Abstract
High-resolution image guidance for resection of residual tumor cells would enable more precise and complete excision for more effective treatment of cancers, such as medulloblastoma, the most common pediatric brain cancer. Numerous studies have shown that brain tumor patient outcomes correlate with the precision of resection. To enable guided resection with molecular specificity and cellular resolution, molecular probes that effectively delineate brain tumor boundaries are essential. Therefore, we developed a bioinformatics approach to analyze micro-array datasets for the identification of transcripts that encode candidate cell surface biomarkers that are highly enriched in medulloblastoma. The results identified 380 genes with greater than a two-fold increase in the expression in the medulloblastoma compared with that in the normal cerebellum. To enrich for targets with accessibility for extracellular molecular probes, we further refined this list by filtering it with gene ontology to identify genes with protein localization on, or within, the plasma membrane. To validate this meta-analysis, the top 10 candidates were evaluated with immunohistochemistry. We identified two targets, fibrillin 2 and EphA3, which specifically stain medulloblastoma. These results demonstrate a novel bioinformatics approach that successfully identified cell surface and extracellular candidate markers enriched in medulloblastoma versus adjacent cerebellum. These two proteins are high-value targets for the development of tumor-specific probes in medulloblastoma. This bioinformatics method has broad utility for the identification of accessible molecular targets in a variety of cancers and will enable probe development for guided resection.
Introduction
Effective surgical resection of brain tumors aims to remove as much diseased tissue as possible while sparing the critical neural tissue immediately adjacent to the malignancy. Whereas some cancers have clearly visible margins, others, such as medulloblastoma, have margins that can be difficult to identify [1]. While preoperative magnetic resonance imaging (MRI) with stereotactic positioning is commonly used to guide resection, poor contrast and loss of position registration due to intraoperative tissue deformation limit this image guidance technique. In addition, the few open-configuration intraoperative MRI units that exist have insufficient resolution, sensitivity, and contrast to delineate tumor margins accurately. Recent advances in micro-optical technologies have enabled the development of miniaturized microscopes to image tumor cells during surgery [2–4]. These devices have been shown to provide high-resolution information that complements wide-field fluorescence image-guided surgery techniques that have been gaining popularity among neurosurgical researchers [3–6]. For instance, Sanai et al. [4] have reported that, in low-grade gliomas, wide-field imaging based on 5-aminolevulinic acid-induced tumor fluorescence is unable to distinguish between tumor and normal regions in human patients but that individual tumor cells are identified and quantified using a real-time surgical confocal microscope. Among this research community, there is consensus about the potential benefits from the use of molecularly specific optical contrast agents that are capable of highlighting tumor tissues relative to normal brain to improve tumor resection while minimizing the destruction of healthy brain tissue.
The development of molecular probes with sufficient contrast to delineate tumor margins requires the identification of targets highly enriched in tumor tissues compared with adjacent, normal tissue. Unlike pharmaceutical development, where drug targets must ideally be present specifically in diseased tissue only, optical probe targets may be highly expressed in other tissues so long as expression in nontumor tissues occurs far from the surgical site. Therefore, bioinformatics techniques that compare gene transcript levels in tumor versus adjacent normal tissue can be used to mine the rich data sets available from human tumors and from animal models as a means of identifying molecular targets for optical probe development.
One attractive disease target for optical probe development is medulloblastoma, the most common brain tumor in children. Medulloblastoma is a solid tumor whose effective resection is both key to prolonged disease-free survival, yet its margins are difficult to visualize [7,8]. Consequently, 20% of children who are cured of the disease develop severe, sometimes irreversible neurologic deficits from overaggressive tumor resection [9,10]. Because of these risks, aggressive resections are often avoided. Real-time and precise image guidance is therefore necessary to allow for complete and accurate resection of medulloblastoma.
Here we describe the use of a novel method to identify transcripts highly enriched in medulloblastoma. Because there is no publicly available data set containing both medulloblastoma samples and normal cerebellum samples drawn from the same patient, we compared medulloblastoma studies published by the Versteeg and the Gilbertson groups [11,12] to normal cerebellum arrays (GSE3526 and GSE13162). Because cell surface or extracellular proteins are preferred targets for an injected or topically applied optical probe, we selected genes coding for proteins with known localization to the plasma membrane or extracellular space. To validate these results, we performed immunohistochemistry on human tissue micro-arrays to determine protein expression of the top 10 candidates coded by the transcripts, and identified two genes, fibrillin-2 and Eph receptor A3, with the gene products of these genes have significantly enhanced immunoreactivity in the medulloblastoma versus that in the cerebellum. Although immunohistochemistry is not the method to be used for image-guided resection, it is a method of target validation. These targets can be used for the development of specific probes, either the antibodies shown here or other molecules, for use in intraoperative image guidance over a range of scales from high-resolution to wide-field imaging.
Materials and Methods
Meta-analysis of Medulloblastoma Gene Expression Microarray Experiments
Gene expression microarray samples of medulloblastoma were obtained from NCBI Gene Expression Omnibus (GEO) experiment GSE10327 and also from the supplementary data accompanying Thompson et al. [12], which was downloaded from the St. Jude Research Data Web site (http://www.stjuderesearch.org/data/medulloblastoma). Microarray samples of normal cerebellum were obtained from NCBI GEO experiment GSE13162. The probe set annotations for micro-array platforms were updated using AILUN [13].
Meta-analysis of genes differentially expressed between normal cerebellum and medulloblastoma was conducted using the Rank Products method [14] implemented in the RankProd package for the R statistical language (http://r-project.org). To compensate for the fact that disease and normal control samples did not come from the same experiment, each set of disease samples was matched with the combined set of normal samples to form a pseudomatched case versus control experiment. Each of these parings was assigned a unique origin identifier in the RankProd analysis to enable aggregate rank-based meta-analysis compensating for per-experiment variance. False discovery rates were estimated by the software using 1000 randomization rounds and differentially expressed genes exhibiting a fold change greater than 2 in medulloblastoma at a false discovery rate less than 5% were selected. The remaining genes were mapped to Gene Ontology cellular component categories using DAVID [15], and genes annotated to be localized on the cell membrane were retained as putative surface biomarkers for medulloblastoma.
Immunohistochemistry
Human tissue microarrays containing human medulloblastoma and normal human cerebellum (GL2082 and GL1001 from US Biomax, Inc, Rockville, MD) were removed from paraffin and rehydrated in water. Slides were treated with 3% hydrogen peroxide to block endogenous peroxidase for 10 minutes. Slides were treated with 0.1 M citric acid at 100°C for 5 minutes and then allowed to cool to room temperature for 40 minutes. Slides were then washed with phosphate-buffered saline (PBS) and then blocked with 2% bovine serum albumin for 30 minutes. Slides were washed with PBS and incubated with primary antibody for 2 hours at room temperature. Slides were washed with PBS and incubated in a biotinylated goat anti-rat or rabbit antibody at 1:500 for 30 minutes at room temperature. Slides were washed with PBS and incubated with streptavidin-HRP at 1:1000 for 30 minutes at room temperature. Slides were washed in PBS and treated with diaminobenzidine. Slides were washed with PBS and counterstained with hematoxylin. Slides were washed in water, dehydrated, cleared, and mounted in permanent mounting media.
Primary antibodies were rabbit anti-Eph Receptor A3 (PAB3005; Abnova, Cambridge, MA), rabbit anti-Fibrillin (HPA012853; Sigma-Aldrich, Seelze, Germany) and rat monoclonal anti-Laminin beta 1 antibody (Abcam, Cambridge, MA), anti-Frizzled-2 (181-46110749; GenWay, San Diego, CA), mouse anti-protocadherin 8 (H00005100; Abnova), rabbit anti-transforming growth factor beta receptor I (PAB3503; Abnova), anti-connective tissue growth factor (PAB3503; Abnova), anti-villin 2 (MAB3822; Millipore, Billerica, MA), anti-poliovirus receptor-related 3 (HPA011038; Sigma-Aldrich), anti-F2R (PAB11822; Abnova). Secondary antibodies were purchased from Jackson Immunoresearch (West Grove, PA).
Images were acquired with a Zeiss AxioVert microscope (Zeiss, Oberkochen, Germany) with a 60x 1.4 NA objective.
Results
To identify candidate cell surface biomarkers for medulloblastoma and building from a previously published method [16], we developed a meta-analysis method called Public Microarray Integration of Cases and Control (PuMiCC) to find disparate microarray data sets for cases and controls, integrate these using rank products, and filter the differentially expressed genes based on the localization of their coded proteins within the cell. For cases of medulloblastoma, we used GSE10327 from NCBI GEO (62 samples) and a medulloblastoma data set obtained from St. Jude Hospital (40 samples) for a total of 102 human medulloblastoma microarray samples. Normal human cerebellum arrays were derived from GSE3526 (10 samples) and GSE13162 (6 samples).
The results identified 380 genes that exhibited a greater-than-twofold change in expression in the medulloblastoma relative to the normal cerebellum at a false-positive rate less than 0.05. This list was then filtered using Gene Ontology cellular component annotations to derive a list of 95 genes known to be located on or within the cell membrane. The top 30 candidate medulloblastoma cell surface markers are shown in Table 1.
Table 1.
Top 30 Most Enriched Transcripts in Medulloblastoma Verse Cerebellum.
| Gene Name | Symbol | Fold Change (Medulloblastoma) |
| Actin-like 6A | ACTL6A | 3.9 |
| Centromere protein F, 350/400KA (mitosin) | CENPF | 3.6 |
| Laminin, beta 1 | LAMB1 | 3.3 |
| CDC42 effector protein (rho GTPase binding) 3 | CDC42EP3 | 3.2 |
| Transmembrane protein 45A | TMEM45A | 3.1 |
| Protocadherin 8 | PCDH8 | 3.1 |
| GTP binding protein overexpressed in skeletal muscle | GEM | 3.0 |
| Connective tissue growth factor | CTGF | 3.0 |
| Calmegin | CLGN | 2.9 |
| Down syndrome critical region gene 2 | PSMG1 | 2.9 |
| DNAJ (HSP40) homolog, subfamily C, member 10 | DNAJC10 | 2.7 |
| Villin 2 (ezrin) | EZR | 2.7 |
| Solute carrier family 25, member 13 (citrin) | SLC25A13 | 2.7 |
| Transmembrane protein 123 | TMEM123 | 2.7 |
| Nucleoporin 37 kDa | NUP37 | 2.7 |
| Thymopoietin | TMPO | 2.7 |
| SEC22 vesicle trafficking protein-LIKE 2 (Saccharomyces cerevisiae) | SEC22A | 2.7 |
| v-HA-ras Harvey rat sarcoma viral oncogene homolog | NRAS | 2.7 |
| Coagulation factor II (thrombin) receptor | F2R | 2.6 |
| Mitochondrial carrier family protein | SLC25A40 | 2.6 |
| Fat tumor suppressor homolog 4 (Drosophila) | FAT4 | 2.6 |
| Fibrillin 2 (congenital contractural arachnodactyly) | FBN2 | 2.6 |
| Frizzled homolog 2 (Drosophila) | FZD2 | 2.6 |
| Eph receptor A3 | EPHA3 | 2.6 |
| Transforming growth factor, beta receptor i | TGFBR1 | 2.5 |
| Kiaa1479 protein (sema6D) | SEMA6D | 2.5 |
| Proline-serine-threonine phosphatase interacting protein 2 | PSTPIP2 | 2.5 |
| Trophoblast glycoprotein | TPBG | 2.5 |
| Poliovirus receptor-related 3 | PVRL3 | 2.4 |
| Solute carrier organic anion transporter family, member 2A1 | SLCO2A1 | 2.4 |
| Chromosome 14 open reading frame 109 | C14orf109 | 2.4 |
| Zinc finger protein 643 | ZNF643 | 2.4 |
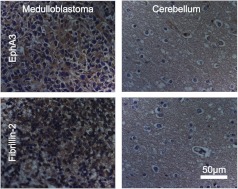
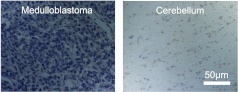
Ten candidate biomarkers were selected for validation by immunohistochemistry (Table 2). We focused on candidate genes coding for proteins with known localization in the outer leaflet of the plasma membrane or in the extracellular space. Genes that had commercially available antibodies reactive against human antigen were selected for immunohistochemistry analysis. These antibodies were used to label commercially available human tissue microarrays (US Biomax, Inc) containing biopsies from 10 cases of medulloblastoma, 10 cases of normal cerebellum, and 80 cases of other brain disorders. Staining intensity was scored on a 0 to 3+ scale. Of the 10 candidate biomarkers, fibrillin 2 was deeply stained (3+) in 4 of 10 medulloblastoma cases and EphA3 was highly stained in 3 of 10 medulloblastoma cases, whereas normal cerebellum was nonreactive (Figure 1). Isotype controls did not stain the medulloblastoma tissue (Figure 2).
Table 2.
Candidate Biomarkers Selected for Immunohistochemistry Validation.
| Gene | Name | Fold Change (Medulloblastoma) |
| LAMB1 | Laminin, beta 1 | 9.68 |
| PCDH8 | Protocadherin 8 | 8.59 |
| CTGF | Connective tissue growth factor | 7.95 |
| EZR | Villin 2 (ezrin) | 6.71 |
| F2R | Coagulation factor II (thrombin) receptor | 6.03 |
| FBN2 | Fibrillin 2 | 5.93 |
| FZD2 | Frizzled homolog 2 (Drosophila) | 5.90 |
| EPHA3 | Eph receptor A3 | 5.85 |
| TGFBR1 | Transforming growth factor, beta receptor I | 5.71 |
| PVRL3 | Poliovirus receptor-related 3 | 5.41 |
Figure 1.
Medulloblastoma but not cerebellum expresses the Eph receptor A3 and fibrillin 2. Bright-field micrographs show immunohistochemical staining of the human medulloblastoma and the normal cerebellum. An anti-Eph A3 antibody (top) and an anti-fibrillin 2-antibody stained medulloblastoma (left), but not cerebellum.
Figure 2.
Medulloblastoma and cerebellum stain lightly with rabbit IgG antibody serum labeled with biotinylated goat anti-rabbit secondary antibody. Bright-field micrographs show immunohistochemical staining of the human medulloblastoma and the normal cerebellum.
Discussion
Taken together, we have demonstrated a novel bioinformatics approach that successfully identified cell surface and extracellular candidate markers enriched in medulloblastoma versus adjacent cerebellum. Furthermore, these candidate markers were validated using immunohistochemistry on human tissue microarrays, revealing two genes, fibrillin 2 and Eph receptor A3.
Fibrillins interact with integrins and heparin-sulphated proteoglycans and likely play important roles in cell migration, adhesion, signaling, and cell differentiation—all important processes in tumor growth and metastasis [17]. Fibrillin 2 transcript and protein is also densely present in rhabdomyosarcoma [18]. The related protein fibrillin 1 is present in some glioblastomas [19], suggesting that fibrillins could be a common marker of invasive tumors. Whereas fibrillin 2 has been shown to have a widespread distribution in the extracellular matrix of healthy skin, lung, liver, and other tissues, it is not highly expressed in the brain [20], making it an excellent specific target for medulloblastoma marker development.
Eph receptors are tyrosine kinases that play a role in cell-cell interaction and cell migration [21]. Both Ephrin and Eph receptor over-expression may promote tumor growth, survival, angiogenesis, and metastasis. Mutations in Eph receptors have also been identified in breast cancer (EPHA1, EPHA6, EPHA10) [22] and in lung cancer (EPHA3, EPHA5, EPHA6, EPHB2, EPHB3, EPHB4) [23]. Interestingly, soluble forms of Eph A receptors (EPHA2 and EPHA3) appear to inhibit tumor angiogenesis and tumor progression [24], suggesting that EphA3 may serve as both an optical imaging agent and a therapeutic target for medulloblastoma.
Because Fibrillin 2 and Eph receptor A3 have previously implicated in tumor progression and metastasis, it is possible that optical probes for these targets will be effective against other brain tumors as well. In addition, as there are many tumor resection surgeries that would benefit from improved surgical visualization, the techniques described here have broad application to the identification of targets for molecular probe development.
Footnotes
This work was funded in part through grants from the National Institutes of Health (U54CA136465 to C.H.C., R01GM079719 to A.J.B., and K99EB008557 to J.T.C.L.) and support from the Center for Children's Brain Tumors at Stanford University. J.T.D. was supported by the NLM Biomedical Informatics Training Grant (T15 LM007033) to Stanford University. H.H. was a fellow in the Stanford Molecular Imaging Scholars Program.
References
- 1.Tonn J-C, Grossman SA, Rutka JT, Westphal M. Neuro-oncology of CNS Tumors. Berlin, Germany: Springer; 2006. [Google Scholar]
- 2.Liu JT, Mandella MJ, Loewke NO, Haeberle H, Ra H, Piyawattanametha W, Solgaard O, Kino GS, Contag CH. Micromirror-scanned dual-axis confocal microscope utilizing a gradient-index relay lens for image guidance during brain surgery. J Biomed Opt. 2010;15:026029. doi: 10.1117/1.3386055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner JD, Sanai N. A brain tumor stem cell origin for glioblastoma endothelium. World Neurosurg. 2011;75:574–575. doi: 10.1016/j.wneu.2011.03.030. [DOI] [PubMed] [Google Scholar]
- 4.Sanai N, Eschbacher J, Hattendorf G, Coons SW, Preul MC, Smith KA, Nakaji P, Spetzler RF. Intraoperative confocal microscopy for brain tumors: a feasibility analysis in humans. Neurosurgery. 2011;68(2):282–290. doi: 10.1227/NEU.0b013e318212464e. [DOI] [PubMed] [Google Scholar]
- 5.Koc K, Anik I, Cabuk B, Ceylan S. Fluorescein sodium-guided surgery in glioblastoma multiforme: a prospective evaluation. Br J Neurosurg. 2008;22:99–103. doi: 10.1080/02688690701765524. [DOI] [PubMed] [Google Scholar]
- 6.Liao H, Shimaya K, Wang K, Maruyama T, Noguchi M, Muragaki Y, Kobayashi E, Iseki H, Sakuma I. Combination of intraoperative 5-aminolevulinic acid-induced fluorescence and 3-D MR imaging for guidance of robotic laser ablation for precision neurosurgery. Med Image Comput Comput Assist Interv. 2008;11:373–380. doi: 10.1007/978-3-540-85990-1_45. [DOI] [PubMed] [Google Scholar]
- 7.Grill J, Sainte-Rose C, Jouvet A, Gentet JC, Lejars O, Frappaz D, Doz F, Rialland X, Pichon F, Bertozzi AI, et al. Treatment of medulloblastoma with postoperative chemotherapy alone: an SFOP prospective trial in young children. Lancet Oncol. 2005;6:573–580. doi: 10.1016/S1470-2045(05)70252-7. [DOI] [PubMed] [Google Scholar]
- 8.Chargari C, Feuvret L, Levy A, Lamproglou I, Assouline A, Hemery C, Ghorbal L, Lopez S, Tep B, G GB, et al. Reappraisal of clinical outcome in adult medulloblastomas with emphasis on patterns of relapse. Br J Neurosurg. 2010;24:460–467. doi: 10.3109/02688691003739881. [DOI] [PubMed] [Google Scholar]
- 9.Zeltzer PM, Boyett JM, Finlay JL, Albright AL, Rorke LB, Milstein JM, Allen JC, Stevens KR, Stanley P, Li H, et al. Metastasis stage, adjuvant treatment, and residual tumor are prognostic factors for medulloblastoma in children: conclusions from the Children's Cancer Group 921 randomized phase III study. J Clin Oncol. 1999;17:832–845. doi: 10.1200/JCO.1999.17.3.832. [DOI] [PubMed] [Google Scholar]
- 10.Nomura Y, Yasumoto S, Yanai F, Akiyoshi H, Inoue T, Nibu K, Tsugu H, Fukushima T, Hirose S. Survival and late effects on development of patients with infantile brain tumor. Pediatr Int. 2009;51:337–341. doi: 10.1111/j.1442-200X.2008.02760.x. [DOI] [PubMed] [Google Scholar]
- 11.Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, Troost D, Meeteren NS, Caron HN, Cloos J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM, Kellie SJ, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 13.Chen R, Li L, Butte AJ. AILUN: reannotating gene expression data automatically. Nat Methods. 2007;4:879. doi: 10.1038/nmeth1107-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 15.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 16.English SB, Butte AJ. Evaluation and integration of 49 genome-wide experiments and the prediction of previously unknown obesity-related genes. Bioinformatics. 2007;23:2910–2917. doi: 10.1093/bioinformatics/btm483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritty TM, Broekelmann TJ, Werneck CC, Mecham RP. Fibrillin-1 and -2 contain heparin-binding sites important for matrix deposition and that support cell attachment. Biochem J. 2003;375:425–432. doi: 10.1042/BJ20030649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grass B, Wachtel M, Behnke S, Leuschner I, Niggli FK, Schafer BW. Immunohistochemical detection of EGFR, fibrillin-2, P-cadherin and AP2beta as biomarkers for rhabdomyosarcoma diagnostics. Histopathology. 2009;54:873–879. doi: 10.1111/j.1365-2559.2009.03303.x. [DOI] [PubMed] [Google Scholar]
- 19.Lal A, Glazer CA, Martinson HM, Friedman HS, Archer GE, Sampson JH, Riggins GJ. Mutant epidermal growth factor receptor up-regulates molecular effectors of tumor invasion. Cancer Res. 2002;62:3335–3339. [PubMed] [Google Scholar]
- 20.Dubuisson L, Lepreux S, Bioulac-Sage P, Balabaud C, Costa AM, Rosenbaum J, Desmouliere A. Expression and cellular localization of fibrillin-1 in normal and pathological human liver. J Hepatol. 2001;34:514–522. doi: 10.1016/s0168-8278(00)00048-9. [DOI] [PubMed] [Google Scholar]
- 21.Surawska H, Ma PC, Salgia R. The role of ephrins and Eph receptors in cancer. Cytokine Growth Factor Rev. 2004;15:419–433. doi: 10.1016/j.cytogfr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 23.Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, Stevens C, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 24.Cheng N, Brantley D, Fang WB, Liu H, Fanslow W, Cerretti DP, Bussell KN, Reith A, Jackson D, Chen J. Inhibition of VEGF-dependent multistage carcinogenesis by soluble EphA receptors. Neoplasia. 2003;5:445–456. doi: 10.1016/s1476-5586(03)80047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]