Abstract
Genetic manipulation of carotenoid biosynthesis has become a recent focus for the alleviation of vitamin A deficiency. However, the genetically modified phenotypes often challenge the expectation, suggesting the incomplete comprehension of carotenogenesis. Here, embryogenic calli were engineered from four citrus genotypes as engineered cell models (ECMs) by over-expressing a bacterial phytoene synthase gene (CrtB). Ripe flavedos (the coloured outer layer of citrus fruits), which exhibit diverse natural carotenoid patterns, were offered as a comparative system to the ECMs. In the ECMs, carotenoid patterns showed diversity depending on the genotypes and produced additional carotenoids, such as lycopene, that were absent from the wild-type lines. Especially in the ECMs from dark-grown culture, there emerged a favoured β,β-pathway characterized by a striking accumulation of β-carotene, which was dramatically different from those in the wild-type calli and ripe flavedos. Unlike flavedos that contained a typical chromoplast development, the ECMs sequestered most carotenoids in the amyloplasts in crystal form, which led the amyloplast morphology to show a chromoplast-like profile. Transcriptional analysis revealed a markedly flavedo-specific expression of the β-carotene hydroxylase gene (HYD), which was suppressed in the calli. Co-expression of CrtB and HYD in the ECMs confirmed that HYD predominantly mediated the preferred carotenoid patterns between the ECMs and flavedos, and also revealed that the carotenoid crystals in the ECMs were mainly composed of β-carotene. In addition, a model is proposed to interpret the common appearance of a favoured β,β-pathway and the likelihood of carotenoid degradation potentially mediated by photo-oxidation and vacuolar phagocytosis in the ECMs is discussed.
Keywords: Amyloplast, carotenogenesis, chromoplast, citrus, CrtB, crystal, engineered cell model, flavedo, phytoene synthase
Introduction
Carotenoids and their derivatives play essential physiological and ecological roles in plant growth, development, and reproduction (Cazzonelli and Pogson, 2010). They are also essential health-protecting compounds involved in human vision, immunity, embryonic development, and reproduction (von Lintig, 2010). Carotenoids in nature originate from phytoene biosynthesis via the condensation of geranylgeranyl pyrophosphate (GGPP), which is the crucial rate-controlling step mediated by phytoene synthase (PSY) (Welsch et al., 2010) (see Supplementary Fig. S1 at JXB online). Enzymatic desaturation and isomerization, as well as a light-mediated photoisomerization, convert non-coloured phytoene into coloured lycopene (Chen et al., 2010). The lycopene flux then branches into two pathways via cyclization reaction. Lycopene β-cyclase (LCYB) adds two β-rings to the ends of lycopene molecule to form β-carotene, while the dual action of lycopene ε-cyclase (LCYE) and LCYB results in the formation of α-carotene with one β-ring and one ε-ring (Bai et al., 2009). Subsequently, carotenes are converted into various xanthophylls, which is mediated by carotene hydroxylases and epoxidase as well as de-epoxidase (Vallabhaneni et al., 2009).
Carotenoids are assembled and accumulated in nearly all types of plastids (Howitt and Pogson, 2006), while the deposition of massive carotenoids requires plastid modification for enhancing the storage capacity. Chromoplasts have been postulated to be the most important plastid type for striking carotenoid sequestration in higher plants (Kim et al., 2010). They are generally derived from pre-existing plastids like chloroplasts and amyloplasts during the ripening of carotenoid-containing organs such as fruits and flowers (Egea et al., 2010). For example, amyloplast to chromoplast conversion is observed in developing ornamental tobacco floral nectaries (Horner et al., 2007). Amyloplasts are starch-storing plastids, and are distinguished by the starch-granule-filled profile (Wise, 2006). During amyloplast conversion, starch breakdown begins, the starch granules diminish then disappear, while carotenoid sequestering structures such as plastoglobules and carotenoid crystals form and chromoplasts appear (Horner et al., 2007). Genes including fibrillin, CHRC, and OR have been identified to participate in the regulation of chromoplast development and carotenoid sequestration (Li and Van Eck, 2007).
In recent decades, genetic manipulation has been successfully used to modify carotenogenesis in many plants (Giuliano et al., 2008; Fraser et al., 2009; Farré et al., 2011). In particular, over-expression of PSY leads to a higher carotenoid generation in carotenoid-poor organs, such as canola seeds (Shewmaker et al., 1999), potato tubers (Ducreux et al., 2005), white carrot root (Maass et al., 2009), and cassava root (Welsch et al., 2010). These engineered crops have exhibited an effective approach to human health promotion, especially the alleviation of vitamin A deficiency. However, many physiological and biochemical alterations associated with modification of the carotenoid pathway that occur in genetically engineered plants remain unexplainable. For example, an altered carotenoid profile, particularly an increased β,β-carotenoid flux, is commonly observed (Ducreux et al., 2005; Paine et al., 2005; Schaub et al., 2005; Zhu et al., 2008; Maass et al., 2009; Diretto et al., 2010). In addition, it is difficult to obtain the desired phenotypes for some important carotenoids, such as lycopene (Schaub et al., 2005). Moreover, engineered plants with enhanced carotenoid biosynthesis require sink formation for effective carotenoid storage (Li and Van Eck, 2007). Morphological changes in plastids have been observed in PSY over-expressed tomato fruits and canola endosperm (Shewmaker et al., 1999; Fraser et al., 2007), which implies the possibility of plastid modification for carotenoid deposition. Over-expression of PSY can also drive crystalline-type carotenoid sequestering in Arabidopsis callus (Maass et al., 2009). However, the crucial mechanism governing carotenoid metabolic sink formation is still unclear, and needs to be further clarified.
Genetic manipulation of carotenogenesis in higher plants has primarily been focused on fruits (tomato), seeds (rice, maize, and canola), and roots or tubers (potato, carrot, and cassava). However, inherent deficiencies in these systems, including time-consuming cultivation and management due to long growth cycles and poorly controlled growth conditions, challenge the further study on carotenogenesis. Plant cell culture offers an attractive option for producing natural active compounds. Despite the fact that plant cells in vitro often exhibit an inconsistent yield of natural products owing to the dedifferentiation process, it could be circumvented partially through selection of the genotype and explant source for deriving cell lines (Engelmann et al., 2010; Lee et al., 2010), as well as through metabolic engineering (Hellwig et al., 2004). Interestingly, the carotenoid accumulation patterns in OR mutant cauliflower curd (Li et al., 2006), engineered potato tuber (Diretto et al., 2007, 2010), and carrot root (Jayaraj et al., 2008) are similar to those in the homologous cell lines in vitro. Besides, maize callus provides an efficient model for identifying the most productive PSY gene from different plant species (Paine et al., 2005). Arabidopsis callus has been used as a non-green system in investigating carotenoid metabolic sink formation driven by the over-expression of AtPSY (Maass et al., 2009). Thus, it is particularly important to construct a plant cell model cultured in vitro that could facilitate the study on the molecular, biochemical and cytological processes related to carotenoid accumulation.
Citrus is one of the most important and widely grown fruit crops with great economic significance and value for humans (Talon and Gmitter, 2008). Citrus fruits have diverse carotenoid patterns, highly regulated by the co-ordinated expression of carotenogenic genes (Kato et al., 2004). Over the years, citrus fruits have served as a unique system to provide new insights into the understanding of carotenogenesis. A novel chromoplast-specific LCYB gene has been found to be associated with lycopene accumulation in the fruits of Star Ruby grapefruit (C. paradise Macf.) (Alquezar et al., 2009). Recent studies using comparative ‘omics’ revealed some processes, such as post-transcriptional regulation, partial impairment of lycopene downstream flux, enhanced photosynthesis, and miRNA-directed molecular process, in conferring the red-fleshed phenotype in the ‘Honganliu’ sweet orange (Pan et al., 2009; Xu et al., 2009, 2010). In addition, an in vitro culture system of citrus juice sacs provides a new approach to the comprehension of carotenogenesis in response to controlled environmental stimuli (Zhang et al., 2012).
By applying genetic manipulation of carotenoid accumulation as well as using carotenoid-rich citrus fruit, it is possible to understand the basis of carotenoid biosynthesis further. Therefore, engineered cell models (ECMs) were established by over-expressing CrtB (phytoene synthase from Erwinia herbicola) or co-expressing CrtB and DSM2 (a non-haem-diiron β-carotene hydroxylase from rice) in various non-coloured citrus embryogenic callus cells. To compare with the ECMs, the ripe flavedos from four genotypes were offered as natural carotenoid accumulation systems. Further analysis using comparative biochemistry, molecular biology, and cytology contributed to a better understanding of crystalline β-carotene accumulation in the amyloplasts of the ECM cells.
Materials and methods
Plant materials
Abortive ovule embryogenic calli from four citrus genotypes were used in this study. They were derived from Star Ruby grapefruit (C. paradise Macf.), Pink Marsh grapefruit (C. paradise Macf.), Cara Cara navel orange [C. sinensis (L.) Osb.], and Sunburst mandarin [Citrus reticulata Blanco×(C. paradisi Macf.×C. reticulata)], designated as RB, M, HQC, and SBT, respectively. The specimens were supplied by the National Key Laboratory of Crop Genetic Improvement at Huazhong Agricultural University (HZAU), Wuhan, China.
Fruits were obtained from citrus trees grown at the National Center for Citrus Breeding, Huazhong Agricultural University. Marsh grapefruit (M) (C. paradise Macf.) was selected instead of Pink Marsh grapefruit because Pink Marsh grapefruit is a periclinal chimera whose abortive ovules and flavedos are identical to those of the Marsh grapefruit in terms of their genotype (Cameron et al., 1964). Ripe fruits were collected at the same time in November. Flavedos of the fruits were separated with scalpels. A part of flavedo tissues were used for cytology, and others were immediately frozen in liquid nitrogen, and stored at –80 °C until analysis. The samples used for carotenoid extraction were lyophilized at –55 °C in a Lyolab 3000 (Heto, Denmark) and then stored at –80 °C.
Plasmid construction and transformation
The CrtB gene (a bacterial phytoene synthase gene, GenBank No. M90698) from Erwinia herbicola was PCR-amplified from plasmid vector PSL525, which was kindly provided by Professor Shih-Tung Liu (Taiwan, China). Pea rbcS transit peptide cDNA (GenBank No. X00806) was amplified by RT-PCR from the pea leaf pool. To create the chimeric gene (tp–rbcS–CrtB) construct, the PCR products were gel-purified, digested with XhoI/BamHI, and ligated into the corresponding sites in the PMV vector with a kanamycin resistance marker and the CaMV 35S promoter (modified pBI121) (Zhang et al., 2009). The primer pairs used were as listed in Supplementary Table S1 at JXB online. The plasmid pCB2004H with DSM2 [a non-haem-diiron β-carotene hydroxylase gene isolated from rice (Du et al., 2010)] over-expression construct was kindly provided by Professor Lizhong Xiong, Huazhong Agricultural University, China. These constructs and empty vectors were electroporated into an Agrobacterium tumefaciens strain (EHA105).
Explant preparation and transformation were performed according to Duan et al. (2007). To reduce the occurrence of chimeric calli, a very small piece of each recovered callus, as an independent cell line, was transferred onto selective culture media and subcultured for several cycles. To generate CrtB/DMS2 double transgetic lines, the DSM2 over-expression construct was introduced into a stable CrtB transformed callus line (M-33) and the transgenic calli were selected with hygromycin (50 mg l−1). Each independent line was propagated in the dark or under 35 μmol m−2 s−1 illumination for 16 h daily and kept at 25±1 °C.
Twenty-day-old calli were harvested for cellular analysis or stored at –80 °C for molecular analysis. The calli used for carotenoids extraction were lyophilized and stored at –80 °C until use.
DNA analysis
Genomic DNA for PCR and Southern blot analysis was extracted from stored calli according to Cheng et al. (2003). A PCR-derived CrtB fragment (see Supplementary Table S1 at JXB online) was labelled with a Dig-DNA labelling kit and used as a probe (Roche Diagnostics GmbH). Prehybridization, hybridization, and membrane washing and detection were conducted according to the manufacturer’s instructions (Roche Diagnostics GmbH).
Quantitative analysis of gene expression
First-strand cDNA was synthesized from 1 μg of total RNA isolated from calli and flavedos using the RevertAid M-MuLV KIT (MBI, Lithuania) as previously described by Liu et al. (2006). The primer pairs used were as listed (Liu et al., 2007) or designed using the Primer Express software (Applied Biosystems, Foster City, CA, USA) ((see Supplementary Table S2 at JXB online)). Actin was used as an endogenous control to normalize expression in different samples (Liu et al., 2007). Quantitative real-time PCR was performed using ABI 7500 Real Time System (PE Applied Biosystems; Foster City, CA, USA) according to Liu et al. (2007).
Generation of anti-CrtB antibodies and western blot analysis
The sequence of CrtB encoding 117 amino acids in the C-terminus was amplified, cloned into pGEX-4T, and transformed into E. coli strain BL21 (see Supplementary Table S1 at JXB online). After induction and purification, inclusion bodies were dissolved in SDS sample buffer and separated by SDS-PAGE. Gel slices containing the peptide were used to immunize rabbits to generate anti-CrtB antibodies.
Total callus and flavedo proteins were prepared as described previously (Isaacson et al., 2006; Pan et al., 2009), and quantified using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA) based on the Lowry method using bovine serum albumin (BSA) as standard. Protein samples (50 μg) were separated by SDS-PAGE and blotted onto PVDF membranes (Millipore, USA). Membranes were blocked with a TBST solution, containing 5% (w/v) not-fat dry milk power overnight at 4 °C and were incubated with anti-CrtB antibodies (1:2500) in the same solution for 2 h at room temperature. After rinsing five times for 10 min each with TBST solution, the membranes were incubated with secondary antibodies (peroxidase-conjugated immunopure goat anti-rabbit IgG [H+L], Pierce) at 1:20 000 in TBST solution, containing 5% (w/v) not-fat dry milk power and rinsed six times for 10 min each with TBST solution. The signal was detected using SuperSignal West Pico system (Thermo Scientific, USA) according to the manufacturer’s instructions. Two rabbit polyclonal antibodies (Agrisera@, Sweden) against the Rubisco large subunit (RubcL, 53 kDa) and PsbA protein (D1 protein of Photosystem II, 32 kDa) were used to detect the plastid marker proteins.
Carotenoid profile analysis
Carotenoid extraction and analysis using reversed-phase high-performance liquid chromatography (RP-HPLC) was conducted as previously described (Paine et al., 2005; Liu et al., 2007). For the calli, a 0.2 g homogeneous lyophilized sample was extracted. Because of the abundant carotenoid esters in citrus fruits, the extracts from 0.3 g of lyophilized flavedos were saponified with 15% (w/v) KOH:methanol. The RP-HPLC analysis was performed in a Waters liquid chromatography system equipped with a model 600E solvent delivery system, a model 2996 photodiode array detection (PAD) system, a model 717 plus autosampler and an Empower chromatography manager. A C30 carotenoid column (150×4.6 mm; YMC, Japan) was used to elute the carotenoids. The carotenoids were identified by their characteristic absorption spectra and typical retention time based on the literature and standards from CaroNature Co. (Bern, Switzerland). The quantification of the carotenoids was achieved using calibration curves for standards including violaxanthin, lutein, phytoene, α-carotene, β-carotene, β-cryptoxanthin, and lycopene; phytofluene was quantified as phytoene, and α-cryptoxanthin was quantified as β-cryptoxanthin.
Microscopy analysis
Protoplasts from the calli were isolated as described by Grosser and Gmitter (1990), after which protoplast suspensions were plated onto microscope slides to observe modes of carotenoid deposition. Microcellular light microscopy of fresh flavedo cells were performed using a frozen sectioning technique with a Leica CM1900 (Leica, Germany). More detailed inspections of the carotenoid accumulative structures in vitro were performed based on a protocol for isolating chromoplasts from fresh tissues (Lopez et al., 2008). An optical microscope (BX61, Olympus) equipped with a DP70 camera was used in tandem with differential interference contrast (DIC) technique.
The same callus and flavedo samples were also selected for transmission electron microscopy (TEM) analysis. Samples were prepared using a normal fixation process with 2.5% glutaraldehyde adjusted to pH 7.4, and 0.1 M phosphate buffer with 2% OsO4. They were dehydrated and embedded in epoxy resin (Luft, 1961) and SPI-812, respectively. Ultrathin sections obtained with a Leica UC6 ultramicrotome were stained with uranyl acetate and subsequently with lead citrate. Recording of the images was performed with a HITACHI H-7650 transmission electron microscope at 80 kV and a Gatan 832 CCD camera. Images were obtained at various magnifications along with a corresponding a waffle-pattern diffraction grafting replica with line spacing being 0.463 μm (Product No. 607, Ted Pella, Inc., Redding. CA).
Statistical analysis
The data presented in this study were analysed using SAS statistical software. Analysis of variance (ANOVA) was used to determine Pearson correlation (r) and to compare the statistical difference based on Student–Newman–Keuls’ multiple range test at the significance levels of P <0.05 (*), P <0.01 (**), and P <0.001 (***), respectively.
Results
Construction of carotenoid engineered cell models (ECMs)
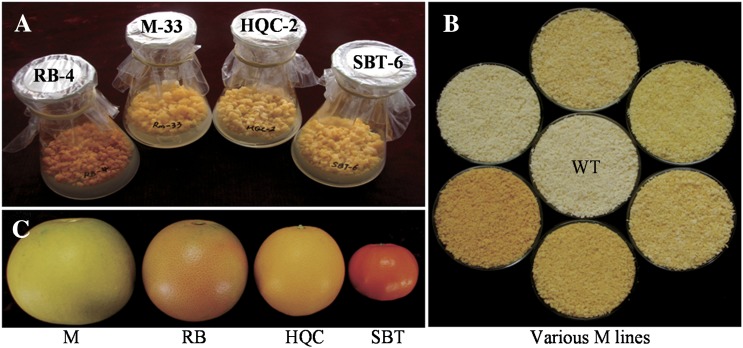
CrtB was first transformed into embryogenic calli from four citrus genotypes. Positive transformants of 58 independent lines were recovered, including five lines from Star Ruby grapefruit (RB), five lines from Sunburst mandarin (SBT), six lines from Cara Cara navel orange (HQC), and 32 lines from Pink Marsh grapefruit (M). Most of the transgenic lines had a yellow or orange-red colour that was not observed in the empty-vector controls and wild types. Coloured transgenic callus lines were selected as engineered cell models (ECMs). Four engineered genotypes presented distinctive colour characteristics where RB ECMs appeared red and the others orange or yellow (Fig. 1A).
Fig. 1.
Various ECMs and ripe citrus fruits used in this study. (A) The representative ECMs from four genotypes (from left to right: RB-4, M-33, HQC-2, and SBT-6). (B) Various M lines [wild type (WT) is in the middle]. (C) Ripe fruits [from left to right: Marsh grapefruit (M), Ruby Star grapefruit (RB), Cara Cara navel orange (HQC), and Sunburst mandarin (SBT)].
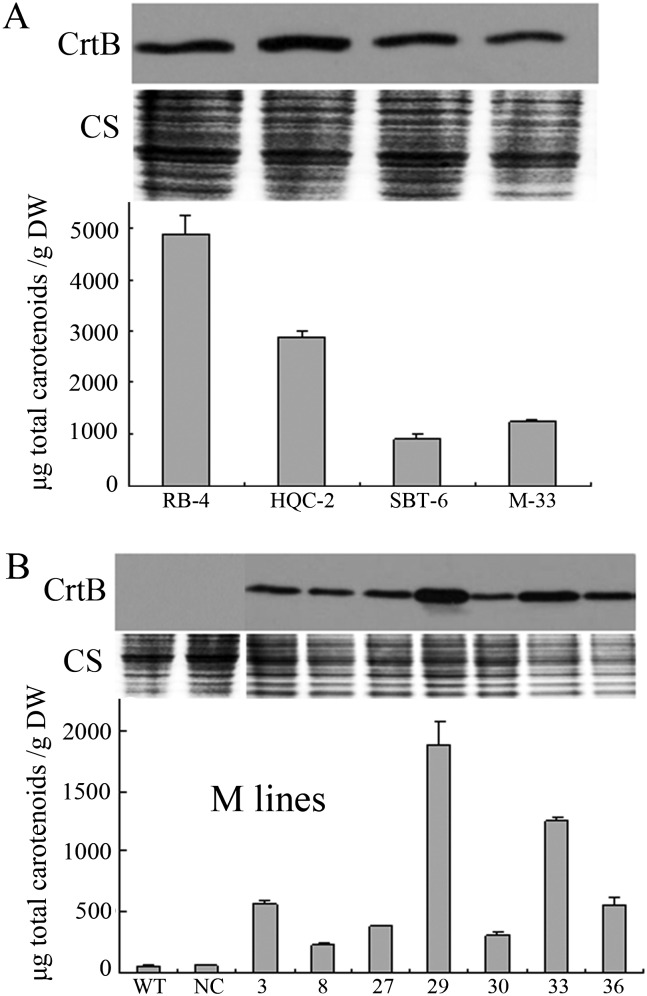
Southern blot analysis confirmed the independent transformation events in the representative M ECMs (see Supplementary Fig. S2 at JXB online). However, no correlation was observed between transgene copy number and carotenoid content (data not shown). Western blot analysis using anti-CrtB antibodies indicated the presence of CrtB proteins in the representative ECMs but an absence in the wild type and negative control line (Fig. 2A, B). Furthermore, correlation between CrtB protein levels and carotenoid contents was observed in seven M ECMs (Fig. 2B). The ECMs showed normal growth, and their pigment expression was found to be stable in subcultures for at least three years.
Fig. 2.
CrtB protein levels and their correlation with carotenoid contents in various ECMs. (A) CrtB protein levels and carotenoid contents in the representative ECMs from four genotypes. (B) Positive correlation between CrtB levels and carotenoid contents in various M lines, including the ECMs, wild type (WT), and negative line (NC). CS, Coomassie blue staining for loading control. Values and bars represent the means and ±SD (n=3 replicate experiments), respectively.
Carotenoid accumulation patterns in the ECMs
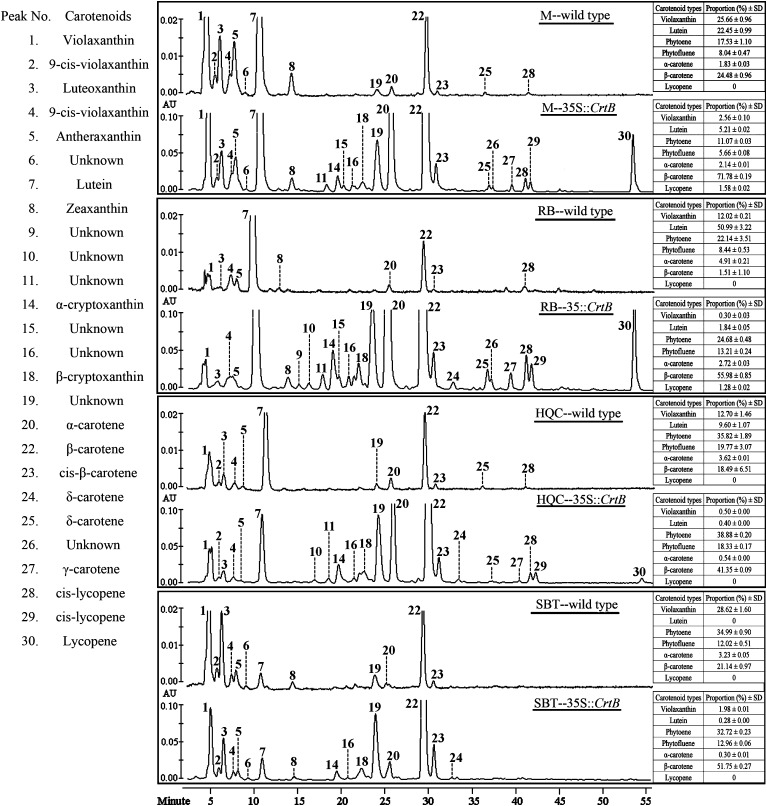
The carotenoid profile was investigated in various callus lines from dark-grown culture using HPLC. Low amount of carotenoids was observed in wild-type embryogenic calli (Table 1). They mainly consisted of violaxanthin, lutein, β-carotene, and phytoene, and their accumulation patterns were markedly diverse among the genotypes (Fig. 3). In particular, RB contained predominantly lutein, and SBT had significantly suppressed lutein biosynthesis. Other important carotenoids, such as β-cryptoxanthin, lycopene, and esterified xanthophylls, were all at negligible levels or undetected in these calli.
Table 1.
Carotenoid content and composition in wild-type callus lines, representative ECMs and flavedos
Vio, violaxanthin; α-Cry, α-cryptoxanthin, β-Cry. β-cryptoxanthin; Phy, phytoene; Phytof, phytofluene; α-Car, α-carotene; β-Car, β-carotene; Lyc, lycopene; β/ε ratio represents the β-cycle to ε-cycle derivatives ratio. Xan./Tol. represents the xanthophylls to total carotenoids ratio. (–) Not detected or trace; (– –) infinite. Values are means ±SD from three replicate experiments and are expressed as μg g−1 dry weight.
| Event | Vio | Lutein | α-Cry | β-Cry | Phy | Phytof | α-Car | β-Car | Lyc | Total | β/ε ratio | Xan./Tol. |
| Wild-type callus lines from dark-grown culture (non-saponified samples) | ||||||||||||
| M | 13.78±0.46 | 12.05±0.56 | – | – | 9.41±0.89 | 4.32±0.48 | 0.98±0.042 | 13.14±1.26 | – | 53.68±3.13 | 2.07±0.086 | 0.48±0.019 |
| RB | 2.16±0.14 | 9.17±0.45 | – | – | 3.98±0.82 | 1.52±0.17 | 0.88±0.008 | 0.27±0.17 | – | 17.98±0.94 | 0.24±0.015 | 0.63±0.03 |
| HQC | 3.48±0.14 | 2.63±0.19 | – | – | 9.80±0.57 | 5.41±0.45 | 0.99±0.079 | 5.06±2.26 | – | 27.37±2.18 | 2.36±0.55 | 0.22±0.024 |
| SBT | 7.75±0.45 | – | – | – | 9.48±0.14 | 3.26±0.18 | 0.88±0.007 | 5.73±0.31 | – | 27.09±0.39 | 15.32±0.47 | 0.29±0.016 |
| ECMs from dark-grown culture (non-saponified samples) | ||||||||||||
| M-33 | 31.97±0.89 | 65.14±0.73 | 3.49±0.027 | 3.13±0.14 | 138.51±2.16 | 70.79±0.95 | 26.76±0.32 | 897.82±16.2 | 19.73±0.54 | 1257.34±19.64 | 9.78±0.056 | 0.083±0.001 |
| RB-4 | 14.84±1.81 | 89.73±5.63 | 15.34±1.48 | 9.51±0.70 | 1204.39±72.54 | 644.66±37.75 | 132.53±6.97 | 2732.07±78.20 | 62.22±4.47 | 4935.30±204.85 | 11.60±0.39 | 0.026±0.0009 |
| HQC-2 | 14.29±0.59 | 11.27±0.44 | 5.69±0.79 | 3.89±0.72 | 1108.63±32.79 | 523.00±8.98 | 15.26±0.37 | 1179.28±31.11 | – | 2861.30±73.02 | 37.16±1.09 | 0.012±0.002 |
| SBT-6 | 18.10±0.98 | 2.59±0.21 | 2.26±0.46 | 3.94±0.44 | 298.47±15.03 | 118.21±6.24 | 2.75±0.18 | 472.10±29.39 | – | 918.42±51.55 | 65.02±4.96 | 0.029±0.001 |
| Flavedos (saponified samples) | ||||||||||||
| M | 10.37±1.29 | 1.64±0.22 | – | 0.76±0.074 | 8.77±1.06 | 6.12±0.68 | 0.32±0.01 | 2.31±0.38 | – | 30.28±3.62 | 6.89±0.23 | 0.42±0.009 |
| RB | 9.86±0.78 | 2.19±0.37 | 1.62±0.036 | – | 167.75±7.92 | 177.38±9.09 | 2.23±0.13 | 177.19±13.32 | 225.71±13.86 | 763.92±45.22 | 30.99±0.63 | 0.018±0.0005 |
| HQC | 97.49±5.91 | – | – | 3.66±0.17 | 72.23±5.03 | 25.87±1.35 | – | 0.60±0.11 | – | 199.86±10.09 | – – | 0.51±0.017 |
| SBT | 163.46±13.03 | – | – | 24.20±2.38 | 79.95±7.85 | 101.78±6.89 | – | 7.04±1.02 | – | 376.44±28.77 | – – | 0.50±0.011 |
Fig. 3.
HPLC chromatograms of carotenoids (non-saponified) in the wild types and representative ECMs (35S::CrtB) monitored at 450 nm. The peaks are numbered according to the elution time as detailed in Supplementary Table S3 at JXB online. Inserted tables show the proportion (%) and ±SD (n=3 replicate experiments) of the representative carotenoids relative to total carotenoid amount.
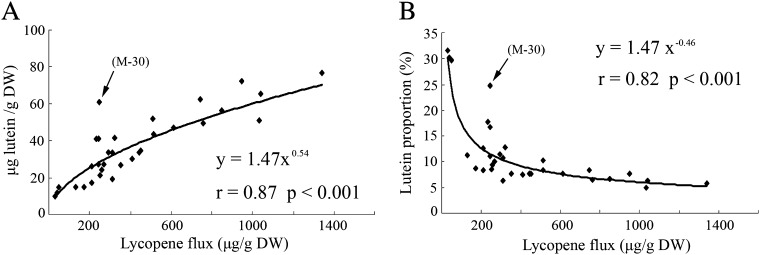
By contrast, the levels of total carotenoids markedly increased in all ECMs compared with the wild types (Table 1 shows the carotenoid profiles in four representative ECMs). Phenotypes with a darker orange colour in the RB ECMs were associated with a larger amount of carotenoids, ranging from 3052 to 4880 μg g−1 dry weight. Although ECMs’ carotenoid patterns were diverse among genotypes, up-regulation of the β,β-pathway was favoured (Table 1; Fig. 3). For example, in wild-type RB and M, the proportion of β-carotene in the total carotenoids was only 1.51% and 24.48%, respectively, and lutein accounted for 50.99% and 22.45%; but in the corresponding ECMs, β-carotene rose to 55.98% and 71.78% while lutein decreased to 1.84% and 5.21%, respectively (Fig. 3). It was also noted that violaxanthin increased slightly, so its proportion decreased in the ECMs. To characterize the altered balance of carotenoid patterns in the ECMs further, lutein levels were analysed in 32 M callus lines, including the wild type, empty-vector control, negative control, and 29 ECMs (35S::CrtB). Statistical analysis showed that the content/proportion change of lutein relative to lycopene flux followed a power function (Fig. 4). Herein, lycopene flux represented the total content of violaxanthin, lutein, α-carotene, β-carotene, and lycopene. This power functional relationship showed the dynamic change of branch flux, suggesting that the β,ε-branch was less competitive when the upstream flux become strong. Noticeably, a novel ECM (M-30) was found not to correlate well with the power function (Fig. 4). M-30 showed a dark yellow colour and contained a higher lutein proportion (19.7%) in relation to other ECMs (see Supplementary Fig. S3A at JXB online).
Fig. 4.
Correlation between lutein content (A) or lutein proportion (B) and lycopene flux. 32 M callus lines, including wild type, empty-vector control, negative control, and 29 ECMs, are used for analysis. Lycopene flux is represented by the total content of violaxanthin, lutein, α-carotene, β-carotene, and lycopene, and lutein proportion (%) is relative to this total content. Pearson correlation (r) and statistical significance (P) are determined using SAS statistical software. The arrows show a significantly deviating line (M-30).
In addition, a large number of carotenoids under the detection threshold in wild types were detected in the ECMs (Fig. 3; see Supplementary Fig. S4 and Table S3 at JXB online). In particular, a low level of lycopene was observed in the ECMs of RB and M. Furthermore, identical HPLC chromatography profiles for saponified and unsaponified ECM samples demonstrated that non-esterified carotenoids were predominant in the ECMs (see Supplementary Fig. S5A at JXB online).
Effect of illumination on carotenoid accumulation in the ECMs
To determine the carotenoid accumulation in the ECMs from light-grown culture, illumination (35 μmol m−2 s−1 illumination for 16 h daily) treatments were performed. As shown in Supplementary Fig. S6 at JXB online, after excitation with 35 μmol m−2 s−1 irradiance (light/dark, 16/8 h), the visual inspection of the calli revealed that M-33 and RB-4 were both yellow instead of dark orange. HPLC analysis showed that the levels of carotenoids except lutein decreased under illumination. This was especially the case for β-carotene, which showed a 56% and an 85% reduction in M-33 and RB-4, respectively.
Carotenoid accumulation patterns in the ripe flavedos from four genotypes
To compare the ECMs with the natural carotenoid-rich system, carotenoid profiles in the ripe flavedos from four corresponding genotypes were characterized (Fig. 1C; Table 1). The flavedos of SBT and HQC chiefly contained violaxanthin, cryptoxanthin, phytoene, and phytofluene. RB accumulated high level of lycopene as well as β-carotene, phytoene, and phytofluene in the flavedo. There was a low level of carotenoids consisting predominantly of violaxanthin, phytoene, and phytofluene in the flavedo of Marsh grapefruit (M). In addition, carotenoid esterification is a typical character of carotenogenesis in the ripe flavedos (see Supplementary Fig. S5B at JXB online).
Light microscopic observations of carotenoid storage structures
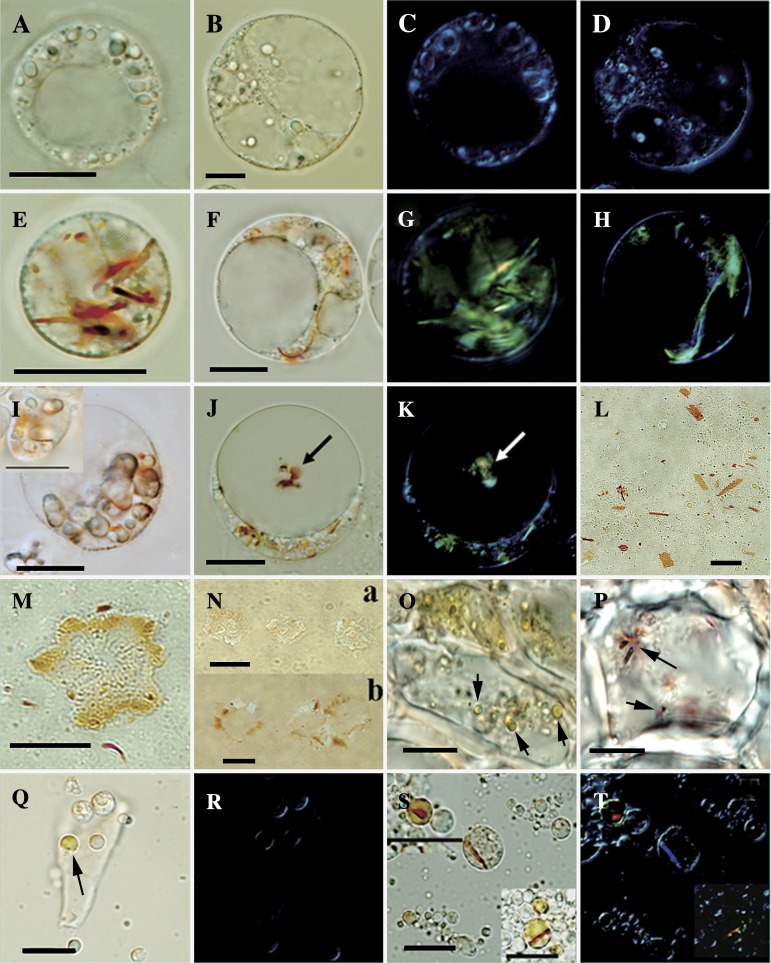
Significant carotenoid accumulation is accommodated by chromoplast development. To examine the formation of carotenoid sequestering structures in the ECMs, light microscopy of the protoplasts was performed. Unlike wild-type controls, visible red-orange carotenoid sequestering structures were found in the ECM cells (Fig. 5A, B, E, F), and polarization microscopy confirmed their crystal nature (Fig. 5C, D, G, H). Our further observation showed that the crystals were formed in a circular pattern localizing to the periphery of the amyloplasts (Fig. 5I). Some orange conglomerates, which were undergoing Brownian motion, were observed inside the vacuoles of the ECM cells (Fig. 5J). The conglomerates exhibited birefringence (Fig. 5K), indicating their crystal nature and the likelihood of containing carotenoids. According to a protocol for isolating plastids (Lopez et al., 2008), abundant carotenoid crystals with the size from 1 μm to 10 μm were detected free in the isotonic buffer (Fig. 5L), and no surrounding membrane envelope was observed. Coloured plastid structures containing plastoglobules peripherally were observed in abundance in isotonic buffer (Fig. 5M). Noticeably, compared with the wild types (Fig. 5N-a), these structures in the ECMs loaded more carotenoids (Fig. 5N-b).
Fig. 5.
Light microscopic inspection. (A) Protoplasts from wild-type RB under normal light field microscope. (B) Protoplasts from wild-type M under normal light field microscope. (C, D) Polarization micrographs for (A) and (B), respectively. (E, F) Protoplasts exhibiting the orange-coloured structures in the ECM cells of RB and M, respectively. (G, H) Carotenoid crystals confirmed by polarization microscopy in the ECM cells of RB and M, respectively. (I) Crystals located around the amyloplasts. (J) Coloured conglomerate (shown with arrows) observed in the vacuole of the ECM cells. (K) Conglomerate in the vacuole containing carotenoid crystals. (L) Flattened sheet (carotenoid crystals) in the lysate sample of the ECM cells. (M) Coloured plastid structure containing plastoglobules peripherally. (N) Coloured plastid structures from the wild-type cells (a) and ECM cells (b). (O) Inspection of the flavedo cells from Marsh; arrows show the chromoplasts. (P) Inspection of the flavedo cells from Ruby Star; arrows show the chromoplasts. (Q) Globular chromoplast (shown with arrows) in the lysate samples of the flavedo cells from Marsh. (R) Polarization microscopy for (Q). (S) Chromoplasts in the lysate samples of the flavedo cells from RB. (T) Polarization microscopy for (S), confirming the carotenoid crystals inside the chromoplasts of RB. The bar in each figure represents 10 μm.
Carotenoid sequestering structures in the flavedo cells were also inspected using frozen sectioning and plastid isolation techniques. As shown in Fig. 5O, Q, and R (and Supplementary Fig. S7 at JXB online), the flavedo cells of ripe fruits from all four genotypes contained abundant globular chromoplasts. Interestingly, the globular chromoplasts in the RB flavedo cells showed a distinct morphology characterized by visible carotenoid crystals inside (Fig. 5P, S, T). The crystals exhibited red colour, implicating the association with the accumulation of lycopene and β-carotene in the RB flavodo cells. This type of chromoplast has not been reported in citrus, while its morphology is similar to the crystalline chromoplast in tomato fruit and carrot root (Ben-Shaul and Naftali, 1969; Kim et al., 2010).
Transmission electron microscopic observations of carotenoid storage structures
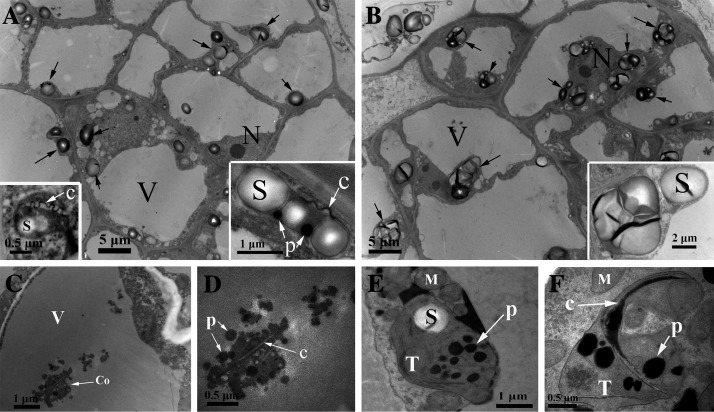
Transmission electron microscopy (TEM) was used to study the carotenoid sequestering structures of our callus and flavedo specimens further. Amyloplasts were the main plastid type observed in all callus cells (Fig. 6A, B). By comparison, there were slightly more plastoglobules in the amyloplasts of the ECM cells, which also had remnant carotenoid crystals and characteristic internal membranes exclusively, showing chromoplast-like structures (Fig. 6A). Conglomerate structures were noted in the vacuoles of the ECM cells under the TEM field (Fig. 6C), and high resolution image demonstrated that the conglomerate consisted of carotenoid plastoglobules and crystals without a surrounding membrane structure (Fig. 6D). The results of extensive TEM investigation indicated the frequent appearance of conglomerate structure in the vacuoles (data not shown), which provided further confirmation of the vacuolar location of carotenoid sequestering structures.
Fig. 6.
Cellular ultrastructure of the callus and flavedo. (A) Ultrastructure of ECM cells (M) containing conspicuous amyloplasts (showed with arrows). Inserts in the lower corner showed the detail of amyloplasts, which contained a small number of plastoglobules, carotenoid crystal remnant, and characteristic internal membrane. (B) Ultrastructure of wild-type callus cells. (C) Conglomerate in the vacuole of the ECM cell. (D) A detailed image of the conglomerate containing plastoglobules and crystals without a surrounding membrane structure. (E, F) Aspects of the young chromoplasts in the flavedo cells from ripe fruits of Marsh (M) and Ruby Star (RB). Crystal structure is observed in the chromoplast of RB (F). Remnant thylakoids are always observed in all chromoplasts. S, starch granules; P, plastoglobules; M, mitochondrion; C, crystal; V, vacuole; Co, conglomerate; T, remnant thylakoids.
In contrast to the ECM cells, plastids containing massive plastoglobules and remnant thylakoid membranes were detected in the flavedo cells of all four genotypes (Fig. 6E, F; see Supplementary Fig. S7 at JXB online), which defined the young chromoplasts derived from chloroplasts. Moreover, the chromoplasts in the flavedo cells of RB contained crystals and were almost devoid of starch granules (Fig. 6F).
Western blot analysis of two plastid proteins
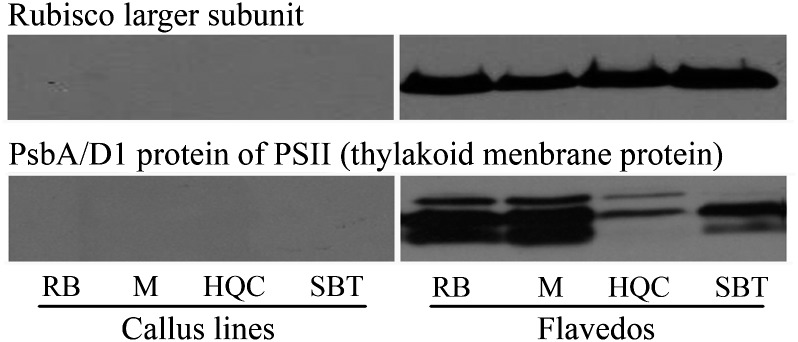
Comparative microscopy indicated distinctive plastid formation in flavedos and calli. To characterize the plastids further, Western blot analysis was performed to detect two plastid proteins, a Rubisco large subunit and PsbA/D1, both of which are photosynthetic proteins and typical of chloroplasts, and are also detected in the chromoplasts arising from chloroplasts (Barsan et al., 2010). The result revealed that both proteins were missing in all calli, both light-grown and dark-grown cultures, although they were expressed actively in all flavedos (Fig. 7).
Fig. 7.
Western blot analysis of two plastid proteins: a Rubisco large subunit and PsbA/D1 protein. Western blot result of only one sample in each genotype was demonstrated here, representing the results of wild type and ECMs cultured in the light and dark, as they all show the same blot response. Total proteins from all samples were loaded in 50 μg, and separated by SDS-PAGE.
Transcript profiles of isoprenoid and carotenoid metabolism in the ECMs and flavedos
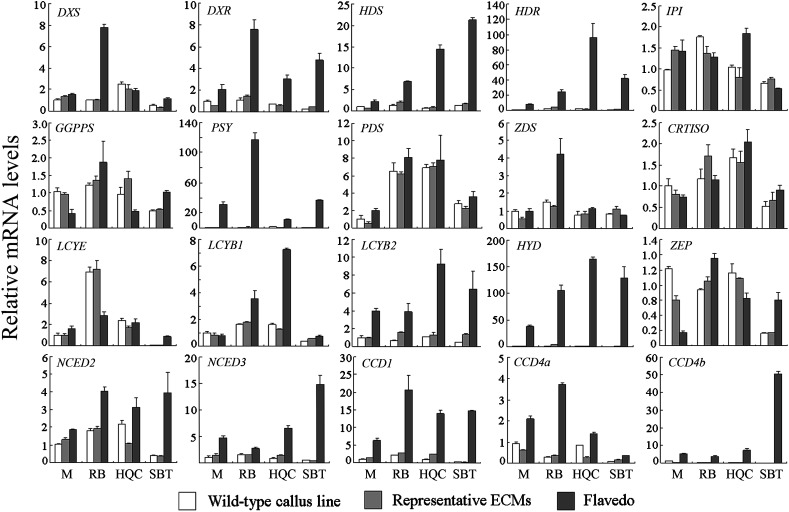
Expression data showed that most genes encoding enzymes for isoprenoid and carotenoid metabolism were expressed at a similar level in all callus lines, including four wild types and four representative ECMs (M-33, RB-4, SBT-6, HQC-2) (Fig. 8; see Supplementary Fig. S8 at JXB online). However, some exceptions were observed. The gene encoding phytoene desaturase (PDS) was expressed approximately 6-fold higher in HQC and RB compared with M. LCYE was expressed 7-fold higher and approximately 15-fold lower in RB and SBT, respectively, compared with M. Transcriptional analysis of all tested M callus lines showed that M-30 exhibited the highest transcript level of LCYE gene (P <0.001) (see Supplementary Fig. S3B at JXB online).
Fig. 8.
Transcriptional analysis of genes encoding enzymes for isoprenoid and carotenoid metabolism. Transcript levels are expressed relative to M wild type. For one genotype at least five ECMs were analysed, and data of only one ECM was shown due to their similar expression trend. All data are presented as means ±SD from three replicate experiments.
Furthermore, comparison of the expression patterns of the calli and the corresponding flavedos, it was possible to group these genes into three categories. The first group exhibited a flavedo-specific expression pattern, including the genes encoding DXR (1-deoxy-D-xylulose 5-phosphate reductoisomerase), HDS (hydroxymethylbutenyl 4-diphosphate synthase), HDR (hydroxymethylbutenyl 4-diphosphate reductase), PSY, LCYB2 (lycopene β-cyclase 2), HYD (β-carotene hydroxylase), NCEDs (9-cis-epoxycarotenoid dioxygenases), and CCDs (carotenoid cleavage dioxygenases). In particular, HYD was the most flavedo-specific gene, reflected by the striking up-regulation of its expression in the flavedo of almost all four genotypes whereas suppression in all calli (see Supplementary Fig. S8B at JXB online). The second group showed a constitutive expression pattern in the calli and corresponding flavedos, including the genes encoding IPI (isopentenyl diphosphate isomerase), GGPPS (geranylgeranyl diphosphate synthase), PDS, and CRTISO (carotene isomerase). Other genes were assigned to the third category due to their unique expression patterns. In particular, DXS (encoding 1-deoxy-D-xylulose 5-phosphate synthase) and ZDS (encoding ζ-carotene desaturase) were expressed at higher levels in the flavedos of RB as compared to other lines, while LCYE was expressed at unusually low levels only in the callus of SBT.
Over-expression of CrtB in combination with DSM2 in callus tissue
Transcriptional analysis suggested HYD plays a key role in regulating the preferred carotenoid patterns between the ECMs and flavedos. To prove this, one non-haem-diiron β-carotene hydroxylase gene (DMS2, also named as HYD3) from rice (Du et al., 2010) was introduced into an ECM of M. This co-transformation system was also expected to be used to examine whether β-carotene was the major component of the crystals in the ECMs.
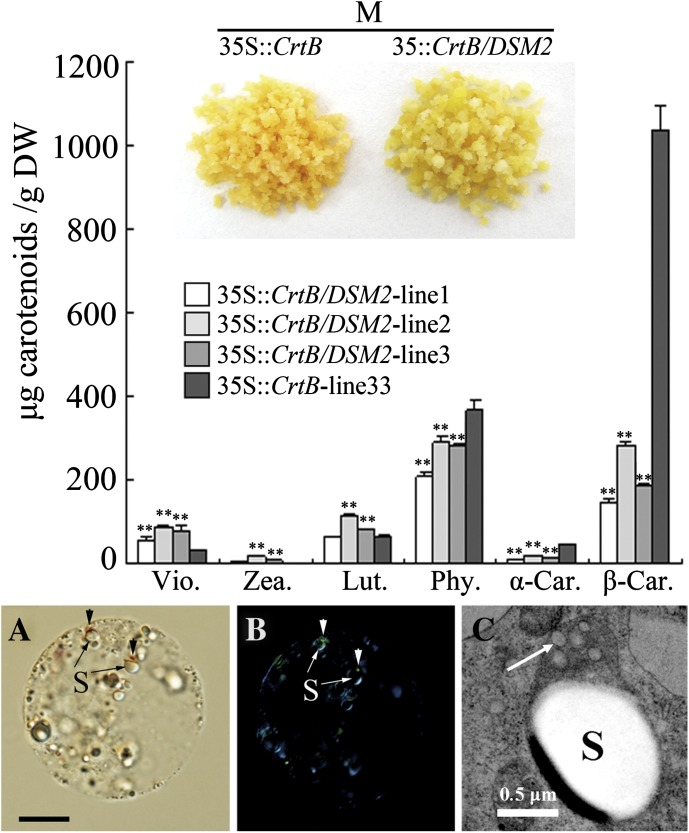
As shown in Fig. 9, co-transformed lines showed an altered phenotype from orange to gold. HPLC analysis revealed an 80.4% reduction of β-carotene on average and a 67.7% reduction of α-carotene in the co-transformed lines. As expected, xanthophyll content was noticeably increased in the co-transformed lines, particularly violaxanthin content, which exhibited a 2.4-fold increase. However, unlike flavedos, the co-transformed lines accumulated unesterified xanthophylls. Furthermore, carotenoid crystals became smaller in size, as shown by small dots attaching to the amyloplasts of co-transformed cells, or even being invisible (Fig. 9A, B). Subsequently, the ultrastructural inspection of the amyloplasts in the co-transformed cells showed an increased number of plastoglobules (Fig. 9C).
Fig. 9.
Carotenoid analysis and cellular inspection of co-transformed lines (35S::CrtB/DSM2) and M-33 (35S::CrtB). Via., violaxanthin; Zea., zeaxanthin; Lut., lutein; Phy., phytoene; α-Car., α-carotene; β-Car., β-carotene. Values and bars represent the means and ±SD (n=3 replicate experiments), respectively. ** Indicates that the values are significantly different compared with M-33 at the significance level of P <0.01. (A) Normal light microscopy. (B) Polarization microscopy (arrows show the dot-crystals attaching to the amyloplasts). (C) Ultrastructure of amyloplast (arrow shows the electron-transparent plastoglobules; S, starch granule). The bar in picture (A) represents 10 μm.
Discussion
Comparative analysis of various ECMs with the natural carotenoid-rich system (flavedo) provides a powerful tool for gaining insight into the key regulatory patterns and metabolic sink formation, particularly those with regard to β-carotene accumulation. More importantly, the ECMs from dark-grown culture were found to be excellent accumulators of carotenoids consisting predominantly of β-carotene. In a representative RB ECM, the carotenoid content, with about 56% β-carotene, was even higher than those of carotenoid-rich crops, such as carrot roots (2000–4000 μg g−1 dry weight) and tomatoes (2000 μg g−1 dry weight) (Maass et al., 2009). Therefore, an improved understanding of the mechanisms underlying β-carotene accumulation in the ECMs could promote effective carotenoid engineering in non-green tissues, which represent major food sources for humans (Howitt and Pogson, 2006).
ECMs possess a favoured β,β-pathway
CrtB created a considerable carotenogenic flux in the callus tissues of various genotypes, including a red-fruit variety (RB) which accumulated lycopene in ripe fruits. However, in the ECMs, no abundant lycopene accumulation was found, except for a low level in that of RB and M (Table 1; Fig. 3). Supposedly, lycopene accumulation in fruits is a tissue-specific developmental process that is difficult to assess from non-fruit tissues, conforming to the report that a unique chromoplast-specific lycopene β-cyclase gene (LCYB2) determines the lycopene accumulation in the RB fruits (Alquezar et al., 2009). The ECMs contained a substantial favoured β,β-branch pathway, consistent with the observations in other engineered plants, such as canola seeds (Shewmaker et al., 1999), golden rice (Schaub et al., 2005), and golden potatoes (Diretto et al., 2010).
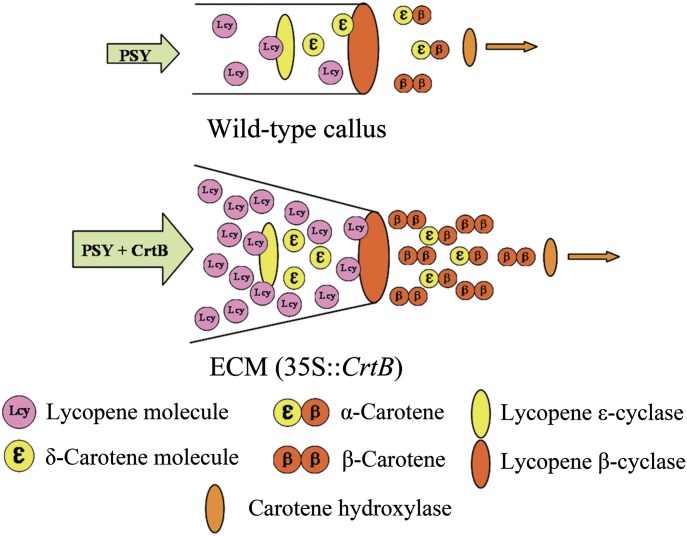
However, in most cases, the β,β-branch pathway is not predominant in wild-type tissues (or organs), this is also confirmed by the result of our experiment in which β-carotene only accounted for 1.5% of total carotenoids in wild-type RB calli, while lutein took up a substantial proportion (51.1%). Thus, the balance of carotenoid accumulation was altered as a result of genetic manipulation. It is noted that, in the present study, a suppressed transcript level for LCYE could explain the scarce accumulation of lutein in SBT callus, and M-30, which probably contained additional variation, exhibited increased LCYE transcription, yielding high lutein accumulation (see Supplementary Fig. S3 at JXB online). Based on these data and the power function of the content/proportion change of lutein relative to lycopene flux (Fig. 4), as well as a suggestion of metabolic channelling from LCYE to LCYB (Bai et al., 2009), a model was established to explain the favouring of the β,β-pathway in the ECMs here (Fig. 10) and is predicted to be valid in other carotenoid engineered plants where the phenomena is observed. In the wild-type calli of the model, a small amount of synthesized lycopene was sufficiently captured by LCYE and converted to δ-carotene, the substrate for LCYB, and formed a high proportion of the β,ε-carotenoids, such as lutein. However, in this model, LCYE was regarded as a cyclase with limited activity relative to LCYB, based on the fact that LCYB is the effective bi-cyclic cyclase, in the absence of which, lycopene accumulates mainly in plant tissues, whereas LCYE is generally the mono-cyclic cyclase, and less lycopene accumulates when LCYE is absent (Pogson et al., 1996; Alquezar et al., 2009; Bai et al., 2009). Accordingly, in the ECMs abundant lycopene molecules that were not captured by LCYE diffused to LCYB and were converted to β-carotene. Subsequently, β-carotene was predominantly accumulated, but less leaked to xanthophylls, which implicated the low carotenoid hydroxylase activity in the ECMs.
Fig. 10.
A hypothetical schematic model of carotenoid metabolic channels toward the β-carotene branch point in callus tissues. In wild-type callus, most of lycopene molecules can be captured by LCYE because of low carotenoid metabolism flux. In the ECMs, exogenous CrtB engineers a striking carotenoid metabolism flux. The activity of LCYE is unaltered. A mass of lycopene molecules diffuses to LCYB and is converted into β-carotene.
Therefore, this model suggests that LCYE has a bottleneck role in the presence of abundant lycopene substrates leading to an altered carotenoid composition. This is supported by the reports that diverse LCYE enzymatic activity determines alternative β-carotene and lutein compositions in maize and wheat (Harjes et al., 2008; Howitt et al., 2009), as well as by the fact that over-expression of CrtI, a bacterial phytoene desaturase that converts phytoene to lycopene, leads to substantially increased β,β-carotenoid accumulation in tobacco leaves (Misawa et al., 1993) and tomato fruits (Romer et al., 2000).
In addition, a β- and ε-cyclase complex has once been proposed to explain the branch control of carotenoid pathway (Bai et al., 2009). In this complex, it is speculated that there is competition for substrate between LCYB and LCYE, probably relying on their different cyclase affinity to substrate. This proposed system suggested a feedback regulation mechanism, mediated by the altered lycopene substrate preference by cyclase, which may be acting to influence the proportion of β,β-carotenoids and β,ε-carotenoids (Cazzonelli and Pogson, 2010).
Amyloplasts serve as crystalline-type β-carotene sequestering sink in the ECMs
As revealed by cellular inspection and Western blot analysis, plastid types differed in the callus and flavedo cells. In flavedo cells, chromoplasts transformed from chloroplasts served as a carotenoid metabolic sink. By comparison, almost all the observed carotenoids in the ECM cells were sequestered within amyloplasts; despite the fact that amyloplasts showed chromoplast-like profiles owing to visible crystals and plastoglobules inside, starch granules were still the most predominant (Fig. 6A). β-Carotene content was correlated with crystal development in CrtB/DSM2 co-transformed lines, which reveals that the carotenoid crystals predominantly contained β-carotene. Other carotenoids, such as xanthophylls, might be deposited in amyloplasts without crystallization, which was suggested by the multiplication of plastoglobules observed in the amyloplasts (Figs 6A, 9C), and also by the coloured plastid structures (Fig. 5M), which were probably the broken amyloplasts losing most starch granules, as previously observed in potato tubers (Lopez et al., 2008). In agreement with the previous description of crystal formation in proplastids driven by over-expression of PSY (Maass et al., 2009), the observations in the present study demonstrate that carotenoid sequestering sink can also be formed in the amyloplasts.
Chromoplast development requires the controlled changes of the pre-existing plastids, such as the internal membrane remodelling (Egea et al., 2010), which may be mediated by some crucial factors. For example, OR functions in directing the transition from non-coloured plastids to chromoplasts, over-expression of OR drives β-carotene crystal formation in potato tubers and cauliflowers (Lopez et al., 2008). In addition, metabolite-induced plastid transition is found in the engineered tomato fruits over-expressing PSY (Fraser et al., 2007). A previous study of tobacco floral nectaries has clearly shown that amyloplasts could be converted into chromoplasts (Horner et al., 2007). However, amyloplasts are sustained in the ECMs, whether cultured in the light or in the dark. Unlike Arabidopsis, citrus has a chromoplast developmental programme (Alquezar et al., 2009; Maass et al., 2009). Probably, such a programme in the cell models is suppressed by the predominant starch accumulation, or the cell models lack some factors that initiate this progress. Further investigation is needed to find out the causes repressing the transition of amyloplasts to chromoplasts, particularly the impact of starch metabolism on chromoplast development and carotenogenesis.
Hypothetical carotenoid degradation in the ECMs
In the present study, abundant carotenoid crystals without surrounding membranes were found in the isotonic buffer (Fig. 5L), suggesting that the crystals were not firmly attached to the amyloplast membranes. As previously discussed by Maass et al. (2009), carotenoid crystals are free from the lipophilic phase of the plastid membrane, and the crystalline form may protect carotenoids from enzymatic catabolism. However, crystallization seems not to protect β-carotene from photo-oxidative degradation in the ECMs. It is noted that the ECMs under irradiation showed a marked decrease in β-carotene level (see Supplementary Fig. S6 at JXB online). This result is not due to a conversion of β-carotene to β,β-xanthophylls as in the cyanobacterium (Schäfer et al., 2006), because violaxanthin levels decreased in the light-grown ECMs. Light can generate oxidative stress, which is also confirmed by our experiment in the citrus callus (data not shown). Thus, it is possible that the lost carotenoids in the light-grown ECMs are involved in an oxidative reaction. Micrographs of the ECM cells revealed carotenoid sequestering structures in the vacuoles, specially plastoglobules and crystals without a membrane structure (Figs 5J, K, 6C, D). The structures appear to be similar to those of starch granules without an intact amyloplast membrane found inside the vacuoles of cultured rice cells during sucrose starvation through vacuolar phagocytosis (Chen. et al., 1994). This provides evidence of an additional site for carotenoids and implies that there is an underlying catabolism mechanism that governs carotenoids in the vacuoles. Further studies are required to discover new pathways related to carotenoids degradation.
HYD transcription predominantly determines the variation of carotenoid accumulation in calli and flavedos
Transcriptional analysis confirmed the crucial rate-controlling role of PSY in the ECMs, given an over-expressed CrtB driving the abundant accumulation of carotenoid. However, the major accumulation pattern of β-carotene in the ECMs is very different from that of esterified β,β-xanthophylls in common fruits (Table 1). In the ECMs, the proportion of violaxanthin was lower than in the wild types (Fig. 3). This phenomenon seemed like a result of feedback inhibition from striking metabolic flux. Furthermore, in the flavedos of the present study, a co-ordinated transcriptional up-regulation of most genes encoding enzymes for isoprenoid and carotenoid metabolism, especially the HYD gene, reflects a fruit-specific regulation of carotenogenesis (Fig. 8; see Supplementary Fig. S8 at JXB online) (Kato et al., 2004). As reported previously, HYD is associated with β-carotene accumulation in the endosperm of maize (Vallabhaneni et al., 2009; Yan et al., 2010). Therefore, the over-expression of an HYD gene from rice was performed in the ECM, which resulted in a significantly decreased β-carotene content and increased violaxanthin and other xanthophylls (Fig. 9). This study identifies the crucial role of HYD in determining the diversified carotenoid accumulation patterns between the ECMs and flavedos, and also suggests that HYD can be a key target for β-carotene metabolic engineering in citrus fruits. However, it is noteworthy that the β,β-xanthophylls in CrtB/DSM2 co-transformed lines were all non-esterified forms, which is presumed to be associated with an inactive fatty acid synthesis in the amyloplasts of the ECM cells. In general, esterification of xanthophylls with fatty acids is a tissue-specific behaviour which is more likely to occur in ripe fruits (Hornero-Mendez and Minguez-Mosquera, 2000; D’Ambrosio et al., 2011).
Conclusion
In summary, this paper reports diverse ECMs depending on the genotypes of citrus used for the construction. However, the carotenoid pattern in the ECMs of one genotype seems not to correlate with that in its flavedo. Comparative analysis of various ECMs with flavedos probes into the β-carotene accumulation mechanism with new insights. The present study correlates striking β-carotene accumulation with the suppressed HYD transcription and a channelling control of alternative lycopene flux in the ECMs. It also shows that, in the ECM cells, β-carotene is synthesized and sequestered in the amyloplasts in crystalline form. Furthermore, carotenoid crystals consisting mainly of β-carotene are confirmed. The likelihood of carotenoid degradation mediated by photo-oxidation and vacuolar phagocytosis in the ECMs is also proposed. Utilization of the ECMs is not limited to comprehension of β-carotene accumulation. The ECMs may facilitate the investigations into more potential gene targets associated with carotenogenesis and into the regulation metabolism of carotenoid accumulation in response to various environmental stimuli.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1 . Primers used in this study.
Supplementary Table S2. Specific primers used in real-time reverse transcriptase-PCR.
Supplementary Table S3. Analysis of carotenoid profiles detected in the wild-type and ECM lines.
Supplementary Fig. S1. Carotenoid biosythesis pathway in higher plants.
Supplementary Fig. S2. A DNA gel blot analysis indicating inserted patterns of T-DNA in the representative lines.
Supplementary Fig. S3. A novel 35S::CrtB ECM (M-30) containing a high proportion of lutein due to the highest expression of the LCYE gene.
Supplementary Fig. S4. Absorbance spectra of the peaks detected in ECMs (peak identification in Supplementary Table S3).
Supplementary Fig. S5. HPLC chromatograms of carotenoids extracted from the RB ECM (A) and HQC flavedo (B) at 450 nm.
Supplementary Fig. S6. Comparative analysis of carotenoids contents in light/dark-grown engineered lines.
Supplementary Fig. S7. Additional photographs of the visual inspection of the flavedo cells and plastid isolation.
Supplementary Fig. S8. Comparative transcriptional analysis of genes encoding enzymes for isoprenoid and carotenoid metabolism among wild-type calli, ECMs, and flavedos.
Acknowledgments
This research was supported by the National Basic Research Program of China (No. 2011CB100600) and NSFC (30830078, 30921002). We thank Professor Shih-Tung Liu (Taiwan, China) for providing the Erwinia herbicola CrtB gene, and Professor Lizhong Xiong and Hao Du (Huazhong Agricultural University, China) for the gift of the DSM2 over-expressing construct. We thank Professor Yuanhuai Han (Shanxi Agricultural University, China), Professor Jihong Liu (Huazhong Agricultural University, China), Professor Hanhui Kuang (Huazhong Agricultural University, China), Zhiyong Pan and Jinzhi Zhang (Huazhong Agricultural University, China) for their critical reading of this paper. We also thank Jianbo Cao (Huazhong Agricultural University, China) and Baoping Chen (Medical College, Wuhan University) for technical assistance.
References
- Alquezar B, Zacarias L, Rodrigo MJ. Molecular and functional characterization of a novel chromoplast-specific lycopene beta-cyclase from Citrus and its relation to lycopene accumulation. Journal of Experimental Botany. 2009;60:1783–1797. doi: 10.1093/jxb/erp048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Kim E, DellaPenna D, Brutnell T. Novel lycopene epsilon cyclase activities in maize revealed through perturbation of carotenoid biosynthesis. The Plant Journal. 2009;59:588–599. doi: 10.1111/j.1365-313X.2009.03899.x. [DOI] [PubMed] [Google Scholar]
- Barsan C, Sanchez-Bel P, Rombaldi C, Egea I, Rossignol M, Kuntz M, Zouine M, Latche A, Bouzayen M, Pech JC. Characteristics of the tomato chromoplast revealed by proteomic analysis. Journal of Experimental Botany. 2010;61:2413–2431. doi: 10.1093/jxb/erq070. [DOI] [PubMed] [Google Scholar]
- Ben-Shaul Y, Naftali Y. The development and ultrastructure of lycopene bodies in chromoplasts of Lycopersicon esculentum . Protoplasma. 1969;67:333–344. [Google Scholar]
- Cameron J, Soost R, Olson E. Chimeral basis for color in pink and red grapefruit. Journal of Heredity. 1964;55:23–28. [Google Scholar]
- Cazzonelli C, Pogson B. Source to sink: regulation of carotenoid biosynthesis in plants. Trends in Plant Science. 2010;15:266–274. doi: 10.1016/j.tplants.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Chen MH, Liu LF, Chen YR, Wu HK, Yu SM. Expression of α-amylases, carbohydrate metabolism, and autophagy in cultured rice cells is coordinately regulated by sugar nutrient. The Plant Journal. 1994;6:625–636. doi: 10.1046/j.1365-313x.1994.6050625.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Li F, Wurtzel ET. Isolation and characterization of the Z-ISO gene encoding a missing component of carotenoid biosynthesis in plants. Plant Physiology. 2010;153:66–79. doi: 10.1104/pp.110.153916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YJ, Guo WW, Yi HL, Pang XM, Deng XX. An efficient protocol for genomic DNA extraction from Citrus species. Plant Molecular Biology Reporter. 2003;21:177–178. [Google Scholar]
- D’Ambrosio C, Stigliani AL, Giorio G. Overexpression of CrtR-b2 (carotene beta hydroxylase 2) from S. lycopersicum L. differentially affects xanthophyll synthesis and accumulation in transgenic tomato plants. Transgenic Research. 2011;20:47–60. doi: 10.1007/s11248-010-9387-4. [DOI] [PubMed] [Google Scholar]
- Diretto G, Al-Babili S, Tavazza R, Papacchioli V, Beyer P, Giuliano G. Metabolic engineering of potato carotenoid content through tuber-specific overexpression of a bacterial mini-pathway. PLoS One. 2007;2:350. doi: 10.1371/journal.pone.0000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diretto G, Al-Babili S, Tavazza R, Scossa F, Papacchioli V, Migliore M, Beyer P, Giuliano G. Transcriptional-metabolic networks in beta-carotene-enriched potato tubers: the long and winding road to the Golden phenotype. Plant Physiology. 2010;154:899–912. doi: 10.1104/pp.110.159368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Wang NL, Cui F, Li XH, Xiao JH, Xiong LZ. Characterization of the β-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiology. 2010;154:1304–1318. doi: 10.1104/pp.110.163741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan YX, Guo WW, Meng HJ, Tao NG, Li DD, Deng XX. High efficient transgenic plant regeneration from embryogenic calluses of Citrus sinensis. Biologia Plantarum. 2007;51:212–216. [Google Scholar]
- Ducreux LJ, Morris WL, Hedley PE, Shepherd T, Davies HV, Millam S, Taylor MA. Metabolic engineering of high carotenoid potato tubers containing enhanced levels of beta-carotene and lutein. Journal of Experimental Botany. 2005;56:81–89. doi: 10.1093/jxb/eri016. [DOI] [PubMed] [Google Scholar]
- Egea I, Barsan C, Bian WP, Purgatto E, Latche A, Chervin C, Bouzayen M, Pech JC. Chromoplast differentiation: current status and perspectives. Plant and Cell Physiology. 2010;51:1601–1611. doi: 10.1093/pcp/pcq136. [DOI] [PubMed] [Google Scholar]
- Engelmann NJ, Campbell JK, Rogers RB, Rupassara SI, Garlick PJ, Lila MA, Erdman JW. Screening and selection of high carotenoid producing in vitro tomato cell culture lines for [13C]-carotenoid production. Journal of Agricultural and Food Chemistry. 2010;58:9979–9987. doi: 10.1021/jf101942x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré G, Bai C, Twyman RM, Capell T, Christou P, Zhu CF. Nutritious crops producing multiple carotenoids: a metabolic balancing act. Trends in Plant Science. 2011;16:532–540. doi: 10.1016/j.tplants.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Fraser P, Enfissi E, Bramley P. Genetic engineering of carotenoid formation in tomato fruit and the potential application of systems and synthetic biology approaches. Archives Biochemistry and Biophysics. 2009;483:196–204. doi: 10.1016/j.abb.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Fraser PD, Enfissi EM, Halket JM, Truesdale MR, Yu D, Gerrish C, Bramley PM. Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. The Plant Cell. 2007;19:3194–3211. doi: 10.1105/tpc.106.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Tavazza R, Diretto G, Beyer P, Taylor MA. Metabolic engineering of carotenoid biosynthesis in plants. Trends in Biotechnology. 2008;26:139–145. doi: 10.1016/j.tibtech.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Grosser J, Gmitter F. Protoplast fusion and citrus improvement. Plant Breeding Reviews. 1990;8:339–374. [Google Scholar]
- Harjes CE, Rocheford TR, Bai L, et al. Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science. 2008;319:330–333. doi: 10.1126/science.1150255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig S, Drossard J, Twyman RM, Fischer R. Plant cell cultures for the production of recombinant proteins. Nature Biotechnology. 2004;22:1415–1422. doi: 10.1038/nbt1027. [DOI] [PubMed] [Google Scholar]
- Horner HT, Healy RA, Ren G, Fritz D, Klyne A, Seames C, Thornburg RW. Amyloplast to chromoplast conversion in developing ornamental tobacco floral nectaries provides sugar for nectar and antioxidants for protection. American Journal of Botany. 2007;94:12–24. doi: 10.3732/ajb.94.1.12. [DOI] [PubMed] [Google Scholar]
- Hornero-Mendez D, Minguez-Mosquera MI. Xanthophyll esterification accompanying carotenoid overaccumulation in chromoplast of Capsicum annuum ripening fruits is a constitutive process and useful for ripeness index. Journal of Agricultural and Food Chemistry. 2000;48:1617–1622. doi: 10.1021/jf9912046. [DOI] [PubMed] [Google Scholar]
- Howitt CA, Cavanagh CR, Bowerman AF, Cazzonelli C, Rampling L, Mimica JL, Pogson BJ. Alternative splicing, activation of cryptic exons and amino acid substitutions in carotenoid biosynthetic genes are associated with lutein accumulation in wheat endosperm. Functional and Integrative Genomics. 2009;9:363–376. doi: 10.1007/s10142-009-0121-3. [DOI] [PubMed] [Google Scholar]
- Howitt CA, Pogson BJ. Carotenoid accumulation and function in seeds and non-green tissues. Plant, Cell and Environment. 2006;29:435–445. doi: 10.1111/j.1365-3040.2005.01492.x. [DOI] [PubMed] [Google Scholar]
- Isaacson T, Damasceno CM, Saravanan RS, He Y, Catala C, Saladie M, Rose JK. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nature Protocols. 2006;1:769–774. doi: 10.1038/nprot.2006.102. [DOI] [PubMed] [Google Scholar]
- Jayaraj J, Devlin R, Punja Z. Metabolic engineering of novel ketocarotenoid production in carrot plants. Transgenic Research. 2008;17:489–501. doi: 10.1007/s11248-007-9120-0. [DOI] [PubMed] [Google Scholar]
- Kato M, Ikoma Y, Matsumoto H, Sugiura M, Hyodo H, Yano M. Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiology. 2004;134:824–837. doi: 10.1104/pp.103.031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Rensing K, Douglas C, Cheng K. Chromoplasts ultrastructure and estimated carotene content in root secondary phloem of different carrot varieties. Planta. 2010;231:549–558. doi: 10.1007/s00425-009-1071-7. [DOI] [PubMed] [Google Scholar]
- Lee EK, Jin YW, Park JH, Yoo YM, Hong SM, Amir R, Yan Z, Kwon E, Elfick A, Tomlinson S. Cultured cambial meristematic cells as a source of plant natural products. Nature Biotechnology. 2010;28:1213–1217. doi: 10.1038/nbt.1693. [DOI] [PubMed] [Google Scholar]
- Li L, Lu S, Cosman KM, Earle ED, Garvin DF, O’Neill J. β-Carotene accumulation induced by the cauliflower Or gene is not due to an increased capacity of biosynthesis. Phytochemistry. 2006;67:1177–1184. doi: 10.1016/j.phytochem.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Li L, Van Eck J. Metabolic engineering of carotenoid accumulation by creating a metabolic sink. Transgenic Research. 2007;16:581–585. doi: 10.1007/s11248-007-9111-1. [DOI] [PubMed] [Google Scholar]
- Liu Q, Xu J, Liu Y, Zhao X, Deng X, Guo L, Gu J. A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck) Journal of Experimental Botany. 2007;58:4161–4171. doi: 10.1093/jxb/erm273. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu Q, Tao NG, Deng XX. Efficient isolation of RNA from fruit peel and pulp of ripening navel orange (Citrus sinensis Osbeck) Journal of Huazhong Agricultural University. 2006;25:300–304. [Google Scholar]
- Lopez AB, Van Eck J, Conlin BJ, Paolillo DJ, O’Neill J, Li L. Effect of the cauliflower Or transgene on carotenoid accumulation and chromoplast formation in transgenic potato tubers. Journal of Experimental Botany. 2008;59:213–223. doi: 10.1093/jxb/erm299. [DOI] [PubMed] [Google Scholar]
- Luft J. Improvements in epoxy resin embedding methods. The Journal of Biophysical and BiochemicalCcytology. 1961;9:409–414. doi: 10.1083/jcb.9.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass D, Arango J, Wust F, Beyer P, Welsch R. Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS One. 2009 doi: 10.1371/journal.pone.0006373. 4, e6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa N, Yamano S, Linden H, de Felipe MR, Lucas M, Ikenaga H, Sandmann G. Functional expression of the Erwinia uredovora carotenoid biosynthesis gene crtl in transgenic plants showing an increase of beta-carotene biosynthesis activity and resistance to the bleaching herbicide norflurazon. The Plant Journal. 1993;4:833–840. doi: 10.1046/j.1365-313x.1993.04050833.x. [DOI] [PubMed] [Google Scholar]
- Paine JA, Shipton CA, Chaggar S, et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nature Biotechnology. 2005;23:482–487. doi: 10.1038/nbt1082. [DOI] [PubMed] [Google Scholar]
- Pan ZY, Liu Q, Yun Z, Guan R, Zeng WF, Xu Q, Deng XX. Comparative proteomics of a lycopene-accumulating mutant reveals the important role of oxidative stress on carotenogenesis in sweet orange (Citrus sinensis [L.] Osbeck) Proteomics. 2009;9:5455–5470. doi: 10.1002/pmic.200900092. [DOI] [PubMed] [Google Scholar]
- Pogson B, McDonald KA, Truong M, Britton G, DellaPenna D. Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. The Plant Cell. 1996;8:1627–1639. doi: 10.1105/tpc.8.9.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer S, Fraser PD, Kiano JW, Shipton CA, Misawa N, Schuch W, Bramley PM. Elevation of the provitamin A content of transgenic tomato plants. Nature Biotechnology. 2000;18:666–669. doi: 10.1038/76523. [DOI] [PubMed] [Google Scholar]
- Schäfer L, Sandmann M, Woitsch S, Sandmann G. Coordinate up-regulation of carotenoid biosynthesis as a response to light stress in Synechococcus PCC7942. Plant, Cell and Environment. 2006;29:1349–1356. doi: 10.1111/j.1365-3040.2006.01515.x. [DOI] [PubMed] [Google Scholar]
- Schaub P, Al-Babili S, Drake R, Beyer P. Why is golden rice golden (yellow) instead of red? Plant Physiology. 2005;138:441–450. doi: 10.1104/pp.104.057927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiggert RM, Steingass CB, Heller A, Esquivel P, Carle R. Characterization of chromoplasts and carotenoids of red-and yellow-fleshed papaya (Carica papaya L.) Planta. 2011;234:1031–1044. doi: 10.1007/s00425-011-1457-1. [DOI] [PubMed] [Google Scholar]
- Shewmaker CK, Sheehy JA, Daley M, Colburn S, Ke DY. Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. The Plant Journal. 1999;20:401–412. doi: 10.1046/j.1365-313x.1999.00611.x. [DOI] [PubMed] [Google Scholar]
- Talon M, Gmitter FG. Citrus genomics. International Journal of Plant Genomics. 2008;2008:528361. doi: 10.1155/2008/528361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhaneni R, Gallagher CE, Licciardello N, Cuttriss AJ, Quinlan RF, Wurtzel ET. Metabolite sorting of a germplasm collection reveals the Hydroxylase3 locus as a new target for maize provitamin A biofortification. Plant Physiology. 2009;151:1635–1645. doi: 10.1104/pp.109.145177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lintig J. Colors with functions: elucidating the biochemical and molecular basis of carotenoid metabolism. Annual Review of Nutrition. 2010;30:35–56. doi: 10.1146/annurev-nutr-080508-141027. [DOI] [PubMed] [Google Scholar]
- Welsch R, Arango J, Bar C, Salazar B, Al-Babili S, Beltran J, Chavarriaga P, Ceballos H, Tohme J, Beyer P. Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. The Plant Cell. 2010;22:3348–3356. doi: 10.1105/tpc.110.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RR. The diversity of plastid form and function. Advances in Photosynthesis and Respiration. 2006;23:3–26. [Google Scholar]
- Xu Q, Liu YL, Zhu AD, Wu XM, Ye JL, Yu KQ, Guo WW, Deng XX. Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genomics. 2010;11:246. doi: 10.1186/1471-2164-11-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Yu KQ, Zhu AD, Ye JL, Liu Q, Zhang JC, Deng XX. Comparative transcripts profiling reveals new insight into molecular processes regulating lycopene accumulation in a sweet orange (Citrus sinensis) red-flesh mutant. BMC Genomics. 2009;10:540. doi: 10.1186/1471-2164-10-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kandianis CB, Harjes CE, Bai L, Kim EH, Yang X, Skinner DJ, Fu Z, Mitchell S, Li Q. Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nature Genetics. 2010;42:322–327. doi: 10.1038/ng.551. [DOI] [PubMed] [Google Scholar]
- Zhang JC, Tao NG, Xu Q, Zhou WJ, Cao HB, Xu J, Deng XX. Functional characterization of Citrus PSY gene in Hongkong kumquat (Fortunella hindsii Swingle) Plant Cell Reports. 2009;28:1737–1746. doi: 10.1007/s00299-009-0774-3. [DOI] [PubMed] [Google Scholar]
- Zhu C, Naqvi S, Breitenbach J, Sandmann G, Christou P, Capell T. Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proceedings of the National Academy of Sciences, USA. 2008;105:18232–18237. doi: 10.1073/pnas.0809737105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ma G, Kato M, Yamawaki K, Takagi T, Kiriiwa Y, Ikoma Y, Matsumoto H, Yoshioka T, Nesumi H. Regulation of carotenoid accumulation and the expression of carotenoid metabolic genes in citrus juice sacs in vitro. Journal of Experimental Botany. 2012;63:871–886. doi: 10.1093/jxb/err318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.