Abstract
The phytohormone ethylene plays important roles in regulating plant responses to phosphate (Pi) starvation. To date, however, no molecular components have been identified that interact with ethylene signalling in regulating such responses. In this work, an Arabidopsis mutant, hps4, was characterized that exhibits enhanced responses to Pi starvation, including increased inhibition of primary root growth, enhanced expression of Pi starvation-induced genes, and overproduction of root-associated acid phosphatases. Molecular cloning indicated that hps4 is a new allele of SABRE, which was previously identified as an important regulator of cell expansion in Arabidopsis. HPS4/SABRE antagonistically interacts with ethylene signalling to regulate plant responses to Pi starvation. Furthermore, it is shown that Pi-starved hps4 mutants accumulate more auxin in their root tips than the wild type, which may explain the increased inhibition of their primary root growth when grown under Pi deficiency.
Keywords: Antagonistic interaction, auxin accumulation, ethylene signalling, hps4 mutant, HPS4/SABRE, phosphate starvation responses
Introduction
Plant growth and development are regulated by both internal genetic programmes and external signals. When grown in soil, plants are exposed to various environmental stimuli, such as light, temperature, water, nutrients, pathogens, and mechanical forces. Among these environmental factors, the nutrient level has a profound effect on plant growth and development. Because plants are sessile organisms, those experiencing nutrient scarcity must display a set of responses to cope with this environmental stress. An essential nutrient that is often scarce is phosphorus (P). In soil, inorganic phosphate (Pi) is the major form of P that is taken up by plants through phosphate transporters on the root surface. In most soils, however, the Pi level is <10 μM, which is below the concentration required for optimal plant growth (Schachtman et al., 1998; Raghothama, 1999). The responses of plants to Pi starvation include changes in root architecture (i.e. inhibition of primary root growth and formation of more lateral roots and root hairs), increased expression of Pi transporter genes, induction and secretion of acid phosphatases (APases) and RNases, and accumulation of anthocyanin (Yuan and Liu, 2008). Although these responses have been well documented in many plant species, the underlying signalling mechanism is poorly understood.
The regulation of plant responses to Pi starvation involves the plant hormone ethylene (Nagarajan and Smith, 2012). The level of ethylene was twice as high in Pi-starved than in non-starved common bean plants (Borch et al., 1999). The change of ethylene level has also been found in tomato and Medicago falcate plants grown under Pi deficiency (P–) (Kim et al., 2008; Li et al., 2009). When grown under P– conditions, maize roots exhibited an enhanced sensitivity to ethylene (He et al., 1992). In addition, Ma et al. (2003) found that treatment of Arabidopsis seedlings with the ethylene biosynthesis inhibitor aminoethoxyvinylglycine (AVG) enhanced primary root growth under Pi-sufficient (P+) conditions but inhibited primary root growth under P– conditions. Ethylene is also involved in Pi starvation-mediated root hair development. In the ethylene-insensitive Arabidopsis mutant ein2, the induction of root hair formation was partially blocked. Further anatomical examination indicated that P availability and ethylene have interacting but distinct effects on root hair development (Ma et al., 2001; Zhang et al., 2003). Several genomic studies have demonstrated that, in Pi-starved root tissues, the expression of some genes related to both ethylene biosynthesis and signalling is altered (Mission et al., 2005; Thibaud et al., 2010; Chacon-Lopez et al., 2011). All these results indicate that ethylene is an important mediator of Pi starvation-regulated root development. Previous findings regarding the Arabidopsis mutant hps2 (hypersensitive to Pi starvation 2), however, demonstrated that in addition to regulating root growth when plants are exposed to Pi starvation, ethylene is also involved in regulating Pi starvation-induced (PSI) gene expression, APase production, and anthocyanin accumulation (Lei et al., 2011). hps2 was identified based on its enhanced expression of the high affinity Pi transporter gene Pht1;4. It was shown to be a new allele of the CTR1 gene. Functional disruption of the CTR1 gene causes plants to display a constitutive ethylene response. Under P– conditions, treatment of wild-type (WT) plants with the ethylene signalling inhibitor Ag+ suppressed expression of Pht1; 4, whereas addition of the ethylene biosynthesis precursor 1-aminocyclopropane-1-carboxylic acid (ACC) dramatically enhanced its expression. Similarly, the expression of the Pht1;4 gene is partially suppressed in the ethylene-insensitive mutant ein2-5 but is enhanced in the ethylene-overproducing mutant eto1. A similar expression pattern was also observed for several other PSI genes in hps2 and ein2 mutants. In addition, production of PSI APase is enhanced in hps2/ctr1 but is partially suppressed in ein2. Li et al. (2011) also found that induction of APase in M. falcate roots could be stimulated by ACC under P+ conditions but that induction of APase in roots was blocked by the ethylene biosynthesis inhibitor AVG under P– conditions. These results demonstrated that ethylene is a positive regulator of PSI gene expression and APase production. Ethylene, however, is a negative regulator of PSI anthocyanin accumulation because, under P– conditions, ein2 produces more anthocyanin but hps2 produces less anthocyanin (Lei et al., 2011). Recently, another Arabidopsis mutant hps3 which is hypersensitive to Pi starvation has been characterized (Wang et al., 2012). hps3 was identified as a new allele of the ETO1 gene which, when mutated, led to the overproduction of ethylene in Arabidopsis seedlings (Wang et al., 2004). Although these results have clearly defined the role of ethylene in regulating multiple plant responses to Pi starvation, the molecular components that interact with ethylene signalling in regulating plant Pi responses have yet to be identified.
In this work, an Arabidopsis mutant, hps4, that exhibits enhanced sensitivity to Pi starvation was characterized. The HPS4 gene encodes SABRE, which was previously identified as an important regulator of cell expansion in Arabidopsis (Aeschbacher et al., 1995). It is demonstrated that HPS4 antagonistically interacts with ethylene signalling to regulate plant responses to Pi starvation. Furthermore, it is shown that Pi-starved hps4 accumulates more auxin in its root tip than the WT, which may explain the increased inhibition of its primary root growth under Pi starvation.
Materials and methods
Plant materials and growth conditions
All plants used in this study were of the Columbia ecotype background. The Pi-sufficient medium (P+) used was half-strength MS medium (Murashige and Skoog, 1962) with 1% (w/v) sucrose and 1.2% (w/v) agar (Sigma catalogue no. A1296). The Pi-deficient medium (P–) was made by replacing the 1.25 mM KH2PO4 in the P+ medium with 0.65 mM K2SO4. Seeds were surface sterilized with 20% (v/v) bleach for 20 min. After three washes in sterile distilled water, seeds were sown on Petri plates containing P+ or P– medium. After the seeds were stratified at 4 °C for 2 d, the agar plates were placed vertically in a growth room with a photoperiod of 16 h of light and 8 h of darkness at 22–24 °C. The light intensity was 100 μmol m−2 s−1.
Mutant isolation
About 80 000 ethylmethane sulphonate (EMS)-mutagenized M2 seeds representing ∼5000 M1 plant lines were used for mutant screening. The EMS-mutagenized lines were generated according to Weigel and Glazebrook (2002). The roots of seedlings that had grown vertically for 7 d were overlaid with a 0.5% agar solution containing 0.01% BCIP (5-bromo-4-chloro-3-indoxyl phosphate) for 24 h at 23 °C (Lloyd et al., 2001). The seedlings with dark-blue BCIP staining were identified as putative mutants and were transferred to soil. The plants were self-pollinated, and the mutant phenotypes were confirmed in the next generation. The mutants were back-crossed to the WT plants twice before they were characterized further.
Genetic mapping of the HPS4 gene
The mapping population was generated by crossing the mutant hps4 to a plant of the Ler ecotype. The F2 progeny that displayed the mutant phenotype were selected, and DNAs from these seedlings were isolated for molecular mapping. A set of simple sequence length polymorphism (SSLP) and cleaved amplified polymorphic sequence (CAPS) markers were used to map the HPS4 gene. The sequences and chromosomal positions of the molecular markers are listed in Supplementary Table S1 available at JXB online.
Quantitative real-time PCR analysis
Total RNAs were extracted from 8-day-old seedlings with the TIANGEN RNAprep pure plant kit with on-column DNase I digestion (Tiangen Co., Beijing). The first-strand cDNA was synthesized using oligo(dT) and Takara MLV-Reverse transcriptase. Real-time PCR analysis was carried out on the Applied Biosystems 7500 real-time PCR detection system. UBC mRNA was used as an internal control. The genes and the primers used for detection of their mRNA expression are listed in Supplementary Table S2 at JXB online.
In-gel assays of the APase profile
The protein extraction and in-gel assay for APase profiles were performed essentially as described (Trull and Deikman, 1998).
Quantitative analysis of cellular Pi and total P content
Cellular Pi content was determined using the method described by Ames (1966). Basically, the pre-weighed fresh shoot and root tissues were submerged in 1 ml of 1% glacial acetate and freeze–thawed eight times. A 100 μl volume of the extract was mixed with 200 μl of H2O and 700 μl of Pi reaction buffer containing a mixture of 0.48% NH4MoO4+2.85% (v/v) H2SO4 and 10% (w/v) ascorbic acid in a ratio of 6:1. The reaction was allowed to proceed at 37 °C for 1 h. The Pi content was determined at A 820 according to a pre-made standard curve and was expressed as μmol g−1 fresh weight (FW). For determination of total P content, ∼50 mg of fresh tissue was oven-dried at 500 °C for 3 h and flamed to ash. The ashes were dissolved in 100 μl of 30% (v/v) HCl and 10% (v/v) HNO3. A 10 μl volume of dissolved sample was mixed with 290 μl of ddH2O and 700 μl of Pi reaction buffer, and Pi was quantified by Ames’s method. The total P content of plant tissues was determined and expressed as Pi content extracted from flamed ashes.
Measurement of anthocyanin content
Anthocyanins were extracted with propanol:HCl:H2O (18:1:81, v/v/v) at room temperature overnight. Absorbance was measured at 530 nm and 650 nm. Anthocyanin content was expressed as A 530–A 650 g−1 FW.
Quantitative analysis of total APase activity
About 50 mg of shoot or root tissue was ground in liquid N2, and the total protein was extracted in protein extraction buffer [0.1 M KAc, 20 mM CaCl2, 2 mM EDTA, 0.1 mM phenylmethylsulphonyl fluoride (PMSF)]. For quantitative analysis of total APase activity, 10 μl of extracted proteins was mixed with 620 μl of reaction buffer (10 mM MgCl2, 50 mM NaAc, pH 4.9), and 10 μl of p-nitrophenol phosphate (10 mg ml−1) (pNPP; Sigma, N-2770). After incubation at 37 °C for 1 h, the reaction was terminated with 120 μl of 2% SDS. The colour that developed was measured spectrophotometrically at 410 nm, and APase activity was expressed as A 410 mg−1 protein.
Quantitative analysis of root-associated APase activity
Root-associated APase activity was measured according to Boutin et al. (1981) with some modifications. Roots were excised from two 8-day-old seedlings of uniform size and transferred to a 2 ml Eppendorf tube containing 620 μl of reaction buffer (10 mM MgCl2, 50 mM NaAc, pH 4.9). A 50 μl aliquot of pNPP (5 mg ml−1) was added to the tubes, which were incubated at 37 °C for 1 h. Then, 120 μl of 2% SDS was added to terminate the reaction, and absorbance was determined spectrophotometrically at 410 nm. The APase activity was expressed as A 410 cm−1 root.
Histochemical analysis of GUS activity
The histochemical analysis of β-glucuronidase (GUS) activity was performed according to Jefferson et al. (1989).
Measurement of IAA contents
Free indole-3-acetic acid (IAA) was extracted and purified from apical 5 mm root sections of 9-day-old hps4 and WT seedlings, and analysed by gas chromatography–mass spectrometry as described (Edlund et al., 1995), except that an Agilent 7890/7000 gas chromatographer–mass spectrometer was used, with the separation performed in a DB-5ms column (Agilent, http://www.agilent.com). The internal standard [13C]IAA was purchased from Cambridge Isotope Laboratories (http://www.isotope.com).
Method for statistical analysis
The two-sample t-test function of Origin software (OriginLab Corporation, Northampton, USA) was used to perform statistical analysis of the data generated in this work.
Results
hps4 exhibits enhanced production of root surface-associated APase, reduced anthocyanin accumulation, and increased inhibition of primary root growth under Pi-deficient conditions
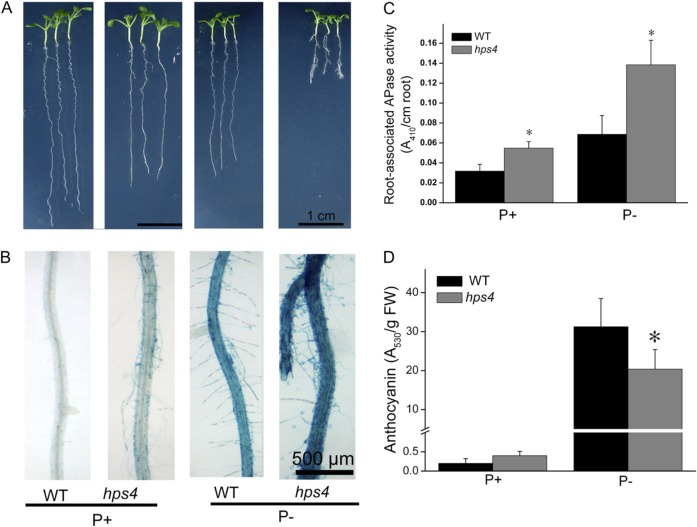
APase production is a universal response of plants to Pi starvation (Tran et al., 2010). To identify novel molecular components involved in plant responses to Pi starvation, APase was used as a biomarker in a large-scale screen for Arabidopsis mutants with altered sensitivity to Pi starvation. M2 seeds from EMS-mutagenized M1 plants were directly sown on half-strength MS P+ or P– medium. Nine days after germination (DAG), APase production was examined. APase activity on the root surface can be detected by application of the APase substrate BCIP; cleavage of BCIP by APases produces a blue precipitate (Lloyd et al., 2001). Using this method, an Arabidopsis mutant, hps4 (hypersensitive to Pi starvation 4), was identified with enhanced APase activity on its root surface under P– conditions as indicated by a dark-blue staining compared with the WT (Fig. 1B; Supplementary Fig. S1A at JXB online). On P+ medium, hps4 also displayed a light-blue staining, whereas no blue staining was evident on the WT (Fig. 1B; Supplementary Fig. S1A). Under both P+ and P– conditions, total APase activity in shoots and roots did not differ between hps4 and the WT as analysed by both quantitative measurement and in-gel assays (Supplementary Fig. S1B, C); however, the secreted, root surface-associated APase activity was much higher for hps4 than for the WT (Fig. 1C).
Fig. 1.
Growth characteristics, APase activities, and anthocyanin contents of the WT and the hps4 mutant. (A) Morphology of 9-day-old seedlings of the WT and hps4 grown on Pi-sufficient (P+) and Pi-deficient (P–) medium. (B) Detection of APase activity by BCIP staining on the root surfaces of WT and hps4 seedlings shown in A. (C) Root-associated APase activity in 9-day-old seedlings of the WT and hps4 grown on P+ and P– medium. (D) Anthocyanin contents in 12-day-old seedlings of the WT and hps4. For C and D, values represent the mean and SE of three replicates. Means with asterisks are significantly different from the WT (P < 0.05, two-sample t-test).
Accumulation of anthocyanin is another hallmark response of plants to Pi starvation and is thought to protect chloroplast membranes. Thus, the levels of anthocyanin were compared in the hps4 mutant and the WT. On P+ medium, anthocyanin levels were low and did not differ between hps4 and WT plants. Under P– conditions, anthocyanin accumulation was dramatically increased in both the hps4 mutant and the WT, but the level was ∼30% lower in hps4 than in the WT (Fig. 1D; Supplementary Fig. S2 at JXB online).
hps4 was also more sensitive to PSI inhibition of primary root growth. At 9 DAG, the hps4 primary root was ∼15% shorter than that of the WT on P+ but 70% shorter on P– medium (Fig. 1A). In addition, hps4 formed lateral roots earlier than the WT under P– conditions (Fig. 1A).
Expression of PSI genes is enhanced in hps4
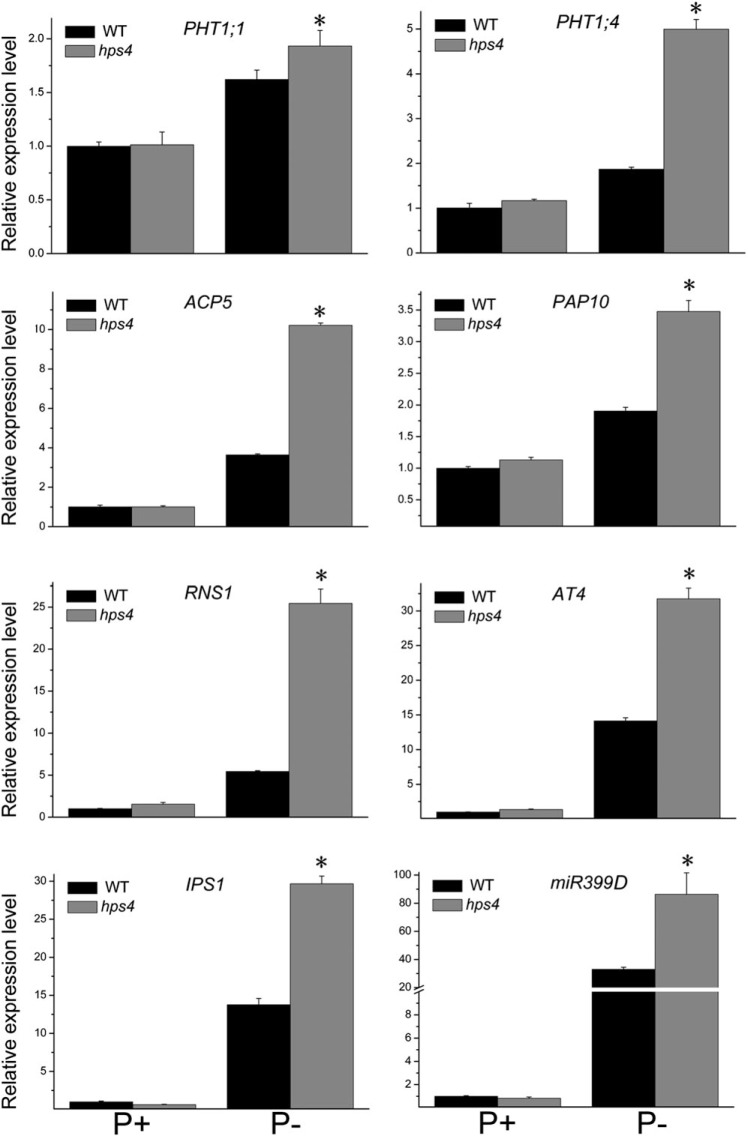
To examine whether PSI gene expression was affected in hps4, the expression of eight PSI genes in hps4 and the WT was analysed. The PSI genes examined included two high-affinity Pi transporters, Pht1;1 and Pht1;4 (Muchhal et al., 1996); two non-coding transcripts, At4 and IPS1 (Burleigh and Harrison, 1999; Martín et al., 2000); an RNase, RNS1 (Bariola et al., 1994); an miR399D (Fujii et al., 2005); and two APases, ACP5 (del Pozo et al., 1999) and AtPAP10 (Wang et al., 2011). At 4 DAG, the induction of all eight PSI genes was significantly higher in hps4 than in the WT under Pi starvation, while induction did not differ when plants were grown on P+ medium (Fig. 2). At 9 DAG, the expression of six PSI genes was still higher in hps4 than in the WT, but the expression of the two non-coding transcripts was similar in hps4 and the WT (Supplementary Fig. S3 at JXB online).
Fig. 2.
Analysis of PSI gene expression in the WT and the hps4 mutant. Four-day-old seedlings of the WT and hps4 grown on P+ or P– medium were used for real-time PCR analysis. The names of the genes examined are indicated on the top of each panel. Values are the means and SD of three biological replicates and represent fold changes normalized to transcript levels of the WT on P+ medium. Means with asterisks are significantly different from the WT (P < 0.05, two-sample t-test).
The contents of total P and cellular Pi were then analysed in 9-day-old WT and hps4 seedlings. As shown in Supplementary Fig. S4 at JXB online, total P and cellular Pi contents did not differ significantly between hps4 and WT shoot and root tissues under either P+ or P– conditions.
Genetic and molecular analysis of hps4
When hps4 was backcrossed to the WT, all F1 plants showed WT phenotypes, and F2 progeny derived from selfed F1 plants segregated into mutant and WT phenotypes in a ratio of 1:3 (55:165). These results indicated that the hps4 mutant phenotypes were caused by a single recessive mutation. A map-based cloning approach was then used to identify the molecular lesion in hps4. hps4 (Columbia ecotype) was crossed to a plant with Landsberg erecta background to establish a mapping population. Using F2 progeny derived from this cross, the HPS4 gene was mapped to a 91 kb region on chromosome 1 (Supplementary Fig. S5 at JXB online). After sequencing all the annotated genes in this region, a point mutation was found within the SABRE gene (At1g58250) (Fig. 3A). This mutation caused a transition of nucleotide G to C, which converted an alanine to a proline at position of 2118 on the SABRE protein. To confirm that mutated SABRE was linked to the mutant phenotypes, regions encompassing the mutated site in 10 WT and 10 mutant F2 progeny derived from the cross between hps4 and the WT (Columbia ecotype) were sequenced. All 10 of the mutant progeny were homozygous for the mutation, and the 10 progeny with the WT phenotype either had no mutation or were heterozygous for the mutation, indicating that the homozygote mutation in the SABRE gene was linked to the hps4 mutant phenotypes; this was consistent with the genetic analysis, which had indicated that hps4 is caused by a single recessive mutation. The phenotypes of a previously characterized sabre mutant (sab1-1) (Aeschbacher et al., 1995) and six additional SALK T-DNA insertion lines of the SABRE gene were then further examined. The T-DNA insertions in sab1-1 and the six SALK lines disrupted expression of the SABRE gene. On the P+ medium, similar to hps4, sab1-1 and the six SALK lines showed light-blue BCIP staining, while on the P– medium, the roots of the sab1-1 and six SALK lines had darker blue BCIP staining (Fig. 3B, C). This provided additional evidence that hps4 is a new allele of the SABRE gene. The length of primary roots of sab1-1 and the six SALK lines on both P+ and P– media were, however, much shorter than those of hps4. The stronger root phenotypes of sab1-1 and the six SALK lines were probably due to the complete disruption of transcription of the SABRE gene because of T-DNA insertion. In contrast, the transcription level of the SABRE gene in hps4 was not affected by the point mutation, suggesting that hps4 is only a weak mutant allele (see the results later in the text).
Fig. 3.
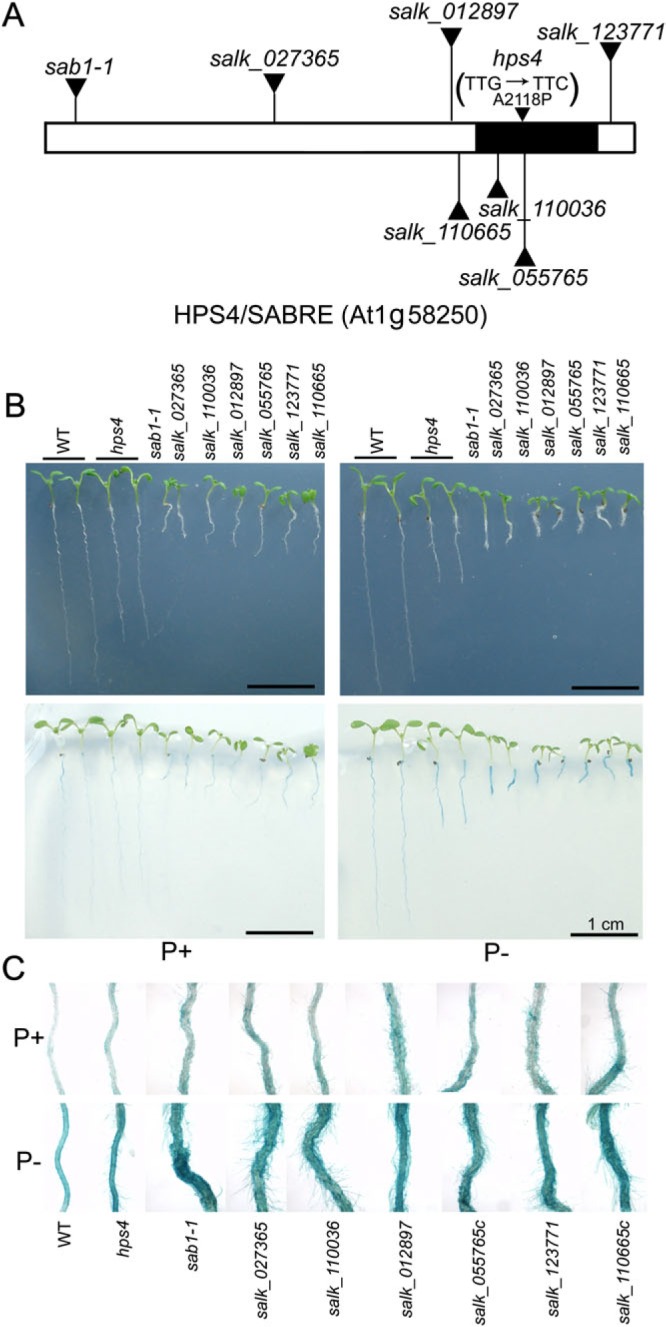
Molecular cloning of the HPS4 gene. (A) A diagram of the structure of the HPS4/SABRE protein. The AGI code of the HPS4 gene is indicated. The filled region indicates the segment that shares sequence homology with a group of Golgi-localized plant proteins. The positions of T-DNA insertions in the sab1-1mutant line and six SALK lines, and the position of the point mutation in hps4 are indicated. The changes in nucleotide and amino acid in the hps4 mutant are shown in parentheses. (B) Morphologies and BCIP staining of 9-day-old seedlings of the WT, hps4, sab1-1, and six SALK T-DNA insertion lines grown on P+ and P– medium. (C) Close-up view of BCIP staining of the seedlings shown in B.
Expression patterns of the HPS4/SABRE gene
SABRE was previously identified as a gene that is required for normal cell expansion in Arabidopsis (Aeschbacher et al., 1995). Alhough analysis of its protein sequence indicated that its C-terminal part (residues 1900–2450, Fig. 4A) shares homology with the sequences presented in a group of plant Golgi-localized proteins (Procissi et al., 2003; Xu and Dooner, 2006), the exact biochemical function of the SABRE protein is still unknown. For the maize homologue of SABRE, APT1, this sequence is required for protein localization to Golgi bodies (Xu and Dooner, 2006).
Fig. 4.
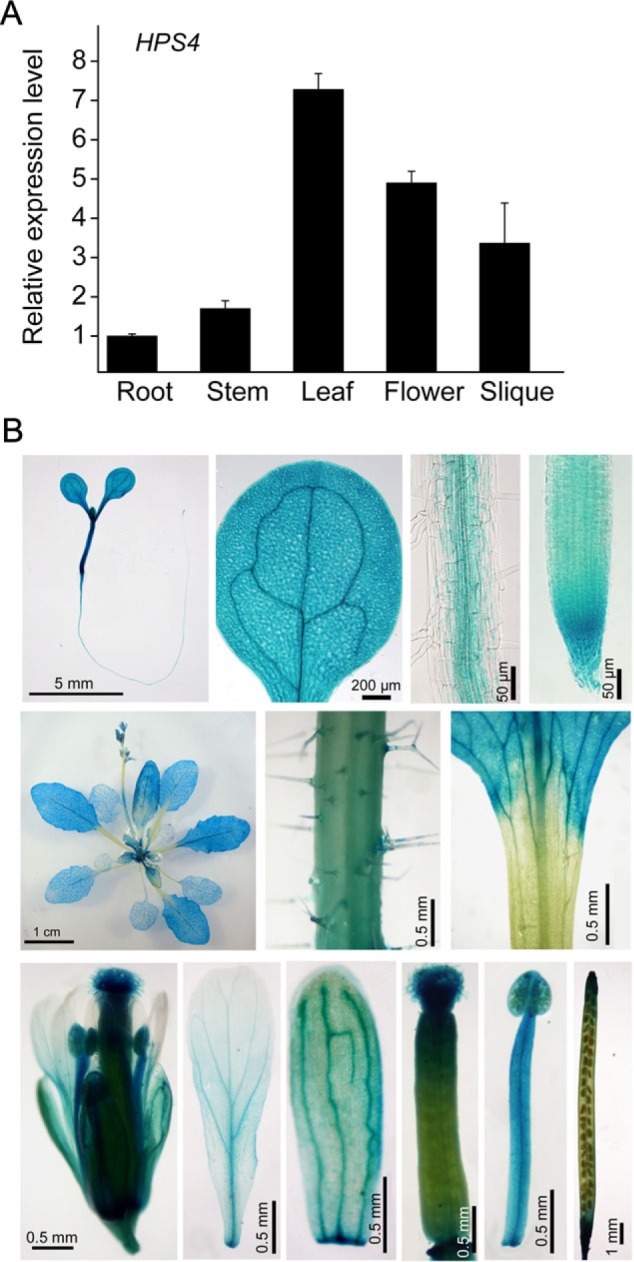
Expression patterns of the HPS4 gene. (A) Relative expression of the HPS4 gene in different plant organs determined by q-PCR. (B) Tissue-specific expression patterns of the HPS4::GUS gene. Top row, from left to right: a 9-day-old seedling, a cotyledon, part of the root elongation zone, and root apex. Middle row, from left to right: a 20-day-old mature plant, stem, and a junction between leaf blade and leaf petiole. Bottom row, from left to right: a fully opened flower, petal, sepal, gynoecium, stamen, and silique.
A previous study showed that the expression of the SABRE gene was so low that it could not be detected by northern blot, even when poly(A) RNA was used (Aeschbacher et al., 1995). In this work, quantitative real-time PCR (q-PCR) was used to analyse SABRE expression in different plant organs. The results showed that HPS4/SABRE was expressed in all plant organs but that the expression was lower in roots and stem than in leaves, flowers, and siliques, and that expression was highest in leaves (Fig. 4A). In the hps4 mutant, the expression level of HPS4 was similar to that of the WT (data not shown), indicating that the point mutation in HPS4 did not affect its RNA stability. To determine further the tissue-specific expression patterns of HPS4, a 2 kb DNA sequence was fused upstream of its transcription start site with a GUS reporter gene and this gene construct was transformed into WT plants. Twenty-five independent HPS4::GUS transgenic plants were generated, and the GUS expression pattern of one representative line is shown in Fig. 4B. In a 9-day-old HPS4::GUS seedling, GUS expression was observed in all types of cells in the root apex and was restricted to vascular tissue in the upper part of the root. GUS expression was strong in the hypocotyl and the entire cotyledon. In a mature plant, GUS expression was evident in all leaves, with stronger expression in young leaves than in old leaves. GUS expression in the stem was relatively weak, and no GUS expression was detected in leaf petioles. In addition, GUS expression was evident in all flower organs, including the sepal, petal, stamen, and gynoecium. In the silique, GUS expression was high at both ends but weak in the middle. The expression pattern of GUS (Fig. 4B) was consistent with the q-PCR analysis (Fig. 4A). Furthermore, it was found that the expression of the HPS4 gene was not affected by Pi starvation (data not shown).
HPS4 antagonistically interacts with ethylene signalling in regulating plant responses to Pi starvation
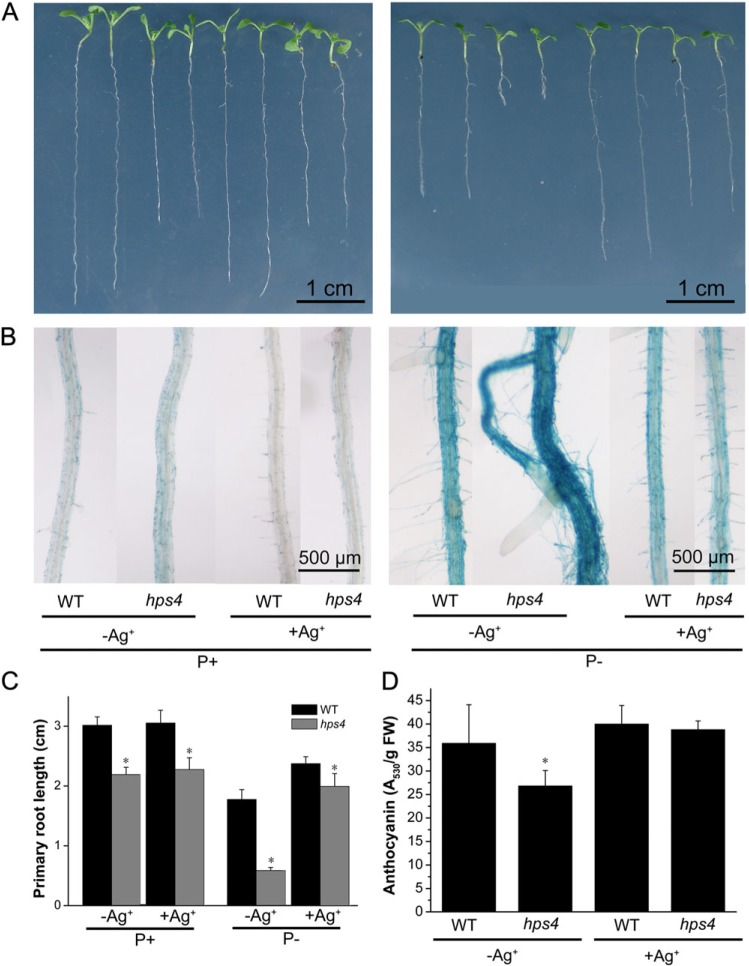
Previous studies indicated that SABRE interacts with ethylene signalling to regulate cell expansion (Aeschbacher et al., 1995). Therefore, in order to determine whether HPS4/SABRE was also involved in regulating plant responses to Pi starvation by interacting with ethylene signalling, WT and hps4 seeds were directly sown on P+ medium. At 5 DAG, the germinated seedlings were transferred to P+ or P– medium with or without addition of the ethylene action inhibitor Ag+. Addition of Ag+ to P+ medium had no effect on primary root growth of either the WT or hps4 but it suppressed the APase activity on the root surface of hps4 (Fig. 5A–C). On P– medium, however, addition of Ag+ partially suppressed the PSI inhibition of primary root growth and APase activity of the WT (Fig. 5A–C), and the hypersensitivity of hps4 in PSI inhibition of primary root growth and production of APase was reduced (Fig. 5A–C). The length of the primary root of hps4 was restored to 85% of that of the WT; that is, the difference in root length became similar to that on P+ medium. When Ag+ was added to P– medium, the APase activity on the root surface of the WT and hps4 also became similar, and APase activities for both were even lower than that of the WT without the addition of Ag+. This result was consistent with a previous finding that ethylene is a positive regulator of PSI production of APase (Lei et al., 2011).
Fig. 5.
Effects of the ethylene perception inhibitor Ag+ on root growth, APase activity, and anthocyanin accumulation of WT and hps4 seedlings. (A) Morphology of 9-day-old seedlings of WT and hps4 grown on P+ and P– medium with or without addition of 10 μM Ag+ (labels are provided below panel B). (B) Close-up view of APase activities detected by BCIP staining on the root surfaces of the seedlings shown in A. (C) Primary root length of 9-day-old seedlings of the WT and hps4 grown on P+ and P– medium with or without addition of 10 μM Ag+. (D) Anthocyanin accumulation in 9-day-old seedlings of the WT and hps4 grown on P– medium with or without addition of 10 μM Ag+. For C and D, values represent the mean and SE of three replicates. Means with asterisks are significantly different from the WT (P < 0.05, two-sample t-test).
In a previous work, it was found that enhanced ethylene signalling suppresses anthocyanin accumulation in Pi-starved plants (Lei et al., 2011). Therefore, the effect of Ag+ on anthocyanin accumulation in hps4 was examined. On P– medium, addition of Ag+ increased anthocyanin accumulation in both hps4 and the WT, and levels of anthocyanin became similar in WT and hps4 plants (Fig. 5D). This indicated that the low accumulation of anthocyanin in Pi-starved hps4 seedlings was caused by enhanced ethylene signalling.
The effect of the ethylene biosynthesis inhibitor AVG on primary root growth and root-associated APase activity was then tested. The seeds of the WT and hps4 were directly sown on P+ medium. At 5 DAG, the seedlings were transferred to P+ and P– medium with or without addition of 0.2 μM AVG. After another 7 d, the primary root growth and root-associated APase activity were examined. The results showed that AVG treatment did not suppress these two mutant phenotypes in hps4 (Supplementary Fig. S6 at JXB online). This indicated that the mutant phenotypes in hps4 were not due to enhanced ethylene biosynthesis.
Root tips of Pi-starved hps4 accumulate more auxin than do those of the WT
Several previous studies have shown that ethylene inhibits root growth by up-regulating auxin biosynthesis and interfering with its transport process (Stepanova et al., 2005, 2007; Růzicka et al., 2007; Swarup et al., 2007; Negi et al., 2008). Thus, it was of interest to determine whether the hps4 mutant accumulates more auxin in its root tissues than the WT under P– conditions. To test this hypothesis, the free IAA contents of apical 5 mm root sections of 9-day-old hps4 and WT seedlings grown on P– medium were directly measured. As shown in Fig. 6A, the IAA content was about twice as great in the hps4 mutant than in the WT. Accordingly, the expression of the auxin-responsive genes GH3.4, IAA2, and IAA14 was significantly higher in hps4 than in the WT under Pi starvation (Fig. 6B). The expression of 10 auxin biosynthetic genes (Zhao, 2010) was further compared between the WT and hps4. Expression of all these genes, except YUC5, was significantly higher in hps4 than in the WT (Fig. 6C). These results indicated that transcriptional control was involved in the enhanced auxin production in the hps4 mutant.
Fig. 6.
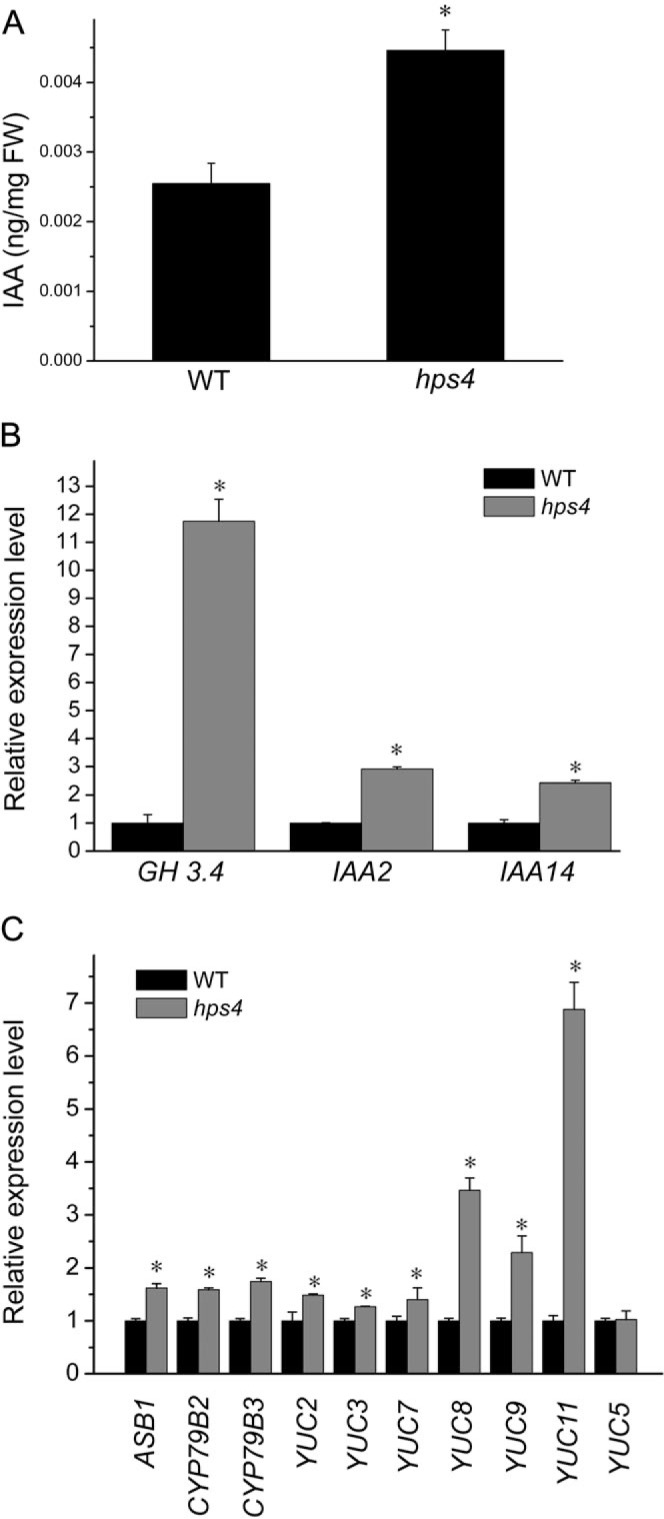
IAA content, and expression of auxin-responsive genes and auxin biosynthetic genes between Pi-starved hps4 and Pi-starved WT. (A) Free IAA contents in apical 5 mm root sections of 9-day-old seedlings of Pi-starved hps4 and Pi-starved WT. (B) Relative expression of four auxin-responsive genes in the roots of 9-day-old Pi-starved hps4 and Pi-starved WT. (C) Relative expression of 10 auxin biosynthetic genes in the roots of 9-day-old Pi-starved hps4 and Pi-starved WT. In B and C, the expression of all the genes in the WT was set to 1. In A–C, values represent the mean and SE of three replicates. Means with asterisks are significantly different from the WT (P < 0.05, two-sample t-test).
Discussion
Researchers have long recognized that ethylene plays an important role in regulating primary root growth and root hair development under Pi starvation (He et al., 1992; Borch et al., 1999; Ma et al., 2003). Recently, it has been shown in several studies that ethylene is involved in regulating PSI gene expression, APase production, and anthocyanin accumulation when plants are subjected to P– conditions (Lei et al., 2011; Li et al., 2011; Wang et al., 2012). However, how ethylene signalling interacts with other protein factors to coordinate plant responses to Pi starvation remains unknown.
In this work, an Arabidopsis mutant, hps4, wasidentified that exhibited enhanced expression of PSI genes and production of root-associated APases, increased inhibition of primary root growth, and reduced accumulation of anthocyanin when grown under P– conditions. Molecular cloning of HPS4 indicated that it is a new allele of the SABRE gene. In hps4, the point mutation in the SABRE gene caused a conversion of an alanine to a proline. Proline is generally thought to be a strong disruptor of protein secondary structure, thus such an amino acid conversion might have a strong impact on the conformation of the SABRE protein. So, although the mRNA level of HPS4 is not altered in hps4, the function of the HPS4 protein may still be affected. So far, the biochemical function of SABRE remains unknown. A SABRE homologue, APT1 (aberrant pollen transmission 1), has been identified in maize and proven to be localized in Golgi bodies (Xu and Dooner, 2006). A SABRE-like gene, KIP (Kinky Pollen), was also found in Arabidopsis (Procissi et al., 2003). Functional disruption of the APT1 and KIP genes caused defects in pollen tube growth in maize and Arabidopsis. Because membrane vesicle trafficking at the tip of pollen tube is critical for pollen tube growth, researchers have proposed that the APT1 and KIP genes are required to meet the high secretory demands of tip growth in pollen tubes (Procissi et al., 2003; Xu and Dooner, 2006). In hps4, the mutated SABRE protein may also affect the protein secretory process because it only increased the activity of secreted root-associated APase (Fig. 1B, C), but not the total activity of APase in roots (Supplementary Fig. S1B, C at JXB online).
In Arabidopsis, SABRE has been suggested to interact with ethylene signalling to regulate cell expansion during root development. In the sab1-1 mutant, there is a shift in the orientation of expansion in root cortex cells. Aeschbacher et al. (1995) found that sab1-1 had a normal level of ethylene production and ethylene responsiveness; however, treatment of sab1-1 with an inhibitor of ethylene action (Ag+) partially rescued its mutant phenotypes. So, they inferred that the extent or direction of cell expansion is determined by the antagonistic interaction between the activity of the SABRE protein and ethylene signalling. According to this explanation, when SABRE function was lost, the equilibrium shifted toward radial expansion under a normal ethylene level (Aeschbacher et al., 1995).
The phenotypes of enhanced PSI gene expression, increased production of APase, and reduced accumulation of anthocyanin in hps4 resemble that of another mutant, hps2 (Lei et al., 2011). hps2 contains a T-DNA insertion within the CTR1 gene that caused plants to display a constitutive ethylene response (Kieber et al., 1993). Thus, it was decided to determine whether the mutant phenotypes in hps4 also resulted from enhanced ethylene signalling (caused by a reduction in HPS4/SABRE activity). The results showed that, when plants were grown on P– medium supplemented with Ag+, the large difference in the root-associated APase activity between the WT and hps4 disappeared. Furthermore, the root-associated APase activity in both Ag+-treated WT and hps4 plants was even lower than that of untreated WT plants, which was similar to that observed in the ethylene-insensitive mutant ein2-5 (Lei et al., 2011). In addition, the reduced anthocyanin in hps4 was reversed to the level of that in the WT by Ag+ treatment, suggesting that this mutant phenotype was also caused by enhanced ethylene signalling. Under P– conditions, the Ag+-treated WT primary roots were longer than untreated roots, but they were still shorter than those produced by the WT grown under the P+ condition. This indicated that ethylene is partially involved in PSI inhibition of primary root growth. Similarly, Ag+ treatment abolished the hypersensitivity of hps4 to PSI inhibition of primary root growth, suggesting that the hypersensitivity of hps4 to PSI inhibition of primary roots was caused by enhanced ethylene signalling. In contrast to treatment with Ag+, treatment with the ethylene biosynthesis inhibitor AVG did not suppress the hps4 mutant phenotypes, further indicating that the hypersensitivity of hps4 to Pi starvation is not caused by enhanced ethylene biosynthesis.
Several studies have indicated that auxin plays an important role in PSI changes in root architecture. A maximal concentration of auxin at the root tip and polar auxin transport in root tissues are believed to be critical for primary root growth and formation of lateral roots. Arabidopsis grown under P– conditions has enhanced sensitivity to auxin, which is required for development of lateral roots (Lopez-Bucio et al., 2002; Perez-Torres et al., 2008). Nacry et al. (2005) found that Pi-starved root tissues of Arabidopsis accumulated twice as much auxin as non-starved root tissues and proposed that Pi starvation changed auxin distribution, which led to the inhibition of primary root growth and the increased development of lateral roots. Miura et al. (2011) further indicated that SIZ1, a SUMO E3 ligase, may be a negative regulator of enhanced accumulation of auxin in the root tip of Pi-starved Arabidopsis plants. In the siz1 mutant, more auxin may accumulate at the root tip and thereby enhance inhibition of primary root growth under Pi deficiency. Several recent reports have demonstrated cross-talk between auxin and ethylene in regulating root growth in Arabidopsis. Specifically, ethylene inhibits primary root elongation by up-regulating auxin biosynthesis and by altering auxin polar transport (Stepanova et al., 2005, 2007; Růzicka et al., 2007; Swarup et al., 2007; Negi et al., 2008).
Based on the results reviewed in the previous paragraph and the results generated in the current research, it is proposed that Pi starvation increases both ethylene biosynthesis and responsiveness in plant cells. Increased ethylene biosynthesis and/or enhanced ethylene responses would then up-regulate auxin biosynthesis, leading to inhibition of primary root growth. The degree of inhibition of primary root growth induced by Pi starvation would be determined by the antagonistic interactions between ethylene signalling and the activity of HPS4/SABRE proteins. When the activity of HPS4/SABRE is reduced in the hps4 mutant, the equilibrium between the HPS4/SABRE and ethylene signalling would be broken and the effect of ethylene signalling would be enhanced. This would cause the Pi-starved hps4 plant to accumulate more auxin in its root tissues. As a consequence, the hps4 mutant would become more sensitive than the WT to the inhibition of primary root growth triggered by Pi starvation. However, it is also necessary to point out that though this model is supported by direct measurement of free IAA content in hps4 and the WT, the possibility that enhanced ethylene signalling and an increased auxin level in hps4 are two independent events triggered by Pi starvation cannot be excluded.
In summary, genetic and molecular approaches have been combined to identify a molecular component, HPS4/SABRE, that interacts with ethylene signalling to regulate multiple plant responses to Pi starvation. This is the first such component to be identified. The biochemical function of HPS4/SABRE, however, remains unknown. To increase understanding of how HPS4/SABRE and ethylene signalling interact to regulate plant responses to Pi starvation, the direct targets or interacting proteins of HPS4/SABRE must now be identified and the biochemical function of the HSP4/SABRE protein should be elucidated.
Supplementary data
Supplementary data are available at JXB online.
Figure S1 . APase activity in 9-day-old WT and hps4 seedlings grown on P+ and P– medium.
Figure S2 . Anthocyanin contents in 14-day-old WT and hps4 seedlings grown on P+ and P– medium.
Figure S3 . Analysis of PSI gene expression in 9-day-old WT and hps4 seedlings.
Figure S4 . Total phosphorus and cellular Pi contents in 9-day-old WT and hps4 seedlings grown on P+ and P– medium.
Figure S5 . The strategy for fine mapping of the HPS4 gene.
Figure S6 . The effect of AVG treatment on primary root growth and APase activity in WT and hps4 seedlings.
Acknowledgments
We thank Dr Philip Benfey of Duke University for providing the seeds of the sab1-1 mutant, the Arabidopsis Biological Resource Centre for providing seeds of six SALK T-DNA lines, Ms Zhen Xue of the Institute of Botany, Chinese Academy of Sciences for analysis of IAA contents, and Dr Jia-Wei Wu of Tsinghua University for help with analysis of the HPS4/SABRE protein sequence. This work was supported by the National Natural Science Foundation of China (grant no. 31170238), the Ministry of Science and Technology of China (grant no. 2009CB119100), and the Ministry of Agriculture of China (grant no. 2009ZX08009-123B).
References
- Aeschbacher RA, Hauser M, Feldmann KA, Benfey P. The SABRE gene is required for normal cell expansion in Arabidopsis. Genes and Development. 1995;9:330–340. doi: 10.1101/gad.9.3.330. [DOI] [PubMed] [Google Scholar]
- Ames BN. Assay of inorganic phosphate, total phosphate and phosphatases. Methods in Enzymology. 1966;8:115–118. [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. The Plant Journal. 1994;6:673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- Borch K, Bouma TJ, Lynch JP, Brown KM. Ethylene: a regulator of root architectural responses to soil phosphorus availability. Plant, Cell and Environment. 1999;22:425–431. [Google Scholar]
- Boutin JP, Provot M, Roux L. Effect of cycloheximide and renewal of phosphorus supply on surface acid-phosphatase-activity of phosphorus deficient tomato roots. Physiologia Plantarum. 1981;51:353–360. [Google Scholar]
- Burleigh SH, Harrison MJ. The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiology. 1999;119:241–248. doi: 10.1104/pp.119.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon-Lopez A, Ibarra-Laclette E, Sanchez-Calderon L, Gutierrez-Alanis D, Herrera-Estrella L. Global expression pattern comparison between low phosporus insensitive 4 and WT Arabidopsis reveals an important role of reactive oxygen species and jasmonic acid in the root tip responses to phosphate starvation. Plant Signaling and Behavior. 2011;6:638–392. doi: 10.4161/psb.6.3.14160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. The Plant Journal. 1999;19:579–589. doi: 10.1046/j.1365-313x.1999.00562.x. [DOI] [PubMed] [Google Scholar]
- Edlund A, Eklöf S, Sundberg B, Moritz T, Sandberg G. A microscale technique for gas chromatography–mass spectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissues. Plant Physiology. 1995;108:1043–1047. doi: 10.1104/pp.108.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. A miRNA involved in phosphate-starvation response in Arabidopsis. Current Biology. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- He CJ, Morgan PW, Drew MC. Enhanced sensitivity to ethylene in nitrogen-starved or phosphate-starved roots of Zea mays L during aerenchyma formation. Plant Physiology. 1992;98:137–142. doi: 10.1104/pp.98.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. The GUS reporter gene system. Nature. 1989;342:837–838. doi: 10.1038/342837a0. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Lynch JP, Brown KM. Ethylene insensitivity impedes a subset of responses to phosphorus deficiency in tomato and petunia. Plant, Cell and Environment. 2008;31:1744–1755. doi: 10.1111/j.1365-3040.2008.01886.x. [DOI] [PubMed] [Google Scholar]
- Lei M, Zhu C, Liu Y, Karthikeyan AS, Bressan RA, Raghothama KG, Liu D. Ethylene signalling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New Phytologist. 2011;189:1084–1095. doi: 10.1111/j.1469-8137.2010.03555.x. [DOI] [PubMed] [Google Scholar]
- Li YS, Gao Y, Tian QY, Shi FL, Li LH, Zhang WH. Stimulation of root acid phosphatase by phosphorus deficiency is regulated by ethylene in Medicago falcate. Environmental and Experimental Botany. 2011;71:114–120. [Google Scholar]
- Li YS, Mao XT, Tian QY, Li LH, Zhang WH. Phosphorus deficiency-induced reduction in root hydraulic conductivity in Medicago falcata is associated with ethylene production. Environmental and Experimental Botany. 2009;67:172–177. [Google Scholar]
- Lloyd JC, Zakhleniuk OV, Raines CA. Identification of mutants in phosphorus metabolism. Annals of Applied Biology. 2001;138:111–115. [Google Scholar]
- Lopez-Bucio J, Hernandez-Abreu E, Sanchez-Calderon L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiology. 2002;129:244–256. doi: 10.1104/pp.010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Baskin TI, Brown KM, Lynch JP. Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiology. 2003;131:1381–1390. doi: 10.1104/pp.012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Bielenberg DG, Brown KM, Lynch JP. Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant, Cell and Environment. 2001;24:459–467. [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de La Peña A, Leyva A, Paz-Ares J. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. The Plant Journal. 2000;24:559–567. doi: 10.1046/j.1365-313x.2000.00893.x. [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proceedings of the National Academy of Sciences, USA. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Gong Q, Ma S, Jin JB, Yoo CY, Miura T, Sat A, Bohnert HJ, Hasegawa PM. SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation. Plant Physiology. 2011;155:1000–1012. doi: 10.1104/pp.110.165191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG. Phosphate transporters from the higher plant Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1996;93:10519–10523. doi: 10.1073/pnas.93.19.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Nacry P, Canivenc G, Muller B, Azmi A, Van Onclelen H, Rossignol M, Doumas A role of auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiology. 2005;138:2061–2074. doi: 10.1104/pp.105.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan VK, Smith AP. Ethylene’s role in phosphate starvation signaling: more than just a root growth regulator. Plant and Cell Physiology. 2012;53:277–286. doi: 10.1093/pcp/pcr186. [DOI] [PubMed] [Google Scholar]
- Negi S, Ivanchenko MG, Muday GK. Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. The Plant Journal. 2008;552:175–187. doi: 10.1111/j.1365-313X.2008.03495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Torres CA, Lopez-Bucio J, Cruz-Ramirez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L. Phosphate availability alters lateral root development in arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. The Plant Cell. 2008;20:3258–3272. doi: 10.1105/tpc.108.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procissi A, Guyon A, Pierson ES, Giritch A, Knuiman B, Grandjean O, Tonelli C, Derksen J, Pelletier G, Bonhomme S. KINKY POLLEN encodes a SABRE-like protein required for tip growth in Arabidopsis and conserved among eukaryotes. The Plant Journal. 2003;36:894–904. doi: 10.1046/j.1365-313x.2003.01933.x. [DOI] [PubMed] [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- Růzicka K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. The Plant Cell. 2007;197:2197–2212. doi: 10.1105/tpc.107.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Reid RJ, Ayling SM. Phosphorus uptake by plants: from soil to cell. Plant Physiology. 1998;116:447–453. doi: 10.1104/pp.116.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. The Plant Cell. 2005;178:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM. Multilevel interactions between ethylene and auxin in Arabidopsis roots. The Plant Cell. 2007;197:2169–2185. doi: 10.1105/tpc.107.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Perry P, Hagenbeek D, Van Der Straeten D, Beemster GT, Sandberg G, Bhalerao R, Ljung K, Bennett MJ. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. The Plant Cell. 2007;197:2186–2196. doi: 10.1105/tpc.107.052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. The Plant Journal. 2010;64:775–789. doi: 10.1111/j.1365-313X.2010.04375.x. [DOI] [PubMed] [Google Scholar]
- Tran HT, Hurley BA, Plaxton WC. Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Science. 2010;179:14–27. [Google Scholar]
- Trull MC, Deikman D. An Arabidopsis mutant missing one acid phosphatase isoform. Planta. 1998;20:544–550. doi: 10.1007/s004250050431. [DOI] [PubMed] [Google Scholar]
- Wang K, Yoshida H, Lurin C, Ecker JR. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature. 2004;428:945–950. doi: 10.1038/nature02516. [DOI] [PubMed] [Google Scholar]
- Wang L, Dong J, Gao Z, Liu D. The Arabidopsis gene HYPERSENSITIVE TO PHOSPHATE STARVATION 3 encodes ETHYLENE OVERPRODUCTION 1. Plant and Cell Physiology. 2012 doi: 10.1093/pcp/pcs072. (in press) [DOI] [PubMed] [Google Scholar]
- Wang L, Li Z, Qian W, Guo W, Gao X, Huang L, Wang H, Zhu H, Wu J, Wang DW, Liu D. The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiology. 2011;157:1283–1299. doi: 10.1104/pp.111.183723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. Arabidopsis: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. pp. 24–36. [Google Scholar]
- Xu Z, Dooner HK. The maize aberrant pollen transmission 1 gene is a SABRE/KIP homolog required for pollen tube growth. Genetics. 2006;172:1251–1261. doi: 10.1534/genetics.105.050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Liu D. Signaling components involved in plant responses to phosphate starvation. Journal of Integrative Plant Biology. 2008;50:849–859. doi: 10.1111/j.1744-7909.2008.00709.x. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Lynch JP, Brown KM. Ethylene and phosphorus availability have interacting yet distinct effects on root hair development. Journal of Experimental Botany. 2003;54:2351–2361. doi: 10.1093/jxb/erg250. [DOI] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis and its role in plant development. Annual Review of Plant Biology. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.