Abstract
The resistance of methicillin-resistant Staphylococcus aureus (MRSA) to all β-lactam classes limits treatment options for serious infections involving this organism. Our goal is to discover new agents that restore the activity of β-lactams against MRSA, an approach that has led to the discovery of two classes of natural product antibiotics, a cyclic depsipeptide (krisynomycin) and a lipoglycopeptide (actinocarbasin), which potentiate the activity of imipenem against MRSA strain COL. We report here that these imipenem synergists are inhibitors of the bacterial type I signal peptidase SpsB, a serine protease that is required for the secretion of proteins that are exported through the Sec and Tat systems. A synthetic derivative of actinocarbasin, M131, synergized with imipenem both in vitro and in vivo with potent efficacy. The in vitro activity of M131 extends to clinical isolates of MRSA but not to a methicillin-sensitive strain. Synergy is restricted to β-lactam antibiotics and is not observed with other antibiotic classes. We propose that the SpsB inhibitors synergize with β-lactams by preventing the signal peptidase-mediated secretion of proteins required for β-lactam resistance. Combinations of SpsB inhibitors and β-lactams may expand the utility of these widely prescribed antibiotics to treat MRSA infections, analogous to β-lactamase inhibitors which restored the utility of this antibiotic class for the treatment of resistant Gram-negative infections.
INTRODUCTION
Staphylococcus aureus is a human pathogen that causes diseases of varied severity ranging from minor skin infections to life-threatening conditions such as bacteremia, endocarditis, necrotizing pneumonia, toxic shock syndrome, and septicemia. The synthetic derivatives of penicillin, such as methicillin, were once the front line agents to treat S. aureus infections, but resistance is now prevalent in hospital and community settings and is global in its distribution (31). There are few treatment options for methicillin-resistant Staphylococcus aureus (MRSA), and the development of new agents remains a priority to ensure the availability of efficacious therapies. We describe in the manuscript an approach to developing combination therapies for MRSA infections through the potentiation of β-lactam antibiotics by inhibiting signal peptidase type I.
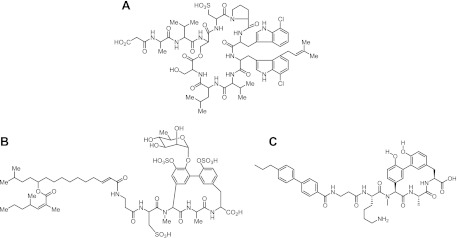
Resistance to β-lactams in MRSA is associated with the acquisition and expression of PBP2A, a member of the penicillin binding protein (PBP) family, which is a group of enzymes responsible for the cross-linking of the peptidoglycan cell wall. Resistance is due to the lower acylation rate of PBP2A by β-lactams compared to other PBPs (12) and requires cooperativity between PBP2A and PBP2, which respectively provide the transpeptidase and glycosyltransferase activities required for peptidoglycan cross-linking (26). Genetic experiments have identified a number of other factors beyond PBP2 that are involved in cell wall biosynthesis and that, when inactivated, restore β-lactam susceptibility to MRSA (2, 9, 20, 34). These genetic findings are confirmed pharmacologically by the observation that antibiotics that target peptidoglycan synthesis pathways also restore β-lactam susceptibility to MRSA (18, 26, 34). We therefore reasoned that a search for compounds which synergize with β-lactams against MRSA would identify novel inhibitors of cell wall biosynthesis and lead to the development of improved agents for combination therapies (16). Natural product screening at Merck led to the discovery of two classes of compounds derived from microbial extracts, a cyclic depsipeptide (krisynomycin) and a lipoglycopeptide (actinocarbasin), which potentiate the activity of carbapenems against MRSA (K. E. Wilson et al., unpublished data). We report here the mechanism of action studies that demonstrate that these two molecules, as well as a synthetic derivative of actinocarbasin, M131, are potent inhibitors of the signal peptidase of Staphylococcus type IB (SpsB) (Fig. 1).
Fig 1.
β-Lactam synergists. Structure of krisynomycin (A), actinocarbasin (B), and M131 (C), a synthetic derivative of actinocarbasin.
SpsB is a conserved essential cell surface protease that cleaves the amino-terminal signal peptide of secreted proteins and whose activity is required for protein secretion through the Sec and Tat protein export systems (24). SpsB has long been considered an attractive antibacterial target, and inhibitors of the enzyme have been described previously in the literature (5, 19). However, the compounds have a rather narrow spectrum of antistaphylococcal activity and relatively low potency against methicillin-resistant Staphylococcus aureus. Here we show that the novel inhibitors of SpsB exhibit intrinsic antibacterial activity against MRSA and significantly synergize with β-lactams (penicillins, cephalosporins, and carbapenems), but not other classes of antibiotics. Furthermore, synergy with imipenem (a carbapenem) was observed in murine models of MRSA infection. We surmise that the robust synergy with β-lactams is due to the obstruction of secreted proteins that are essential for the β-lactam resistance phenotype of MRSA and normally play a role in bacterial cell wall biosynthesis as described above. The β-lactam-sensitizing effects of signal peptidase inhibitors could lead to the development of a combination therapy that would expand the already broad activity spectrum of β-lactam antibiotics to include resistant strains of S. aureus.
MATERIALS AND METHODS
Compounds and medium.
Krisynomycin was isolated from Streptomyces fradiae strain MA7310, and actinocarbasin was isolated from Actinoplanes ferrugineus strain MA7383 (Wilson et al., unpublished). M131 is a synthetic derivative of actinocarbasin that was synthesized by following the approach of Roberts et al. (30; Kevin et al., unpublished data). M131(R,S-lysine) is an early preparation of M131 consisting of a 1:1 mixture of diastereomers at the lysine stereogenic center, of which the single (S,S,S,S) diastereomer (M131) is more active (4-fold). Imipenem was supplied by the Merck compound repository. Other antibiotics were purchased from Sigma and were analytical grade. Cation-adjusted Mueller-Hinton II broth (CAMHB; BBL) was used for all studies unless otherwise stated.
MRSA strains.
MRSA COL was a generous gift from Alexander Tomasz. The MRSA isolates were obtained from various types of infection: bloodstream, skin and soft tissues, pulmonary, and urinary tract. Organisms from studies 1 to 3 (Table 1) are from the Merck culture collection. Studies 2 and 3 include the vancomycin-resistant MRSA strain of Hiramatsu et al. (Mu50) and an epidemic clone of community-acquired MRSA, USA300 (ATCC BAA1556) (10, 15). Eurofins-Medinet collected the set used in study 4 (Table 1), which include organisms isolated from 34 U.S. hospitals (18 states) between 2001 and 2007, with some additional organisms from Japan, France, Greece, and Germany.
Table 1.
Antibacterial activity of SpsB inhibitors and synergy with imipenem in clinical isolates of MRSA
| Study | Compound | No. of organisms | IPM MIC range | MIC range (μg/ml)a with: |
FICI rangeb | |
|---|---|---|---|---|---|---|
| No IPM | 4 μg/ml IPM | |||||
| 1 | Krisynomycin | 24 | 16–≥128 | 16–≥128 | ≤0.06–8 | 0.15–0.26 |
| 2 | Actinocarbasin | 10 | 0.5–128 | 1–8 | 0.25–0.5 | 0.19–0.42 |
| 3 | M131(R,S-lysine) | 10 | 1–128 | 1–2 | ≤0.06–0.13 | 0.27–0.45 |
| 4 | M131 | 59 | 8–≥32 | 0.5–8 | 0.03–1 | NDc |
Range of MICs for MRSA strains resistant to imipenem (IPM) (MICs > 4 μg/ml).
FICI values were calculated from results of checkerboard combination studies .
ND, not determined; checkerboard analysis was not performed.
S. aureus fitness test.
The S. aureus fitness test was performed by following the method of Donald et al. (11). Briefly, the assay consists of 245 strains, derivatives of the methicillin-sensitive strain RN4220, where each strain is individually modified to inducibly express antisense (AS) oligonucleotides specific for a distinct essential gene to reduce expression (11, 16, 25). DNA sequences encoding the antisense RNA were expressed from an expression vector under the control of a xylose-regulatable promoter. Strain sensitivities were tested in a competitive growth assay for approximately 20 population doublings. Quantitative PCR was used to measure the abundances of strains at the end of the experiment, and results of compound-treated cells were compared with mock-treated controls. The assay yielded antisense-induced strain sensitivity profiles.
SpsB overexpression analysis.
The plasmid pTET10::spsB, containing the spsB gene under the control of a tetracycline-regulatable promoter, was introduced into strain MB5393 (MRSA COL) by electroporation to produce the recombinant strain MB6281. The control, strain MB6280, is MRSA COL with the empty pTET10 vector (38). The strains were cultured in the absence or presence of various concentrations of the tetracycline analog anhydrotetracycline (ATc) for 2 to 3 h at 35°C to induce gene expression. MICs of compound M131 were measured in the presence and absence of added ATc by following the broth dilution susceptibility protocol of the Clinical and Laboratory Standards Institute (CLSI) (7).
Preparation of membrane fractions.
Membranes were prepared from MRSA COL (MB5393), MRSA COL pTET10::spsB (MB6281), or MRSA COL pTET10 (MB6280) as follows. Cells from 500-ml overnight cultures were harvested by centrifugation (12,000 × g for 10 min at 4°C) and lysed by treatment with lysostaphin (20 U/ml, 37°C for 1 h), followed by sonication. Intact cells and debris were removed by centrifugation (12,000 × g for 10 min at 4°C), and membranes were collected by further centrifugation (39,000 × g for 75 min at 4°C). Membrane-containing pellets were resuspended in cold sodium phosphate buffer (50 mM, pH 7.5) and stored in aliquots at −80°C. Membranes were used for SpsB enzyme assays and Western blotting as described below.
Western blotting.
Membrane fractions (10 μg/well) of MB6280 and MB6281 were separated by polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes for immunoblotting according to standard methods. Polyclonal anti-SpsB rabbit antiserum (raised against a peptide corresponding to residues 173 to 191 of S. aureus SpsB; prepared by Open Biosystems, Lafayette, CO) was used as the primary antibody, and horseradish peroxidase-labeled goat anti-rabbit immunoglobulin was used as the secondary antibody. SpsB bands were detected using the ECL kit (Amersham).
Signal peptidase I enzymatic assay.
Enzyme activity was measured using a fluorescence-based assay and membrane fraction of MRSA strain COL as the source of the SpsB enzyme (4). The SpsB substrate was a synthetic lipopeptide, decanoyl-k(DABCYL)-TPTAKA↓ASKKD-D(EDANS)-NH2 (prepared by JPT Peptide Technologies, Berlin, Germany). Assays were performed with 10 μM peptide substrate (Km > 10 μM), and reactions were initiated with enzyme addition (final protein concentration of 0.75 mg/ml). SpsB-mediated cleavage of the peptide substrate was detected as an increase in fluorescence (excitation, 340 nm; emission, 460 nM).
β-Lactamase secretion assay.
The whole-cell protein secretion assay measured the generation of type I β-lactamase activity in the culture supernatant. The assay used strain MB6361, a derivative of the methicillin-sensitive strain RN4220 which carries plasmid pM200 encoding type I β-lactamase and a chloramphenicol resistance gene. Strain MB6361 was grown in Miller's LB medium (Invitrogen) containing chloramphenicol (17 μg/ml) at 37°C to an optical density at 595 nm (OD595) of 0.3. Cells were washed twice with fresh medium and then diluted to an OD595 of 0.1. Cell suspension (196 μl) was distributed to wells of a 96-well plate (clear, flat bottom), and then 4 μl compound dilution or vehicle (dimethyl sulfoxide [DMSO[) was added to each well. Cells were incubated for 90 to 120 min until they reached an OD595 of 0.4 to 0.5. OD595 measurements were taken at the beginning of the incubation and prior to assay termination by filtration to determine changes in cellular density. Cultures were filtered using 96-well filtration plates (Millipore MultiScreen, 0.45 um), and the culture filtrate was retained. To determine secreted β-lactamase activity, nitrocefin reagent (0.5 mg/ml in 100 mM phosphate buffer [pH 7.0], 5% DMSO) was added to the culture filtrate (170 μl); absorbance (482 nm) was measured at 0 and 5 min to yield rates of nitrocefin hydrolysis.
Broth susceptibility assays and checkerboard analysis of antibiotic combinations.
The broth dilution method was used to determine MIC values by following the protocol of the CLSI (7). Pairwise combinations of two antimicrobial agents were tested using the checkerboard dilution technique by following the method of Bonapace et al. (3). At least seven concentrations of one agent were tested in combination with seven concentrations of a second test agent to determine fractional inhibitory concentrations (FIC values). The FIC value for a test agent in a particular combination is the MIC of that agent in the combination divided by the MIC of that agent tested alone. Isobologram plots were generated from the two FIC values for each pairwise combination. Synergy is demonstrated when the sum of the two FIC values for each combination (FIC index or FICI) is ≤0.5 and is indicated in the isobologram plots as data points that lie below the dashed diagonal line.
Time-kill assay.
An 18-h culture of MRSA strain COL, grown in tryptic soy broth, was diluted into flasks containing 20 ml CAMHB to achieve a cellular density of 5 × 105 CFU/ml. Cultures were incubated 1 h at 37°C, and then test compounds were added to the exponentially growing cultures. Treated and mock-treated cultures were incubated at 37°C, with shaking (85 rpm). Aliquots were taken at time intervals and plated onto brain heart infusion agar (Difco) to determine colony counts.
In vivo efficacy.
The MRSA COL deep-thigh infection model was performed with female BALB/c mice (Charles River Laboratories, St. Constant, Canada) that were rendered neutropenic with cyclophosphamide intraperitoneal (i.p.) treatment (150 and 100 mg/kg i.p., given 4 days and 1 day prior to infection, respectively; cyclophosphamide was from Baxter Corporation, Toronto, Canada). Mice were then inoculated by intramuscular injection (∼1.6 × 104 CFU/thigh) and, 2 h later, were administered doses of M131, intraperitoneally, with or without concomitant imipenem (10 mg/kg, coadministered with 50 mg/kg cilastatin). Thigh homogenates were prepared 24 h posttreatment to enumerate bacterial counts per gram of tissue. The MRSA COL systemic infection model also used female BALB/c mice that were rendered neutropenic by cyclophosphamide treatment (250 mg/kg, i.p.) 4 days prior to infection. Mice were inoculated by i.p. injection (∼1.0 × 104 CFU/mouse) and 0.5 h later were administered increasing doses of M131 with or without concomitant imipenem (10 mg/kg, coadministered with 50 mg/kg cilastatin). Kidney homogenates were prepared 24 h after initiation of therapy to enumerate bacterial CFU per gram of tissue. In this systemic model, MRSA COL bacterial burden is typically 6.5 to 7 log10 CFU/g kidney at 24 h postinfection. Both infection models involved five mice per study group. Statistical significance was determined by one-way analysis of variance (ANOVA) with Bonferroni posttest.
Resistance analysis.
The agar dilution procedure was used to determine the MIC of M131 in the presence and absence of imipenem by following the CLSI agar dilution protocol (7). To determine frequency of resistance, aliquots of overnight cultures were plated onto selective CAMHB medium containing M131 (various concentrations) in the absence or presence of imipenem (4 μg/ml). Approximately 1.5 × 109 CFU of each culture was plated onto selective medium, and the experiment was performed with three independent cultures. Dilutions of cultures were also plated onto nonselective medium to determine the bacterial counts. Plates were incubated for 48 h at 35°C. Frequencies were determined as the total number of resistant colonies that formed on selective medium divided by the total number of CFU plated.
RESULTS
Discovery of imipenem potentiators.
In order to identify molecules that potentiate the activity of β-lactams against MRSA, a high-throughput screen was conducted to detect molecules, derived from microbial extracts, that synergize with imipenem in an MRSA agar growth assay (Wilson et al., unpublished). This approach yielded two compounds, krisynomycin and actinocarbasin, that were identified from culture extracts of Streptomyces fradiae and Actinoplanes ferrugineus, respectively (Fig. 1A and B). Krisynomycin and actinocarbasin enhance the activity of imipenem against MRSA strain COL at concentrations at which each agent is inactive alone. The MIC of imipenem against MRSA strain COL is 16 to 32 μg/ml. Krisynomycin (at 4 μg/ml) and actinocarbasin (at 0.5 μg/ml) restored susceptibility of MRSA COL to imipenem, where imipenem susceptibility is defined as an MIC value equal to 4 μg/ml. The potentiating concentrations of krisynomycin and actinocarbasin are 16-fold below the native MIC values of 64 and 8 μg/ml, respectively.
Synthetic derivatives of the actinocarbasin structure were generated to improve the microbiological and pharmacological properties of this comparatively large natural compound (Kevin et al., unpublished). Chemical optimization yielded compound M131 (Fig. 1C), which has improved antibacterial potency (MIC = 1 μg/ml). M131 at 0.125 μg/ml restored imipenem susceptibility to MRSA strain COL.
Identification of SpsB as the cellular target.
Structural similarity of actinocarbasin and M131 to previously described lipoglycopeptide and arylomycin inhibitors of signal peptidase type IB, termed SpsB, pointed to this enzyme as the potential target (19, 23). SpsB is a membrane-localized serine protease that cleaves the amino-terminal signal peptide from most secreted proteins (24). However, krisynomycin is a new structural class with no precedent to draw on for the mechanism of action.
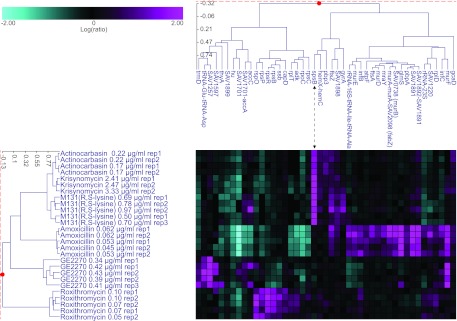
S. aureus fitness test profiling was conducted to identify the cellular target of actinocarbasin, krisynomycin, and M131(R,S-lysine), an early preparation of M131 consisting of a mixture of diastereomers of M131 (see Materials and Methods). In this assay, regulated expression of antisense RNA was used to repress the gene expression. A set of 245 antisense strains corresponding to the majority of the S. aureus essential genes were included in the profiling set. Growth inhibition by a target-specific inhibitor should be exacerbated under conditions where that target's activity is genetically reduced by antisense expression. Growth inhibition was tested in a competitive growth assay and was measured as depletion of a PCR marker for each strain relative to the other strains in the assay. Prior reports validated the fitness test through the profiling of mechanistically diverse antibacterials and by applying the method to determine the targets of novel antibacterial compounds (11, 16, 25). Actinocarbasin, krisynomycin, and M131(R,S-lysine) caused a dose-dependent inhibition of the SpsB antisense strain in the fitness test, ranging from 16- to nearly 200-fold. No other strains were as significantly inhibited, indicating that the activity is specific for the SpsB target. Two-dimensional hierarchal clustering showed that the profiles of actinocarbasin, krisynomycin, and M131(R,S-lysine) cluster tightly together, confirming that they act by the same mechanism (Fig. 2).
Fig 2.
Two-dimensional cluster analysis of S. aureus fitness test profiles. Actinocarbasin, krisynomycin, and M131(R,S-lysine) were compared to three compounds of known mechanism of action, amoxicillin, the translation inhibitor GE2270, and roxithromycin. Antisense strains that are significantly growth inhibited by the compound treatment are shown in magenta, and strain resistances are shown in cyan. The thresholds for including strains in the cluster analysis are a >5-fold reduction in strain abundance and a P value of ≤0.01 in three or more experiments. The profiles of strains that are reduced by actinocarbasin, krisynomycin, or M131(R,S-lysine) treatment cluster tightly together and are characterized by significant depletions of the spsB antisense strain at all compound concentrations tested.
While reduced expression of a drug target can cause hypersensitivity, overexpression may also cause resistance. Indeed, resistance to M131 was observed by increasing the expression of the SpsB enzyme with a tetracycline inducible expression plasmid, pTET10::spsB, relative to the control strain which carried the empty pTET10 vector (38). The plasmid-bearing strains were incubated in the presence of anhydrotetracycline (ATc) to induce gene expression. The M131 MIC of the SpsB overexpression strain (MB6281) is 16-fold higher than the vector control strain (MB6280) in the presence of ≥10 ng/ml ATc (see Fig. S1 in the supplemental material). The reduced susceptibility to M131 correlated with increased SpsB protein levels and activity, as measured by Western blotting and enzyme assays.
Inhibition of SpsB enzyme activity and SpsB-mediated protein secretion.
To directly demonstrate that the compounds inhibit SpsB, we measured their ability to inhibit SpsB enzymatic activity using membrane fractions of MRSA strain COL as the source of the SpsB enzyme and a fluorescent synthetic lipopeptide substrate of SpsB (4). Krisynomycin, actinocarbasin, and M131 were potent inhibitors of SpsB enzymatic activity with 50% inhibitory concentrations (IC50s) of 120, 50, and 10 nM, respectively (Table 2).
Table 2.
Inhibition of SpsB enzymatic activity and β-lactamase secretion by SpsB inhibitors
| Compound | IC50 (nM) |
|
|---|---|---|
| SpsB enzymatic activity | β-Lactamase secretiona | |
| Krisynomycin | 120 | 7 |
| Actinocarbasin | 50 | 3 |
| M131 | 10 | 2 |
Values are on average more than three orders of magnitude lower than IC50s for inhibition of bacterial growth, as determined side by side in the same experiment.
We also assessed the effects of our compounds on SpsB-dependent secretion of β-lactamase type A with a whole-cell assay similar to that described by Kulanthaivel and coworkers (19). The assay used a methicillin-sensitive strain of S. aureus RN4220 expressing a plasmid-borne β-lactamase reporter gene (MB6361). Strain MB6361 was incubated in the presence of the test compound, and then the β-lactamase present in the culture supernatant was detected by hydrolysis of the chromogenic substrate nitrocefin. Bacterial cell density was measured in parallel to differentiate a reduction in β-lactamase production from a reduction in cell viability. Krisynomycin, actinocarbasin, and M131 were potent inhibitors of β-lactamase secretion (IC50s < 10 nM) (Table 2). Inhibition was observed at concentrations that did not impact cellular density. IC50s for secretion inhibition were on average more than three magnitudes lower than IC50s for growth (data not shown).
Synergy with β-lactam antibiotics against MRSA determined by checkerboard analysis.
Initial experiments described above suggested a strong synergy between imipenem and the SpsB inhibitors; that is, the antibacterial effects of the imipenem combinations were significantly greater than expected compared to simply adding up the effects of either of the compounds and imipenem on their own. To determine whether the chemical interaction met the definition of synergy, in vitro studies were performed where krisynomycin, actinocarbasin, and M131(R,S-lysine) were each tested in combination with imipenem against at least 10 MRSA clinical isolates using the checkerboard method (3). Significant synergy was observed with FIC indices below 0.5 against all of the strains (Table 1).
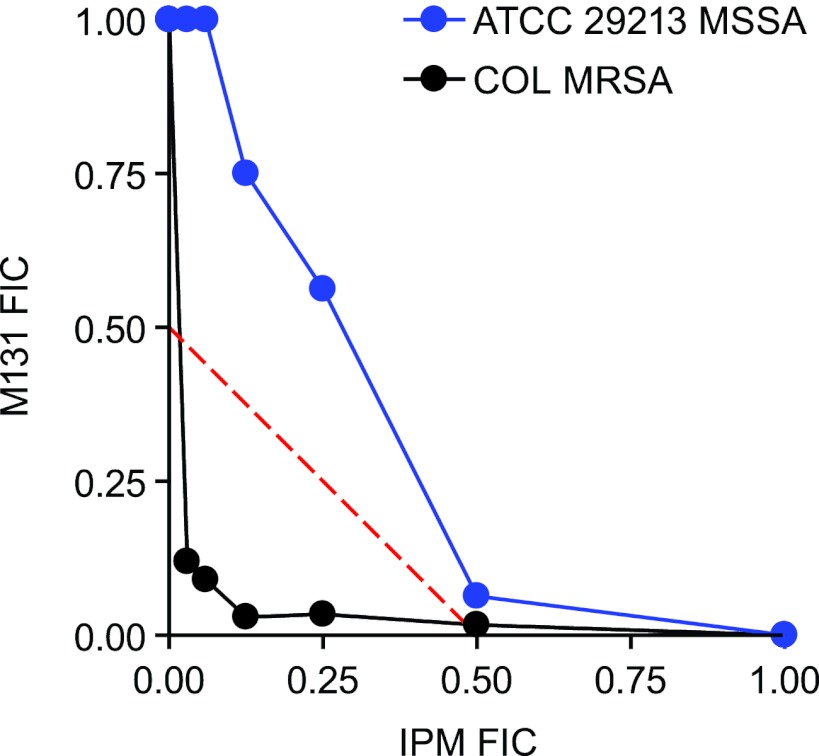
The optimized compound M131 showed synergy with imipenem against MRSA COL (Fig. 3) and was further tested against a set of 59 geographically diverse clinical isolates of MRSA, including vancomycin-intermediate and -resistant S. aureus (VISA and VRSA) strains. MIC values of M131 ranged from 0.5 to 8 μg/ml (Table 1) with an MIC90 value of 4 μg/ml. Addition of sub-MIC imipenem (4 μg/ml) reduced the M131 MIC value of all strains (range, 0.03 to 1 μg/ml; MIC90 = 0.5 μg/ml). The synergy of M131 appeared specific for MRSA: synergy was not observed with the methicillin-susceptible S. aureus (MSSA) strain ATCC 29213, as seen in the isobologram plot (Fig. 3).
Fig 3.
Combinations of imipenem with the SpsB inhibitor M131. Combinations were tested using the checkerboard technique against MRSA strain COL and MSSA strain ATCC 29213. Fractional inhibitory concentrations (FIC) were determined for each combination by dividing the MIC in the presence of the other compound by MIC in its absence. Synergy is achieved when the sum of the FIC values for each agent (FIC index or FICI) is ≤0.5. Data points below the dashed diagonal line have an FICI of ≤0.5, demonstrating synergy.
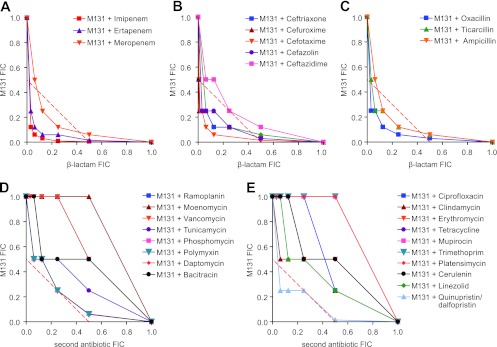
Interestingly, the synergy described above for M131 is specific to the β-lactam class of antibiotics. Synergy (mean FICI ≤ 0.5) was observed with 13/14 of the β-lactams tested, including cephalosporins, penicillins, and two carbapenems beyond imipenem, the sole exception being ceftazidime for which the mean FICI was borderline at 0.56 (Fig. 4A to C). In contrast, 19/20 antibiotics of other classes did not synergize with M131 (FICI > 0.5), including membrane disruptors, inhibitors of nucleic acid and protein synthesis, and non-β-lactam cell wall inhibitors (Fig. 4D and E).
Fig 4.
Combinations of M131 with antibiotics of different classes and mechanisms. Combinations with the following were tested using the checkerboard technique with MRSA strain COL: carbapenems (A), cephalosporins (B), penicillins (C), cell wall inhibitors that are not β-lactams (D), and antibiotics that act by other mechanisms (i.e., inhibition of DNA replication, protein synthesis, RNA synthesis, fatty acid synthesis, folate metabolism, and membrane function) (E).
Synergy with imipenem against MRSA COL determined using the time-kill method.
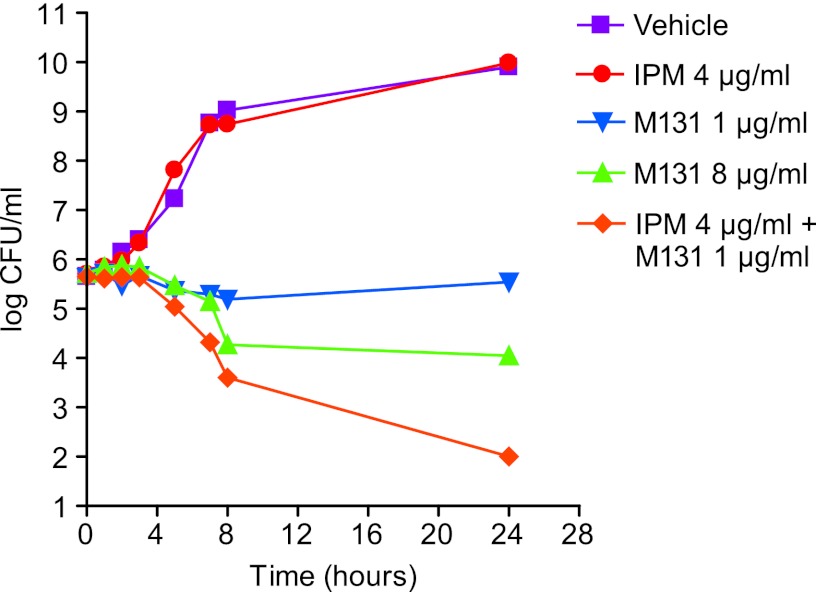
The combination of M131 with imipenem caused a bactericidal effect in vitro against MRSA strain COL (Fig. 5). Imipenem (4 μg/ml; 0.25× MIC) in combination with M131 (1 μg/ml; 1× MIC) caused a 4-log reduction in the viable cell count over the 24-h time course. In contrast, M131, tested alone at 1 μg/ml, did not significantly reduce the cellular viability. M131 at 8 μg/ml (8× MIC) caused only a ∼2-log reduction in viable counts. Similar results were observed with actinocarbasin and krisynomycin (see Fig. S2 in the supplemental material).
Fig 5.
Killing curve analysis of imipenem-M131 combinations. Cultures of MRSA COL were treated with M131 at 1 μg/ml (1× MIC) with or without imipenem (4 μg/ml; 0.25× MIC), and viable bacterial counts (CFU/ml) were monitored over 24 h. M131 was also tested alone at 8 μg/ml (8× MIC).
In vivo efficacy.
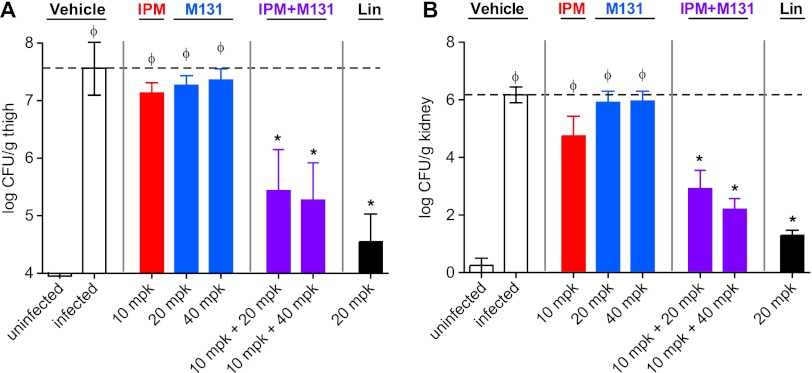
Synergy of M131 with imipenem was observed in two murine models of MRSA infection: a localized deep-tissue infection model and a disseminated infection model where bacterial counts were monitored in the thigh tissue and kidney, respectively. M131 significantly reduced the bacterial burden at 24 h postinfection in both models at 20 and 40 mg/kg doses, but only in the presence of subefficacious doses of imipenem (Fig. 6). Treatment with M131 on its own failed to decrease the bacterial burden in either model, likely due to inadequate exposure relative to the MIC. Indeed, free M131 concentrations in the blood did not reach the MIC level (1 μg/ml) but were well above the MIC observed in the presence of imipenem (0.125 μg/ml) (not shown).
Fig 6.
In vivo activity of M131 combinations with imipenem on MRSA infection. (A) MRSA COL deep-thigh infection model. (B) MRSA COL systemic infection model. Both assays are performed with neutropenic female BALB/c mice. A single intraperitoneal dose of test agents were applied 2 h postinfection for the thigh model and 30 min postinfection for the bacteremia model. Linezolid (Lin) was employed as a positive control and dosed orally (20 mg/kg) at 2 h and 8 h postinoculation. Results are plotted as means of bacterial counts in infected tissue (n = 5); the error bars represent standard deviations. The dashed lines indicate the bacterial counts recovered from vehicle-treated MRSA-infected mice. Statistical significance was determined by one-way ANOVA with Bonferroni posttest: ϕ, P values of <0.05 for comparisons with uninfected mice; *, P values of <0.05 for comparisons with vehicle-treated mice.
Reduced resistance emergence with M131-imipenem combinations.
A low rate of resistance emergence is an important characteristic of all clinically useful antibiotics. The frequency of spontaneous mutants of MRSA COL that are resistant to M131 was measured by plating cultures onto agar medium containing M131 at various concentrations with or without added imipenem (4 μg/ml). The addition of sub-MIC imipenem resulted in a reduced number of resistant M131 mutants at all M131 concentrations (Table 3). With medium containing M131 alone at 4.0 μg/ml (4× MIC, no imipenem), the frequency of resistance emergence was 3 × 10−8 to 4 × 10−8 resistant mutants per CFU. No resistant mutants were detected when the medium contained both M131 (4 μg/ml) and sub-MIC imipenem (4 μg/ml), and the limit of detection was 7 × 10−10 resistant variants per CFU. This finding indicates that combining imipenem with M131 suppresses the emergence of M131 resistance.
Table 3.
Frequency of resistance emergence for M131 alone and in combination with imipenem
| [M131] (μg/ml) | Fold MIC (no IPM) | Fold MIC (combined with 4 μg/ml IPM) | Frequencya |
|
|---|---|---|---|---|
| No IPM | 4 μg/ml IPM | |||
| 0.06 | 1/8 | 1 | NDb | 6.3 × 10−7 |
| 0.13 | 1/4 | 2 | ND | 1.3 × 10−8 |
| 0.25 | 1/2 | 4 | ND | 1.9 × 10−8 |
| 0.5 | 1 | 8 | 1.1 × 10−6 | 1.2 × 10−9 |
| 1.0 | 2 | 16 | 1.8 × 10−7 | <6.7 × 10−10c |
| 2.0 | 4 | 32 | 1.0 × 10−7 | <6.7 × 10−10 |
| 4.0 | 8 | 64 | 3.5 × 10−8 | <6.7 × 10−10 |
| 8.0 | 16 | 128 | 5.6 × 10−8 | <6.7 × 10−10 |
Frequency of resistant variants, the number of colonies on selective medium per CFU plated.
ND, not determined; concentration is lower than effective MIC in the absence of imipenem (IPM).
No resistant colonies were isolated under these conditions (total number of CFU plated, 1.5 × 109 ).
DISCUSSION
There is a growing clinical need for the development of novel therapeutics for the treatment of life-threatening infections involving MRSA. Our approach to this problem has been to identify compounds which rescue the activity of β-lactams, including piperacillins, cephalosporins, and carbapenems, against these normally resistant organisms (16, 34). We hypothesized that compounds interfering with cell wall biosynthesis would potentially sensitize MRSA to β-lactams on the premise that reliance on PBP2A, the inefficient transpeptidase enzyme that evades inhibition by β-lactams, causes the cell wall of β-lactam-treated MRSA to be significantly weakened (28, 36), albeit insufficiently for cell growth to be fully inhibited. We therefore designed a whole-cell synergy assay in which our natural product library was screened in the presence of a carbapenem antibiotic at subinhibitory concentrations (Wilson et al., unpublished). Of the multiple hits identified, we focused on two structurally unrelated natural products, krisynomycin and actinocarbasin, which we determined to be potent inhibitors of the S. aureus type I signal peptidase SpsB. An optimized synthetic analog of actinocarbasin, M131, has potent antibacterial activity against laboratory and clinical strains of MRSA and synergizes with β-lactams. Indeed, synergy was observed with 13/14 of the β-lactams tested, including cephalosporins, penicillins, and carbapenems, the sole exception being ceftazidime. The reason for the lower level of synergy observed with ceftazidime is not clear but could be related to distinct PBP inhibition patterns by different β-lactams which may differentially alter cell wall cross-linking in subtle but significant ways. The combination of M131 with imipenem is bactericidal, has a low frequency of resistance emergence, and is efficacious in vivo in two murine models of infection, indicating that development of such a combination is a valid therapeutic approach for tackling MRSA infections. The synergistic effects of M131 appear to be specific to the β-lactam resistance phenotype of MRSA: synergy is observed only with β-lactam antibiotics, is not observed with most other types of antibacterials, and is not detected with methicillin-sensitive S. aureus. Quinupristin-dalfopristin was the only non-β-lactam combination that demonstrated synergy with the SpsB inhibitor, and other protein synthesis inhibitors did not demonstrate synergy. The synergy of quinupristin-dalfopristin is not easily explained, but it should be noted that this analysis is complicated by the fact that three, rather than two, distinct antibacterial agents are interacting in this case.
The intrinsic antibacterial effects of SpsB inhibitors are to be expected since SpsB-dependent protein secretion is essential for S. aureus viability (8). The synergy between SpsB inhibitors and β-lactams is consistent with the observation that genetic repression of SpsB with antisense RNA expression also potentiates β-lactam activity against MRSA (20). We therefore infer that SpsB function is essential for the β-lactam resistance phenotype of MRSA and that the synergy of SpsB inhibitors with β-lactam antibiotics is a consequence of blocking the proteolysis of SpsB substrates that are required for resistance. While the identities of the SpsB substrates involved in resistance are not known, several recent studies offer potential insight into the mechanism of synergy. Previous genetic studies have identified numerous genes which, when inactivated or repressed, restore susceptibility of MRSA to β-lactam antibiotics. Many of these genes (e.g., femA, femB, murE, pbp2, and fmtA) encode functions involved in the biosynthesis, cross-linking, and turnover of the cell wall peptidoglycan (2, 13, 14, 17, 20, 21). Peptidoglycan hydrolases, enzymes involved in reorganizing the cell wall during cell division, are likely to be secreted via type I signal peptidases, as recently demonstrated for Atl, IsaA, SERP0318 (SACOL0723), and SERP2263 (SACOL2666) (1, 27, 29). In addition to being involved in protein secretion of proteins such as peptidoglycan hydrolases, SpsB may play a role in regulating the enzymatic activity of membrane-bound enzymes that contribute to the β-lactam resistance phenotype of MRSA. For example, Powers and colleagues have reported that the β-lactam response sensor (BlaR1) is a substrate of SpsB in Staphylococcus epidermidis (27). BlaR1 is a β-lactam responsive sensor that, along with the MecR-MecI system, regulates PBP2A and β-lactamase (BlaZ) expression in MRSA in response to β-lactam antibiotics (22). As also reported by this group, and confirmed by Wörmann and coworkers (35), SpsB regulates the activity of lipoteichoic acid synthase (LtaS) through cleavage at a noncanonical site. The LtaS product, lipoteichoic acid, is a key component of the Staphylococcus cell envelope and plays a role in, among other processes, peptidoglycan wall turnover through its positive regulation of autolysis (37). Although a role of SpsB in the biosynthesis of the structurally related wall teichoic acids has thus far not been described, the recent observation by Campbell and coworkers that inhibitors of wall teichoic acid synthesis synergize with β-lactams against MRSA may also be relevant to β-lactam potentiation by SpsB inhibitors (6). Overall, the aforementioned studies are consistent with the notion that the growth of MRSA in the presence of a β-lactam results in an altered and fragile peptidoglycan wall (due to reliance on the inefficient PBP2A) that causes a greater reliance on proteins that are secreted or regulated by SpsB and that directly or indirectly modulate the synthesis and turnover of cell wall peptidoglycan. Furthermore, our data show that interfering with the activities of one or more of these proteins through SpsB inhibition results in potentiation of β-lactam activity in MRSA.
The bacterial type IB signal peptidase enzyme has attracted significant attention as a drug discovery target, as it is a conserved and essential enzyme and its surface localization makes it accessible to inhibitors without the need for cellular penetration (24). Prior drug discovery efforts generated inhibitors with insufficient potency for clinical utility (5, 19, 23). A synthetic derivative of arylomycin A2 has been shown to exhibit potent antibacterial activity against certain coagulase-negative Staphylococcus species in vitro (MIC90 < 2 μg/ml), but weak activity was observed against S. aureus strains including MRSA (MICs, 12 to >128 μg/ml) (30, 32, 33). The M131 compound reported here is the first example of an SpsB inhibitor with significant antibacterial activity both in vitro and in vivo. We propose that SpsB inhibitors could be used in combination with a β-lactam antibiotic to treat severe bacterial infections suspected to involve MRSA. This approach offers significant advantages over monotherapy with an SpsB inhibitor. The first is the ability to harness the broad spectrum activity of the β-lactam antibiotic, an important consideration for severe hospital infections for which broad-spectrum agents are still the first-line treatment of choice in the absence of a definitive diagnostic test for the infecting agent. Furthermore, the reduced frequency of resistance emergence of the combination, compared to the SpsB inhibitor alone, may translate to reduced problems of future resistance emergence in the clinic. Thus, analogous to the use of β-lactamase inhibitors for β-lactam-resistant Gram-negative infections, SpsB inhibitors in combination with β-lactams have the potential to expand the spectrum of antibacterial activity for treatment of severe infections caused by MRSA.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chris Pillar and colleagues at Eurofins-Medinet (Chantilly, VA) for providing the clinical MRSA isolates and for performing susceptibility testing of these organisms. We also thank Ying Cong Zhang and Doris Cully for plasmid pM200.
This work was supported by the Merck Research Laboratories.
Footnotes
Published ahead of print 18 June 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Becher D, et al. 2009. A proteomic view of an important human pathogen—towards the quantification of the entire Staphylococcus aureus proteome. PLoS One 4:e8176 doi:10.1371/journal.pone.0008176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berger-Bächi B, Rohrer S. 2002. Factors influencing methicillin resistance in staphylococci. Arch. Microbiol. 178:165–171 [DOI] [PubMed] [Google Scholar]
- 3. Bonapace CR, White RL, Friedrich LV, Bosso JA. 2000. Evaluation of antibiotic synergy against Acinetobacter baumannii: a comparison with Etest, time-kill, and checkerboard methods. Diagn. Microbiol. Infect. Dis. 38:43–50 [DOI] [PubMed] [Google Scholar]
- 4. Bruton G, et al. 2003. Lipopeptide substrates for SpsB, the Staphylococcus aureus type I signal peptidase: design, conformation and conversion to α-ketoamide inhibitors. Eur. J. Med. Chem. 38:351–356 [DOI] [PubMed] [Google Scholar]
- 5. Buzder-Lantos P, Bockstael K, Anné J, Herdewijn P. 2009. Substrate based peptide aldehyde inhibits bacterial type I signal peptidase. Bioorg. Med. Chem. Lett. 19:2880–2883 [DOI] [PubMed] [Google Scholar]
- 6. Campbell J, et al. 2011. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 6:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. CLSI 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—8th edition. CLSI document M7-A8. CLSI, Wayne, PA [Google Scholar]
- 8. Cregg KM, Wilding I, Black MT. 1996. Molecular cloning and expression of the spsB gene encoding an essential type I signal peptidase from Staphylococcus aureus. J. Bacteriol. 178:5712–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Lencastre H, Tomasz A. 1994. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 38:2590–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diep BA, et al. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731–739 [DOI] [PubMed] [Google Scholar]
- 11. Donald RG, et al. 2009. A Staphylococcus aureus fitness test platform for mechanism-based profiling of antibacterial compounds. Chem. Biol. 16:826–836 [DOI] [PubMed] [Google Scholar]
- 12. Fuda C, Suvorov M, Vakulenko SB, Mobashery S. 2004. The basis for resistance to β-Lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. J. Biol. Chem. 279:40802–40806 [DOI] [PubMed] [Google Scholar]
- 13. Gardete S, et al. 2004. Role of murE in the expression of β-lactam antibiotic resistance in Staphylococcus aureus. J. Bacteriol. 186:1705–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henze U, Sidow T, Wecke J, Labischinski H, Berger-Bächi B. 1993. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J. Bacteriol. 175:1612–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hiramatsu K, et al. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135–136 [DOI] [PubMed] [Google Scholar]
- 16. Huber J, et al. 2009. Chemical genetic identification of peptidoglycan inhibitors potentiating carbapenem activity against methicillin-resistant Staphylococcus aureus. Chem. Biol. 16:837–848 [DOI] [PubMed] [Google Scholar]
- 17. Komatsuzawa H, Ohta K, Labischinski H, Sugai M, Suginaka H. 1999. Characterization of fmtA, a gene that modulates the expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 43:2121–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Komatsuzawa H, Suzuki J, Sugai M, Miyake Y, Suginaka H. 1994. Effect of combination of oxacillin and non-β-lactam antibiotics on methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 33:1155–1163 [DOI] [PubMed] [Google Scholar]
- 19. Kulanthaivel P, et al. 2004. Novel lipoglycopeptides as inhibitors of bacterial signal peptidase I. J. Biol. Chem. 279:36250–36258 [DOI] [PubMed] [Google Scholar]
- 20. Lee SH, et al. 2011. Antagonism of chemical genetic interaction networks re-sensitize methicillin resistant Staphylococcus aureus to β-lactam antibiotics. Chem. Biol. 18:1379–1389 [DOI] [PubMed] [Google Scholar]
- 21. Maidhof H, Reinicke B, Blumel P, Berger-Bächi B, Labischinski H. 1991. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Bacteriol. 173:3507–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McKinney TK, Sharma VK, Craig WA, Archer GL. 2001. Transcription of the gene mediating methicillin resistance in Staphylococcus aureus (mecA) is corepressed but not coinduced by cognate mecA and β-lactamase regulators. J. Bacteriol. 183:6862–6868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paetzel M, Goodall JJ, Kania M, Dalbey RE, Page MGP. 2004. Crystallographic and biophysical analysis of a bacterial signal peptidase in complex with a lipopeptide-based Inhibitor. J. Biol. Chem. 279:30781–30790 [DOI] [PubMed] [Google Scholar]
- 24. Paetzel M, Karla A, Strynadka NCJ, Dalbey RE. 2002. Signal Peptidases. Chem. Rev. 102:4549–4580 [DOI] [PubMed] [Google Scholar]
- 25. Phillips John W, et al. 2011. Discovery of kibdelomycin, a potent new class of bacterial type II topoisomerase inhibitor by chemical-genetic profiling in Staphylococcus aureus. Chem. Biol. 18:955–965 [DOI] [PubMed] [Google Scholar]
- 26. Pinho MG, de Lencastre H, Tomasz A. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. U. S. A. 98:10886–10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Powers ME, et al. 2011. Type I signal peptidase and protein secretion in Staphylococcus epidermidis. J. Bacteriol. 193:340–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qoronfleh MW, Wilkinson BJ. 1986. Effects of growth of methicillin-resistant and -susceptible Staphylococcus aureus in the presence of beta-lactams on peptidoglycan structure and susceptibility to lytic enzymes. Antimicrob. Agents Chemother. 29:250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ravipaty S, Reilly JP. 2010. Comprehensive characterization of methicillin-resistant Staphylococcus aureus subsp. aureus COL secretome by two-dimensional liquid chromatography and mass spectrometry. Mol. Cell. Proteomics 9:1898–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roberts TC, Smith PA, Cirz RT, Romesberg FE. 2007. Structural and initial biological analysis of synthetic arylomycin A2. J. Am. Chem. Soc. 129:15830–15838 [DOI] [PubMed] [Google Scholar]
- 31. Rosenthal VD, et al. 2010. International Nosocomial Infection Control Consortium (INICC) report, data summary for 2003–2008, issued June 2009. American J. Infect. Control 38:95–104 [DOI] [PubMed] [Google Scholar]
- 32. Smith PA, Powers ME, Roberts TC, Romesberg FE. 2011. In vitro activities of arylomycin natural-product antibiotics against Staphylococcus epidermidis and other coagulase-negative Staphylococci. Antimicrob. Agents Chemother. 55:1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith PA, Roberts TC, Romesberg FE. 2010. Broad-spectrum antibiotic activity of the arylomycin natural products is masked by natural target mutations. Chem. Biol. 17:1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan CM, et al. 2012. Restoring methicillin-resistant Staphylococcus aureus susceptibility to β-lactam antibiotics. Sci. Transl. Med. 4:126ra35. [DOI] [PubMed] [Google Scholar]
- 35. Wörmann ME, Reichmann NT, Malone CL, Horswill AR, Gründling A. 2011. Proteolytic cleavage inactivates the Staphylococcus aureus lipoteichoic acid synthase. J. Bacteriol. 193:5279–5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wyke AW, Ward JB, Hayes MV. 1982. Synthesis of peptidoglycan in vivo in methicillin-resistant Staphylococcus aureus. Eur. J. Biochem. 127:553–558 [DOI] [PubMed] [Google Scholar]
- 37. Xia G, Kohler T, Peschel A. 2010. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int. J. Med. Microbiol. 300:148–154 [DOI] [PubMed] [Google Scholar]
- 38. Xu HH, et al. 2010. Staphylococcus aureus TargetArray: comprehensive differential essential gene expression as a mechanistic tool to profile antibacterials. Antimicrob. Agents Chemother. 54:3659–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.