Abstract
We previously showed that phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] and septin regulation play major roles in maintaining Candida albicans cell wall integrity in response to caspofungin and other stressors. Here, we establish a link between PI(4,5)P2 signaling and septin localization and demonstrate that rapid redistribution of PI(4,5)P2 and septins is part of the natural response of C. albicans to caspofungin. First, we studied caspofungin-hypersusceptible C. albicans irs4 and inp51 mutants, which have elevated PI(4,5)P2 levels due to loss of PI(4,5)P2-specific 5′-phosphatase activity. PI(4,5)P2 accumulated in discrete patches, rather than uniformly, along surfaces of mutants in yeast and filamentous morphologies, as visualized with a green fluorescent protein (GFP)-pleckstrin homology domain. The patches also contained chitin (calcofluor white staining) and cell wall protein Rbt5 (Rbt5-GFP). By transmission electron microscopy, patches corresponded to plasma membrane invaginations that incorporated cell wall material. Fluorescently tagged septins Cdc10 and Sep7 colocalized to these sites, consistent with well-described PI(4,5)P2-septin physical interactions. Based on expression patterns of cell wall damage response genes, irs4 and inp51 mutants were firmly positioned within a group of caspofungin-hypersusceptible, septin-regulatory protein kinase mutants. irs4 and inp51 were linked most closely to the gin4 mutant by expression profiling, PI(4,5)P2-septin-chitin redistribution and other phenotypes. Finally, sublethal 5-min exposure of wild-type C. albicans to caspofungin resulted in redistribution of PI(4,5)P2 and septins in a manner similar to those of irs4, inp51, and gin4 mutants. Taken together, our data suggest that the C. albicans Irs4-Inp51 5′-phosphatase complex and Gin4 function upstream of PI(4,5)P2 and septins in a pathway that helps govern responses to caspofungin.
INTRODUCTION
Candida albicans is the most common fungal pathogen of humans, causing a wide range of superficial, mucosal, and systemic diseases. Candidemia and other types of systemic candidiasis are associated with mortality rates approaching or exceeding 40% (35). The echinocandin antifungals (caspofungin, anidulafungin, and micafungin) have emerged as front-line therapy against systemic candidiasis (40). These agents inhibit β-1,3-d-glucan synthase, an enzyme that synthesizes a major component of the Candida cell wall, and thereby kill cells by disrupting cell wall integrity. Independent of its essential role in maintaining cellular viability, the cell wall is central to the pathogenesis of candidiasis. It forms the interface of the pathogen-host interaction, elaborates virulence factors, and makes complex contributions to cellular morphogenesis and resistance to host defenses (2, 3, 11, 16, 21, 22, 33, 41, 44, 50, 57, 58). Not surprisingly, C. albicans cell wall regulation is among the most dynamic areas of translational and basic research in medical mycology.
The C. albicans cell wall consists of three layers. Echinocandins target the middle layer, which constitutes a backbone comprised largely of cross-linked β-1,3- and β-1,6-glucans. A mannan- and mannoprotein-rich outer layer is the favored target of the innate immune system. Cell wall rigidity is provided by an inner layer of the polymer chitin, which lies in proximity to the plasma membrane. Much of our understanding of C. albicans cell wall synthesis and regulation comes from work with the model yeast Saccharomyces cerevisiae (8, 24–27). There are many examples in which orthologous S. cerevisiae and C. albicans genes involved in cell wall regulation have the same biological function (4). However, de novo gene discovery in C. albicans has increasingly revealed nonconservation of function (7, 19, 29, 32, 39, 51, 61), which has prompted analysis of cell wall-related genes and genetic relationships in C. albicans itself (38, 42). In addition to their clinical implications, studies of C. albicans responses to echinocandin exposure are important for understanding cell wall regulation in general and defining mechanisms of susceptibility and resistance to cell wall stress.
In this study, we focus on the relationship between phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] and septins in the regulation of C. albicans cell wall integrity in response to caspofungin. We began our studies of PI(4,5)P2 regulation after demonstrating that C. albicans Irs4 was among a group of proteins that was recognized by sera from patients with candidiasis (2, 3, 10, 48). We selected Irs4 for further study for two reasons: (i) it has an EH domain, a protein binding motif implicated in cell wall regulation (2, 3, 34), and (ii) it is the sole homologue of S. cerevisiae ScIrs4 and ScTax4, which jointly interact with the 5′-phosphatase ScInp51 to regulate levels of PI(4,5)P2 (2, 3, 34). We established that C. albicans Irs4 and Inp51 physically interact and that disruption of IRS4 or INP51 causes elevated intracellular PI(4,5)P2 levels but does not affect levels of phosphatidylinositol 4-phosphate [PI(4)P], PI(3)P, or PI(3,5)P2 (3). In addition, Irs4 and Inp51 are required for the progression but not the initiation of disseminated candidiasis in a murine model (2, 3).
A connection between PI(4,5)P2 and cell wall organization was first made in S. cerevisiae, in which irs4/tax4 and inp51 mutations have synthetic interactions with known cell wall integrity regulators (34). However, the role of PI(4,5)P2 in cell wall integrity is well buffered in S. cerevisiae (34). Indeed, extremely sensitive competitive growth assays failed to reveal hypersensitivity of an inp51 mutant to caspofungin (18). In contrast, PI(4,5)P2 has a highly prominent role in C. albicans cell wall organization: irs4 and inp51 null mutants are hypersusceptible to caspofungin, do not exhibit paradoxical growth at high caspofungin concentrations, overactivate the protein kinase C–mitogen-activated protein kinase (PKC-MAPK) cell integrity pathway, and exhibit aberrant sites of chitin accumulation (2, 18, 48). Thus, the study of PI(4,5)P2 signaling in C. albicans provides a unique opportunity to advance our understanding of both cell wall integrity and echinocandin susceptibility.
Septins are cytoskeletal proteins that serve as scaffolds for cytokinesis and other dynamic events at the plasma membrane (20, 30, 53). They were first connected to cell wall biogenesis in S. cerevisiae by the finding that they are required for localization of chitin synthase Chs3 (13, 45). Septin-defective mutants display aberrant sites of chitin deposition (13). We were led to C. albicans septins through a screen of protein kinase mutants for cell wall integrity defects (6). Among 24 protein kinase mutants that were hypersensitive to caspofungin, one group had cohesive properties: they displayed increased expression of numerous cell wall integrity genes, and in many cases their S. cerevisiae orthologs were implicated in the function of the septin ring. We extended these observations with the finding that C. albicans septin mutants are hypersensitive to caspofungin and by showing that a newly characterized protein kinase gene of this group, KIN3, is required for septin ring integrity. We observed that the septin ring becomes disorganized and delocalized upon caspofungin treatment. These results suggested that septins may be involved in the natural response to caspofungin and other cell wall inhibitors (6).
S. cerevisiae septins interact specifically with PI(4,5)P2, and the presence of PI(4,5)P2 promotes septin polymerization and organization (5). As such, we hypothesized that the role of PI(4,5)P2 in C. albicans cell wall integrity may be mediated by septins or their regulators. Here we present phenotypic comparisons that reveal close similarities between irs4, inp51 and septin regulatory protein kinase mutants. In addition, we observe that PI(4,5)P2 and septin localization is altered in wild-type C. albicans upon caspofungin treatment. Our findings argue that redistribution of PI(4,5)P2 and septins is part of the natural cell wall integrity response in C. albicans.
MATERIALS AND METHODS
Strains and growth conditions.
C. albicans strains used or constructed in this study are listed in Table 1. C. albicans SC5314 was the wild-type control for irs4, inp51, and gin4 null mutants created using the SAT-flipper method (see below). For our previously created septin regulatory protein kinase mutants, the wild-type control strain was DAY185 (6, 59). Strains were routinely grown in yeast extract-peptone-dextrose (YPD) liquid medium (1% yeast extract, 1% Bacto peptone, 2% [α-35C]glucose) or on YPD or Sabouraud dextrose agar (SDA) at 30°C. For embedded growth, ∼100 cells of overnight-grown C. albicans were mixed into 20 ml of YPD-reverse agar and incubated for 3 days at 30°C or 37°C. Reverse agar (BASF pluronic polyol F-127; kindly provided by Fungal Genetics Stock Center [http://www.fgsc.net]) is a lock polymer of polyoxypropylene and polyoxyethylene that can be used as a replacement for conventional agar in solid medium. It is a liquid in solution at 4°C and solidifies to a viscous solution at room temperature. For invasive growth, cells were grown in liquid medium in 35-mm-diameter glass-bottom dishes (Matek, Ashland, MA) coated with 10 μg/cm2 of Cell-Tak (BD Biosciences, Bedford, MA) to adsorb a thin layer. C. albicans cells immobilized in the layer were covered with ∼100 μl of YPD or YPD plus 5% fetal bovine serum and incubated at 30°C or 37°C.
Table 1.
C. albicans strains
| Strain | Parent | Description | Research purpose | Reference |
|---|---|---|---|---|
| SC5314 | C. albicans reference strain | Wild type | 15 | |
| irs4_2KO | SC5314 | irs4Δ/irs4Δ | irs4 null mutant | This study |
| irs4_2KO_Reins | irs4_2KO | Complemented, irs4Δ/IRS4 | IRS4 reinsertion | This study |
| inp51_2KO | SC5314 | inp51Δ/inp51Δ | inp51 null mutant | 3 |
| inp51_2KO_Reins | Inp51_2KO | Complemented, inp51Δ/INP51 | INP51 reinsertion | 3 |
| gin4_2KO* | SC5314 | gin4Δ/gin4Δ | gin4 null mutant | This study |
| gin4_2KO_Reins* | gin4_2KO | Complemented, irs4Δ/IRS4 | GIN4 reinsertion | This study |
| SC_AGPH | SC5314 | RP10/rp10::CaPHx2-GFP | PH-GFP expression | This study |
| irs4_AGPH | irs4_2KO | irs4Δ/irs4Δ, RP10/rp10::CaPHx2-GFP | PH-GFP expression | This study |
| inp51_AGPH | inp51_2KO | inp51Δ/inp51Δ, RP10/rp10::CaPHx2-GFP | PH-GFP expression | This study |
| gin4_AGC10 | gin4_2KO | gin4Δ/gin4Δ, RP10/rp10::CaPHx2-GFP | PH-GFP expression | This study |
| SC_AGC10 | SC5314 | RP10/rp10::CDC10-GFP | CDC10-GFP expression | This study |
| irs4_AGC10 | irs4_2KO | irs4Δ/irs4Δ, RP10/rp10::CDC10-GFP | CDC10-GFP expression | This study |
| inp51_AGC10 | inp51_2KO | inp51Δ/inp51Δ, RP10/rp10::CDC10-GFP | CDC10-GFP expression | This study |
| SC_AGS7 | SC5314 | RP10/rp10::SEP7-GFP | SEP7-GFP expression | This study |
| irs4_AGS7 | irs4_2KO | irs4Δ/irs4Δ, RP10/rp10::SEP7-GFP | SEP7-GFP expression | This study |
| inp51_AGS7 | inp51_2KO | inp51Δ/inp51Δ, RP10/rp10::SEP7-GFP | SEP7-GFP expression | This study |
| CAI_R5G | CAI4 | ura3::imm434/ura3::imm434 iro1/iro1::imm434, Rbt5sig.GFP.GPI | RBT5-GFP expression | 31 |
| rs4_AGR5 | CAI_R5G | Same as parent, irs4Δ/irs4Δ | RBT5-GFP expression | This study |
| inp51_AGR5 | CAI_R5G | Same as parent, inp51Δ/inp51Δ | RBT5-GFP expression | This study |
| SC_AGPHRC10 | SC_AGPH | RP10/rp10::CaPHx2-GFP, CDC10-RFP | PH-GFP and CDC10-RFP expression | This study |
| irs4_AGPHRC10 | irs4_AGPH | irs4Δ/irs4Δ, RP10/rp10::CaPHx2-GFP, CDC10-RFP | PH-GFP and CDC10-RFP expression | This study |
| inp51_AGPHRC10 | inp51_AGPH | inp51Δ/inp51Δ, RP10/rp10::CaPHx2-GFP, CDC10-RFP | PH-GFP and CDC10-RFP expression | This study |
| gin4_AGPHRC10 | gin4_ AGPH | gin4Δ/gin4Δ, RP10/rp10::CaPHx2-GFP, CDC10-RFP | PH-GFP and CDC10-RFP expression | This study |
| SC_AGPHRS7 | SC_AGPH | RP10/rp10::CaPHx2-GFP, SEP7 | PH-GFP and SEP7-RFP expression | This study |
| irs4_ AGPHRS7 | irs4_2KO | irs4Δ/irs4Δ, RP10/rp10::CaPHx2-GFP, SEP7 | PH-GFP and SEP7-RFP expression | This study |
| inp51_ AGPHRS7 | inp51_2KO | inp51Δ/inp51Δ, RP10/rp10::CaPHx2-GFP, SEP7 | PH-GFP and SEP7-RFP expression | This study |
| SC_AGV4 | SC5314 | RP10/rp10::Vrg4-GFP | VRG4-GFP expression | This study |
| irs4_ AGV4 | irs4_2KO | irs4Δ/irs4Δ, RP10/rp10::Vrg4-GFP | VRG4-GFP expression | This study |
| inp51_ AGV4 | inp51_2KO | inp51Δ/inp51Δ, RP10/rp10::Vrg4-GFP | VRG4-GFP expression | This study |
| SC_S1SPG | SC5314 | RP10/rp10::SAP1_SP-GFP | SAP1-GFP (secretory) expression | This study |
| irs4_ S1SPG | irs4_2KO | irs4Δ/irs4Δ, RP10/rp10::SAP1_SP-GFP | SAP1-GFP (secretory) expression | This study |
| inp51_ S1SPG | inp51_2KO | inp51Δ/inp51Δ, RP10/rp10::SAP1_SP-GFP | SAP1-GFP (secretory) expression | This study |
| SC_AGV4 | SC5314 | RP10/rp10::Arf1-GFP | ARF1-GFP expression | This study |
| irs4_ AGV4 | irs4_2KO | irs4Δ/irs4Δ, RP10/rp10::Arf1-GFP | ARF1-GFP expression | This study |
| inp51_ AGV4 | inp51_2KO | inp51Δ/inp51Δ, RP10/rp10::Arf1-GFP | ARF1-GFP expression | This study |
| SC_S1SPG | SC5314 | RP10/rp10::Arf2-GFP | ARF2-GFP expression | This study |
| irs4_ S1SPG | irs4_2KO | irs4Δ/irs4Δ, RP10/rp10::Arf2-GFP | ARF2-GFP expression | This study |
| inp51_ S1SPG | inp51_2KO | inp51Δ/inp51Δ, RP10/rp10::Arf2-GFP | ARF2-GFP expression | This study |
| BWP17 | C. albicans reference strain (auxotropically marked) | 59 | ||
| DAY185 | BWP17 | C. albicans reference strain (auxotrophies corrected) | Wild-type control | 59 |
| DAY_AGPH | DAY185 | RP10/rp10::CaPHx2-GFP | PH-GFP expression | This study |
| cdc10−/− (YAW7) | BWP17 | cdc10Δ/cdc10Δ | cdc10 null mutant | 55, 56 |
| cdc11−/− (YAW11) | BWP17 | cdc10Δ/cdc10Δ | cdc10 null mutant | 55, 56 |
| ckb2−/− (JJH351) | BWP17 | ckb2Δ/ckb2Δ | ckb2 null mutant | 6 |
| CKB2−/−/+ | ckb2Δ/ckb2Δ | Complemented, ckb2Δ/CKB2 | CKB2 reinsertion | 6 |
| ckb2_AGPH | ckb2Δ/ckb2Δ | ckb2Δ/ckb2Δ, RP10/rp10::CaPHx2-GFP | PH-GFP expression | This study |
| cla4−/− (JJH262) | BWP17 | cla4Δ/cla4Δ | cla4 null mutant | 6 |
| CLA4−/−/+ | cla4Δ/cla4Δ | Complemented, cla4Δ/CLA4 | CLA4 reinsertion | 6 |
| cla4_AGPH | cla4Δ/cla4Δ | cla4Δ/cla4Δ, RP10/rp10::CaPHx2-GFP | PH-GFP expression | This study |
| gin4−/− (JJH276) | BWP17 | gin4Δ/gin4Δ | gin4 null mutant | 6 |
| GIN4−/−/+ | gin4Δ/gin4Δ | Complemented, gin4Δ/GIN4 | GIN4 reinsertion | 6 |
| gin4_AGPH | gin4Δ/gin4Δ | gin4Δ/gin4Δ, RP10/rp10::CaPHx2-GFP | PH-GFP expression | This study |
| hsl1−/− (JJH377) | BWP17 | hsl1Δ/hsl1Δ | hsl1 null mutant | 6 |
| HSL1−/−/+ | hsl1Δ/hsl1Δ | Complemented, hsl1Δ/HSL1 | HSL1 reinsertion | 6 |
| hsl1_AGPH | hsl1Δ/hsl1Δ | hsl1Δ/hsl1Δ, RP10/rp10::CaPHx2-GFP | PH-GFP expression | This study |
| kin3−/− (JJH381) | BWP17 | kin3Δ/kin3Δ | kin3 null mutant | 6 |
| KIN3−/−/+ | hsl1Δ/hsl1Δ | Complemented, kin3Δ/KIN3 | KIN3 reinsertion | 6 |
| kin3_AGPH | hsl1Δ/hsl1Δ | kin3Δ/kin3Δ, RP10/rp10::CaPHx2-GFP | PH-GFP expression | This study |
| tpk1−/− (JJH384) | BWP17 | tpk1Δ/tpk1Δ | tpk1 null mutant | 6 |
| TPK1−/−/+ (JJH233) | tpk1Δ/tpk1Δ | Complemented, tpk1Δ/TPK1 | TPK1 reinseriton | 6 |
| bck1−/− (JJH256) | BWP17 | bck1Δ/bck1Δ | bck1 null mutant | 6 |
| BCK1−/−/+ (JJH246) | bck1Δ/bck1Δ | Complemented, bckΔ/BCK1 | BCK1 reinsertion | 6 |
| ire1−/− (SF008P) | BWP17 | ire1Δ/ire1Δ | ire1 null mutant | 6 |
| IRE1−/−/+ (SF008C) | ire1Δ/ire11Δ | Complemented, ire1Δ/IRE1 | IRE1 reinsertion | 6 |
Gene disruption.
C. albicans irs4 and gin4 null mutant strains were created by the standard SAT-flipper method, as used previously to create our inp51 null mutants (3, 43). In brief, fragments at the 5′ (F1) and 3′ (F2) ends of the targeted genes were amplified by PCR using primers that introduced appropriate restriction sites. These fragments were subcloned into pSFS-2A between the KpnI and XhoI sites and the SacII and SacI sites, respectively. In parallel, the full gene with 1 kb of downstream noncoding sequence was amplified and used as F1 for gene reinsertion. The resulting plasmids were extracted and linearized with KpnI and SacI and introduced into competent C. albicans cells by electroporation. Transformants were grown on selective medium plates containing 200 μg/ml streptothricin (Alexis Biochemicals) for 2 to 3 days and then screened by PCR. Positive transformants were grown overnight in maltose-based medium to induce excision of the plasmid cassette, plated on selective medium plates for 3 to 4 days, and screened for positive excisions to enable the strain for another round of targeted recombination. All positive events of homologous recombination and excision were confirmed by Southern blotting. We verified previously described phenotypes for the irs4 and inp51 mutants, including normal growth in liquid media, impaired invasive growth and hyphal formation in contact with solid agar, increased susceptibility to caspofungin, zymolase, and SDS, and distribution of chitin in patches along the cell surfaces of yeast and filamentous morphologies (2, 3). The irs4 and inp51 mutants were indistinguishable in each of these assays. The gin4 mutant was indistinguishable from gin4 mutants previously created in our lab using C. albicans strain BWP17 (6). In all instances, gene reconstitutions corrected the aberrant phenotypes. Similar results were obtained using independently created null mutant and reconstitution strains.
Caspofungin exposure.
Sensitivity to caspofungin (Merck Research Laboratories) was measured in 48-well microtiter plates. C. albicans strains grown overnight in YPD at 30°C were diluted to an optical density at 600 nm (OD600) of 0.1 in YPD. Caspofungin was added at concentrations ranging from 0.075 to 20 μg/ml and transferred to the microtiter plates (800 μl per well). The plates were incubated at 30°C with shaking at 250 rpm, and the OD620 was measured every hour. The MIC of caspofungin was determined using the broth macrodilution method recommended by CLSI (48, 49). To assess the impact of caspofungin exposure on PI(4,5)P2 and septin localization, we used our previously described methods with the following modifications (6). Cells were exposed to the MIC of caspofungin (0.125 μg/ml) in YPD for 5 min, followed by washing twice with phosphate-buffered saline (PBS). This regimen was verified to be nonlethal to C. albicans cells by colony count enumerations prior to caspofungin exposure, as well as after exposure and washing. PI(4,5)P2 and septin were visualized as described below.
CaPH-GFP and other fluorescent reporters.
Pleckstrin homology (PH) domains bind specifically to PI(4,5)P2 (17, 46). To create a green fluorescent protein (GFP)-tagged C. albicans PH domain, the sequence of human PLCδ1 PH was modified to account for noncanonical CTG codon usage (encoding serine instead of leucine). The C. albicans PH domain (CaPH; 549 bp) was synthesized using the custom gene synthesis service of Integrated DNA Technologies, Inc. (Coralville, IA), and it was cloned into pZEro-2. For the other fluorescently labeled reporters listed in Table 1, the respective open reading frames were amplified from C. albicans SC5314 genomic DNA and fused to yeGFP (C. albicans-optimized GFP, as described in reference 12), CaGFP (C. albicans-optimized GFP, as described in reference 60), or red fluorescent protein (RFP) (47). In the case of RFP, the sequence of the photostable TagRFP-T version of RFP was optimized for expression using the codon preference of C. albicans and was chemically synthesized by Integrated DNA Technologies Inc. The fusion genes were digested with the appropriate restriction enzymes and cloned into pSFS under the control of ADH1p and ADH1t (43). pSFS plasmids carried target fragments for genomic integration by homologous recombination into either the RP10 locus or a noncoding region in chromosome 1 upstream to RP10 (bp 25000 to 27000). For targeted homologous recombination in C. albicans, we used transformation by electroporation (3, 43). C. albicans wild-type strain CAI4 expressing Rbt5-GFP (strain CAI_R5G) was generously provided by Brian Wong. As previously described, CAI_R5G expresses a C. albicans-optimized GFP fused in frame to the signal peptide and GPI sequence from the N and C termini, respectively, of Rbt5 (31). CAI_R5G was used to generate irs4 and inp51 null mutants expressing GFP-Rbt5 by the SAT-flipper method (41).
Chitin staining and endocytosis assays.
For chitin staining, cells were incubated for 5 min at room temperature in 0.1 mg/ml calcofluor white and then washed three times in cold PBS. Since calcofluor white may bind other glycan polymers in addition to chitin, we also localized chitin with white germ agglutinin during electron microscopy experiments (described below). For endocytosis assays, cells were resuspended in either a 50-μg/ml solution of FM4-64 (catalog no. T3166; Molecular Probes) or a 6 mg/ml solution of Lucifer yellow (catalog no. 62642; Fluka). Following incubation for 1 h at room temperature or 30°C, cells were washed 3 times in cold PBS with sodium azide and sodium fluoride (10 mM each).
Microscopy.
For microscopy of fluorescently labeled cells, respective strains were grown in 35-mm-diameter glass-bottom dishes (Matek, Ashland, MA) for 2 to 3 h, as described above for invasive growth. For chitin colocalization, cells were fixed with 2% formaldehyde and stained with calcofluor white. Microscopy was performed at the University of Pittsburgh Center for Biologic Imaging, using established protocols. For fluorescence microscopy, optical sections were captured using an Olympus Fluoview 1000 confocal laser scanning microscope. Images were captured with an exposure time of 1 s on a Coolsnap HQ2 (Photometrics) camera using Axiovision (Zeiss) software. For transmission electron microscopy, cells were collected from within reverse agar (3) and fixed at 4°C in 0.1 M sodium cacodylate buffer (pH 7.2) containing 2% glutaraldehyde and 2% paraformaldehyde (37). The samples were then dehydrated through a graded series of ethanol and embedded in Lowicryl K4M (Electron Microscopy Sciences, Hatfield, PA). Thin sections were imaged with a Zeiss EM902 electron microscope. Wheat germ agglutinin localization of chitin was performed using previously described methods (52).
Quantitative reverse transcription-PCR (6).
Overnight cultures of cells were diluted to an OD600 of 0.2 in 50 ml fresh YPD medium. Cultures were allowed to grow at 30°C with shaking until the culture density reached an OD600 of ∼1. Cells were then harvested by vacuum filtration and flash frozen in a dry ice/ethyl alcohol (EtOH) bath. Cells were kept frozen on filters at −80°C until RNA extraction. RNA was extracted using a RiboPure-Yeast kit (Ambion) following the manufacturer's instructions with the following modifications. Cells were resuspended from filters with 1.5 ml ice-cold distilled water (dH2O) followed by 10 to 15 s of vigorous vortexing. Resuspended cells were transferred to a 1.5-ml tube and spun down following the manufacturer's protocol. Furthermore, during the cell disruption step, cells were beaten with a Next Advance Bullet Blender for 3 min at 4°C to maximize cell lysis. Ten micrograms of the resulting total RNA was treated with the DNA-free kit (Ambion) followed by first-strand cDNA synthesis from half of the DNA-free RNA using the AffinityScript multiple temperature cDNA synthesis kit (Stratagene). Absence of DNA contamination was confirmed using control sets for which reverse transcriptase was omitted from the cDNA reaction. Primer3 software (http://frodo.wi.mit.edu/) was used to design primers for 14 genes (TDH3, DDR48, SOD5, STP4, FDH2, AOX2, STF2, ECM331, RTA4, SKS1, RHR2, ORF19.4445, ALS1, and HGT6) (6). Then, 2× iQ SYBR green Supermix (Bio-Rad), 1 μl of first-strand cDNA reaction mixture, and 0.1 μM primers were mixed in a total volume of 50 μl per reaction, and real-time PCR was performed in triplicate for each sample on an iCycler iQ real-time PCR detection system (Bio-Rad). Product amplification was detected using SYBR green fluorescence, and specificity of the reaction was monitored by melt-curve analysis following the real-time program. TDH3 was used as a reference gene for normalization of gene expression, which was determined using Bio-Rad iQ5 software (ΔΔCT method [CT, threshold cycle]). Cluster analysis was performed with the Multiexperiment Viewer (MeV 4.3) from the J. Craig Venter Institute. Hierarchical clustering was performed with average linkage and a Manhattan distance metric. Each mutant strain was normalized to its respective complement strain to account for strain background differences.
RESULTS
PI(4,5)P2 and chitin are redistributed in C. albicans irs4 and inp51 mutants at sites of plasma membrane and cell wall derangements.
Building upon our observation that C. albicans Irs4 and Inp51 interact to regulate PI(4,5)P2 levels (3), we hypothesized that PI(4,5)P2 distribution would be abnormal in irs4 and inp51 mutants. To localize PI(4,5)P2, we constructed a reporter comprised of a C. albicans-optimized PI(4,5)P2-binding PH domain tagged with GFP (CaPH-GFP). In wild-type C. albicans SC5314, CaPH-GFP was distributed uniformly along the surfaces of yeast and filamentous cells and at septae. In contrast, PI(4,5)P2 was concentrated in discrete patches at the cell surfaces of irs4 and inp51 mutants, which appeared to extend intracellularly (Fig. 1). The aberrant distribution of PI(4,5)P2 resembled the chitin accumulations we observed previously (2, 3). Using the CaPH-GFP reporter and calcofluor white, we demonstrated that PI(4,5)P2 and chitin were colocalized in these patches (Fig. 2). Reconstitution of IRS4 and INP51 in the respective null mutants restored the uniform distribution of PI(4,5)P2 and chitin.
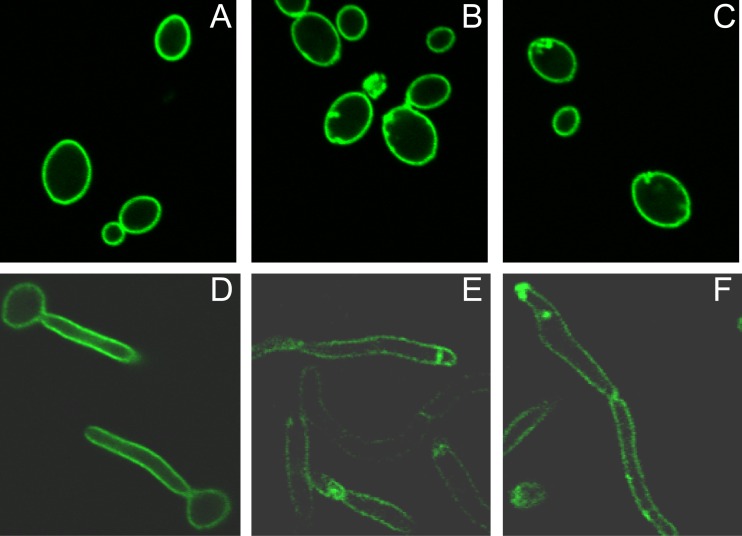
Fig 1.
PI(4,5)P2 accumulates in discrete patches in irs4 and inp51 mutants. C. albicans SC5314 (wild-type) and irs4 and inp51 mutants expressing CaPH-GFP were grown in yeast (A to C, respectively; YPD medium at 30°C) or hyphal (D to F, respectively; YPD medium + 5% fetal bovine serum [FBS] at 37°C) morphologies in 35-mm-diameter glass-bottom dishes and imaged using confocal microscopy. As shown, CaPH-GFP was distributed uniformly in SC5314 (A and D) but concentrated in discrete patches in irs4 (B and E) and inp51 (C and F) mutants.
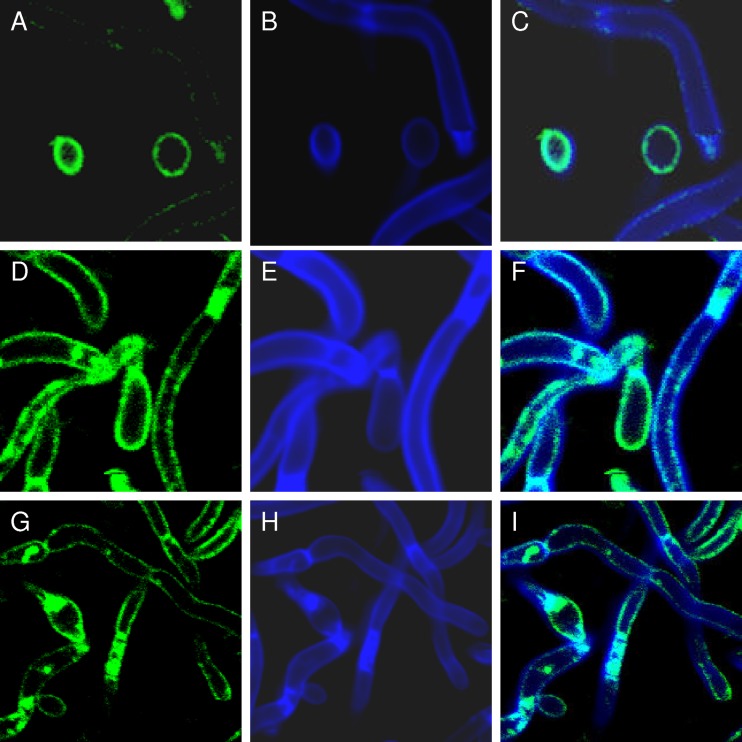
Fig 2.
PI(4,5)P2 and chitin colocalize at aberrant patches in irs4 and inp51 mutants. PI(4,5)P2 was localized in C. albicans SC5314 and irs4 and inp51 mutants using a CaPH-GFP reporter (A, D, and G, respectively), as in the experiments shown in Fig. 1. Chitin was localized in the same cells by calcofluor white staining (B, E, and H, respectively). Overlay images revealed that CaPH-GFP and calcofluor white colocalized to aberrant sites in irs4 (F) and inp51 (I) mutants (cyan color).
Since the null mutants are hypersusceptible to caspofungin (2, 3), we hypothesized that the PI(4,5)P2-chitin accumulations were associated with cell wall derangements. To test this hypothesis, we first visualized the GPI-anchored cell wall protein Rbt5 in mutant cells. Indeed, an Rbt5-GFP fusion colocalized with the chitin accumulations (Fig. 3). Next, we used transmission electron microscopy to assess the cell walls of mutant cells in greater detail. The cells demonstrated plasma membrane and cell wall invaginations into the cytoplasm (Fig. 4). The invaginations concentrated immunogold-labeled wheat germ agglutinin, consistent with the accumulation of chitin.
Fig 3.
The GPI-anchored cell wall protein Rbt5 colocalizes with chitin at aberrant patches in mutant cells. C. albicans CAI4 and irs4 and inp51 mutants expressing an Rbt5-GFP fusion were visualized by confocal microscopy as in earlier experiments. inp51 mutant cells are shown as a representative. Rbt5-GFP was distributed normally in CAI4 cells (A) but was concentrated within aberrant patches (white arrows) in the inp51 mutant (B and C [higher-resolution images]). Rbt5-GFP colocalized with calcofluor white in the inp51 mutant (D, cyan color, arrowhead). The irs4 mutant resembled the inp51 mutant in all experiments.
Fig 4.
Mutant cells exhibit plasma membrane and cell wall abnormalities that correspond to sites of chitin accumulation. C. albicans SC5314 exhibited normal plasma membrane and cell wall (A) and septa (B) by transmission electron microscopy. The inp51 mutant, on the other hand, had striking invaginations of the plasma membrane and cell wall (invaginations in panels C and E are shown at higher power [arrowheads] in panels D and F, respectively). Immunogold-labeled wheat germ agglutinin, which binds chitin, was distributed normally in SC5314 cells (G, black dots) but accumulated within the plasma membrane and cell wall invaginations of the mutant (H). The inp51 mutant is shown as a representative; similar results were obtained for the irs4 mutant.
Septins colocalize with PI(4,5)P2 and chitin in irs4 and inp51 mutants.
Consistent with the observation that PI(4,5)P2 directly interacts with septins (5),we demonstrated that PI(4,5)P2 and chitin colocalized with fluorescently tagged septins Cdc10 and Sep7 in irs4 and inp51 mutant cells (Fig. 5 and 6). To test the hypothesis that PI(4,5)P2 and septins are connected functionally, we studied cdc10 and cdc11 septin mutants for cell wall-related abnormalities previously demonstrated for irs4 and inp51 mutants (2, 3). Similar to irs4 and inp51, cdc10 and cdc11 mutants were hypersusceptible to caspofungin and, as previously shown, impaired in the ability to invade solid agar (2, 3, 55). Along these lines, we previously showed that irs4 and inp51 mutants were impaired in invasive growth into mouse kidneys during the course of hematogenously disseminated candidiasis, similar to observations with cdc10 and cdc11 mutants (55, 56).
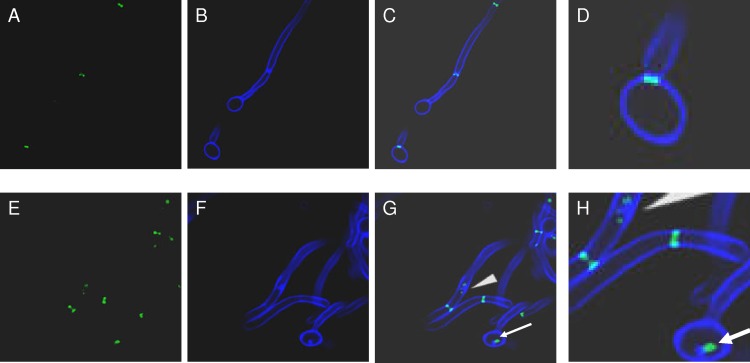
Fig 5.
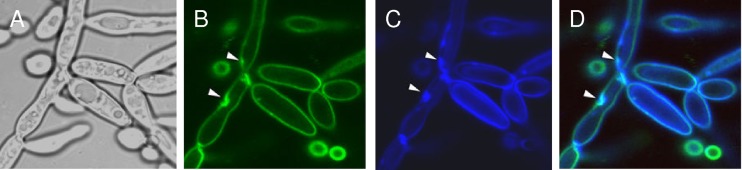
Septins and chitin colocalize in mutant cells. C. albicans SC5314 and irs4 and inp51 mutants expressing Cdc10-GFP were visualized by confocal microscopy, and chitin was localized by calcofluor white staining. Cdc10-GFP and calcofluor white were distributed normally in SC5314 cells (A and B, respectively), but were concentrated within aberrant patches in the inp51 mutant (E and F, respectively). As evident in the overlay images, Cdc10-GFP colocalized with calcofluor white in the inp51 mutant (G and H [higher resolution], cyan color and arrowheads).
Fig 6.
Septins and PI(4,5)P2 colocalize in mutant cells. C. albicans inp51 colocalized Cdc10-RFP and CaPH-GFP (yellow color and arrowheads in overlay images in B; higher-resolution images in C and D). Images of irs4 mutants expressing Cdc10-RFP and CaPH-GFP, and both mutants expressing Sep7-RFP and CaPH-GFP, were similar (not shown).
In addition to their roles in cell wall regulation, EH domain proteins are known to contribute to the regulation of endocytosis and intracellular membrane trafficking and secretion. For this reason, we tested whether the plasma membrane and cell wall derangements of irs4 and inp51 mutant cells were sites of dysregulated endocytosis or secretion. The chitin-containing structures in mutant cells were clearly distinct from endocytic vesicles, as evident by calcofluor white and FM4-64 or Lucifer yellow costaining (see Fig. S1 in the supplemental material). Moreover, we found that mutant cells did not mislocalize GFP-tagged Vrg4 (a cytoplasm-to-Golgi transporter), GFP-Arf1 or GFP-Arf2 (ADP-ribosylation factors, involved in intracellular transport), or GFP-tagged secretory signals from either Rbt5 or the secreted aspartyl protease Sap1 (see Fig. S2 in the supplemental material).
The septin regulatory protein kinase Gin4 acts upstream of Irs4 and Inp51 in regulating cell wall integrity responses.
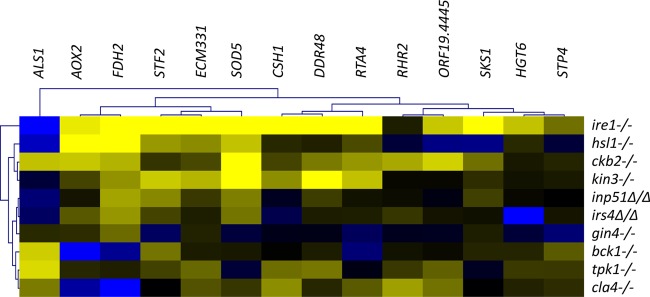
We previously identified several protein kinases with roles in cell wall integrity that also are involved in septin localization, ring function, or signaling (6). In the absence of stress, mutants of these protein kinases displayed a common cell wall damage response gene expression profile. In this study, we used quantitative RT-PCR to assess the expression of 14 damage response genes by irs4 and inp51 mutants, septin regulatory protein kinase mutants (gin4, kin3, ckb2, cla4, and hsl1), and other cell wall-related protein kinase mutants (ire1, bck1, and tpk1). The irs4 and inp51 mutants were firmly positioned with the septin mutants by hierarchical analysis of the expression data (Fig. 7), clustering most tightly with each other and the gin4 mutant. The ire1 mutant, in contrast, was a distinct outlier, consistent with its positioning in our earlier analysis (6).
Fig 7.
C. albicans irs4 and inp51 mutants display cell wall damage response gene expression profiles that are similar to those of septin regulatory protein kinase mutants. The expression of 14 damage response genes was analyzed in mutant strains in the absence of cell wall stress. The expression of TDH3, a housekeeping gene involved in glycolysis, was used to normalize expression between strains. Strains were further normalized to their complemented strain to reduce strain background errors. Resultant values were log base 2 transformed. As shown, irs4 and inp51 mutants were firmly positioned within the septin regulatory protein kinase functional group by hierarchical analysis and clustered most tightly with gin4. The cell wall-related protein kinase mutant ire1 was an outlier from the other mutants. Blue and yellow on heat map refer to log base 2 transformed values of −2 and 2, respectively.
The results suggest that the septin regulatory protein kinases are linked with PI(4,5)P2 and septins within a network that regulates cell wall integrity. If this interpretation is correct, septin regulatory protein kinase and irs4 and inp51 mutants should exhibit a number of similar cell wall-related phenotypes. Indeed, phenotypic analysis of the mutant strains showed variation in the degree of similarity (Table 2; see Fig. S3 in the supplemental material). Of particular note, gin4, irs4, and inp51 mutant strains exhibited similar phenotypes for each assay, consistent with our hierarchical clustering analysis and suggesting a tight correlation between the functions of these genes. In fact, gin4 was the only septin regulatory protein kinase mutant to demonstrate aberrant PI(4,5)P2 and chitin patches (Fig. 8). Taken with our previous demonstration that septins were also delocalized in the gin4 mutant, the data suggest that this protein kinase acts upstream of PI(4,5)P2 and septins in regulating C. albicans responses to cell wall stress imposed by caspofungin.
Table 2.
Phenotypes of C. albicans mutants
| Phenotype | Result for C. albicans mutant |
||||||
|---|---|---|---|---|---|---|---|
| irs4 | inp51 | gin4 | kin3 | ckb2 | cla4 | hsl1 | |
| Caspofungin hypersusceptibility | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Impaired invasive growth | Yes | Yes | Yes | No | Yes | Yes | No |
| Chitin mislocalization | Yes | Yes | Yes | Yes | No | No | No |
| PI(4,5)P2 mislocalization | Yes | Yes | Yes | No | No | No | No |
| Septin mislocalization | Yes | Yes | Yesa | Yesa | Noa | ±a,b | Noa |
Fig 8.
The gin4 mutant (A) demonstrates aberrant chitin and PI(4,5)P2-containing patches. CaPH-GFP (B) and calcofluor white (C) colocalized in aberrant patches in C. albicans gin4 mutant cells (D, cyan color and arrowheads in overlay image), similarly to irs4 and inp51 mutants. gin4 mutants were created using the SAT-flipper method and C. albicans SC5314.
C. albicans rapidly redistributes PI(4,5)P2 and septins in response to caspofungin.
In our earlier study, caspofungin treatment of wild-type C. albicans cells resulted in septin ring delocalization. To determine if PI(4,5)P2 is also mislocalized in response to caspofungin, we performed PI(4,5)P2 and septin colocalization experiments. First, we identified a brief caspofungin exposure that was sublethal to a CaPH-GFP, Cdc10-RFP expressing wild-type C. albicans strain. Indeed, a 5-min exposure to the MIC (0.125 μg/ml) of caspofungin did not kill the reporter strain, as established by colony count enumerations. At the same time, this treatment regimen resulted in redistribution of PI(4,5)P2 and septins to common sites, which was evident in 50 to 75% of C. albicans cells (Fig. 9).
Fig 9.
Brief exposure to caspofungin results in redistribution of PI(4,5)P2 and septins to aberrant patches in wild-type C. albicans cells. A 5-min exposure to caspofungin at the MIC (0.125 μg/ml) did not kill a wild-type C. albicans strain (A) but resulted in redistribution of CaPH-GFP (B) and Cdc10-RFP (C) to aberrant patches (D, overlay image). PI(4,5)P2-septin colocalization was similar to that seen in irs4, inp51, and gin4 mutant cells in the absence of cell wall stress.
Taken together, therefore, our data demonstrate that a network that includes the protein kinase Gin4 and the Irs4/Inp51 phosphatase complex regulates PI(4,5)P2 and septin localization and is essential for maintaining C. albicans cell wall integrity in response to caspofungin. Moreover, C. albicans cells redistribute PI(4,5)P2 and septins rapidly as part of the cell wall stress response to caspofungin.
DISCUSSION
In our previous studies, we demonstrated that PI(4,5)P2 regulation and septin ring formation both play major roles in the maintenance of C. albicans cell wall integrity (2, 3, 6). Here, we present a series of observations that tie PI(4,5)P2 signaling to septin localization, in keeping with in vitro studies showing that PI(4,5)P2 directly interacts with septins and promotes septin filament organization and stability (5). Our most important finding is that PI(4,5)P2 signaling and septin localization are part of the natural response of C. albicans to cell wall inhibition by caspofungin. We propose that a PI(4,5)P2-septin regulatory network relays a signal that helps govern responses to cell wall stress and the inhibition of cell wall synthesis (Fig. 10).
Fig 10.

Model for the PI(4,5)P2-septin cell wall regulatory network. Exposure of C. albicans to caspofungin inhibits β-1,3-d-glucan synthesis and creates cell wall stress (box 1). Upon sensing this stress, cells rapidly redistribute PI(4,5)P2 and septins (box 3). PI(4,5)P2 accumulation activates the PKC-MAPK cell wall integrity pathway, and PI(4,5)P2 and septins direct the deposit of chitin and other cell wall components at sites of colocalization (box 4). The redistribution of PI(4,5)P2 and septins in response to caspofungin is similar to that observed in gin4, irs4, and inp51 mutants, which are hypersusceptible to caspofungin and exhibit highly similar cell wall damage response gene expression profiles. Therefore, the data suggest that (i) Gin4 and the Irs4/Inp51 5′-phosphatase complex function upstream of PI(4,5)P2 and septins in a pathway that relays the cell wall stress signal (box 2), and (ii) an intact Gin4-Irs4/Inp51-PI(4,5)P2-septin pathway is required for the maintenance of cell wall integrity in the face of ongoing caspofungin exposure (box 5). Cell wall regulation (CWR) through the pathway is part of the natural response to caspofungin and, as our earlier mouse data indicated (2), necessary for the progression of invasive candidiasis after the initial stages of tissue colonization (box 6). White and blue boxes represent components of this model that have been established or suggested by our data, respectively.
We have explored PI(4,5)P2 function through deletion mutations of IRS4 and INP51, genes that encode the PI(4,5)P2-specific 5′-phosphatase and its EH domain-containing regulatory subunit, respectively. As predicted, the mutations resulted in the same biochemical consequence—accumulation of PI(4,5)P2, which we demonstrated by high-performance liquid chromatography (HPLC) analysis (3). In the present study, we extend these observations by showing that the excess PI(4,5)P2 accumulates in discrete plasma membrane patches in budding cells and hyphae of inp51 and irs4 mutant cells. These patches do not represent sites of disrupted endocytosis or secretion but, rather, correspond to aberrant deposition of cell wall components, including chitin and the GPI-linked protein Rbt5. We previously observed that inp51 and irs4 mutants were hypersensitive to caspofungin and other stressors that cause cell wall disruption, a phenotype that is consistent with indirect effects on cell wall biogenesis. In this context, our current data indicate that PI(4,5)P2 directs deposition of at least some cell wall components. In fact, upregulation of chitin synthesis is a well-recognized adaptive response of C. albicans to β-1,3-d-glucan synthase inhibition by caspofungin (54). In our model for the PI(4,5)P2-septin network, chitin synthesis may be promoted through PI(4,5)P2 activation of the PKC-MAPK cell wall integrity pathway, as demonstrated in our previous study (3), and by septin-directed deposition of chitin, as suggested in the present study.
The connection of PI(4,5)P2 to septins rests on three lines of evidence. First, septins Sep7 and Cdc10 localize to the sites of aberrant PI(4,5)P2 and chitin accumulation in inp51 and irs4 mutants. This finding supports the functional role of septin-PI(4,5)P2 binding established in vitro (5). Second, inp51 and irs4 mutants have similar damage response gene expression changes to protein kinase mutants that govern septin localization or downstream responses (6). Similarity to the gin4 mutant was particularly noteworthy and also extended to a defect in invasive growth into solid agar. Third, we observed that the gin4 mutation causes accumulation of PI(4,5)P2 and chitin patches similar to those observed in the inp51 and irs4 mutants (2, 3). Gin4 is known to phosphorylate septins directly to govern their localization (1, 28, 36), but it has not previously been connected to PI(4,5)P2. Therefore, our observations suggest that Gin4 may have a second role in regulating PI(4,5)P2 synthesis. Alternatively, there may be a reciprocal septin-PI(4,5)P2 regulatory mechanism, whereby PI(4,5)P2 governs septin distribution, and septins also control PI(4,5)P2 distribution.
Our data supporting the PI(4,5)P2-septin regulatory relationship have two major clinical implications. First, the relationship appears to be an element of the natural response of C. albicans to cell wall inhibition by caspofungin. We demonstrated that altered PI(4,5)P2-septin distribution occurs within 5 min of caspofungin exposure and therefore represents a rapid response to β-1,3-d-glucan synthase inhibition. In principle, this altered localization might result from various indirect effects of cell wall perturbation. However, most caspofungin-hypersensitive mutants that have been tested to date do not affect septin localization (6) or, as we showed here, PI(4,5)P2 localization. The specificity and rapid onset of the PI(4,5)P2-septin response lead us to believe that it mediates a particular rather than generalized signal and that it plays a natural role in cell wall dynamics. The response may serve a protective purpose, such as by preventing accumulation of unbalanced cell wall components on the cell surface, which may lead to cell rupture, or by directing cell wall biogenesis to specific sites of need. Alternatively, PI(4,5)P2-septin redistribution may represent an early step in a mechanism of cell killing. In either case, the phenomenon provides an assay for some of the earliest events in caspofungin-treated C. albicans cells, which should prove useful in unraveling complex cell wall integrity responses.
The second clinically relevant contribution of the PI(4,5)P2-septin response is to the progression of invasive candidiasis within organs following hematogenous dissemination of C. albicans. We previously showed that IRS4 and INP51 were expressed within mouse kidneys after injection of C. albicans into the lateral tail vein (2, 3). The respective null mutants colonized kidneys as well as wild-type C. albicans but thereafter caused lower mortality and tissue burdens and elicited less inflammation. While the mutants initiated hyphal formation at the early stages of invasive disease, they did not form the dense collections of matted hyphae seen with wild-type strains at later time points within the kidneys. Rather, the mutants were largely found as yeast and truncated filaments. Similarly, C. albicans septin mutants have been shown to colonize kidneys but not invade the renal parenchyma (55). Since gin4 mutants exhibit significant growth defects in vitro, we did not test the relative virulence of these strains in the mouse model. Nevertheless, it is logical that mutations that diminish cell wall integrity are associated with decreased ability of C. albicans to invade both solid agar and target organs. Taken together, our data indicate that the C. albicans PI(4,5)P2-septin network is activated upon both caspofungin exposure and tissue invasion and is necessary for successful long-term adaptation to these cell wall stresses.
Finally, our study offers further evidence that C. albicans has more extensive cell wall regulatory circuitry than S. cerevisiae, as well as novel cell wall integrity pathways that are relevant to virulence and drug susceptibility. Studies of S. cerevisiae have defined biogenesis mechanisms for major classes of cell wall components (24), established the connection between cell polarity and cell wall synthesis (8, 27), developed a detailed mechanistic model of cell wall integrity signaling through the PKC-MAPK cascade (27), and defined genetic and chemical-genetic networks that integrate cell wall integrity into other cell biological processes (14, 24–26). Novel C. albicans pathways may include components that do not have clear S. cerevisiae orthologues, such as the transcription factor Cas5 (7, 9). Alternatively, C. albicans pathways may either be rewired for cell wall regulation or adapted to this purpose as a result of increased “current” through them (6, 14). The role of Gin4, for example, suggests rewiring, in which the protein kinase may be at least partially distinct from its S. cerevisiae orthologue in the nature of its target genes and the signals to which it responds. On the other hand, Irs4 and Inp51 may perform comparable basic functions, which are uniquely adapted in C. albicans as a result of greater stress upon or flux (i.e., “current”) through the pathways that regulate cell wall integrity. In this regard, our data are the foundation for future mechanistic investigations into cell wall regulatory responses that distinguish C. albicans as a versatile opportunistic pathogen and have relevance for both the pathogenesis and treatment of invasive candidiasis.
Supplementary Material
ACKNOWLEDGMENTS
This project was funded in part by grant support from the US Department of Veterans Affairs (Merit Review to C. J. Clancy) and the National Institutes of Health (R56DE020831 to C. J. Clancy).
C. J. Clancy has received investigator-initiated research funding for unrelated projects from Pfizer and Merck. M. H. Nguyen has received investigator-initiated research funding for unrelated projects from Pfizer and Merck.
Footnotes
Published ahead of print 11 June 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Altman R, Kellogg D. 1997. Control of mitotic events by Nap1 and the Gin4 kinase. J. Cell Biol. 138:119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Badrane H, et al. 2005. Candida albicans IRS4 contributes to hyphal formation and virulence after the initial stages of disseminated candidiasis. Microbiology 151:2923–2931 [DOI] [PubMed] [Google Scholar]
- 3. Badrane H, et al. 2008. The Candida albicans phosphatase Inp51p interacts with the EH domain protein Irs4p, regulates phosphatidylinositol-4,5-bisphosphate levels and influences hyphal formation, the cell integrity pathway and virulence. Microbiology 154:3296–3308 [DOI] [PubMed] [Google Scholar]
- 4. Berman J, Sudbery PE. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918–930 [DOI] [PubMed] [Google Scholar]
- 5. Bertin A, et al. 2010. Phosphatidylinositol-4,5-bisphosphate promotes budding yeast septin filament assembly and organization. J. Mol. Biol. 404:711–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. 2010. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 6:e1000752 doi:10.1371/journal.ppat.1000752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bruno VM, et al. 2006. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog. 2:e21 doi:10.1371/journal.ppat.0020021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cabib E, Roh DH, Schmidt M, Crotti LB, Varma A. 2001. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276:19679–19682 [DOI] [PubMed] [Google Scholar]
- 9. Chamilos G, et al. 2009. Candida albicans Cas5, a regulator of cell wall integrity, is required for virulence in murine and toll mutant fly models. J. Infect. Dis. 200:152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheng S, et al. 2003. Identification of Candida albicans genes induced during thrush offers insight into pathogenesis. Mol. Microbiol. 48:1275–1288 [DOI] [PubMed] [Google Scholar]
- 11. Cheng SC, et al. 2011. The dectin-1/inflammasome pathway is responsible for the induction of protective T-helper 17 responses that discriminate between yeasts and hyphae of Candida albicans. J. Leukoc. Biol. 90:357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cormack BP, et al. 1997. Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology 143(Pt 2):303–311 [DOI] [PubMed] [Google Scholar]
- 13. DeMarini DJ, et al. 1997. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139:75–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finkel JS, et al. 2012. Portrait of Candida albicans adherence regulators. PLoS Pathog. 8:e1002525 doi:10.1371/journal.ppat.1002525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 16. Gow NA, et al. 2007. Immune recognition of Candida albicans beta-glucan by dectin-1. J. Infect. Dis. 196:1565–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harlan JE, Hajduk PJ, Yoon HS, Fesik SW. 1994. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature 371:168–170 [DOI] [PubMed] [Google Scholar]
- 18. Hillenmeyer ME, et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320:362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hogues H, et al. 2008. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol. Cell 29:552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kinoshita M. 2006. Diversity of septin scaffolds. Curr. Opin. Cell Biol. 18:54–60 [DOI] [PubMed] [Google Scholar]
- 21. Kumamoto CA. 2005. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. U. S. A. 102:5576–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumamoto CA, Vinces MD. 2005. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 7:1546–1554 [DOI] [PubMed] [Google Scholar]
- 23. Leberer E, et al. 1997. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr. Biol. 7:539–546 [DOI] [PubMed] [Google Scholar]
- 24. Lesage G, Bussey H. 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:317–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lesage G, et al. 2004. Analysis of beta-1,3-glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics 167:35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lesage G, et al. 2005. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Levin DE. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li CR, Yong JY, Wang YM, Wang Y. 2012. CDK regulates septin organization through cell-cycle-dependent phosphorylation of the Nim1-related kinase Gin4. J. Cell Sci. 125:2533–2543 [DOI] [PubMed] [Google Scholar]
- 29. Lo HJ, et al. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949 [DOI] [PubMed] [Google Scholar]
- 30. Longtine MS, et al. 2000. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:4049–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mao Y, Zhang Z, Wong B. 2003. Use of green fluorescent protein fusions to analyse the N- and C-terminal signal peptides of GPI-anchored cell wall proteins in Candida albicans. Mol. Microbiol. 50:1617–1628 [DOI] [PubMed] [Google Scholar]
- 32. Martchenko M, Levitin A, Hogues H, Nantel A, Whiteway M. 2007. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr. Biol. 17:1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Monge RA, Roman E, Nombela C, Pla J. 2006. The MAP kinase signal transduction network in Candida albicans. Microbiology 152:905–912 [DOI] [PubMed] [Google Scholar]
- 34. Morales-Johansson H, Jenoe P, Cooke FT, Hall MN. 2004. Negative regulation of phosphatidylinositol 4,5-bisphosphate levels by the INP51-associated proteins TAX4 and IRS4. J. Biol. Chem. 279:39604–39610 [DOI] [PubMed] [Google Scholar]
- 35. Morgan J, et al. 2005. Excess mortality, hospital stay, and cost due to candidemia: a case-control study using data from population-based candidemia surveillance. Infect. Control Hosp. Epidemiol. 26:540–547 [DOI] [PubMed] [Google Scholar]
- 36. Mortensen EM, McDonald H, Yates J, III, Kellogg DR. 2002. Cell cycle-dependent assembly of a Gin4-septin complex. Mol. Biol. Cell 13:2091–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nguyen KT, et al. 2010. Characterising the post-antifungal effects of micafungin against Candida albicans, Candida glabrata, Candida parapsilosis and Candida krusei isolates. Int. J. Antimicrob. Agents 35:80–84 [DOI] [PubMed] [Google Scholar]
- 38. Nikolaou E, et al. 2009. Phylogenetic diversity of stress signalling pathways in fungi. BMC Evol. Biol. 9:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nobile CJ, Mitchell AP. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15:1150–1155 [DOI] [PubMed] [Google Scholar]
- 40. Pappas PG, et al. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 48:503–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Phan QT, et al. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5:e64 doi:10.1371/journal.pbio.0050064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rauceo JM, et al. 2008. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol. Biol. Cell 19:2741–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reuss O, Vik A, Kolter R, Morschhauser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127 [DOI] [PubMed] [Google Scholar]
- 44. Richard ML, Plaine A. 2007. Comprehensive analysis of glycosylphosphatidylinositol-anchored proteins in Candida albicans. Eukaryot. Cell 6:119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roh DH, Bowers B, Schmidt M, Cabib E. 2002. The septation apparatus, an autonomous system in budding yeast. Mol. Biol. Cell 13:2747–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roth MG. 2004. Phosphoinositides in constitutive membrane traffic. Physiol. Rev. 84:699–730 [DOI] [PubMed] [Google Scholar]
- 47. Shaner NC, et al. 2008. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5:545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shields RK, et al. 2011. Paradoxical effect of caspofungin against Candida bloodstream isolates is mediated by multiple pathways but eliminated in human serum. Antimicrob. Agents Chemother. 55:2641–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shields RK, Nguyen MH, Press EG, Clancy CJ. 2011. Five-minute exposure to caspofungin results in prolonged postantifungal effects and eliminates the paradoxical growth of Candida albicans. Antimicrob. Agents Chemother. 55:3598–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Spreghini E, Davis DA, Subaran R, Kim M, Mitchell AP. 2003. Roles of Candida albicans Dfg5p and Dcw1p cell surface proteins in growth and hypha formation. Eukaryot. Cell 2:746–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Srikantha T, et al. 2006. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot. Cell 5:1674–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tronchin G, Poulain D, Herbaut J, Biguet J. 1981. Localization of chitin in the cell wall of Candida albicans by means of wheat germ agglutinin. Fluorescence and ultrastructural studies. Eur. J. Cell Biol. 26:121–128 [PubMed] [Google Scholar]
- 53. Versele M, Thorner J. 2005. Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 15:414–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Walker LA, et al. 2008. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 4:e1000040 doi:10.1371/journal.ppat.1000040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Warenda AJ, Kauffman S, Sherrill TP, Becker JM, Konopka JB. 2003. Candida albicans septin mutants are defective for invasive growth and virulence. Infect. Immun. 71:4045–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Warenda AJ, Konopka JB. 2002. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell 13:2732–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wheeler RT, Fink GR. 2006. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2:e35 doi:10.1371/journal.ppat.0020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wheeler RT, Kombe D, Agarwala SD, Fink GR. 2008. Dynamic, morphotype-specific Candida albicans beta-glucan exposure during infection and drug treatment. PLoS Pathog. 4:e1000227 doi:10.1371/journal.ppat.1000227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang C, Konopka JB. 2010. A photostable green fluorescent protein variant for analysis of protein localization in Candida albicans. Eukaryot. Cell 9:224–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zordan RE, Galgoczy DJ, Johnson AD. 2006. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. U. S. A. 103:12807–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.