Abstract
Homeodomain-only protein X (HOPX)-β promoter methylation was recently shown to be frequent in human cancers and was suggested as tumor suppressor gene in esophageal and gastric cancer. The aim of this study was to investigate the mechanistic roles of HOPX-β promoter methylation and its clinical relevance in colorectal cancer (CRC). HOPX-β promoter methylation was assessed in human CRC cell lines and 294 CRC tissues. HOPX mRNA and protein levels were measured in relation to HOPX-β promoter methylation. The effects of forced HOPX expression on tumorigenesis were studied using in vitro and in vivo assays. The association between HOPX-β promoter methylation and clinical relevance of CRC patients was determined. HOPX-β promoter methylation is cancer-specific and frequently found in CRC cell lines and tissues, resulting in the down-regulation of HOPX mRNA and protein levels. In CRC cell lines, forced expression of HOPX suppressed proliferation, invasion, and anchorage-independent growth. DNA microarray analyses suggested critical downstream genes that are associated with cancer cell proliferation, invasion or angiogenesis. In a mouse xenograft model, HOPX inhibited tumorigenesis and angiogenesis. Finally, HOPX-β promoter methylation was associated with worse prognosis of stage III CRC patients (hazard ratio= 1.40, P = .035) and also with poor differentiation (P = .014). In conclusion, HOPX-β promoter methylation is a frequent and cancer-specific event in CRC progression. This epigenetic alteration may have clinical ramifications in the diagnosis and treatment of CRC patients.
Introduction
Colorectal cancer (CRC) is the second most prevalent cancer and the fourth leading cause of cancer death worldwide [1]. Although CRC screening, that is, fecal occult blood test and colonoscopy, has contributed to a reduction in mortality [2], 70% of newly discovered CRC patients are still detected at an advanced stage, resulting in poor prognosis [3]. Recently, sophisticated scientific technologies have emerged, such as DNA microarray, pharmacological unmasking epigenetic profiles, and proteomic techniques, advancing the discovery of novel biomarkers for CRC [4].
Alterations in DNA methylation, an epigenetic process present in mammalian cells, are one of the hallmarks of cancer [5]. The promoter regions of many genes, particularly housekeeping genes, are populated by CpG dinucleotides, which are often underrepresented in the remainder of the genome. These regions have been termed “CpG islands” because they are protected from methylation in normal cells [6]. This protection is critical because methylation of CpG islands is associated with loss of expression of that particular gene. In carcinogenesis, global hypomethylation [6,7] is often accompanied by dense hypermethylation of specific promoters [5]. Many studies have demonstrated that silencing of tumor suppressor genes (TSGs) is associated with promoter hypermethylation in human cancers and serves as an alternative mechanism for loss of function. However, cancer-specific methylation is rather a rare event and is usually evident only in genes with a robust tumor suppressive function.
We previously developed pharmacologic reversal of epigenetic silencing and uncovered a myriad of transcriptionally repressed genes in human cancers [8]. Using this technique, we have identified several novel TSG candidates, such as PGP9.5 [9], NMDAR2A/B [10,11], and NEFH [12]. Among these candidates, homeodomain-only protein X (HOPX) was of particular interest because its methylation pattern was found to be cancer-specific [8]. Indeed, HOPX-β methylation was correlated with tumorigenesis and worse prognosis in gastric and esophageal cancers [13,14]. In the present study, we investigated the importance of HOPX-β promoter methylation in CRC. We assessed the methylation status of HOPX-β promoter in CRC cell lines and tissues and looked for its functional role and clinical relevance.
Materials and Methods
Cell Lines
CRC cell lines (HCT15, DLD1, COLO205, and LoVo) and an esophageal squamous cell carcinoma cell line (TE15) were kindly provided by the Cell Response Center for Biomedical Research Institute of Development, Aging and Cancer, Tohoku University (Sendai, Japan). HCT116 and SW480 were purchased from American Type Culture Collection (Manassas, VA). Other CRC cell lines (COLO320, CW2, and CACO2) were obtained from RIKEN BioResource Center (Tsukuba, Japan). We previously reported that CpG islands of HOPX-β promoter are not methylated in TE15. TE15 served as negative control for HOPX-β promoter methylation [13,14]. HCT15, DLD1, COLO205, COLO320, CW2, CACO2, and TE15 were grown in RPMI-1640 medium (GIBCO, Rockville, MD), supplemented with 10% fetal bovine serum (FBS). LoVo, SW480, and HCT116 were grown in Ham F-12, Leibovitz L15, and McCoy 5A mediums, respectively (GIBCO), all supplemented with 10% FBS.
Mouse Xenograft Assay
Twenty male BALB/cAJcl nude mice, 7 weeks old, obtained from CLEA Japan Inc (Tokyo, Japan), were divided into four groups (n = 5 in each). Mice were injected with 1 x 106 cells on the left flank with either HCT116 or DLD1 cells transfected with a mock vector as control; and on the right flank, with cells transfected with a HOPX-expressing vector. Tumor volumes were calculated from caliper measurements of two orthogonal diameters (larger (x) and smaller (y) diameters) by using the formula: volume = xy2/2. All animal experiments were performed in strict accordance with the guidelines for animal experiments of the Kitasato University School of Medicine.
Human CRC Tissues
Paraffin-embedded primary CRC tissues and paired normal mucosa specimens were obtained from 99 patients diagnosed with stage I to IV CRC (Table W1), and 146 patients diagnosed with stage III CRC (Table W2), used for the quantitative methylation-specific polymerase chain reaction (Q-MSP). An independent set of 22 samples was subjected to immunohistochemistry assays. Snap-frozen tissues were taken from 27 patients diagnosed with stage I–IV CRC, used for the Q-MSP assay coupled with reverse transcription.polymerase chain reaction (RT-PCR). All specimens were taken during either potentially curative resection (stages I–III patients), or palliative resection (stage IV patients). TNM classification was made according to the UICC (Unio Internationalis Contra Cancrum) staging system. All individuals gave written informed consent for pathologic assessment and molecular testing on their samples. This study was performed with approval of the Ethics Committee of the Kitasato University School of Medicine.
Bisulfite Treatment of DNA and Sequencing Analysis
Tissue sections from tumor and corresponding normal mucosa were stained with hematoxylin and eosin and dissected under microscope. Genomic DNA was subsequently extracted using QIAamp DNA FFPE Kit (Qiagen Sciences, Hilden, Germany). Genomic DNA from cell lines was extracted using QIAamp DNA Mini Kit (Qiagen). Bisulfite treatment was carried out with EpiTect Bisulfite Kit (Qiagen) and was subsequently amplified by PCR. Primer sequences for the HOPX-β promoter region were designed to recognize DNA alterations caused by bisulfite treatment (Table W3). The PCR products were either directly sequenced (Figure 1B) or purified with QIAquick Spin (Qiagen) and inserted into a pCR4-TOPO vector using the TOPO TA Cloning Kit for Sequencing (Invitrogen, Carlsbad, CA; Figure 1D).
Figure 1.
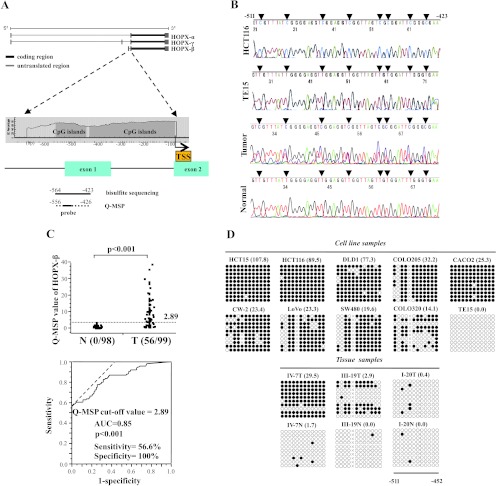
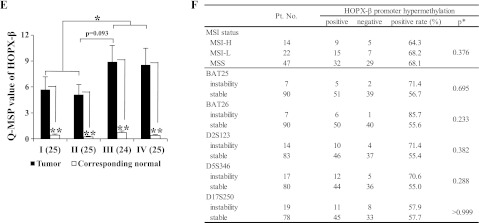
Analysis of HOPX-β promoter methylation and expression in CRC. (A) Schematic diagram of the three spliced transcript variants of HOPX (top), and of CpG islands (gray area) in the 5′-flanking region of HOPX-β (bottom). Solid and dotted lines below the bottom panel indicate the sequences for bisulfite sequencing analysis or TaqMan-MSP (Q-MSP), respectively. The length of the 5′ untranslated region for transcript variants, HOPX-α, β, and γ is 464, 191, and 535 bp of exons, respectively. TSS, translation start site. (B) Representative results of bisulfite sequencing of CRC cell lines, HCT116, TE15 (esophageal cancer cell line, negative control), primary CRC, and its corresponding normal tissue. Arrowhead indicates dinucleotide CpG. N indicates corresponding normal tissue; T, primary tumor. (C) Top: Scatterplots of TaqMan Q-MSP values of HOPX-β in primary CRC and their corresponding normal tissues. Dashed line indicates the optimal cutoff value (2.89). Bottom: ROC curve of HOPX-β promoter methylation for distinguishing between malignant and normal samples. Area under the curve (AUC) is 0.85 and represents the accuracy in distinguishing CRC from normal samples in terms of sensitivity and specificity (P < .001). (D) Methylation status of 12 individual CpG sites (columns) of 10 cloned PCR products (rows), within the HOPX-β promoter region by bisulfite sequencing. Open and filled circles, unmethylated and methylated CpG sites, respectively; x, deletion. Numbers in parenthesis denote Q-MSP value. Results shown for cell lines (top) or tissue samples (bottom). TE15, esophageal cancer cell line served as negative control. (E) Q-MSP values of HOPX-β in each stage (n = 99). Number in parenthesis indicates patient number. Data are expressed as mean ± SEM. *P < .01. **P < .001. (F) Correlation analysis between MSI status and promoter hypermethylation of HOPX-β. H indicates high; L, low; MSS, microsatellite stable. *χ2 test or Fisher exact test where appropriate.
Quantitative Methylation-Specific PCR
For quantitative methylation analysis, TaqMan Q-MSP was carried out using iQ SuperMix (Bio-Rad Laboratories, Hercules, CA) as reported [11]. All reactions were performed in triplicates. Primer sequences and PCR conditions are described in Table W3. The most frequent location of restriction CpG sites for translation has been reported to be around 500 bp upstream of translation start site [11]. Accordingly, we chose the segment for Q-MSP as in our previous report, which regarded HOPX-β methylation in gastric cancer and showed the correlation between HOPX-β methylation and its expression [14]. In addition, because we used FFPE sections, the primers were set for ∼150-bp segments to keep the quality of PCR products. Although the quality was lower, when we analyzed a ∼300-bp segment, results from available samples were similar to those of the 142-bp segment presented in the article (data not shown). CpGenome universal methylated DNA and unmethylated DNA (Chemicon International, Temecula, CA) served as positive and negative controls, respectively. The methylation value was defined as the ratio of fluorescence intensity emitted from the HOPX-β promoter divided by that of β-actin and multiplied by 100 (Q-MSP value). Receiver operating characteristic (ROC) curve was used to determine the optimal cutoff value for Q-MSP.
Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted from the homogenized tissues and harvested cells using the RNeasy Mini Kit (Qiagen) and reverse-transcribed with SuperScript III reverse transcriptase kit (Invitrogen). RT-PCR was carried out for the three spliced variants and for the common transcript core (HOPX-core) of HOPX and normalized to β-actin. Quantitative realtime RT-PCR (qRT-PCR) was performed using SYBR Premix Ex Taq (Takara Bio, Inc, Shiga, Japan) as previously described [15]. Primers' sequences are described in Table W3.
Western Blot Analysis
Total protein was extracted from DLD1 and HCT116 cells and subjected to Western blot analysis using the following antibodies: mouse HOPX monoclonal IgG1κ antibody (3D6; Sigma-Aldrich, Inc, St Louis, MO), mouse V5 IgG2a monoclonal antibody (Invitrogen), or mouse β-actin IgG2a monoclonal antibody (Sigma-Aldrich).
5-Aza-dC and TSA Treatment
Cells were split to low density (1 x 106/T-75 flask) 12 to 24 hours before treatment. Cells were then treated every 24 hours for 4 days with either 1 or 5 µM 5-aza-2′-deoxycytidine (5-Aza-dC; Sigma-Aldrich) dissolved in 50% acetic acid or were mock treated with PBS including the same amount of acetic acid. As indicated, 300 nM of trichostatin A (TSA; Sigma-Aldrich) was added to the medium for the final 24 hours [13].
Immunohistochemistry
For immunostaining, antigen unmasking was performed with Protease K (DakoCytomation, Glostrup, Denmark) or microwave with pressure cooker, endogenous peroxidase activity was blocked by incubation in 3% H2O2/methanol for 5 minutes, and nonspecific antibody binding was blocked by incubation with 1% diluted normal horse serum for 30 minutes. Sections were then incubated at 4°C overnight with the following antibodies: mouse HOPX monoclonal antibody (3D6), rat CD31 monoclonal antibody, rat EphA2 monoclonal antibody, or rabbit CYR61 polyclonal antibody (Abcam, Cambridge, United Kingdom). Immune complexes were detected with a Vectastain Elite ABC kit (Vector Laboratories, Inc, Burlingame, CA) according to the manufacturer's instruction. These immune complexes were detected using the 3,3′-diaminobenzidine substrate with/without nickel ammonium sulfate (Vector) as a chromogen. Sections were counterstained with hematoxylin.
Immunofluorescence
Cells were fixed with 10% formaldehyde at 4°C for 30 minutes. Nonspecific antibody binding was blocked by incubation with 1% diluted normal horse serum/1% Triton X for 30 minutes. The samples were incubated with Alexa Fluor 568-conjugated phalloidin (Molecular Probes, Inc, Eugene, OR) for 1 hour at room temperature. Then, the samples were incubated with 4′,6-diamidino-2-phenylindole (Molecular Probes, Inc) for 5 minutes at room temperature. Samples were analyzed using a confocal scanning laser microscope (LSM710; Carl Zeiss MicroImaging GmbH, Oberkochen, Germany). Serial optical sections (collected at 1-µm intervals) in the z-axis were collected and overlaid using ZEN-2008 software (Carl Zeiss MicroImaging).
Microvessel Density and Microvessel Area
Microvessel density (MVD) and microvessel area (MVA) in the stroma were assessed as a parameter of tumor-associated angiogenesis, according to the established methods described previously [15]. Areas of highest neovascularization were identified by scanning the stroma around the tumor at low power (40-fold and 100-fold magnification). After identifying the most vascularized area, individual microvessels were counted on a 400-fold magnification field. MVD was expressed in terms of the number of vessels per unit area (mm2). MVA was calculated using the ImageJ software (National Institutes of Health Research Service Branch, Bethesda, MD) and was expressed as the area of vessels per observed area (mm2/mm2). MVD and MVA were calculated in four different sections, four fields in each.
Stable Transfections
A full-length complementary DNA (cDNA) of HOPX was isolated from TE15 cells using RT-PCR with the following primers (5′ to 3′): forward, caccatgtcggcggagaccgcgagcgg; reverse, gtctgtgacggatctgcactctg. Full-length cDNA of c-Fos (ORH01934) was purchased from Kazusa DNA Research Institute (Chiba, Japan) and amplified with the following primers (5′ to 3′): forward, cccaagcttcaccatgatgttctcgggcttc; reverse, gctctagatcacagggccagcagcgtgggtg. The insert of HOPX was subcloned into pcDNA3.1D/V5-His-TOPO vector (pcDNA 3.1-HOPX) using a pcDNA3.1 Directional TOPO expression kit (Invitrogen). The restricted PCR products of c-Fos were ligated into pcDNA3.1D/V5-His-TOPO vector within the HindIII-XbaI sites (pcDNA 3.1-c-Fos). The sequences of the subcloned cDNA were verified by sequencing analyses. Mock vector with self-ligation (pcDNA3.1-mock) was used as a control. Cells were transfected using Lipofectamine 2000 (Invitrogen) in OPTI-MEM medium (Invitrogen) according to the manufacturer's instructions. Stable clones with HOPX or mock were established by G418 (GIBCO) selection. The expression of each gene was confirmed by RT-PCR and Western blot analysis.
Proliferation Assay
Cell proliferation and viability (2 x 103 cells/well) were measured using the Premix WST-1 Cell Proliferation Assay System (Takara Bio, Tokyo, Japan) in 96-well plates. Data are expressed as an absorbance at 450 nm. Experiments were performed in triplicates.
Invasion Assay
Cells were seeded at a density of 1 x 106 per well in the 24-well BD BioCoat Matrigel Invasion Chamber (BD Biosciences Discovery Labware, Bedford, MA). Serum 10% was used as a chemoattractant. After incubation for 22 hours, the membrane of the upper chamber was fixed and stained by Diff-Quik reagent (Sysmex, Kobe, Japan). Invaded cells were counted in four randomly selected sites per membrane. Simultaneously, an equal number of cells were seeded on 24-well plates and incubated for 22 hours, and WST assay was performed.
Anchorage-Independent Colony Formation Assay
Anchorage-independent cell growth was analyzed by plating 0.36% top agarose (Bacto Agar; Becton Dickinson and Company, Franklin Lakes, NJ) containing 1 x 105 cells on the surface of 0.72% bottom agarose in six-well plates. Cells were fed weekly by overlying fresh soft agar solution containing G418. Colonies were photographed after 3 weeks of incubation, visualized by ethidium bromide staining. Two independent experiments were performed, each in triplicate.
Cell Cycle Assay
Cells (1 x 106 cells/ml) were starved in the appropriate medium supplemented with 0.2% FBS for 48 hours and were then transferred to 10% FBS-containing medium for 24 hours for cell cycle transition. Cells were then fixed in 75% ethanol and stained with propidium iodide (Guava cell cycle reagent; Guava Technologies, Hayward, CA). Cell cycle assay was carried out using the Guava PCA System. The experiment was performed in duplicate and analyzed using CytoSoft 2.1.5 software (Guava Technologies).
Caspase 3 Assay
Caspase 3 activity was measured with CaspaseGlo 3/7 Assay (Promega, Madison, WI) according to the manufacturer's recommendations. The assay was carried out with 50 µg of cellular extracts in triplicate.
TUNEL Assay
Detection of free 3′-OH was done with the DeadEnd Colorimetric TUNEL System (Promega) according to the manufacturer's protocol. Sections were counterstained with hematoxylin. Apoptotic cells were counted in four different sections, four fields in each. Necrotic areas were excluded.
Microarray Processing
Microarray experiments were performed using Affymetrix Human U133 Plus 2.0 Arrays (Affymetrix, Santa Clara, CA), according to the manufacturer's instructions. Briefly, total RNA was used to prepare a biotinylated target complementary RNA (cRNA) according to the manufacturer's recommendation (Affymetrix). Two hundred nanograms of mRNA was used to generate first-strand cDNA using T7-linked oligo(dT) primer. After second-strand synthesis, the cDNA was subjected to in vitro transcription using an IVT labeling kit (Affymetrix). Quantitative analyses of the isolated total RNA and synthesized cRNA were conducted by electropherogram (Experion; Bio-Rad Laboratories). The biotinylated cRNA was fragmented and hybridized for 16 hours at 45°C with the array, which contains the oligonucleotide probe sets for 54,675 full-length transcripts and expressed sequence tags. The arrays were stained with streptavidin-phycoerythrin and scanned using an Affymetrix Model Fluidics Station 450 and GeneChip Scanner 3000 (Affymetrix). The fluorescence intensity of each probe was quantified using the GeneChip operating software, GCOS version 1.4 (Affymetrix). Each microarray was subjected to a standard quality control evaluation; the percentage of probe sets reliably detected (present flag) in each array was between 41% and 45%, and the 3′/5′ ratio of the GAPDH was less than 1.25. All background intensities and noise factors were within the acceptable range of 40.10 to 48.40 and 1.20 to 1.70, respectively.
Microarray Data Analysis
We compared expression levels of genes in HCT116 or DLD1 cells, transfected with either a mock vector or a HOPX-expressing vector. To define genes regulated by HOPX, we first floored to 50 all values less than 50. Upregulated or downregulated genes were defined as genes whose expression was changed by at least two-fold and also defined as “present” in the upregulated or downregulated sample, respectively. To select for more valid genes, we present genes up- or downregulated in both HCT116 and DLD1 cells by at least two-fold (516 and 77 genes, Tables W4 and W5, respectively) or by at least four-fold (2 and 5 genes, respectively; Table 1). All microarray data are MIAME-compliant, and these data have been deposited in CIBEX at DDBJ (Japan) (CBX253).
Table 1.
Genes Differentially Expressed after HOPX Transfection.
| Accession No. | Gene Symbol | Description | GeneChip U133 (Fold Change) | qRT-PCR (Fold Change) | ||
| HCT116 | DLD1 | HCT116 | DLD1 | |||
| Upregulated genes | ||||||
| NM_005809 | PRDX2 | Peroxiredoxin 2 | 4.50 | 4.61 | 1.62 | 1.67 |
| NM_004906 | WTAP | Wilms tumor 1 associated protein | 5.47 | 4.40 | 5.79 | 2.75 |
| Downregulated genes | ||||||
| NM_001554 | CYR61 | Cysteine-rich, angiogenic inducer | -5.56 | -33.33 | -1.56 | -2.70 |
| NM_001423 | EMP1 | Epithelial membrane protein 1 | -7.69 | -20.00 | -2.50 | -2.70 |
| NM_004431 | EPHA2 | Ephrin receptor EphA2 | -4.55 | -5.26 | -1.89 | -1.82 |
| NM_005252 | FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | -11.11 | -33.33 | -33.33 | -50.00 |
| NM_006931 | SLC2A3 | Solute carrier family 2, member 3 | -6.67 | -33.33 | -1.56 | -4.00 |
Microsatellite Instability Assay
DNA was extracted from 98 pairs of FFPE tissue samples as mentioned above. Finally, 97 pairs of samples were analyzed successfully. Microsatellite assay was performed using Bethesda consensus panel (BAT25, BAT26, D2S123, D5S346, and D17S250) [16] by Aoba Genetics, Inc (Yokohama, Japan), as in a previous report [17]. The presence of extra or shifted bands was classified as microsatellite instability (MSI).
Statistical Analysis
χ2 test or Fisher exact test was used for categorical variables, and Student's t test was used for continuous variables. Data are expressed as mean ± SEM. Clinicopathologic characteristics and follow-up data were analyzed in association with 5-year disease-specific survival (DSS). The follow-up time was calculated from the date of surgery. DSS was estimated with Kaplan-Meier method and compared using the log-rank test. A multivariate proportional hazard model was built using the variables that had prognostic potential in the univariate analysis (P < .1). Fisher exact tests and multivariate logistic regression analysis were performed for correlation analysis of HOPX-β promoter methylation status with clinicopathologic parameters. P < .05 was considered significant.
Results
HOPX-β Promoter Structure
The location of the CpG islands in the HOPX promoter genomic sequence is shown in Figure 1A. HOPX has three transcript variants (HOPX-α [GenBank accession number NM_139212.2], HOPX-β [NM_139211.2], and HOPX-γ [NM_032495.4]), of which only HOPX-β promoter harbors CpG islands. Importantly, all three spliced variants share the same open reading frame.
HOPX-β Promoter Methylation Is Frequent and Cancer-Specific in CRC Cell Lines and Human Tissues
We first examined the HOPX-β promoter methylation using bisulfite sequencing in nine CRC cell lines (DLD1, HCT116, HCT15, COLO205, LoVo, SW480, COLO320, CW-2, and CACO2). All nine CRC cell lines harbored HOPX-β promoter methylation (representative results shown in Figure 1B, top).
We next examined the HOPX-β promoter methylation status in human CRC tissues. Using bisulfite sequencing, we detected HOPX-β promoter methylation in 62.5% of the primary CRCs examined (10/16), as opposed to none (0/16) in the corresponding normal mucosa (representative results shown in Figure 1B, bottom). We then performed Q-MSP analysis of 99 primary human CRC samples and 98 corresponding normal mucosa (normal mucosa was not available in one patient). The methylation value was significantly higher in primary CRC (7.04 ± 0.83) than in the corresponding normal tissues (0.43 ± 0.06, P < .001; Figure 1C, top, and E). The optimal cutoff value for distinguishing between malignant and normal tissues was calculated using a ROC analysis (Figure 1C, bottom). No normal tissue had a Q-MSP value higher than 2.89, yielding 100% specificity, whereas 56.6% (56/99) of primary CRC tissues had a value equal to 2.89 or higher. To assess the correlation between Q-MSP values and methylation status of individual CpG islands, we carried out the bisulfite sequencing with cloned PCR products. All cell lines or human samples with Q-MSP values higher than 2.89 showed denser methylation (Figure 1D). To evaluate the correlation between Q-MSP values and different clinical stages, we calculated Q-MSP values for each stage using the same 99 patient cohort. CRC samples from each stage had an averageQ-MSP value higher than 2.89, which was significantly higher than that of their normal counterparts (Figure 1E). When comparing all pairs of stages, we could not detect a significant difference but could detect a trend toward a higher value of patients with stage III compared with that of stage II. Nevertheless, advanced stages (III-IV) displayed significantly higher Q-MSP values than those of early stages (I-II).
Because CpG island methylator phenotype is thought to be associated with MSI [18], we analyzed MSI status in 98 paired tumor and normal samples of the same patient cohort. Of which, we could successfully assayed MSI status in 97 pairs of samples. No correlation was found between HOPX-β promoter methylation and MSI status (Figure 1F).
HOPX-β Promoter Methylation Results in Transcriptional Silencing in CRC Cell Lines and Human Tissues
To investigate whether HOPX-β methylation results in transcriptional silencing, we performed RT-PCR of the HOPX-core (common to all the variants) and of the α, β, and γ variants, separately. HOPX-β mRNA expression was undetectable in both DLD1 and HCT116 cell lines (Figure 2A). Notably, DLD1 expressed low detectable levels of the HOPX-core mRNA, in contrast to undetectable levels in HCT116. The weak expression of HOPX-core mRNA in DLD1 was due to HOPX-α mRNA expression. Nevertheless, Western blot analysis showed that HOPX protein was undetectable in both cell lines (Figure 2B). To further substantiate the hypothesis that HOPX-β silencing is due to its promoter hypermethylation, DLD1 and HCT116 were treated with the demethylating agent 5-Aza-dC alone or in combination with histone deacetylase inhibitor TSA (Figure 2C). HOPX-β and core expression, but not HOPX-α, or γ, were restored by demethylation with or without deacetylation. We next examined mRNA levels of HOPX variants and core in human CRC frozen tissue samples. As expected from the cancer-specific methylation of HOPX-β promoter, lower levels of HOPX-β mRNA were detected in CRC samples compared with their normal counterparts (Figure 2D).
Figure 2.
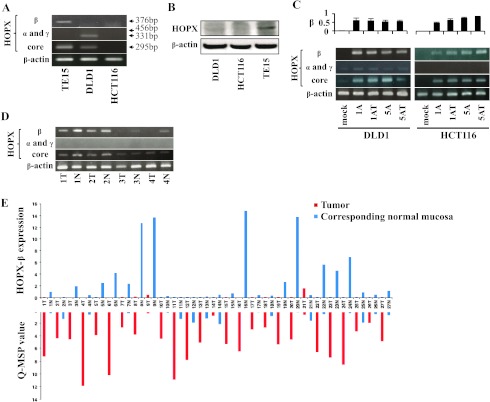
(A) RT-PCR analysis of mRNA expression of HOPX-β, α, and γ and core in DLD1 and HCT116 cell lines. TE15 served as a positive control of HOPX-β and core expression. (B) Western blot analysis of HOPX using whole-cell lysates of DLD1 and HCT116 cell lines. TE15 served as positive control of HOPX protein expression. (C) RT-PCR analysis of mRNA expression of HOPX-β, α, and γ and core in DLD1 and HCT116 after treatment with a demethylating agent, 5-Aza-dC, in the presence or absence of TSA, a histone deacetylase inhibitor. Upper: Densitometric quantification of the bands of HOPX-β mRNA (normalized to β-actin). 1A or 5A indicates 1 or 5 µM 5-Aza-dC; T, TSA. (D) RTPCR analysis of mRNA expression of HOPX-β, α, and γ and core in human CRC samples. N indicates corresponding normal tissue; T, primary tumor. (E) HOPX-β mRNA (top panel) and Q-MSP values of HOPX-β promoter methylation (bottom panel) in 27 pairs of fresh-frozen human samples. HOPX-β mRNA values were normalized to β-actin. (F) Reciprocal relationship between Q-MSP value of HOPX-β (filled markers) and gene expression of HOPX-β (open markers) in 27 patient samples (triangular and circular markers represent tumors and paired normal mucosa, respectively). (G) Correlation between HOPX protein expression and Q-MSP values in independent 22 human primary CRC patients (top). Representative pictures taken from immunohistochemistry of HOPX in methylation-positive or negative CRC samples (bottom left and middle, respectively) and normal mucosa (bottom right). Bars, 100 µm.
In an independent set of 27 patients, HOPX-β promoter hypermethylation was significantly (P = .002) associated with decreased HOPX-β mRNA expression (Figure 2E). Although several samples did not show clear correlation between Q-MSP values and HOPX-β expression (Figure 2E), HOPX-β methylation and gene expression were inversely correlated (Spearman rank correlation coefficient = .0.87, P < .0001), suggesting that the promoter methylation is functionally linked to gene silencing (Figure 2F).
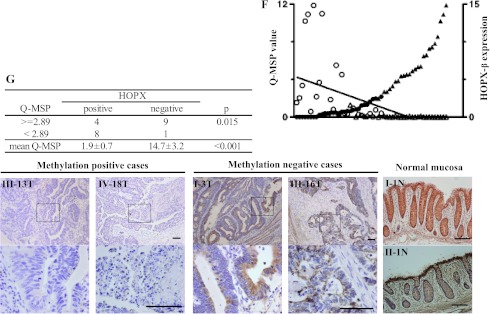
To evaluate the effect of HOPX-β methylation and silencing on HOPX protein expression, we performed immunohistochemistry of the HOPX protein in independent 22 human CRC samples. HOPX protein expression was either present in more than 50% of malignant cells per sample or completely absent in these cells. Therefore, we defined samples as either HOPX-positive or -negative. Primary CRC tissues with Q-MSP value higher than 2.89 were significantly less HOPX-positive compared to tissues with Q-MSP value less than 2.89 (Fisher exact test, P = .015; Figure 2G). In addition, the only four cases that both have a Q-MSP higher than 2.89 and are HOPX-positive had the lowest Q-MSP values among the group with a Q-MSP of 2.89 or higher (data not shown), and therefore, a Q-MSP average of only 5.1, compared to 16.2 in the cases with Q-MSP of 2.89 or higher and HOPX-negative. In addition, HOPX-positive samples had a significantly lower Q-MSP value compared to that of HOPX-negative samples (1.9 and 14.7, respectively; P < .001). Furthermore, immunohistochemistry of the HOPX protein in all 22 normal mucosa counterparts revealed strong HOPX-positive expression (representative results shown in Figure 2G, bottom right). Interestingly, more stromal cells are HOPX-positive in normal tissues.
Tumor-Suppressive Activity Is Increased in CRC Cells Overexpressing HOPX
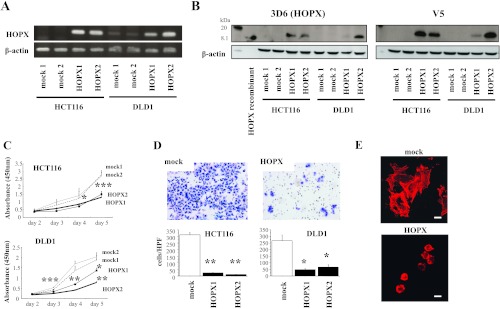
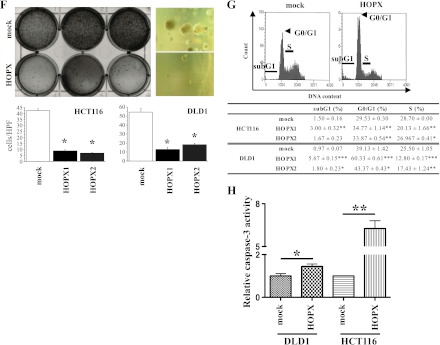
We built a construct containing the full-length cDNA of HOPX, which is common to all three transcript variants, as mentioned before, and used it to stably transfect HCT116 and DLD1 cells. As shown in Figure 3A, parental HCT116 and DLD1 cells express undetectable or low levels of HOPX, respectively (similar results shown also in Figure 2A). As expected, HOPX overexpression resulted in increased levels of HOPX mRNA and protein (Figure 3, A and B). The mRNA levels in the HOPX transfectants were comparable to those in normal human colonic mucosa (data not shown). The specificity of the anti-HOPX antibody (3D6) was validated by blocking the antibody-specific reaction using a recombinant HOPX protein in immunochemistry and Western blot assays (data not shown). To investigate the effects of HOPX reactivation, we performed several tumor aggressiveness assays. Cell proliferation was significantly decreased in HOPX transfectants compared with mock cells using the WST-1 assay (Figure 3C). In addition, Matrigel invasive activity was substantially attenuated in HOPX transfectants (Figure 3D). Interestingly, the morphologic appearance of the mock cells, that is, spindle or spicular shaped, was different from that of HOPX transfectants, which was round shaped. Staining with F-actin showed that this is due to the loss of ability in HOPX transfectants to form filopodia protrusions, manifested by actin aggregations in the cytoplasm (Figure 3E). Moreover, HOPX transfectants showed a dramatic reduction of colony number in an anchorage-independent colony formation assay (Figure 3F). Cell cycle distribution demonstrated that HOPX significantly increased G1 arrest. The sub-G1 fraction (apoptotic cells) and G0/G1 populations were increased (Figure 3G). In contrast, the S population was decreased (Figure 3G). Indeed, HOPX transfectants showed more caspase 3 activity than mock transfectants in vitro (Figure 3H).
Figure 3.
Tumor suppressor functions of HOPX in human CRC cells. (A and B) HOPX mRNA (A) and protein (B) levels in control (mock) and HOPX transfected cells (DLD1 and HCT116). Western blot analysis was performed using a HOPX-specific monoclonal antibody (3D6) and a flag-V5 antibody (V5). Recombinant HOPX served as positive control. (C) Proliferation assay was performed for 5 days. Data are expressed as absorbance levels at 450 nm. Experiments were repeated twice in triplicates. *P < .05. **P < .01. ***P < .001. Error bars, SEM. (D) Matrigel invasion assay. After fixation and staining, invading cells were photographed (top) and counted (bottom) at 100x magnification. Cell growth for 22 hours determined by the WST-1 assay was similar (data not shown). Two independent experiments were done in triplicate, and values indicate means ± SEM. *P < .01. **P < .001. Error bars, SEM. (E) Invading cells in Matrigel invasion assay were stained with phalloidin. F-actin labeling with phalloidin revealed that the mock cells exhibit active filopodia, whereas HOPX-expressing cells exhibit fewer filopodia fibers and F-actins are aggregated in the cytoplasm. (F) Anchorage-independent colony formation assay (soft agar assay) was performed with the indicated transfected cells. Colonies were counted and photographed under a microscope after 3 weeks of cell culture (top right). Colonies were also visualized by ethidium bromide staining (top left). Numbers of colonies were decreased after HOPX overexpression (bottom). *P < .001. (G) Top: Representative images of cell cycle analysis. Bottom: Distribution of cell cycle phases. *P < .05. **P < .01. ***P < .001. Error bars, SEM. (H) Caspase 3 activity was assayed in the transfectants. Values indicate relative activity to each mock transfected cell lines. *P < .01. **P < .001. Error bars, SEM.
DNA Microarray Analysis of Genes Regulated by HOPX in DLD1 and HCT116
To delineate the pathways by which HOPX exerts its tumor suppressor functions, we used DNA microarrays to analyze gene expression profiles of DLD1 and HCT116 cells transfected with HOPX. Using this method, we identified 516 and 77 genes that were either upregulated or downregulated, respectively, at least at a two-fold ratio, both in DLD1 and HCT116 cells on transfection with HOPX (Tables W4 and W5). Several of these genes were upregulated or downregulated by at least a four-fold ratio in both cells, suggesting a higher level of significance (Table 1). The DNA microarray results were validated by qRT-PCR (Table 1). Of the downregulated genes, EPHA2 and CYR61 were previously shown to promote angiogenesis [19–22].
We performed the Matrigel invasion assay using double transfection with FOS and HOPX. As shown in Figure W1A, forced expression of FOS restored invasive ability in HOPX transfectants. However, comparing HOPX-FOS transfectants with mock-mock transfectants, the restoration is not sufficient, suggesting that the suppressive effect of HOPX in invasive ability may not be only due to down-regulation of FOS. We also analyzed expression of CYR61 and EPHA2 by immunohistochemistry in randomly selected 17 CRC patients. Of 12 patients with high Q-MSP, 9 (75%) and 11 (92%) were CYR61- and EPHA2-positive in CRC cells, respectively (Figure W1B).
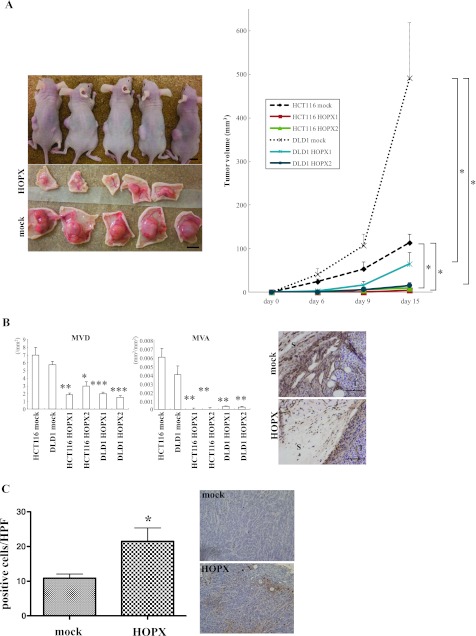
Tumor Suppressive Activity of HOPX In Vivo
To examine the effect of HOPX in vivo, colon cancer cells stably transfected with HOPX were subcutaneously injected into the flanks of nude mice. A remarkable reduction of tumor volume was observed in mice injected with HOPX-transfected cells (Figure 4A). In addition, as the DNA microarray analysis and immunohistochemistry showed down-regulation of the proangiogenic factors EPHA2 and CYR61 by HOPX (Table 1 and Figure W1B), we assessed the effect of HOPX on angiogenesis in the xenograft mouse model. As expected, both MVD and MVA were significantly reduced in tumors derived from HOPX-transfected cells compared with the mock cells (Figure 4B). Furthermore, tumor derived from HOPX transfectants showed increased apoptotic cells by TUNEL assay (Figure 4C).
Figure 4.
Effects of HOPX on tumorigenicity and angiogenesis in vivo. (A) Mock cells were injected into the left flank and HOPX-expressing cells into the right flank of each mouse. Fifteen days after injection, mice were killed, and pictures were taken (left). Time course of tumor growth (right). *P < .05. Error bars, SEM. (B) Left: Stromal angiogenesis on day 15. P values were calculated by comparison with a mock group of each cell line. *P < .05. **P < .01. ***P < .001. Error bars, SEM. Right: Immunohistochemical staining with CD31 was performed to detect endothelial cells. Angiogenesis was significantly suppressed in mice injected with HOPX transfectants. S indicates stroma; T, tumor. Bars, 100 µm. (C) TUNEL assay was performed in tumor derived from HOPX or mock-transfected DLD1 (original magnification, x100). Apoptotic cells were counted in four different sections, four fields in each. Necrotic areas were excluded. *P < .05. Error bars, SEM.
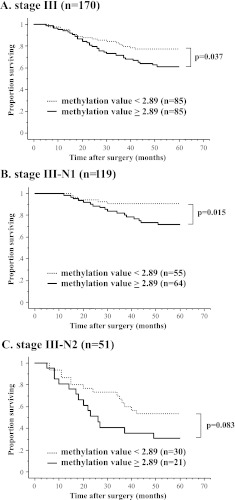
HOPX-β Promoter DNA Hypermethylation in Stage III Patients Is Correlated with Worse Prognosis
Current guidelines published by the National Comprehensive Cancer Network regarding treatment of CRC patients dictate that all patients with stage III disease should receive adjuvant chemotherapy. We therefore focused on this population of patients to identify whether there is a subgroup of patients with stage III disease, with a low value of HOPX-β promoter methylation, which have a favorable prognosis, suggesting reconsideration of the need for adjuvant chemotherapy. We analyzed HOPX-β promoter methylation values in 170 patients with stage III CRC (Figure 5 and Table W2) and looked for a correlation with prognosis, based on low or high values of methylation, previously defined as less or greater than 2.89, respectively (Figure 1C). Indeed, patients with stage III disease with one to three involved regional lymph nodes (N1), with a low Q-MSP value, have a 5-year DSS of 90.6% compared with 71.4% in patients with a high Q-MSP value (Figure 5B). Patients with high Q-MSP values also showed worse prognosis in patients with stage III disease with four or more involved regional lymph nodes (N2; Figure 5C). In a multivariate analysis of all patients with stage III disease, HOPX-β promoter hypermethylation was independently associated with worse prognosis, with a hazard ratio of 1.40 (P = .035; Table W2). Furthermore, HOPX-β promoter hypermethylation was independently correlated with poor differentiation (Table W6 and Figure W2), as a previous report [23], and also with elevated CA19-9 levels (Table W6).
Figure 5.
Kaplan-Meier analysis of 5-year DSS in patients with stage III CRC. (A) Patients with stage III (n = 170). (B) Patients with N1-stage III (n = 119). (C) Patients with N2-stage III (n = 51).
Discussion
We have previously shown that HOPX is a frequently methylated tumor suppressor gene in esophageal and gastric cancers [13,14]. Therefore, in the present study, we investigated the role of HOPX in CRC. Indeed, HOPX-β promoter is frequently hypermethylated in a cancer-specific manner in colon cancer cell lines and human tissues, with 100% specificity in CRC human tissues. Of note, only a few genes that have promoter methylation with 100% cancer specificity were defined so far [24]. This methylation results in the downregulation of HOPX mRNA and protein levels. In human CRC cell lines, forced expression of HOPX increased tumorigenicity. DNA microarray revealed that HOPX downregulates genes involved in angiogenesis and tumor progression. In a mouse xenograft model, HOPX inhibited tumorigenesis and angiogenesis. Importantly, stage III CRC patients show HOPX-β promoter hypermethylation are associated with worse prognosis.
HOPX has three transcript variants, but all variants share the same open reading frame. Only the HOPX-β promoter region harbors CpG islands. Therefore, it was critical to investigate the effect of HOPX-β promoter methylation on HOPX protein levels. Indeed, HOPX-β promoter methylation was correlated with decreased levels of HOPX protein in CRC patients (Figure 2G). Interestingly, HOPX protein was undetectable in DLD1 cells, which do not express HOPX-β mRNA, but do express low levels of HOPX-core mRNA, probably because of the expression of the α variant (Figures 2, A and B, and 3, A and B). This may be due to posttranscriptional regulation of the HOPX-α variant. Collectively, these findings suggest that HOPX protein expression mainly relies on the expression levels of the HOPX-β variant. Nevertheless, high-dose (5 µM) 5-Aza-dC with/without TSA produced higher stimulation of HOPX-β reexpression in HCT116, whereas exposure to 1 µM 5-Aza-dC alone was sufficient to induce reexpression in DLD1 (Figure 2C) as in esophageal and gastric cancer [13,14]. Accordingly, other epigenetic modifications such as histone modifications may be involved in silencing the HOPX-β gene in DLD1, requiring further analysis in the future study.
We used DNA microarrays to explore, for the first time, genes regulated by HOPX. Using this method, we suggest that HOPX upregulates the WTAP and PRDX2 genes and downregulates FOS, EMP1, SLC2A3, CYR61, and EPHA2 genes (Table 1). EPHA2 was shown to promote invasion in prostate cancer and glioma [25] and also play a role in angiogenesis and invasion in ovarian cancer [20]. EPHA2 is significantly overexpressed in CRC [26], and its overexpression is correlated with high microvessel count [19] and reduced survival [27] in CRC. CYR61 is a secreted molecule, involved in many types of cancer. CYR61 was shown to promote angiogenesis [22] and invasion [28] in gastric cancer, proliferation and invasion in esophageal cancer [29], and tumorigenesis in breast cancer [30–33]. In accordance with the suggested down-regulation of EPHA2 and CYR61 by HOPX (Table 1), we showed reduced angiogenesis in vivo by HOPX overexpression (Figure 4B).
Recently, HOPX was shown to play a pivotal role in intestinal stem cell interconversion. HOPX labels a quiescent population of intestinal stem cells at the +4 niche, which can give rise to more rapidly proliferating Lgr5-expressing intestinal stem cells at the crypt base [34]. These findings suggest that HOPX down-regulation is correlated with increased proliferation.
We also found out that more stromal cells are HOPX-positive in normal tissues than in tumors. These HOPX-positive stromal cells are mainly located superficially close to the normal mucosa, suggesting that the expression of HOPX in stromal cells may have immunopathologic roles and might be regulated by other mechanisms. Likewise, HOPX expression on normal epithelial may be affected by other mechanisms, resulting in a strong expression. Indeed, Albrecht et al. [35] reported that the persistence of effector memory TH1 cells is critically regulated by HOPX through T-bet and deletion of HOPX suppressed inflammatory response in colon. However, large-sized stromal cells are also HOPX-positive in the present study (Figure 2G), indicating that other immunocytes may be involved in normal immunopathology. It is consistent with the data in UCSC Genome Bioinformatics (http://genome.ucsc.edu/cgi-bin/hgGateway) in which other immune cells including B, CD8, and NK cells express HOPX, requiring further study.
Further study should address the possible association of HOPX-β promoter hypermethylation with worse prognosis in stage II CRC patients, as a possible marker favoring administration of adjuvant chemotherapy. In addition, further definition and investigation of HOPX-regulated genes are critical to understand the pathways by which HOPX exerts its tumor-suppressive activity.
In summary, we have shown that HOPX-β promoter hypermethylation is a frequent and cancer-specific event in CRC patients, correlated with worse prognosis of patients with stage III disease. Inversely, patients with N1-stage III disease without HOPX-β promoter hypermethylation demonstrated excellent prognosis, similar to that of patients with stage II disease, suggesting reconsideration of the adjuvant chemotherapy currently indicated in these patients with stage III disease. Furthermore, HOPX-β promoter hypermethylation and protein levels may be used as markers for aggressiveness in CRC patients and as tools to screen for CRC.
Supplementary Material
Footnotes
The authors have no current external funding sources for this study. The authors declare no conflict of interest.
This article refers to supplementary materials, which are designated by Tables W1 to W6 and Figures W1 and W2 and are available online at www.neoplasia.com.
Our study showed first that HOPX promoter hypermethylation is cancer-specific and frequent in human colorectal cancer. HOPX was found to have multifunctional tumor suppressive activity and suppressed tumorigenesis and angiogenesis in vivo. HOPX promoter hypermethylation independently shows worse prognosis in patients with stage III disease. Importantly, patients with N1-stage III disease without HOPX promoter hypermethylation demonstrated excellent prognosis, comparable to that of stage II patients, suggesting reconsideration of the adjuvant chemotherapy currently indicated in these patients with stage III disease.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Mandel JS, Bond JH, Church TR, Snover DC, Bradley GM, Schuman LM, Ederer F. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota Colon Cancer Control Study. N Engl J Med. 1993;328:1365–1371. doi: 10.1056/NEJM199305133281901. [DOI] [PubMed] [Google Scholar]
- 3.Yamashita K, Watanabe M. Clinical significance of tumor markers and an emerging perspective on colorectal cancer. Cancer Sci. 2009;100:195–199. doi: 10.1111/j.1349-7006.2008.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, Sato F, Meltzer SJ, Sidransky D. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485–495. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 9.Yamashita K, Park HL, Kim MS, Osada M, Tokumaru Y, Inoue H, Mori M, Sidransky D. PGP9.5 methylation in diffuse-type gastric cancer. Cancer Res. 2006;66:3921–3927. doi: 10.1158/0008-5472.CAN-05-1511. [DOI] [PubMed] [Google Scholar]
- 10.Kim MS, Chang X, Nagpal JK, Yamashita K, Baek JH, Dasgupta S, Wu G, Osada M, Woo JH, Westra WH, et al. The N-methyl-d-aspartate receptor type 2A is frequently methylated in human colorectal carcinoma and suppresses cell growth. Oncogene. 2008;27:2045–2054. doi: 10.1038/sj.onc.1210842. [DOI] [PubMed] [Google Scholar]
- 11.Kim MS, Yamashita K, Baek JH, Park HL, Carvalho AL, Osada M, Hoque MO, Upadhyay S, Mori M, Moon C, et al. N-methyl-d-aspartate receptor type 2B is epigenetically inactivated and exhibits tumor-suppressive activity in human esophageal cancer. Cancer Res. 2006;66:3409–3418. doi: 10.1158/0008-5472.CAN-05-1608. [DOI] [PubMed] [Google Scholar]
- 12.Kim MS, Chang X, LeBron C, Nagpal JK, Lee J, Huang Y, Yamashita K, Trink B, Ratovitski EA, Sidransky D. Neurofilament heavy polypeptide regulates the Akt-β-catenin pathway in human esophageal squamous cell carcinoma. PLoS One. 2010;5:e9003. doi: 10.1371/journal.pone.0009003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashita K, Kim MS, Park HL, Tokumaru Y, Osada M, Inoue H, Mori M, Sidransky D. HOP/OB1/NECC1 promoter DNA is frequently hypermethylated and involved in tumorigenic ability in esophageal squamous cell carcinoma. Mol Cancer Res. 2008;6:31–41. doi: 10.1158/1541-7786.MCR-07-0213. [DOI] [PubMed] [Google Scholar]
- 14.Ooki A, Yamashita K, Kikuchi S, Sakuramoto S, Katada N, Kokubo K, Kobayashi H, Kim MS, Sidransky D, Watanabe M. Potential utility of HOP homeobox gene promoter methylation as a marker of tumor aggressiveness in gastric cancer. Oncogene. 2010;29:3263–3275. doi: 10.1038/onc.2010.76. [DOI] [PubMed] [Google Scholar]
- 15.Katoh H, Hosono K, Ito Y, Suzuki T, Ogawa Y, Kubo H, Kamata H, Mishima T, Tamaki H, Sakagami H, et al. COX-2 and prostaglandin EP3/EP4 signaling regulate the tumor stromal proangiogenic microenvironment via CXCL12-CXCR4 chemokine systems. Am J Pathol. 2010;176:1469–1483. doi: 10.2353/ajpath.2010.090607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi YD, Choi J, Kim JH, Lee JS, Lee JH, Choi C, Choi HS, Lee MC, Park CS, Juhng SW, et al. Microsatellite instability at a tetranucleotide repeat in type I endometrial carcinoma. J Exp Clin Cancer Res. 2008;27:88. doi: 10.1186/1756-9966-27-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O'Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–1387. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- 19.Kataoka H, Igarashi H, Kanamori M, Ihara M, Wang JD, Wang YJ, Li ZY, Shimamura T, Kobayashi T, Maruyama K, et al. Correlation of EPHA2 overexpression with high microvessel count in human primary colorectal cancer. Cancer Sci. 2004;95:136–141. doi: 10.1111/j.1349-7006.2004.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin YG, Han LY, Kamat AA, Merritt WM, Landen CN, Deavers MT, Fletcher MS, Urbauer DL, Kinch MS, Sood AK. EphA2 overexpression is associated with angiogenesis in ovarian cancer. Cancer. 2007;109:332–340. doi: 10.1002/cncr.22415. [DOI] [PubMed] [Google Scholar]
- 21.Brantley-Sieders DM, Fang WB, Hicks DJ, Zhuang G, Shyr Y, Chen J. Impaired tumor microenvironment in EphA2-deficient mice inhibits tumor angiogenesis and metastatic progression. FASEB J. 2005;19:1884–1886. doi: 10.1096/fj.05-4038fje. [DOI] [PubMed] [Google Scholar]
- 22.Babic AM, Kireeva ML, Kolesnikova TV, Lau LF. CYR61, a product of a growth factor-inducible immediate early gene, promotes angiogenesis and tumor growth. Proc Natl Acad Sci USA. 1998;95:6355–6360. doi: 10.1073/pnas.95.11.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harada Y, Kijima K, Shinmura K, Sakata M, Sakuraba K, Yokomizo K, Kitamura Y, Shirahata A, Goto T, Mizukami H, et al. Methylation of the homeobox gene, HOPX, is frequently detected in poorly differentiated colorectal cancer. Anticancer Res. 2011;31:2889–2892. [PubMed] [Google Scholar]
- 24.Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- 25.Miao H, Li DQ, Mukherjee A, Guo H, Petty A, Cutter J, Basilion JP, Sedor J, Wu J, Danielpour D, et al. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell. 2009;16:9–20. doi: 10.1016/j.ccr.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herath NI, Spanevello MD, Doecke JD, Smith FM, Pouponnot C, Boyd AW. Complex expression patterns of Eph receptor tyrosine kinases and their ephrin ligands in colorectal carcinogenesis. Eur J Cancer. 2011;48:753–762. doi: 10.1016/j.ejca.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Baeten CI, Hillen F, Pauwels P, de Bruine AP, Baeten CG. Prognostic role of vasculogenic mimicry in colorectal cancer. Dis Colon Rectum. 2009;52:2028–2035. doi: 10.1007/DCR.0b013e3181beb4ff. [DOI] [PubMed] [Google Scholar]
- 28.Lin MT, Zuon CY, Chang CC, Chen ST, Chen CP, Lin BR, Wang MY, Jeng YM, Chang KJ, Lee PH, et al. Cyr61 induces gastric cancer cell motility/invasion via activation of the integrin/nuclear factor-κB/cyclooxygenase-2 signaling pathway. Clin Cancer Res. 2005;11:5809–5820. doi: 10.1158/1078-0432.CCR-04-2639. [DOI] [PubMed] [Google Scholar]
- 29.Xie JJ, Xu LY, Wu JY, Shen ZY, Zhao Q, Du ZP, Lv Z, Gu W, Pan F, Xu XE, et al. Involvement of CYR61 and CTGF in the fascin-mediated proliferation and invasiveness of esophageal squamous cell carcinomas cells. Am J Pathol. 2010;176:939–951. doi: 10.2353/ajpath.2010.090118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen N, Kuliopulos A, Graham RA, Covic L. Tumor-derived Cyr61 (CCN1) promotes stromal matrix metalloproteinase-1 production and protease-activated receptor 1-dependent migration of breast cancer cells. Cancer Res. 2006;66:2658–2665. doi: 10.1158/0008-5472.CAN-05-2082. [DOI] [PubMed] [Google Scholar]
- 31.Tsai MS, Bogart DF, Castaneda JM, Li P, Lupu R. Cyr61 promotes breast tumorigenesis and cancer progression. Oncogene. 2002;21:8178–8185. doi: 10.1038/sj.onc.1205682. [DOI] [PubMed] [Google Scholar]
- 32.Xie D, Miller CW, O'Kelly J, Nakachi K, Sakashita A, Said JW, Gornbein J, Koeffler HP. Breast cancer. Cyr61 is overexpressed, estrogen-inducible, and associated with more advanced disease. J Biol Chem. 2001;276:14187–14194. doi: 10.1074/jbc.M009755200. [DOI] [PubMed] [Google Scholar]
- 33.Xie D, Nakachi K, Wang H, Elashoff R, Koeffler HP. Elevated levels of connective tissue growth factor, WISP-1, and CYR61 in primary breast cancers associated with more advanced features. Cancer Res. 2001;61:8917–8923. [PubMed] [Google Scholar]
- 34.Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albrecht I, Niesner U, Janke M, Menning A, Loddenkemper C, Kuhl AA, Lepenies I, Lexberg MH, Westendorf K, Hradilkova K, et al. Persistence of effector memory TH1 cells is regulated by Hopx. Eur J Immunol. 2010;40:2993–3006. doi: 10.1002/eji.201040936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.