC-SIGN shapes adaptive immunity. Future Microbiol. 4:879–890 [DOI] [PubMed] [Google Scholar]
- 18. Geijtenbeek TB, Gringhuis SI. 2009. Signalling through C-type lectin receptors: shaping immune responses. Nat. Rev. Immunol. 9:465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geijtenbeek TB, et al. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587–597 [DOI] [PubMed] [Google Scholar]
- 20. Geijtenbeek TB, et al. 2002. Identification of different binding sites in the dendritic cell-specific receptor DC-SIGN for intercellular adhesion molecule 3 and HIV-1. J. Biol. Chem. 277:11314–11320 [DOI] [PubMed] [Google Scholar]
- 21. Geijtenbeek TB, van Kooyk Y. 2003. DC-SIGN: a novel HIV receptor on DCs that mediates HIV-1 transmission. Curr. Top. Microbiol. Immunol. 276:31–54 [DOI] [PubMed] [Google Scholar]
- 22. Guo Y, et al. 2004. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 11:591–598 [DOI] [PubMed] [Google Scholar]
- 23. Gurney KB, et al. 2005. Binding and transfer of human immunodeficiency virus by DC-SIGN+ cells in human rectal mucosa. J. Virol. 79:5762–5773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hansen JE, Lund O, Nielsen JO, Brunak S. 1996. Prediction of the secondary structure of HIV-1 gp120. Proteins 25:1–11 [DOI] [PubMed] [Google Scholar]
- 25. Hijazi K, et al. 2011. DC-SIGN increases the affinity of HIV-1 envelope glycoprotein interaction with CD4. PLoS One 6:e28307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hofmann H, Pohlmann S. 2011. DC-SIGN: access portal for sweet viral killers. Cell Host Microbe 10:5–7 [DOI] [PubMed] [Google Scholar]
- 27. Hong PW, Nguyen S, Young S, Su SV, Lee B. 2007. Identification of the optimal DC-SIGN binding site on human immunodeficiency virus type 1 gp120. J. Virol. 81:8325–8336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu Q, et al. 2004. Blockade of attachment and fusion receptors inhibits HIV-1 infection of human cervical tissue. J. Exp. Med. 199:1065–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu Q, et al. 2005. Restricted variable residues in the C-terminal segment of HIV-1 V3 loop regulate the molecular anatomy of CCR5 utilization. J. Mol. Biol. 350:699–712 [DOI] [PubMed] [Google Scholar]
- 30. Hu Q, Younson J, Griffin GE, Kelly C, Shattock RJ. 2006. Pertussis toxin and its binding unit inhibit HIV-1 infection of human cervical tissue and macrophages involving a CD14 pathway. J. Infect. Dis. 194:1547–1556 [DOI] [PubMed] [Google Scholar]
- 31. Hu QX, et al. 2000. Evolution of the human immunodeficiency virus type 1 envelope during infection reveals molecular corollaries of specificity for coreceptor utilization and AIDS pathogenesis. J. Virol. 74:11858–11872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang X, et al. 2012. Highly conserved HIV-1 gp120 glycans proximal to CD4-binding region affect viral infectivity and neutralizing antibody induction. Virology 423:97–106 [DOI] [PubMed] [Google Scholar]
- 33. Ivey-Hoyle M, et al. 1991. Envelope glycoproteins from biologically diverse isolates of immunodeficiency viruses have widely different affinities for CD4. Proc. Natl. Acad. Sci. U. S. A. 88:512–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jameson B, et al. 2002. Expression of DC-SIGN by dendritic cells of intestinal and genital mucosae in humans and rhesus macaques. J. Virol. 76:1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Labrijn AF, et al. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lagenaur LA, Villarroel VA, Bundoc V, Dey B, Berger EA. 2010. sCD4-17b bifunctional protein: extremely broad and potent neutralization of HIV-1 Env pseudotyped viruses from genetically diverse primary isolates. Retrovirology 7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lambert AA, Gilbert C, Richard M, Beaulieu AD, Tremblay MJ. 2008. The C-type lectin surface receptor DCIR acts as a new attachment factor for HIV-1 in dendritic cells and contributes to trans- and cis-infection pathways. Blood 112:1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee B, et al. 2001. cis expression of DC-SIGN allows for more efficient entry of human and simian immunodeficiency viruses via CD4 and a coreceptor. J. Virol. 75:12028–12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ma X, Hu Q, Cui T, Yang J, Zhang Z. 2004. Expression, purification and biological analysis of soluble DC-SIGN. Chin J. Microbiol. Immunol. 24:67–71 [Google Scholar]
- 40. Marzi A, et al. 2007. Modulation of HIV and SIV neutralization sensitivity by DC-SIGN and mannose-binding lectin. Virology 368:322–330 [DOI] [PubMed] [Google Scholar]
- 41. McDonald D, et al. 2003. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 300:1295–1297 [DOI] [PubMed] [Google Scholar]
- 42. Naarding MA, et al. 2005. Lewis X component in human milk binds DC-SIGN and inhibits HIV-1 transfer to CD4+ T lymphocytes. J. Clin. Invest. 115:3256–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peakman M, Senaldi G, Foote N, McManus TJ, Vergani D. 1992. Naturally occurring soluble CD4 in patients with human immunodeficiency virus infection. J. Infect. Dis. 165:799–804 [DOI] [PubMed] [Google Scholar]
- 44. Plazolles N, et al. 2011. Pivotal advance: the promotion of soluble DC-SIGN release by inflammatory signals and its enhancement of cytomegalovirus-mediated cis-infection of myeloid dendritic cells. J. Leukoc. Biol. 89:329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Poignard P, Saphire EO, Parren PW, Burton DR. 2001. gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253–274 [DOI] [PubMed] [Google Scholar]
- 46. Requena M, et al. 2008. Inhibition of HIV-1 transmission in trans from dendritic cells to CD4+ T lymphocytes by natural antibodies to the CRD domain of DC-SIGN purified from breast milk and intravenous immunoglobulins. Immunology 123:508–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78:1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schutten M, Andeweg AC, Bosch ML, Osterhaus AD. 1995. Enhancement of infectivity of a non-syncytium inducing HIV-1 by sCD4 and by human antibodies that neutralize syncytium inducing HIV-1. Scand. J. Immunol. 41:18–22 [DOI] [PubMed] [Google Scholar]
- 49. Serrano-Gomez D, et al. 2008. Structural requirements for multimerization of the pathogen receptor dendritic cell-specific ICAM3-grabbing non-integrin (CD209) on the cell surface. J. Biol. Chem. 283:3889–3903 [DOI] [PubMed] [Google Scholar]
- 50. Sexton A, et al. 2006. Transgenic plant production of Cyanovirin-N, an HIV microbicide. FASEB J. 20:356–358 [DOI] [PubMed] [Google Scholar]
- 51. Shearer WT, et al. 2000. Recombinant CD4-IgG2 in human immunodeficiency virus type 1-infected children: phase 1/2 study. The Pediatric AIDS Clinical Trials Group Protocol 351 Study Team. J. Infect. Dis. 182:1774–1779 [DOI] [PubMed] [Google Scholar]
- 52. Stamatatos L, Werner A, Cheng-Mayer C. 1994. Differential regulation of cellular tropism and sensitivity to soluble CD4 neutralization by the envelope gp120 of human immunodeficiency virus type 1. J. Virol. 68:4973–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Su SV, et al. 2004. DC-SIGN binds to HIV-1 glycoprotein 120 in a distinct but overlapping fashion compared with ICAM-2 and ICAM-3. J. Biol. Chem. 279:19122–19132 [DOI] [PubMed] [Google Scholar]
- 54. Sullivan N, et al. 1998. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J. Virol. 72:6332–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Turville SG, et al. 2002. Diversity of receptors binding HIV on dendritic cell subsets. Nat. Immunol. 3:975–983 [DOI] [PubMed] [Google Scholar]
- 56. West AP, Jr, et al. 2010. Evaluation of CD4-CD4i antibody architectures yields potent, broadly cross-reactive anti-human immunodeficiency virus reagents. J. Virol. 84:261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wu L. 2008. Biology of HIV mucosal transmission. Curr. Opin. HIV AIDS 3:534–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.