Abstract
Chronic myeloid leukemia (CML) is a hematopoietic neoplasm characterized by the Philadelphia chromosome and the related BCR-ABL1 oncoprotein. Acceleration of CML is usually accompanied by basophilia. Several proangiogenic molecules have been implicated in disease acceleration, including the hepatocyte growth factor (HGF). However, little is known so far about the cellular distribution and function of HGF in CML. We here report that HGF is expressed abundantly in purified CML basophils and in the basophil-committed CML line KU812, whereas all other cell types examined expressed only trace amounts of HGF or no HGF. Interleukin 3, a major regulator of human basophils, was found to promote HGF expression in CML basophils. By contrast, BCR-ABL1 failed to induce HGF synthesis in CML cells, and imatinib failed to inhibit expression of HGF in these cells. Recombinant HGF as well as basophil-derived HGF induced endothelial cell migration in a scratch wound assay, and these effects of HGF were reverted by an anti-HGF antibody as well as by pharmacologic c-Met inhibitors. In addition, anti-HGF and c-Met inhibitors were found to suppress the spontaneous growth of KU812 cells, suggesting autocrine growth regulation. Together, HGF is a BCR-ABL1-independent angiogenic and autocrine growth regulator in CML. Basophils are a unique source of HGF in these patients and may play a more active role in disease-associated angiogenesis and disease progression than has so far been assumed. Our data also suggest that HGF and c-Met are potential therapeutic targets in CML.
Introduction
Chronic myeloid leukemia (CML) is a hematopoietic neoplasm characterized by the reciprocal chromosome translocation t(9;22) [1]. This cytogenetic defect creates the BCR-ABL1 fusion gene [2,3]. The associated oncoprotein, BCR-ABL1, is a cytoplasmic molecule that exhibits constitutive tyrosine kinase activity and triggers key downstream signaling molecules, including RAS, the phosphoinositide 3-kinase, and STAT5 [4–6]. BCR-ABL1 and various BCR-ABL1 downstream signaling molecules have been implicated as major triggering factors in the pathogenesis of CML. In line with this assumption, BCR-ABL1-targeting drugs such as imatinib, are successfully used to suppress the growth of neoplastic cells in patients with CML [7,8].
The clinical course in CML can be divided into a chronic phase (CP), an accelerated phase (AP), and a blast phase (BP), which is the terminal phase and resembles an acute leukemia [8–10]. Whereas in CP, BCR-ABL1 is a major driving force of cell survival and proliferation, additional factors and pro-oncogenic molecules, apart from BCR-ABL1, may play a more important or even decisive pathogenetic role in AP and BP [6–10]. A key feature in AP of CML is basophilia [11,12]. Moreover, basophilia is one of the most significant prognostic factors in CML at diagnosis [12,13].
Although little is known about disease initiation and evolution in CML, several mechanisms and molecules have been implicated as potential mediators of acceleration and drug resistance, including survival-related molecules, cytokine receptors, and various signal transduction pathways [4–10,14,15]. In addition, increased angiogenesis in the bone marrow (BM) and other hematopoietic tissues may contribute to disease progression in CML [16–18].
A number of angiogenic cytokines have been identified in CML cells, including vascular endothelial growth factor (VEGF), basic fibroblast growth factor, angiopoietin 1, and matrix metalloproteinases [17–22]. In addition, hepatocyte growth factor (HGF) has been described to be expressed in CML cells [23,24]. In particular, it has been described that patients with CML exhibit elevated HGF levels in their BM and blood and that HGF expression in the BM correlates with microvessel density [23,24]. Moreover, recent data suggest that increased blood levels of HGF correlate with the prognosis in these patients [25]. However, so far, little is known about the cellular source and function of HGF in CML cells and the exact role this cytokine plays in the pathogenesis of CML.
In the current study, we show that HGF is preferentially produced in CML basophils, and that basophil-derived HGF triggers endothelial cell migration and growth through a specific receptor. These observations point to a novel hitherto unrecognized and more active role of basophils and their products in disease acceleration in CML. In addition, these data suggest that HGF and c-Met may serve as potential targets in CML.
Materials and Methods
Antibodies and Reagents
The basophil-specific PE-labeled monoclonal antibody (mAb) 97A6 (CD203c) [26] was purchased from Immunotech (Marseille, France), a polyclonal rabbit anti-HGF antibody (H-145) from Santa Cruz Biotechnology (Santa Cruz, CA), rabbit anti-phospho-c-Met mAb D26 (Tyr1234/1235) from Cell Signaling (Danvers, MA), biotinylated anti-rabbit IgG and Vectastain Universal ABC-AP Kit from Vector Laboratories (Burlingame, CA), and biotinylated goat anti-rabbit IgG from Biocare Medical (San Diego, CA). The basophil-specific mAb BB1 [27] was produced at the University of Southampton, United Kingdom. A specification of antibodies is shown in Table W1. The c-Met inhibitors PF-2341066 and SU11274 were purchased from Selleck Chemicals (Houston, TX). The protoscript first-strand complementary DNA (cDNA) synthesis kit was from New England Biolabs (Beverly, MA), RPMI-1640 medium, Iscove modified Dulbecco medium (IMDM) and fetal calf serum (FCS) from PAA Laboratories (Pasching, Austria), recombinant human (rh) VEGF, and medium 199 from Invitrogen (Camarillo, CA), rhHGF from Sigma-Aldrich (St Louis, MO), endothelial cell growth supplement from Technoclone (Vienna, Austria), and rh granulocyte-macrophage colony stimulating factor (GM-CSF), and murine interleukin 3 (IL-3) from PeproTech (Rocky Hill, NJ). Imatinib was kindly provided by Drs E. Buchdunger and P.W. Manley (Novartis Pharma AG, Basel, Switzerland).
Isolation and Culture of Primary CML Cells
Primary leukemic cells were obtained from 25 patients with untreated CPCML and 7 with APCML. BM aspirate samples (iliac crest) and peripheral blood (PB) cells were collected in heparinized tubes. Normal BM cells were obtained from three patients with lymphoma (routine staging) without BM involvement. Informed consent was obtained from each patient before blood donation or BM puncture. BM mononuclear cells (MNCs) and PB MNC were isolated using Ficoll. The study was approved by the institutional review board of the Medical University of Vienna. Isolated MNCs were cultured in RPMI-1640 medium containing 10% FCS with or without cytokines. In four patients with CML with marked basophilia, PB MNCs were separated into two fractions by cell sorting: a CD203c+ fraction (>98% basophils) and a CD203c. fraction (<1% basophils) using the CD203c mAb 97A6 as described [26]. In three patients with CML, CD34+/CD38- stem cells and CD34+/CD38+ progenitor cells were enriched to greater than 98% purity by cell sorting using a PE-labeled CD34 mAb (clone 581) and an APC-conjugated mAb against CD38 (HIT2) (Table W1). Cell sorting was performed on a FACS-Aria (BD Biosciences, San Jose, CA). After sorting, cell viability was more than 95% in all experiments.
Cell Lines and Culture Conditions
Ton.B210-X is an IL-3-dependent Ba/F3-derived cell line, in which BCR-ABL1 can be induced conditionally through addition of doxycycline (1 µg/ml) [21]. Ton.B210-X cells were grown in RPMI-1640 medium with 10% FCS and IL-3 (1 ng/ml) at 37°C. For starvation, cells were cultured in the absence of IL-3 for up to 24 hours [21]. The BCR-ABL1+ cell lines K562 (multilineage) and KU812 (basophil-committed), the BCR-ABL1- cell lines HL60, KG1, U937, and MO7e, as well as lung fibroblasts (LUFs), were maintained in RPMI-1640 medium and 10% FCS. MO7e cells were transfected with BCR-ABL1 by lentiviral-mediated gene transfer as described [28]. Whereas control MO7e cells (nontransfected and empty vector transfected) were cultured in GM-CSF (100 ng/ml), BCR-ABL1-transfected MO7e cells (MO7e-p210) were maintained without GM-CSF. HMC-1.1 and HMC-1.2 cells were maintained in Iscove modified Dulbecco medium plus 10% FCS. To inhibit BCR-ABL1 activity, KU812 cells were cultured in imatinib (1 µM) for up to 8 hours. After exposure to imatinib, cells were subjected to quantitative polymerase chain reaction (qPCR) and ELISA measurements. Human umbilical vein-derived endothelial cells (HUVECs) were purchased from Technoclone and cultured at 37°C and 5% CO2 in gelatin-coated plastic flasks in Medium 199 supplemented with endothelial cell growth supplement (20 µg/ml).
Northern Blot Analysis and qPCR
Northern blot analysis was performed as described previously [21]. Total RNA was extracted from leukemic cell lines using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Twenty micrograms of RNA was size fractionated on 1.0% formaldehyde agarose gels and transferred to nylon membranes (Hybond N; Amersham, Aylesbury, United Kingdom) [21]. Hybridization was performed with 32P-labeled cDNA probes (Table W2), and labeling was conducted using MegaPrime kit (Amersham). Blots were washed in 0.2 x SSC (1 x SSC = 150 mM NaCl and 15 mM sodium citrate, pH 7.0) with 0.1% sodium dodecyl sulfate [21]. Bound radioactivity was visualized by exposure to Biomax MS films (Kodak, Rochester, NY) at -80°C using intensifying screens. For PCR analysis, total RNA was isolated from cell lines and primary leukemic cells (patients with CML CP: BM, n = 14 and PB, n = 20; AP: BM, n = 8; and PB, n = 12) using the RNeasy MinElute Cleanup Kit (Qiagen, Hilden, Germany). Complementary DNA was synthesized using Moloney murine leukemia virus reverse transcriptase (Invitrogen), random primers, first-strand buffer, dNTPs (100 mM), and RNasin according to the manufacturer's instructions (Invitrogen). Quantitative PCR was performed as described [28] using iTaq SYBR Green SuperMix with ROX (Bio-Rad, Hercules, CA) and primers specific for HGF, c-Met, histidine decarboxylase (HDC), VEGF, and ABL (Table W3).
Immunohistochemistry and Immunocytochemistry
In 16 patients with CP CML, 14 with AP, 6 with BP, and 5 with normal BM, expression of HGF was analyzed by IHC on serial sections (2 µm) prepared from formalin-fixed, paraffin-embedded, BM specimens. IHC was performed by the indirect immunoperoxidase staining technique as described [29] using a polyclonal anti-HGF antibody (work dilution, 1:50) and the anti-basophil mAb BB1 (1:300). After incubation (overnight) slides were washed, incubated with biotinylated second step goat anti-rabbit or horse anti-mouse antibodies (30 minutes), washed, and stained using 3-amino-9-ethyl-carbazole (Sigma-Aldrich). Slides were counterstained in Meyer hematoxylin. For ICC, primary CML cells (CML CP: BM, n = 17, and PB, n = 17; AP: BM, n = 8, and PB, n = 10) and cell lines were spun on cytospin slides and incubated with the anti-HGF antibody (1:50) or with mAb BB1 (1:300) overnight. Then, slides were washed and incubated with a biotinylated goat anti-rabbit antibody for 30 minutes. Slides were again washed and incubated with streptavidin alkaline phosphatase complex (30 minutes), washed, and stained with New Fuchsin (Nichirei Corporation, Tokyo, Japan).
Flow Cytometry
To investigate expression of c-Met on cell lines, and HUVEC, flow cytometry was performed using fluorochrome-conjugated mAb (Table W1). Multicolor flow cytometry was performed on a FACSCalibur (BD Biosciences) using FlowJo software (TreeStar, Ashland, OR). Antibody reactivity was controlled by isotype-matched antibodies. In a separate set of experiments, CML cells were incubated (37°C, 5% CO2) with the c-Met inhibitor PF-2341066 (1 µM, 24 or 48 hours) and were then examined for signs of apoptosis by combined staining for surface antigens and Annexin V-fluoresceine isothiocyanate and caspase 3, following the instructions of the manufacturer (Bender MedSystems, Vienna, Austria).
Evaluation of Proliferation of CML Cells and Endothelial Cells
CML cells (primary CMLMNC and cell lines: K562, KU812) were incubated in control medium, HGF (100 ng/ml), VEGF (100 ng/ml), or KU812 supernatants in the absence or presence of the c-Met inhibitors PF-2341066 or SU11274 (each 0.01-10 µM) for 48 hours (37°C, 5% CO2). After incubation, proliferation was determined by measuring 3H-thymidine incorporation in 96-well microtiter plates (CML cells: 5 x 104 cells per well). After incubation, 3H-thymidine (0.5 µCi) was added to each well. Sixteen hours later, cells were harvested on filter membranes (Packard Bioscience, Meriden, CT) in a Filtermate 196 harvester (Packard Bioscience). Filters were then air-dried, and the bound radioactivity was measured in a β-counter (Top-Count NXT; Packard Bioscience). All experiments were performed in triplicates.
Measurement of HGF and VEGF by ELISA
HGF and VEGF were measured in lysates and supernatants of cultured primary CML cells and KU812 cells (1 x 107 cells per well) after incubation in control medium in the presence or absence of cytokines (IL-3; 100 ng/ml). In a separate set of experiments, KU812 cells were incubated in control medium or in medium containing various concentrations of imatinib (0.1–1 µM) for 24 or 48 hours. HGF and VEGF concentrations were determined by ELISA (R&D Systems, Minneapolis, MN).
Endothelial Cell Migration (Scratch Wound) Assay
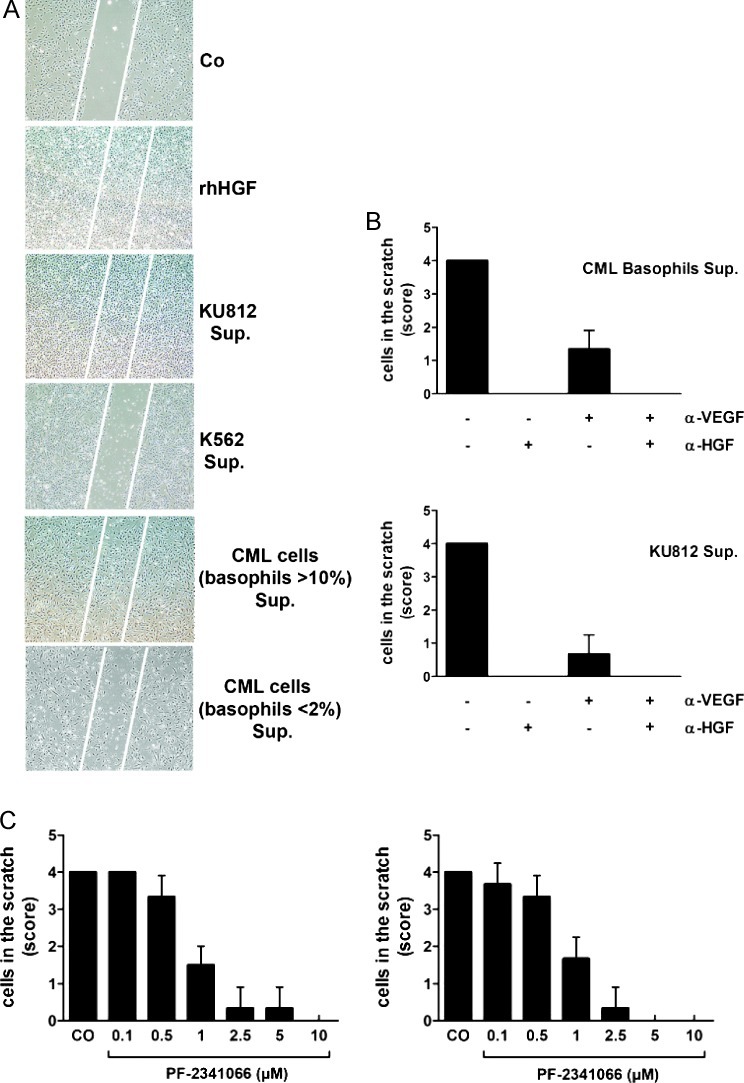
To demonstrate functional activity of basophil-derived HGF, we applied basophil supernatants and KU812 supernatants as well as K562 supernatants (control) in a scratch assay. In this assay, HUVEC were seeded in six-well plates and cultured at 37°C in 5% CO2 until confluence was reached. A linear scratch wound (100 µm diameter) was produced by a pipette tip. Endothelial migration was induced by adding rhVEGF (100 ng/ml) and/or rhHGF (100 ng/ml). In addition, cell supernatants were applied at various dilutions (1:1, 1:4, 1:8, 1:16, and 1:32) with or without rhVEGF (100 ng/ml), a polyclonal blocking anti-HGF antibody (60 ng/ml), anti-VEGF antibody (100 ng/ml), or the c-Met inhibitor PF-2341066 (1 µM). After 24 and 48 hours, endothelial cell migration was examined in an inverted microscope (Olympus, Hamburg, Germany), photographed (Eclipse TE 300; Nikon, Tokyo, Japan) and graded using the following scores: 0 = scratched areas empty, 1 = few single endothelial cells in scratch wound, 2 = multiple isolated endothelial cells in the scratch lesion, 3 = endothelial cells form aggregates and bridges in scratch wound, and 4 = scratch wound completely “healed” with confluent endothelial layer (Figure W1).
Statistical Analysis
Data were analyzed by Student's t test, Mann-Whitney U test, oneway analysis of variance, and Kruskal-Wallis test using GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA). The Student's t test was used to compare differences between two experimental groups, whereas one-way analysis of variance (followed by Bonferroni adjustment) was applied to analyze differences between three or more groups. Results were considered to be significantly different when P < .05.
Results
Expression of HGF in Primary CML Cells
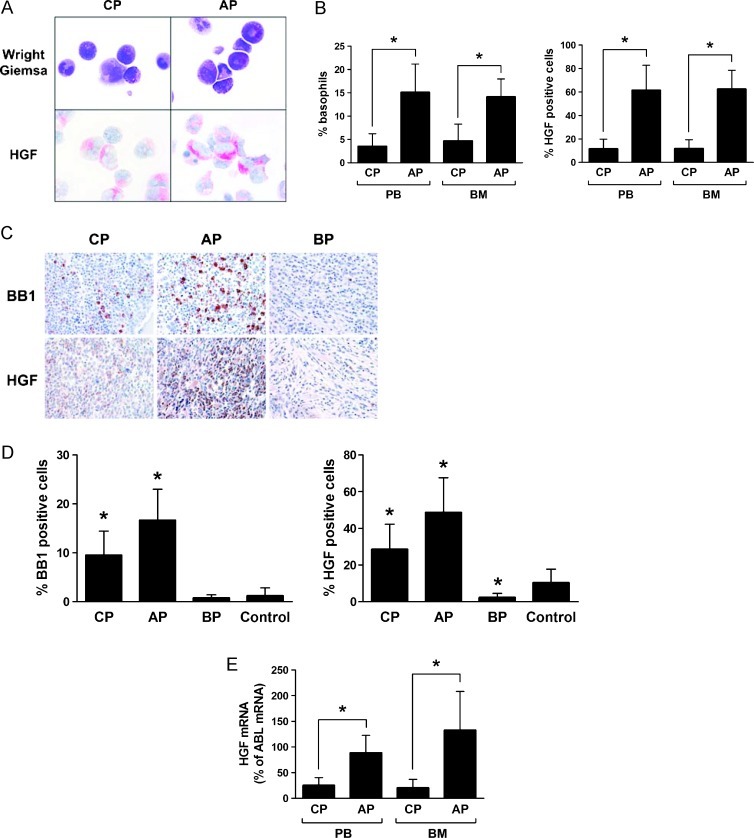
As assessed by immunocytochemistry (ICC) and immunohistochemistry (IHC), primary CML cells were found to express the HGF protein (Figure 1, A–D). In both staining protocols (ICC and IHC), only a subset of leukemic cells were found to react with the anti-HGF antibody (Figure 1, A–D). The percentage of HGF-positive cells varied from patient to patient and was higher in patients with AP compared to patients with CP or BP (Figure 1, A–D; P < .05). The difference in HGF expression in BM cells in the various phases of CML was clearly demonstrable in our ICC staining experiments as well as in the IHC protocol. Preincubation of the anti-HGF antibody with a HGF-specific blocking peptide resulted in a negative stain (Figure W2). We were also able to demonstrate expression of HGF mRNA in primary CML cells by qPCR. As expected, HGF transcript levels were found to be higher in CML cells in AP compared to patients with CP (Figure 1E; P < .05).
Figure 1.
Detection of HGF in primary CML cells. (A) Ficoll-isolated PB cells obtained from patients with CML in CP or AP were spun on cytospin slides and stained by Wright-Giemsa (upper panels) or with an antibody against HGF (lower panels) by indirect ICC (magnification, 100x/1.35). (B) Percentage of basophils (left panel) and HGF-positive cells (right panel) counted in PB and BM samples obtained from patients with CML CP (n = 17) and CML AP (n = 10) by ICC. Results represent the mean ± SD from all patients. *P < .05. (C) Immunohistochemical (IHC) detection of BB1 and HGF in BM cells. Serial sections were prepared from paraffin-embedded BM (iliac crest) in a patient in CML CP (left panels), one with CML AP (middle panels), and one in BP (right panels). Sections were stained with antibodies against BB1 (upper panels) or HGF (lower panels). Magnification, 40x/0.85. (D) Percentage of BB1-positive basophils (left panel) and HGF-positive cells (right panel) in BM sections obtained from patients with CML CP (n = 16), CML AP (n = 14), CML BP (n = 6), or control marrow (n = 5). Expression of BB1 and HGF was examined by IHC. Results represent the mean ± SD from all donors in each group. *P < .05 compared with control (normal BM). (E) Expression of HGF mRNA in primary CML cells. Ficoll-isolated cells (PB: CP, n = 20; AP, n = 12; BM: CP, n = 14; AP, n = 8) were subjected to RNA isolation and qPCR using primers specific for HGF and ABL. Results show HGFmRNA expression levels as percent of ABL mRNA levels. Results represent the mean ± SD from all donors in each group. *P < .05.
Identification of Basophils as a Major Source of HGF in CML
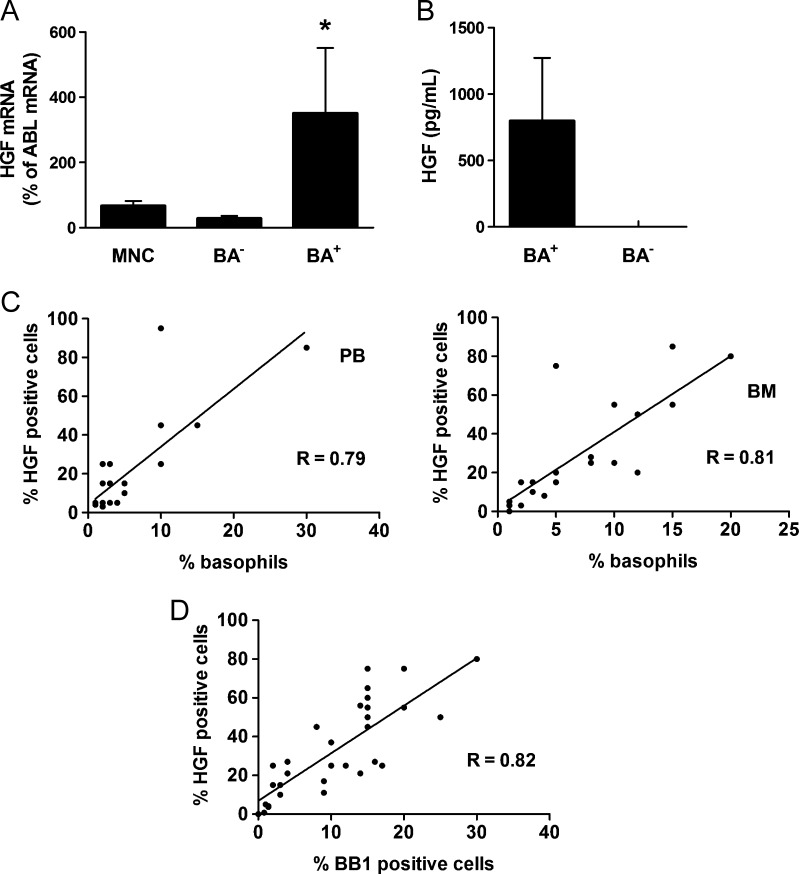
On the basis of the observation that HGF is expressed at high levels in leukemic cells in AP, we examined the expression of HGF in basophils, known to increase substantially in AP [11,12]. As shown in Figure 2A, sorted CD203c+ CML basophils were found to express substantial amounts of HGF mRNA, whereas basophil-depleted cell fractions contained only low amounts of HGF mRNA. Moreover, we were able to show that supernatants of basophil-rich cell fractions contain substantial amounts of the HGF protein, whereas supernatants of CML samples with low numbers of basophils, exhibited no detectable HGF (Figure 2B). In a next step, we established correlations between HGF+ cells and basophil numbers in our immunostaining experiments. On cytospin slides, basophils were identified and counted after Wright-Giemsa staining, and the numbers of HGF+ cells were determined by ICC. In BM biopsy sections, basophils were quantified by BB1 staining, and the numbers of HGF+ cells were determined on adjacent BM sections by IHC. In both analyses, we found a good correlation between the numbers (percentage) of basophils and the numbers (percentage) of HGF+ cells (Figure 2, C and D), confirming that basophils are a primary source of HGF in the BM and PB in CML.
Figure 2.
Identification of basophils as a source of HGF in CML. (A) Expression of HGF mRNA in unfractionated PB MNCs, highly purified sorted CD203c+ basophils (BA+), and basophil-depleted (BA-) cells. MNC were prepared from PB of three CML donors. mRNA levels were quantified by qPCR using primers specific for HGF and ABL. Results show HGF mRNA levels as percent of ABL mRNA levels and represent the mean ± SD from three donors. *P < .05 compared with MNC. (B) Measurement of HGF protein levels in supernatants of cultured PB cells (5-day culture) obtained from three patients with CML with marked basophilia (>10%) and three patients with CML with low basophil counts (≤2%). HGF levels were determined by ELISA. Results represent the mean ± SD of three donors. (C) Correlations between HGF+ cells and basophils (Wright-Giemsa stain) in BM MNC samples (n = 20, left panel) and PB MNC (n = 17, right panel) of patients with CML. The numbers (percentage) of HGF+ cells were determined by IHC. R indicates the correlation coefficient. (D) Correlation between HGF+ cells and BB1+ cells (percentage of nucleated cells) in BM sections in patients with CML (n = 29). Adjacent BM sections were stained with antibodies against HGF and BB1 by IHC. R indicates the correlation coefficient.
The Basophil-Committed CML Cell Line KU812 Produces HGF
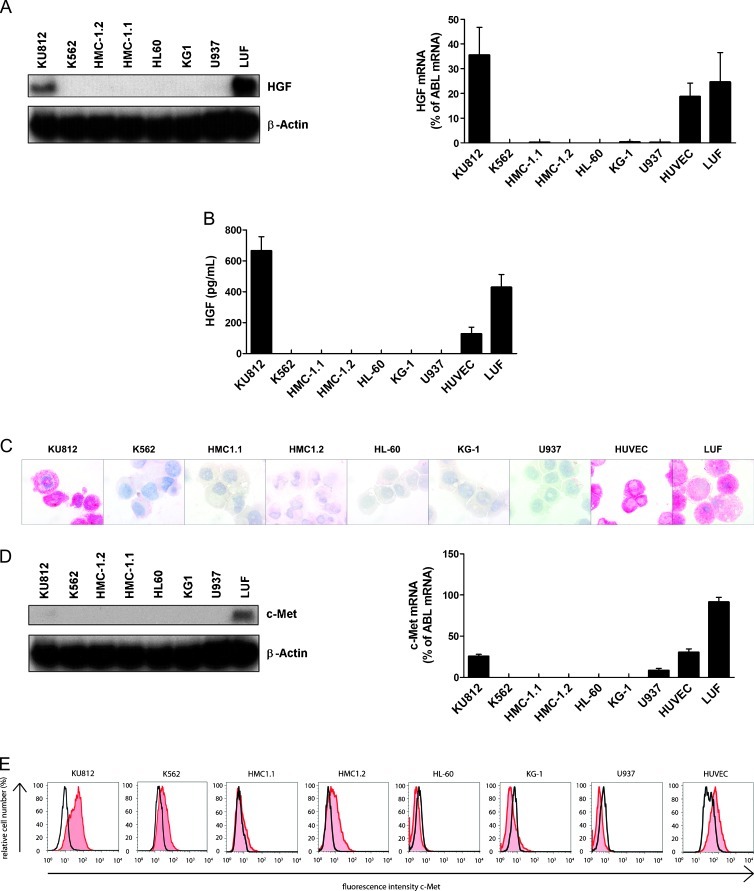
In a next step, we examined HGF expression in various leukemic cell lines including the basophil-committed CML line KU812, the immature (uncommitted) CML line K562, HUVECs, and LUFs. As shown in Figure 3A, KU812 cells were found to express substantial amounts of HGF mRNA, whereas K562 cells and the other leukemic cell lines tested did not express substantial amounts of HGF mRNA. Correspondingly, KU812 cells, but neither K562 cells nor the other leukemic cell lines tested, were found to express and release the HGF protein (Figure 3B). Moreover, KU812 cells, but not the other leukemic cell lines tested, were found to stain positive for HGF by ICC (Figure 3C). HUVEC and LUF, known to express HGF, served as a positive control. Preincubation of the anti-HGF antibody with a HGF-specific blocking peptide resulted in a negative stain (Figure W2). We also examined expression of c-Met in various cell lines. As assessed by Northern blot analysis and qPCR, KU812 cells and LUFs were found to express c-Met mRNA, whereas the other cell lines tested expressed only low or undetectable amounts of c-Met mRNA (Figure 3D). Similar results were obtained by flow cytometry. Again, KU812 cells were found to display substantial amounts of c-Met (Figure 3E). Unexpectedly, K562 cells were also found to express c-Met on their surface, although expression levels were lower compared with KU812 cells. As expected, HUVEC expressed c-Met on their surface (positive control; Figure 3E).
Figure 3.
Expression of HGF and c-Met in various leukemic cell lines. (A) Evaluation of expression of HGF mRNA in various leukemic cell lines, and LUFs by Northern blot analysis (NB left side) and qPCR (right side). β-Actin (NB) and ABL (qPCR) served as control. (B) Measurement of HGF in supernatants of leukemic cell lines, HUVECs, and LUF. Supernatants were obtained after culturing cells in medium with 10% FCS for 5 days. HGF concentrations were determined by ELISA. Results represent the mean ± SD of three independent experiments. (C) ICC evaluation of HGF expression in leukemic cell lines, LUFs and HUVECs. Cells were spun on cytospin slides and stained with an antibody against HGF. Magnification, 100x/1.35. (D) Evaluation of expression of c-Met mRNA in various leukemic cell lines and LUFs by Northern blot analysis (NB; left panel) and qPCR (right panel). β-Actin (NB) and ABL (qPCR) served as control. (E) Surface expression of c-Met on KU812, K562, HMC-1.1, HMC-1.2, HL60, KG-1, U937, LUF, and HUVEC. Cells were analyzed for expression of c-Met by flow cytometry. Expression of c-Met (red histograms) was controlled by an isotype-matched antibody (black open histograms).
Expression of HGF in CML Cells Is Independent of BCR-ABL1
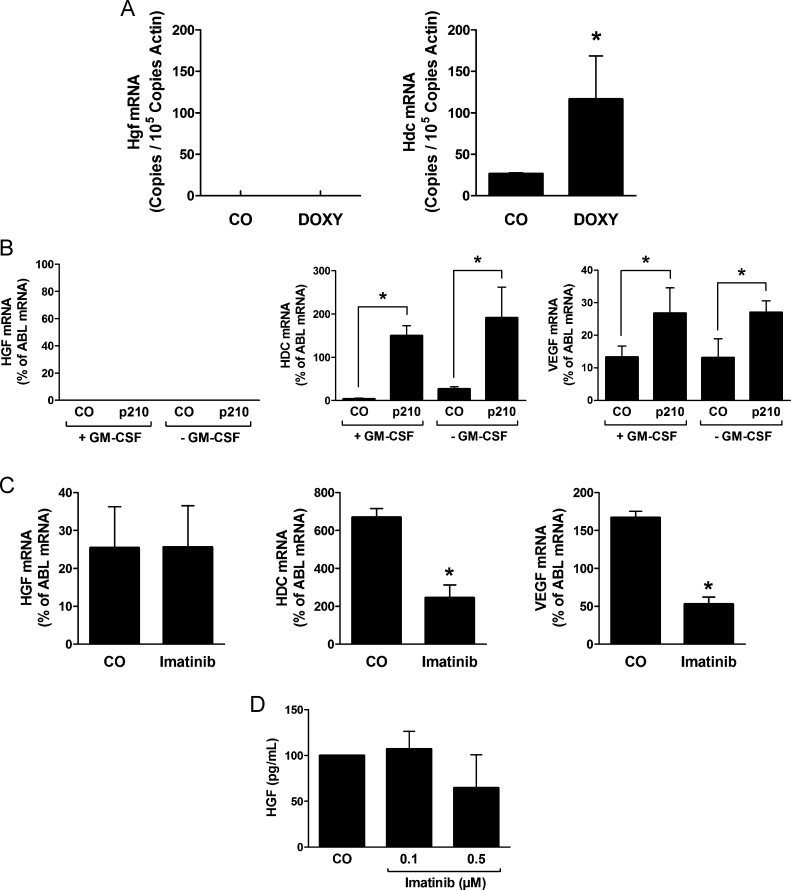
The CML-specific oncoprotein BCR-ABL1 supposedly contributes to the expression of various angiogenic growth factors in leukemic cells [18,19,21]. In the present study, we asked whether BCR-ABL1 is involved in the regulation of expression of HGF. In a first step, we examined the expression of Hgf mRNA in Ton.B210-X cells in which BCR-ABL1 can conditionally be induced by adding doxycycline. However, as shown in Figure 4A, BCR-ABL1 did not induce the expression of Hgf in Ton.B210-X cells, whereas BCR-ABL1 induced the expression of Hdc mRNA in the same experiment. In line with this observation, we were not able to detect any differences in expression of HGF mRNA levels or HGF protein levels when comparing untransfected (or control vector transfected) MO7e cells with MO7e cells stably transfected with BCR-ABL1 (Figure 4B). We also asked whether the BCR-ABL1 inhibitor imatinib would modulate the expression of HGF mRNA (or the HGF protein) in KU812 cells. However, no effect of imatinib on expression of HGF mRNA was found, whereas imatinib caused down-regulation of expression of VEGF mRNA and HDC mRNA in these experiments (Figure 4C). Moreover, although inducing rapid apoptosis in KU812 cells, imatinib did not substantially suppress HGF expression or HGF release in KU812 cells (Figure 4D). All in all, these data suggest that HGF production in CML cells is regulated independent of BCR-ABL1.
Figure 4.
Expression of HGF in CML cells is independent of BCR-ABL1. (A) Expression of Hdc mRNA and Hgf mRNA in Ton.B210-X cells determined by qPCR. Cells were cultured in the presence (DOXY) or absence (CO) of doxycycline (1 µg/ml) for 24 hours to induce BCR-ABL1 and were then subjected to RNA isolation, cDNA synthesis, and qPCR using primers specific for Hdc, Hgf, and Actin. Results show Hdc mRNA expression levels and Hgf mRNA expression levels as copies/105 copies of Actin. Results represent the mean ± SD of three independent experiments. *P < .05. (B) Expression of HGF, HDC and VEGF mRNA in MO7e cells stably transfected with BCR-ABL1 (p210) or with a control vector (CO) determined by qPCR. Cells were cultured in the absence (-GM-CSF) or presence (+GM-CSF) of GM-CSF (100 ng/ml) for 24 hours and were then subjected to RNA isolation, cDNA synthesis, and qPCR using primers specific for HGF, HDC, VEGF, and ABL. Results show HDC and VEGF mRNA levels as percent of ABL mRNA levels and represent the mean ± SD of three independent experiments. *P < .05. (C) Effects of imatinib on expression of HGF, HDC, and VEGF mRNA in KU812 cells. Cells were cultured in the absence (CO) or presence of imatinib (1 µM) for 8 hours and were then subjected to qPCR analysis using primers specific for HGF, HDC, VEGF, and ABL. Results show HGF, HDC, and VEGF mRNA levels as percent of ABL mRNA, and represent the mean ± SD of three independent experiments. (D) Measurement of HGF in supernatants of KU812 cell. Supernatants were obtained after culturing cells in medium with 10% FCS in the absence (CO) or presence of imatinib at 0.1 and 0.5 µM for 24 hours. HGF concentrations were determined by ELISA. Results represent the mean ± SD of three independent experiments.
IL-3 Regulates the Expression and Release of HGF in CML Cells
We next asked whether IL-3, a major regulator of basophil differentiation and function [30,31], would promote the production and expression of HGF in CML cells. Indeed, IL-3 was found to upregulate the expression of HGF mRNA in purified CML basophils (Figure W3). Moreover, IL-3 was found to promote the expression and release of the HGF protein in unfractionated primary CML MNC (Figure W3). By contrast, IL-3 failed to upregulate the expression or release of VEGF in CML cells (not shown).
Basophil-Derived HGF Promotes Endothelial Cell Migration In Vitro
To define a functional role for basophil-derived HGF, we measured the effects of supernatants derived from KU812 cells or primary CML cells on endothelial cell growth and migration using HUVEC and a wound healing (scratch) assay. In these experiments, recombinant HGF as well as supernatants from KU812 cells, induced endothelial growth and migration, whereas supernatants of K562 cells showed no effects (Figure 5A). In addition, we found that supernatants of basophil-rich primary CML samples induced endothelial cell migration, whereas supernatants of primary samples containing low numbers of (or no) basophils did not induce wound healing (Figure 5A). CML supernatant-induced migration of HUVEC was completely blocked by a neutralizing anti-HGF antibody and partly by the addition of an anti-VEGF antibody (Figure 5B). In addition, we were able to show that the CML supernatant-induced endothelial cell migration was blocked by the c-Met inhibitor PF-2341066 (Figure 5C). As expected, migration of HUVEC induced by recombinant HGF was also blocked by the c-Met inhibitor PF-2341066 and by an anti-HGF antibody but not by an anti-VEGF antibody (Figure W4). Together, these data show that basophil-derived HGF and VEGF are functionally active proangiogenic molecules.
Figure 5.
Effects of basophil-derived HGF on endothelial cell migration. (A) Effects of rhHGF and CML-derived HGF on migration of HUVECs in a scratch wound assay. Confluent HUVEC layers were prepared in six-well plates. A linear scratch wound (≈ 100 µm diameter) was produced by a pipette tip. Then, HUVECs were cultured in control medium (CO), rhHGF (100 ng/ml), or 5-day supernatants (Sup.) of KU812 cells, K562 cells (HGF-negative), and primary CML cells (CML-enriched fractions, > 10% basophils; and CML fractions containing < 2% basophils= negative control). After 24 hours, migration of HUVEC was examined under an inverted microscope magnification: 4x/0.13. The scratch wound is marked by white bars. (B) Effects of a neutralizing anti-HGF antibody (αHGF) and a neutralizing anti-VEGF antibody (αVEGF) on migration of HUVECs determined by scratch wound migration assay. After introducing a scratch wound, HUVECs were incubated with supernatants (Sup.) of primary basophil-rich CML cells (>10% basophils) or KU812 cells in the absence (-) or presence (+) of αHGF (60 ng/ml) or/and αVEGF (100 ng/ml) as indicated. After 24 hours (37°C), migration of HUVECs was examined under an inverted microscope and photographed. Cell density in the scratch was scored from 0 to 4 as shown in Figure W1. Results represent the mean ± SD of three independent experiments. (C) Effects of the c-Met inhibitor PF-2341066 on migration of HUVECs induced by KU812 cell supernatant (left panel) or supernatant of primary basophil-rich cells (right panel). After a linear scratch was produced, HUVEC monolayers were incubated with supernatants of CML cells in the absence (CO) or presence of various concentrations of PF-2341066 as indicated. After 24 hours, migration of HUVECs was examined under an inverted microscope and photographed. Cell density in the scratch was scored from 0 to 4 as shown in Figure W1. Results represent the mean ± SD of three independent experiments.
Expression of c-Met mRNA in CML Basophils and CML Progenitor Cells
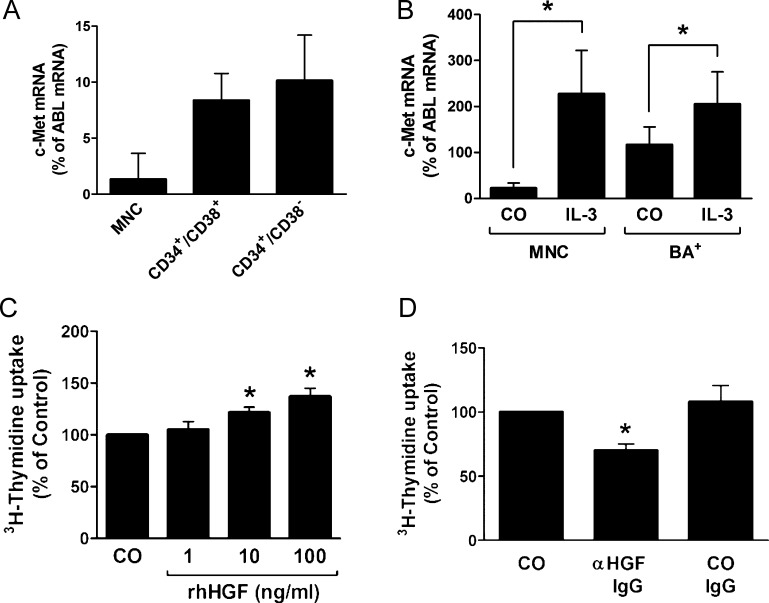
We next examined whether the HGF receptor c-Met is expressed in primary CML cells. In a first step, we examined highly enriched sorted CD34+/CD38- stem cells and CD34+/CD38+ progenitor cells in patients with CML AP. As assessed by qPCR, CML stem cells and CML progenitor cells were found to express low but detectable levels of c-Met mRNA (Figure 6A). In addition, we found that highly enriched CML basophils (CP CML) express low levels of c-Met mRNA (Figure 6B). IL-3 was found to promote c-Met mRNA expression in unfractionated CML MNC as well as in highly enriched CML basophils (Figure 6B). We were also able to show that KU812 cells express c-Met mRNA (Figure 3D). As assessed by flow cytometry, c-Met was found to be expressed on KU812 cells and K562 cells as well as on HUVEC (positive control; Figure 3E). However, unexpectedly, we were unable to detect substantial amounts of c-Met on the surface of unstimulated or IL-3-exposed (CD34+/CD38-) CML stem cells or CML basophils (Figure W5).
Figure 6.
Expression of c-Met on CML progenitor cells and CML basophils. (A) Expression of c-Met mRNA in highly enriched (sorted) CD34+/CD38- stem cells and CD34+/CD38+ progenitor cells obtained from patients with CML in AP as determined by qPCR. Cells were subjected to RNA isolation, cDNA synthesis, and qPCR using primers specific for c-Met and ABL. Results show c-Met mRNA levels as percent of ABL mRNA levels and represent the mean ± SD of three independent experiments (three patients). (B) Expression of c-Met mRNA in PB MNCs and highly purified CD203c+ basophils (BA+) from three patients with CML. MNCs and basophils were incubated in the absence (CO) or presence of IL-3 (100 ng/ml) for 30 minutes and were then subjected to RNA isolation, cDNA synthesis, and qPCR using primers specific for c-Met and ABL. Results show HGF mRNA expression levels as percent of ABLmRNA levels and represent the mean ± SD of three independent experiments (three donors). *P < .05. (C) KU812 cells were incubated in control medium (CO) or in medium containing various concentrations of rhHGF at 37°C for 48 hours. Then, uptake of 3H-thymidine was measured. Results are expressed as percent of control and represent the mean ± SD of three independent experiments. *P < .05. (D) KU812 cells were incubated in control medium (CO) or in medium containing an anti-HGF antibody (αHGF; 1 µg/ml) or a control antibody (CO IgG; 1 µg/ml) at 37°C for 48 hours. Then, uptake of 3H-thymidine was measured. Results are expressed as percent of CO and represent the mean ± SD of three independent experiments. *P < .05.
Effects of HGF on Growth of CML Cells
A number of recent data suggest that angiogenic growth factors, apart from their angiogenic activity, may also act as autocrine growth regulators in leukemic cells. To ask whether HGF can act as a growth regulator in CML cells, we applied HGF and a blocking anti-HGF antibody on CML cells. We found that HGF slightly promotes the proliferation of primary CML cells (Figure W6) and KU812 cells (Figure 6C). In addition, we found that a blocking anti-HGF antibody counteracts spontaneous proliferation of KU812 cells (Figure 6D). These data suggest that HGF is a regulator of autocrine growth of (basophil-committed) CML cells.
Effects of c-Met Inhibitors on Growth of CML Cells
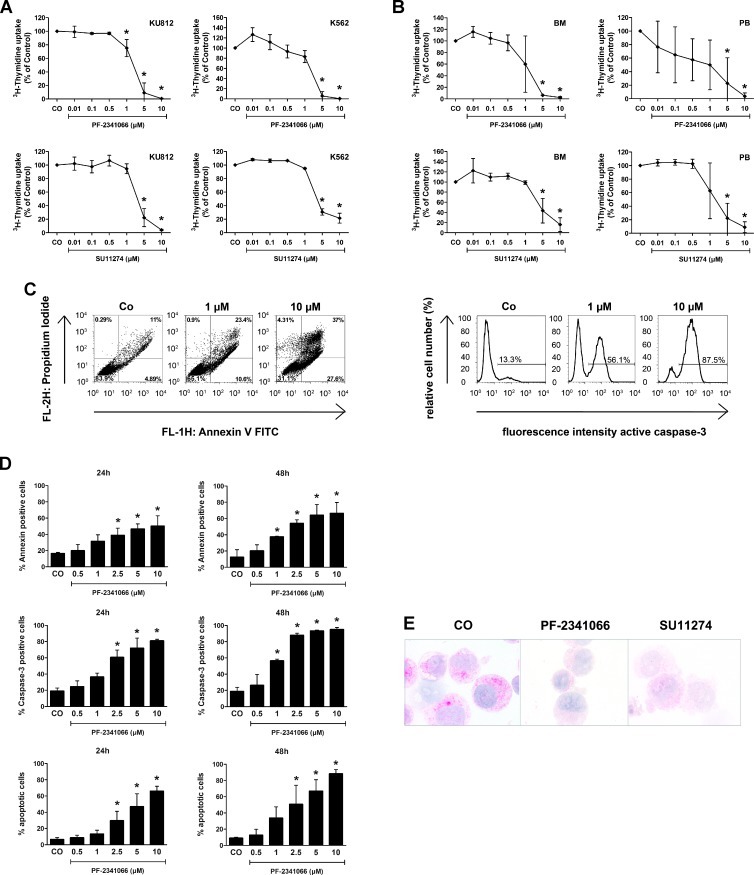
A number of different pharmacologic c-Met inhibitors have been developed during the past few years. In this study, we examined the effects of the c-Met inhibitors PF-2341066 and SU11274 on growth of CML cells. Both inhibitors were found to suppress spontaneous proliferation of KU812 cells and K562 cells in a dose-dependent manner (IC50 = 1–5 µM; Figure 7A). In addition, PF-2341066 and SU11274 were found to inhibit 3H-thymidine uptake in primary CML cells (BM or PB MNC) in all patients tested, with an IC50 of about 1 µM (Figure 7B). The growth-inhibitory effects of the c-Met inhibitor PF-2341066 (in KU812 cells) was found to be accompanied by signs of apoptosis (Figure 7C, D). SU11274 also induced apoptosis in CML cells. However, SU11274-exposed cells also showed signs of necrosis after 24 hours, so that the exact numbers (percentage) of apoptotic cells could not be determined. To confirm drug effects on c-Met kinase activity, ICC was performed using an antibody specific for phospho-c-Met. In these experiments, both PF-2341066 and SU11274 were found to inhibit expression of p-c-Met in KU812 cells (Figure 7E).
Figure 7.
Effects of c-Met inhibitors on growth of CML cells. (A) KU812 cells (left panels) and K562 cells (right panels) were incubated in control medium (CO) or in medium containing various concentrations of the c-Met inhibitors PF-2341066 (upper panels) or SU11274 (lower panels) at 37°C and 5% CO2 for 48 hours. After incubation, uptake of 3H-thymidine was measured. Results are expressed as the percentage of control (CO) and represent the mean ± SD of three independent experiments. (B) Ficoll-isolated mononuclear BM cells (left panels) and PB cells (right panels) were incubated in control medium (CO) or in medium containing various concentrations of the c-Met inhibitors PF-2341066 (upper panels) or SU11274 (lower panels) at 37°C and 5% CO2 for 48 hours. After incubation, 3H-thymidine uptake was measured. Results are expressed as percentage of control and represent the mean ± SD of three independent experiments (three patients). (C and D) KU812 cells were incubated in the absence (CO) or presence of various concentrations of the c-Met inhibitor PF-2341066 at 37°C for 24 and 48 hours. After incubation, apoptosis was measured by light microscopy, Annexin V/PI staining, and active caspase 3 staining by flow cytometry. In C, one typical experiment is shown for Annexin V/PI staining (left panel) and active caspase 3 staining (right panel) after 24 hours. In D, results show the percentage of Annexin V/PI-positive cells (upper panels: left panels, 24 hours; right panels, 48 hours), the percentage of active caspase 3-positive cells (middle panels: left panels, 24 hours; right panels, 48 hours), and the numbers of apoptotic cells by light microscopy (lower panels: left panels, 24 hours; right panels, 48 hours). Results represent the mean ± SD of three independent experiments. *P < .05 compared with control (CO). (E) KU812 cells were incubated in control medium (CO) or with the c-Met inhibitors PF-2341066 or SU11274 (each 1 µM) at 37°C for 4 hours. Then, cells were spun on cytospin slides and stained with an antibody against phospho c-Met (magnification, 100x/1.35).
Discussion
HGF is an established mediator of angiogenesis and is considered to play an important role in the pathogenesis of various myeloid neoplasms [16–22]. In CML, increased levels of HGF have been described as a prognostic variable that correlates with survival [23–25]. It has also been described that HGF levels correlate with BM angiogenesis. However, so far, little is known about the regulation of HGF expression and the cellular source of HGF in CML. We here describe that basophils are a major source of HGF in CML and that basophilderived HGF promotes endothelial cell migration in vitro. In addition, our data show that basophil-derived HGF acts on endothelial cells through a specific receptor, c-Met, and that the effects of HGF can be blocked by anti-HGF antibodies as well as by c-Met inhibitors. Together, these data suggest that basophils may play a more active role in disease acceleration in CML than has so far been assumed. In addition, our data suggest that basophil-derived HGF as well as c-Met may serve as novel potential targets in CML.
Recent data suggest that HGF expression correlates with the phase of disease and the prognosis in CML [23–25]. In the current study, we were able to confirm these data. In particular, we were able to show that HGF mRNA levels are higher in leukemic cells in patients with AP compared to those in patients with CP. Because progression of CML is usually accompanied by an increase in basophils [11–13], we asked whether basophils are a particular source of HGF in CML. Although these cells are well known to produce an array of vascular and angiogenic mediators [32–34], they have not been analyzed for expression of angiogenic growth factors in CML so far. In the present study, we were able to show that basophils are a unique source of HGF in CML. In particular, primary isolated CML basophils were found to express substantial amounts of HGF mRNA as well as the HGF protein, whereas basophil-depleted cell fractions contained only low amounts of HGF. Moreover, we found that the basophil-committed CML cell line KU812 expresses substantial amounts of HGF mRNA and the HGF protein, whereas other leukemic cell lines tested, including the immature uncommitted CML cell line K562, expressed only low amounts or no detectable HGF.
A number of different angiogenic factors, including VEGF and HGF, have been implicated in the pathogenesis of CML [23–25]. The expression and release of most of these growth regulators in CML cells may be triggered by the disease-specific oncoprotein, BCR-ABL1 [19,21,22]. Therefore, we were interested to learn whether BCR-ABL1 would also promote the expression of HGF in leukemic cells. However, unexpectedly, BCR-ABL1 failed to induce expression of HGF in Ba/F3 cells and MO7e cells. Moreover, imatinib failed to inhibit the production of HGF in CML cells. By contrast, we were able to show that BCR-ABL1 promotes expression of VEGF and HDC in Ba/F3 cells and that imatinib inhibits expression of VEGF and HDC in CML cells, confirming previous studies [21]. These data suggest that, in contrast to other angiogenic factors, HGF is expressed in CML cells independent of BCR-ABL1. This is of particular interest because it has been postulated that, in accelerated CML, BCR-ABL1-independent factors play a particular pathogenetic role [6,8–10].
We next asked what factors, apart from BCR-ABL1, could play a role in the production of HGF in leukemic cells. Because IL-3 is known to induce differentiation and activation of human basophils [30,31], we asked whether IL-3 would promote the expression of HGF in leukemic cells. Our data show that IL-3 promotes the expression of HGF in primary CML cells as well as in highly enriched (sorted) CML basophils. This observation suggests that apart from BCR-ABL1, cytokine effects may play a role in the generation of angiogenic factors in CML.
Although several studies have pointed at a potential prognostic value of HGF in CML [23–25], only little is known about the functional role of this angiogenic molecule in CML. So far, HGF levels have been shown to correlate with BM angiogenesis in patients with CML. In the present study, we asked whether basophil-derived HGF would induce endothelial cell migration and growth. Indeed, recombinant HGF as well as basophil-derived HGF were found to induce migration and growth of endothelial cells in a scratch wound assay. The effects of HGF in this assay were blocked by an anti-HGF antibody but not by an anti-VEGF antibody, suggesting that the effects of HGF on endothelial cells were specific and probably mediated through a specific receptor.
HGF exerts effects on its targets cells through a specific receptor, c-Met. We therefore asked whether endothelial cells express c-Met. Indeed, cultured HUVECs were found to express c-Met mRNA as well as surface c-Met protein. We also applied a pharmacologic c-Met inhibitor to show that the effects of CML-derived and recombinant HGF are specific. In these experiments, the c-Met inhibitor PF-2341066 was found to block endothelial cell migration induced by recombinant HGF or CML-derived HGF. Together, these data show that CML-derived HGF is a functionally active molecule that may contribute to CML-associated angiogenesis.
A number of previous studies have shown that HGF regulates growth and function of hematopoietic progenitor cells [35–39]. In the present study, we asked whether CML cells express c-Met and are responsive to HGF. As assessed by qPCR, CML basophils as well as CD34+ CML cells were found to express c-Met mRNA. In addition, KU812 cells and K562 cells were found to express c-Met mRNA. However, whereas KU812 cells expressed substantial amounts of c-Met on their surface, K562 cells expressed only low amounts of c-Met in our flow cytometry experiments, and primary CML cells were found to express very low levels or even stained negative for c-Met. In functional analyses, HGF induced a slight increase in proliferation of KU812 cells and primaryCML cells above control. In addition, a blocking anti-HGF antibody was found to suppress spontaneous growth of KU812 cells, suggesting that HGF acts as an autocrine factor in these cells. Moreover, the c-Met inhibitors PF-2341066 and SU11274 were found to suppress the growth of primary CML cells as well as the growth of KU812 cells and K562 cells. These results suggest that the low amounts of c-Met in CML cells may be sufficient for mediating biologic activity. Alternatively, the c-Met inhibitor exerted “off target effects” on CML cells and thereby introduced growth inhibition. An interesting aspect is that relatively high drug concentrations were required to produce growth inhibition in CML cell lines. However, in primary CML cells, drug effects were within a pharmacologically meaningful range, that is, around or below 1 µM.
In summary, our data show that basophils are a major source of HGF in CML and that basophil-derived HGF acts as a potent paracrine factor promoting endothelial cell migration and growth. Basophils may play a more active role in disease acceleration in CML than has so far been assumed. Whether basophils, basophil-derived HGF, or c-Met could serve as therapeutic targets in CML remains to be elucidated.
Supplementary Material
Acknowledgments
The authors thank Miriam Klauser, Gabriele Stefanzl, and Michaela Seiser for skillful technical assistance. The authors also thank Gunther Hofbauer and Andreas Spittler (Cell Sorting Core Unit of the Medical University of Vienna) for important technical support.
Abbreviations
- AP
accelerated phase
- BM
bone marrow
- BP
blast phase
- c-Met
c-mesenchymal epithelial transition factor (HGF receptor)
- CML
chronic myeloid leukemia
- CP
chronic phase
- IL-3
interleukin 3
- FCS
fetal calf serum
- HDC
histidine decarboxylase
- HGF
hepatocyte growth factor
- HUVEC
human umbilical vein-derived endothelial cells
- LUF
lung fibroblast
- mAb
monoclonal antibody
- MNC
mononuclear cell
- PB
peripheral blood
- PCR
polymerase chain reaction
- rh
recombinant human
- VEGF
vascular endothelial growth factor
Footnotes
This study was supported by the Austrian Science Fund (FWF) grant no. SFB-046-11 and a Cancer Stem Cell Grant from the Medical University of Vienna. Peter Valent received a Research Grant from Novartis and a research grant from BMS. The authors declare no other conflict of interest.
This article refers to supplementary materials, which are designated by Tables W1 to W3 and Figures W1 to W6 and are available online at www.neoplasia.com.
References
- 1.Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 2.de Klein A, van Kessel AG, Grosveld G, Bartram CR, Hagemeijer A, Bootsma D, Spurr NK, Heisterkamp N, Groffen J, Stephenson JR. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982;300:765–767. doi: 10.1038/300765a0. [DOI] [PubMed] [Google Scholar]
- 3.Heisterkamp N, Groffen J. Molecular insights into the Philadelphia translocation. Hematol Pathol. 1991;5:1–10. [PubMed] [Google Scholar]
- 4.Skorski T, Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi JK, Trotta R, Wlodarski P, Perrotti D, Chan TO, et al. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–3356. [PubMed] [Google Scholar]
- 6.Melo JV, Deininger MW. Biology of chronic myelogenous leukaemia—signaling pathways of initiation and transformation. Hematol Oncol Clin North Am. 2004;18:545–568. doi: 10.1016/j.hoc.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Goldman JM. Treatment strategies for CML. Best Pract Res Clin Haematol. 2009;22:303–313. doi: 10.1016/j.beha.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Giles FJ, Cortes JE, Kantarjian HM, O'Brien SM. Accelerated and blastic phases of chronic myelogenous leukemia. Hematol Oncol Clin North Am. 2004;18:753–774. doi: 10.1016/j.hoc.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 10.Perrotti D, Jamieson C, Goldman J, Skorski T. Chronic myeloid leukemia: mechanisms of blastic transformation. J Clin Invest. 2010;120:2254–2264. doi: 10.1172/JCI41246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denburg JA, Wilson WE, Bienenstock J. Basophil production in myeloproliferative disorders: increases during acute blastic transformation of chronic myeloid leukemia. Blood. 1982;60:113–120. [PubMed] [Google Scholar]
- 12.Hasford J, Pfirrmann M, Hehlmann R, Allan NC, Baccarani M, Kluin-Nelemans JC, Alimena G, Steegmann JL, Ansari H. A new prognostic score for survival of patients with chronic myeloid leukemia treated with interferon alfa. Writing Committee for the Collaborative CML Prognostic Factors Project Group. J Natl Cancer Inst. 1998;90:850–858. doi: 10.1093/jnci/90.11.850. [DOI] [PubMed] [Google Scholar]
- 13.Steegmann JL, Odriozola J, Rodriguez-Salvanés F, Giraldo P, Garcïa-Laraña J, Ferro MT, Benïtez E, Pérez-Pons C, Giralt M, Escribano L, et al. Stage, percentage of basophils at diagnosis, hematologic response within six months, cytogenetic response in the first year: the main prognostic variables affecting outcome in patients with chronic myeloid leukemia in chronic phase treated with interferon-α. Results of the CML89 trial of the Spanish Collaborative Group on interferon-α2a and CML. Haematologica. 1999;84:978–987. [PubMed] [Google Scholar]
- 14.Deininger M. Resistance and relapse with imatinib in CML: causes and consequences. J Natl Compr Canc Netw. 2008;6:11–21. [PubMed] [Google Scholar]
- 15.Quintás-Cardama A, Kantarjian HM, Cortes JE. Mechanisms of primary and secondary resistance to imatinib in chronic myeloid leukemia. Cancer Control. 2009;16:122–131. doi: 10.1177/107327480901600204. [DOI] [PubMed] [Google Scholar]
- 16.Aguayo A, Kantarjian H, Manshouri T, Gidel C, Estey E, Thomas D, Koller C, Estrov Z, O'Brien S, Keatina M, et al. Angiogenesis in acute and chronic leukemias and myelodysplastic syndromes. Blood. 2000;96:2240–2245. [PubMed] [Google Scholar]
- 17.Verstovsek S, Kantarjian H, Manshouri T, Cortes J, Giles FJ, Rogers A, Albitar M. Prognostic significance of cellular vascular endothelial growth factor expression in chronic phase chronic myeloid leukemia. Blood. 2002;99:2265–2267. doi: 10.1182/blood.v99.6.2265. [DOI] [PubMed] [Google Scholar]
- 18.Krauth MT, Simonitsch I, Aichberger KJ, Mayerhofer M, Sperr WR, Sillaber C, Schneeweiss B, Mann G, Gadner H, Valent P. Immunohistochemical detection of VEGF in the bone marrow of patients with chronic myeloid leukemia and correlation with the phase of disease. Am J Clin Pathol. 2004;121:473–481. doi: 10.1309/3JLT-FNNE-DQHB-4A0P. [DOI] [PubMed] [Google Scholar]
- 19.Janowska-Wieczorek A, Majka M, Marquez-Curtis L, Wertheim JA, Turner AR, Ratajczak MZ. Bcr-abl-positive cells secrete angiogenic factors including matrix metalloproteinases and stimulate angiogenesis in vivo in Matrigel implants. Leukemia. 2002;16:1160–1166. doi: 10.1038/sj.leu.2402486. [DOI] [PubMed] [Google Scholar]
- 20.Müller A, Lange K, Gaiser T, Hofmann M, Bartels H, Feller AC, Merz H. Expression of angiopoietin-1 and its receptor TEK in hematopoietic cells from patients with myeloid leukemia. Leuk Res. 2002;26:163–168. doi: 10.1016/s0145-2126(01)00110-2. [DOI] [PubMed] [Google Scholar]
- 21.Mayerhofer M, Valent P, Sperr WR, Griffin JD, Sillaber C. BCR/ABL induces expression of vascular endothelial growth factor and its transcriptional activator, hypoxia inducible factor-1α, through a pathway involving phosphoinositide 3-kinase and the mammalian target of rapamycin. Blood. 2002;100:3767–3775. doi: 10.1182/blood-2002-01-0109. [DOI] [PubMed] [Google Scholar]
- 22.Sillaber C, Mayerhofer M, Aichberger KJ, Krauth MT, Valent P. Expression of angiogenic factors in chronic myeloid leukaemia: role of the bcr/abl oncogene, biochemical mechanisms, and potential clinical implications. Eur J Clin Invest. 2004;34:2–11. doi: 10.1111/j.0960-135X.2004.01365.x. [DOI] [PubMed] [Google Scholar]
- 23.Hino M, Inaba M, Goto H, Nishizawa Y, Tatsumi N, Nishino T, Morii H. Hepatocyte growth factor levels in bone marrow plasma of patients with leukaemia and its gene expression in leukaemic blast cells. Br J Cancer. 1996;73:119–123. doi: 10.1038/bjc.1996.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JG, Sohn SK, Kim DH, Baek JH, Lee NY, Suh JS, Chae SC, Lee KS, Lee KB. Clinical implications of angiogenic factors in patients with acute or chronic leukemia: hepatocyte growth factor levels have prognostic impact, especially in patients with acute myeloid leukemia. Leuk Lymphoma. 2005;46:885–891. doi: 10.1080/10428190500054491. [DOI] [PubMed] [Google Scholar]
- 25.Zhelyazkova AG, Tonchev AB, Kolova P, Ivanova L, Gercheva L. Prognostic significance of hepatocyte growth factor and microvessel bone marrow density in patients with chronic myeloid leukaemia. Scand J Clin Lab Invest. 2008;68:492–500. doi: 10.1080/00365510701854991. [DOI] [PubMed] [Google Scholar]
- 26.Bühring HJ, Simmons PJ, Pudney M, Muller R, Jarrossay D, van Agthoven A, Willheim M, Brugger W, Valent P, Kanz L. The monoclonal antibody 97A6 defines a novel surface antigen expressed on human basophils and their multipotent and unipotent progenitors. Blood. 1999;94:2343–2356. [PubMed] [Google Scholar]
- 27.McEuen AR, Buckley MG, Compton SJ, Walls AF. Development and characterization of a monoclonal antibody specific for human basophils and the identification of a unique secretory product of basophil activation. Lab Invest. 1999;79:27–38. [PubMed] [Google Scholar]
- 28.Hoermann G, Cerny-Reiterer S, Perné A, Klauser M, Hoetzenecker K, Klein K, Müllauer L, Gröger M, Nijman SM, Klepetko W, et al. Identification of oncostatin M as a STAT5-dependent mediator of bone marrow remodeling in KIT D816V-positive systemic mastocytosis. Am J Pathol. 2011;178:2344–2356. doi: 10.1016/j.ajpath.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agis H, Krauth MT, Böhm A, Mosberger I, Mullauer L, Simonitsch-Klupp I, Walls AF, Horny HP, Valent P. Identification of basogranulin (BB1) as a novel immunohistochemical marker of basophils in normal bone marrow and patients with myeloproliferative disorders. Am J Clin Pathol. 2006;125:273–281. doi: 10.1309/M9FQ-MQGF-6616-7N2X. [DOI] [PubMed] [Google Scholar]
- 30.Valent P, Schmidt G, Besemer J, Mayer P, Zenke G, Liehl E, Hinterberger W, Lechner K, Maurer D, Bettelheim P. Interleukin-3 is a differentiation factor for human basophils. Blood. 1989;73:1763–1769. [PubMed] [Google Scholar]
- 31.Valent P, Besemer J, Muhm M, Majdic O, Lechner K, Bettelheim P. Interleukin 3 activates human blood basophils via high-affinity binding sites. Proc Natl Acad Sci USA. 1989;86:5542–5546. doi: 10.1073/pnas.86.14.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marone G, Triggiani M, Genovese A, De Paulis A. Role of human mast cells and basophils in bronchial asthma. Adv Immunol. 2005;88:97–160. doi: 10.1016/S0065-2776(05)88004-6. [DOI] [PubMed] [Google Scholar]
- 33.de Paulis A, Prevete N, Fiorentino I, Rossi FW, Staibano S, Montuori N, Ragno P, Longobardi A, Liccardo B, Genovese A, et al. Expression and functions of the vascular endothelial growth factors and their receptors in human basophils. J Immunol. 2006;177:7322–7331. doi: 10.4049/jimmunol.177.10.7322. [DOI] [PubMed] [Google Scholar]
- 34.Crivellato E, Travan L, Ribatti D. Mast cells and basophils: a potential link in promoting angiogenesis during allergic inflammation. Int Arch Allergy Immunol. 2010;151:89–97. doi: 10.1159/000235998. [DOI] [PubMed] [Google Scholar]
- 35.Kmiecik TE, Keller JR, Rosen E, Vande Woude GF. Hepatocyte growth factor is a synergistic factor for the growth of hematopoietic progenitor cells. Blood. 1992;80:2454–2457. [PubMed] [Google Scholar]
- 36.Mizuno K, Higuchi O, Ihle JN, Nakamura T. Hepatocyte growth factor stimulates growth of hematopoietic progenitor cells. Biochem Biophys Res Commun. 1993;194:178–186. doi: 10.1006/bbrc.1993.1801. [DOI] [PubMed] [Google Scholar]
- 37.Galimi F, Bagnara GP, Bonsi L, Cottone E, Follenzi A, Simeone A, Comoglio PM. Hepatocyte growth factor induces proliferation and differentiation of multipotent and erythroid hemopoietic progenitors. J Cell Biol. 1994;127:1743–1754. doi: 10.1083/jcb.127.6.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishino T, Hisha H, Nishino N, Adachi M, Ikehara S. Hepatocyte growth factor as a hematopoietic regulator. Blood. 1995;85:3093–3100. [PubMed] [Google Scholar]
- 39.Ratajcak MZ, Marlicz W, Ratajcak J, Wasik M, Machalinski B, Carter A, Gewirtz AM. Effect of hepatocyte growth factor on early human haematopoietic development. Br J Haematol. 1997;99:228–236. doi: 10.1046/j.1365-2141.1997.3563170.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.