Abstract
Carotenoid accumulation confers distinct colouration to plant tissues, with effects on plant response to light and as well as health benefits for consumers of plant products. The carotenoid pathway is controlled by flux of metabolites, rate-limiting enzyme steps, feed-back inhibition, and the strength of sink organelles, the plastids, in the cell. In apple (Malus × domestica Borkh), fruit carotenoid concentrations are low in comparison with those in other fruit species. The apple fruit flesh, in particular, begins development with high amounts of chlorophylls and carotenoids, but in all commercial cultivars a large proportion of this is lost by fruit maturity. To understand the control of carotenoid concentrations in apple fruit, metabolic and gene expression analysis of the carotenoid pathway were measured in genotypes with varying flesh and skin colour. Considerable variation in both carotenoid concentrations and compound profile was observed between tissues and genotypes, with carotenes and xanthophylls being found only in fruit accumulating high carotenoid concentrations. The study identified potential rate-limiting steps in carotenogenesis, which suggested that the expression of ZISO, CRTISO,and LCY-ε, in particular, were significant in predicting final carotenoid accumulation in mature apple fruit.
Key words: Apple, carotenoid isomerase, carotenoids, gene expression, Malus × domestica, Apple
Introduction
Carotenoids are isoprenoid compounds responsible for the yellow, orange, or red colouration exhibited by flowers, fruits, and vegetables (Southon, 2000; Alquezar et al., 2008). These colour compounds are important in plants for photosynthetic purposes, being involved in light harvesting, and photoprotection (Ducreux et al., 2005; Cazzonelli et al., 2009). Carotenoids accumulate to varying concentrations in plant species and organ types and their synthesis mainly occurs in the plastids, such as chromoplasts, which are usually present in ripe tissues, and chloroplasts, which are predominant in green tissues. Carotenoids are well known for their health benefits, because of their pro-vitamin A activity. β-Carotene is the main precursor for vitamin A synthesis (Diretto et al., 2007), while α-carotene and β-cryptoxanthin are also precursors of vitamin A (Bai et al., 2009). Lutein and zeaxanthin are important for human vision as they have the ability to slow down damage to the retina brought about by age (Hammond et al., 2001).
Apples (Malus × domestica Borkh) are consumed in large quantities globally partly because of the presence of healthy metabolites such as flavonoids and vitamin C. Importantly in dessert apples, carotenoid pigments in the skin contribute to fruit colouration and therefore their attractiveness, but in the flesh their concentrations are low. Fruit of commercial apple cultivars show relatively low concentrations of carotenoids [<2.5 μg (g fresh weight)–1, this study] in comparison with such fruit as citrus [25 μg (g fresh weight)–1], papaya [60 μg (g fresh weight)–1], and persimmons [15 μg (g fresh weight)–1] (Kato et al., 2004; Schweiggert et al., 2011; Zhou et al., 2011). However, there are non-commercial apples such as the rootstock cultivar ‘Aotea’ that show relatively high fruit carotenoid concentrations. An understanding of the causes of these genetic variations in carotenoid accumulation in apple will provide knowledge for breeding of new dessert cultivars with high fruit carotenoid concentrations.
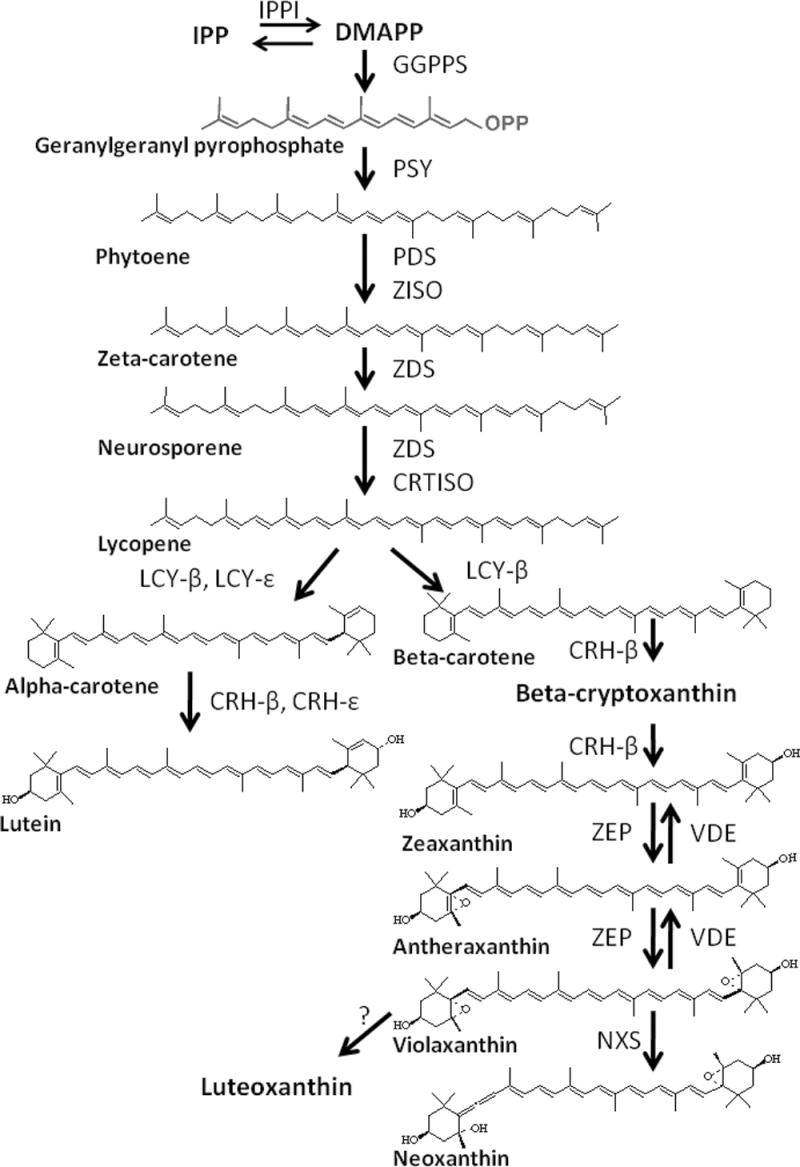
Carotenoids are synthesized in the plastids using isopentenyl diphosphate (IPP), a common precursor for all isoprenoids, which is formed through the methylerythritol-4-phosphate (MEP) pathway (Estevez et al., 2001; Hoeffler et al., 2002; Botella-Pavia et al., 2004). The conversion of the ubiquitous geranylgeranyl pyrophosphate (GGPP), by condensation of two molecules to form phytoene by phytoene synthase (PSY), is the first committed step in the carotenoid biosynthesis (Fig. 1). The activity of PSY, in plants such as Arabidopsis and tomato, has been reported to be the step that influences the extent of carotenoid biosynthesis (Fraser et al., 2007; Rodriguez-Villalon et al., 2009; Cazzonelli and Pogson, 2010). Phytoene is subsequently converted to all-trans lycopene by the activities of two desaturases, phytoene desaturase (PDS) and ζ-carotene desaturase (ZDS), and two isomerase enzymes, ζ-carotene isomerase (ZISO) and carotene isomerase (CRTISO) (Chen et al., 2010). The conversion of the linear lycopene to either β-carotene or α-carotene represents a bifurcation of the pathway and a major regulatory junction in plants (Fig. 1). The conversion to β-carotene requires the addition of two beta rings by lycopene β-cyclase (LCY-β), while the addition of an epsilon ring by lycopene ε-cyclase (LCY-ε) followed by addition of a β-ring by LCY-β results in α-carotene. The flux through the β-carotene and α-carotene pathway branches is controlled by the gene expression of LCY-β and LCY-ε, respectively. In tomato, reduced expression of the lycopene cyclases accounts for the accumulation of lycopene (Fraser et al., 1994; Ronen et al., 1999).
Fig. 1.
. The apple carotenoid biosynthetic pathway with arrows showing enzymatic conversions of substrates from the isoprene biosynthesis pathway. CRH-β, β-carotene hydroxylase; CRH-ε, epsilon-carotene hydroxylase; CRTISO, carotene isomerase; DMAPP, dimethylallyl diphosphate; GGPPS, geranyl geranyl pyrophosphate synthase; IPP, isopentenyl pyrophosphate; IPPI, isopentenyl pyrophosphate isomerase; LCY-β, β-lycopene cyclase; LCY-ε, epsilon lycopene cyclase; NXS, neoxanthin synthase; PDS, phytoene desaturase; PSY, phytoene synthase; VDE, violaxanthin de-expoxidase; ZDS, φ-carotene desaturase; ZEP, zeaxanthin epoxidase; ZISO, φ-carotene isomerase.
Carotenoid accumulation could be potentially controlled by at least four different mechanisms. Firstly, the flux through the carotenoid pathway is controlled by rate-limiting steps. In various plants, different steps have been identified that control the biosynthetic pathway in this manner. In Arabidopsis, PSY enzyme appears to be responsible for the regulation of carotenoid biosynthesis (Rodriguez-Villalon et al., 2009). PSY was found to regulate the levels of 1-deoxy-d-xylulose 5-phosphate-synthase that produces precursors necessary for downstream metabolites of the pathway. Feedback regulation may therefore be an important process in carotenoid synthesis.
Secondly, the carotenoid pathway could be affected by other interacting pathways that divert substrate. For example, the MEP pathway produces GGPP, the substrate required for the first committed step of the carotenoid biosynthetic pathway. However, the biosynthesis of chlorophylls, tocopherols, phylloquinones, and plastoquinones also uses GGPP as substrate, thereby serving as competition. As another example, abscisic acid (ABA) biosynthesis utilizes the epoxidized xanthophylls, therefore affecting their accumulation (Sauret-Gueto et al., 2006).
Thirdly, carotenoid accumulation can be affected by sink capacity because of the presence or absence of chloroplasts and/or chromoplasts. It has been shown recently, through the identification of a cauliflower Orange (Or) mutant, that increasing the metabolic sink capacity could increase carotenoid accumulation (Li et al., 2001; Lu et al., 2006). In most fruits, the ripening process, which is accompanied by the conversion of chloroplasts to chromoplasts, also coincides with the biosynthesis of carotenoids, making chromoplasts the major storage structures of these metabolites. This makes the sink capacity of these plastids an important factor in carotenoid accumulation. In Arabidopsis, a species in which this plastid conversion to chromoplasts does not occur, carotenoid crystals form as an alternate sink for accumulating carotenoids (Maass et al., 2009). Lastly, carotenoid degradation has been shown to impact final carotenoid concentrations; in particular the activity of a group of enzymes, the carotenoid cleavage dioxygenases (CCDs). Down-regulating CCD activity affects carotenoid levels in potato tubers and flowers (Campbell et al., 2010) and roots of Medicago (Floss et al., 2008), while in peach lower expression of CCD4 is correlated with accumulation of carotenoids and carotenoid-derived volatiles (Brandi et al., 2011).
Most commercial apple cultivars are distinct in having white flesh with little or no pigmentation. At maturity, carotenoid and chlorophyll are typically restricted to cells of the skin. This study shows that flesh colour, which varies from white to yellow in different apple genotypes, varies because of differences in carotenoid accumulation. Plastids are present in white flesh, but fail to accumulate carotenoids to any extent in these apples. Analysis of the apple carotenoid pathway genes suggested carotenoid concentration in fruit is controlled by expression of several rate-limiting biosynthetic steps.
Materials and methods
Plant material
Apple genotypes used in the study included parents of a breeding population (Bus et al., 2008), the rootstock cultivars ‘Aotea’ (Malus sieboldii (Regel) and Malling ‘M. 9’ (Malus × domestica Borkh); two hybrid individuals from the ‘Aotea’ × ‘M. 9’ cross, YAM9 (Rosalie R1T16), and WAM9 (Rosalie R1T82) segregating for colour; and two Malus × domestica commercial cultivars, ‘Granny Smith’ and ‘Royal Gala’. All genotypes were grown as single mature trees on an orchard at the Hawkes Bay Research Centre, Plant and Food Research, New Zealand. Fruit samples were harvested on different dates to correspond to four fruit growth stages: 20, 50, and 90 days after full bloom and at tree ripe. At least seven fruit were sampled for large fruited trees, while the ‘Aotea’ sample represented over 50 fruits per time point. Harvested fruit were separated into peel (skin) and flesh (cortex) and snap frozen into liquid nitrogen for storage at –80 °C.
Microscopy
Samples of fruit (5 × 5 × 5 mm) containing skin were fixed in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) under vacuum for 1 h, dehydrated in an ethanol series and embedded in LR White Resin (London Resin, Reading, UK). Structural observations were carried out on 1 μm sections dried on to poly-l-lysine. Sections were viewed using a Vanox AHT3 (Olympus Optical, Tokyo, Japan) and images captured by a CoolSnap colour digital camera system (Roper Scientific, Arizona, USA).
Carotenoid and chlorophyll extraction
Carotenoid and chlorophyll were extracted from samples using methods described earlier by Ampomah-Dwamena et al. (2009) with some modifications. Fruit tissue (1–2 g) was freeze dried and homogenized in 5 ml acetone with 0.1% butylated hydroxytoluene. Homogenates were stored in the dark at 4 °C overnight. One ml of 10% KOH was added and each sample incubated at room temperature for 10 min. The supernatant was extracted twice with 2 ml diethyl ether and washed with 8 ml of 10% (w/v) NaCl. The combined ether phases were taken to dryness by flushing with nitrogen.
HPLC analysis
Samples, supplied as dried extracts, were redissolved in 700 μl of 0.8% BHT/acetone (Ampomah-Dwamena et al., 2009) and analysed by high-performance liquid chromatography (HPLC). HPLC analyses were performed on a Dionex Ultimate 3000 solvent delivery system with a YMC RP C30 column (5 μm, 250 × 4.6 mm), coupled to a 20 × 4.6 C30 guard column (YMC, Wilmington, North Carolina, USA) (column temperature 25 °C) and a Dionex 3000 PDA detector. Elution rate was 1.0 ml min–1 and column temperature 25 °C. Elution was performed using a solvent system comprising solvent A (MeOH), solvent B [H2O/MeOH, 20:80) containing 0.2% ammonium acetate], and solvent C (tert-butyl methyl ether) and a linear gradient starting with 95% A/5% B, decreasing to 80% A/5% B/15% C between 2 and 10 min, decreasing to 30% A/5% B/65% C by 30 min, decreasing to 25% A/5% B/70% C at 40 min, and returning to 95% A/5% B at 45 min, a modified version of the elution gradient described by Fraser et al. (2000). A 25–50 μl aliquot was injected on the HPLC. Carotenoids were detected at 450 nm and the concentrations of carotenoids were determined as β-carotene equivalents (g fresh weight)–1 of tissue. β-Carotene and lutein were identified in the extracts by comparison of retention times and on-line spectral data with standard samples. trans-β-Carotene was purchased from Sigma Chemicals (St Louis, Missouri, USA). Other carotenoids were putatively identified by comparison with reported retention times and spectral data ((Fraser et al., 2000; Lee et al., 2001; Burns et al., 2003; Xu et al., 2006; Kamffer et al., 2010). No esterified carotenoids were detected in any apple samples, which have previously been shown to elute from this column after 25 min of this gradient. Total carotenoid and chlorophyll content of the fruit tissue were also estimated using the methods previously described (Wellburn, 1994).
RNA extraction and cDNA synthesis
Total RNA was extracted from frozen tissues by homogenization in CTAB buffer (Chang et al., 1993). Homogenized tissues were incubated at 65 °C in CTAB buffer for 10 min and extracted twice with chloroform/isoamyl alcohol (24:1). LiCl was added to a final concentration of 3 M and samples were stored at 4 °C overnight and then centrifuged. Pellets were dissolved in SSTE buffer (1 M NaCl, 0.5% sodium dodecyl sulfate, 10 mM TRIS-HCl pH 8.0, and 1 mM sodium EDTA pH 8.0) and further extracted with chloroform followed by precipitation with ethanol. The RNA pellet was redissolved and treated with DNase. cDNA was synthesized from total RNA (0.5–1 μg) using Superscript III reverse transcriptase (Invitrogen) following the manufacturer’s protocol. Reaction components were 50 μM oligo-dT (12) primer, 500 μM dNTPs, 1 × reverse transcription buffer, 5 mM MgCl2, 10 mM DTT, 40 U RNaseOUT, and 200 U reverse transcriptase. Reaction was incubated at 50 °C for 50 min.
Quantitative real-time PCR analysis
Candidate genes for expression analysis were chosen based on best BLAST match, of published carotenoid biosynthetic steps, to expressed sequence tag databases and the apple genome (Velasco et al., 2010). Primers were designed to the genes using PRIMER3 software (Rozen and Skaletsky, 2000) to a stringent set of criteria and are listed in Supplementary Table S1 (available at JXB online). Quantitative real-time PCR (qRT-PCR) was performed under conditions described previously (Lin-Wang et al., 2010). First-strand cDNA products were diluted 1:25 times and used as templates for the PCR reaction. PCR analysis was performed using the LightCycler system (Lightcycler 1.5, Roche). The SYBR Green master mix (Roche) was used following manufacturer’s protocol. Each reaction sample was prepared in four technical replicates, with a negative control using water as template. PCR conditions were as follows: preincubation at 95 °C for 5 min followed by 40 cycles each consisting of 10 s at 95 °C, 10 s at 60 °C and 20 s at 72 °C. Amplification was followed by a melting curve analysis with continuous fluorescence measurement during the 65–95 °C melt. The raw data were analysed using LightCycler software version 4 and the expression of each gene was normalized to Malus × domestica actin gene expression because of its consistency across fruit development (Espley et al., 2007).
Correlation analyses between transcript levels and carotenoid content
Transcript levels as measured by qRT-PCR were correlated with total carotenoid content in ripe fruit skin and flesh. In addition, a composite total transcript index was computed by adding the relative expression values for each sample across the four fruit developmental stages. Pearson correlation (r) analysis was performed and tested for statistical significance using Origin version 7.5 (OriginLab, www.originlab.com).
The statistical significance of variations in total carotenoid content was determined by one-way ANOVA analysis of data (significant at P = 0.00001), was followed by multiple comparisons using Tukey’s test for least-squared-difference (LSD at P = 0.05 level).
Results
Carotenoid accumulation in mature apple fruit differs significantly between genotypes
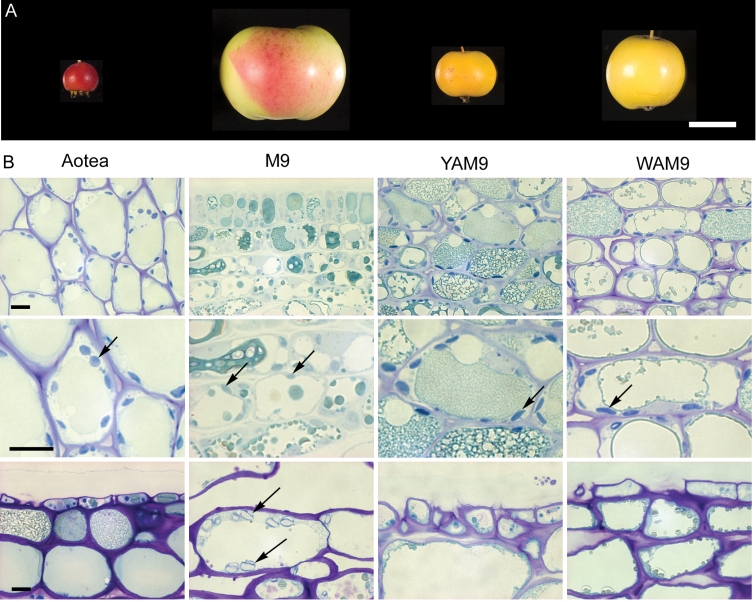
In order to understand the control of carotenoid accumulation in apple, several apple genotypes showing extreme fruit skin and flesh colour were examined, including individuals from a progeny of a cross between ‘Aotea’ and ‘M. 9’ rootstocks (Fig. 2A) that segregate for fruit flesh and skin colour (Bus et al., 2008). All fruit were grown in the same research orchard, thus minimizing environmental effects. Ripe ‘Aotea’ fruit have a red skin and intense yellowish flesh, while ‘M. 9’ fruit have a blush-pink skin with white flesh. Two progeny, YAM9 and WAM9, which showed extremes in pigmentation, were selected. Ripe YAM9 fruit have yellow skin and deep yellowish flesh while ripe WAM9 have a yellowish skin but white flesh. In addition, two widely grown commercial cultivars were also examined: ‘Granny Smith’ (green fruit skin and white flesh) and ‘Royal Gala’ (red skin and cream flesh).
Fig. 2.
. Parents and progeny of a cross between apple ‘Aotea’ and ‘M. 9’. Apple fruit selected based on the pigmentation of their skin and flesh. (A) Ripe fruit from ‘Aotea’, ‘M. 9’ and two progeny from ‘Aotea’ × ‘M. 9’ cross, YAM9 and WAM9, had different fruit sizes. (B) Stained fruit sections of different cultivars showing plastids (arrows) in 20 days after full bloom (top and middle panels) and ripe tissues (bottom panel). Bars, 2 cm (A), 10 μm (B).
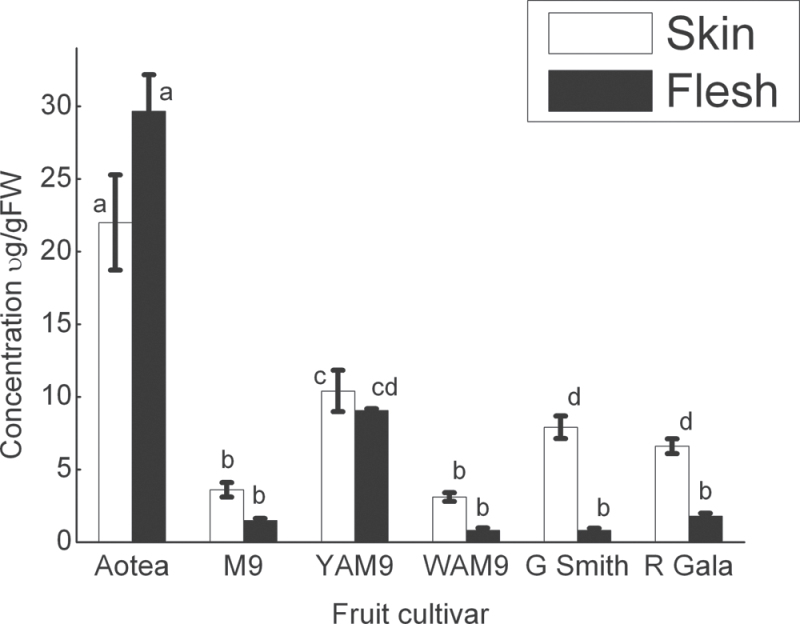
‘Aotea’ had the highest total carotenoid accumulation in both fruit skin and flesh, accumulating more than 10-times total carotenoid concentration than ‘M. 9’ fruit at the ripe fruit stage (Fig. 3). YAM9 fruit accumulated more carotenoids in both skin and flesh than WAM9. The two commercial cultivars, ‘Granny Smith’ and ‘Royal Gala’, were found to have low fruit flesh carotenoids (Fig. 3). ‘Granny Smith’ accumulated high skin carotenoid and chlorophyll concentrations, while flesh carotenoids were the lowest of the apples examined. Overall, total carotenoid concentration was higher in skin than in flesh at the ripe fruit stage, except for ‘Aotea’ (Table 1). Fruit flesh colour was a good indicator of carotenoid concentration, with yellow flesh genotypes accumulating more carotenoids than the white flesh genotypes.
Fig. 3.
. Total carotenoid concentration in ripe fruit skin and flesh of apple cultivars as measured by HPLC analysis. Fruits were harvested ripe from tree and separated into skin (peel) and flesh samples for carotenoid extraction and analysis. Error bars are standard errors of the mean from three technical replicates, with each tissue being the pooled sample of at least seven fruit, at P = 0.05 level. Bars with similar letters are not significantly different (P = 0.05), using one-way ANOVA analysis was followed by a multiple-comparisons T-test.
Table 1.
Carotenoid and chlorophyll concentrations [μg (g fresh weight)–1] in ripe fruit skin and flesh of different apple genotypes
| Compound | ‘Aotea’ | ‘M. 9’ | YAM9 | WAM9 | ‘Granny Smith’ | ‘Royal Gala’ |
| Fruit skin | ||||||
| Antheraxanthin | 1.90 | 0.70 | 1.10 | 0.90 | ||
| Violaxanthin | 1.90 | 1.20 | 1.10 | 1.00 | 1.80 | 1.00 |
| Luteoxanthin | 0.80 | 1.10 | 1.90 | 0.90 | ||
| Lutein | 0.80 | 1.00 | 1.00 | 4.10 | 0.90 | |
| α-Carotene | 4.20 | 1.30 | ||||
| β-Carotene | 7.3 | 1.30 | ||||
| cis-β-Carotene | 2.30 | |||||
| Other carotenoids | 4.40 | 0.10 | 4.60 | 0.1 | 2.90 | |
| Total carotenoid | 22.00 | 3.60 | 10.40 | 3.10 | 7.90 | 6.60 |
| Chlorophyll a | 1.04 | 0.36 | 0.92 | 0.41 | 1.04 | 0.41 |
| Chlorophyll b | 0.99 | 0.42 | 1.24 | 0.29 | 0.92 | 0.61 |
| Fruit flesh | ||||||
| iso-Neoxanthin | 3.40 | 0.35 | 2.35 | 0.51 | ||
| Neoxanthin | 2.35 | 0.31 | 1.66 | 0.16 | 0.17 | 0.29 |
| Violaxanthin | 3.13 | 1.96 | 0.16 | 0.54 | ||
| Luteoxanthin | 1.93 | 0.36 | 0.18 | 0.20 | ||
| Lutein | 0.28 | 0.28 | ||||
| β-Cryptoxanthin | 6.42 | 0.32 | ||||
| Zeaxanthin | 0.47 | |||||
| β-Carotene | 5.06 | 0.17 | 0.60 | 0.15 | 0.17 | |
| cis-β-Carotene | 0.46 | 0.19 | ||||
| Other carotenoids | 6.44 | 0.38 | 1.31 | 0.16 | 0.19 | 0.25 |
| Total carotenoid | 29.66 | 1.48 | 9.06 | 0.81 | 0.81 | 1.79 |
No inhibitory activity (NI) was observed at 5 mM concentration of the cystatin.
There are considerable fruit size differences at maturity between ‘Aotea’ (3.2 g), ‘M. 9’ (80 g), YAM9 (9.7 g), WAM9 (10.2 g), ‘Royal Gala’ (175 g), and ‘Granny Smith’ (180 g) (Fig. 2A). Thus, the lack of carotenoid in genotypes with larger fruit, on a fresh weight basis, may reflect either dilution of carotenoid-containing plastids during fruit expansion or their near absence. Plastids in skin and flesh were examined during early fruit development and mature fruit tissues; stained fruit sections revealed the presence of plastids capable of storing carotenoids in fruit of all the genotypes, but plastid number per cell reduced as the fruit matured (Fig. 2B). Although fruit size appears to affect plastid abundance per tissue area, the progeny YAM9 and WAM9 had comparable fruit size but showed significantly different concentrations of carotenoids. This suggests a genetic basis for carotenoid differences in these fruit that is independent of fruit size.
At fruit maturity, the different apple genotypes had different carotenoid profiles in skin and flesh (Table 1), suggesting a genotypic influence of which particular carotenoid pigment accumulated. In the skin of ripe ‘Aotea’ fruit, β-carotene was the predominant compound accounting for a third of total skin carotenoids, while β-cryptoxanthin (22%) and β-carotene (17%) were the predominant compounds found in ripe ‘Aotea’ flesh. These compounds were present in both skin and flesh of the YAM9 fruit but generally absent in its white-fleshed sibling WAM9. On the other hand, ripe ‘Royal Gala’ fruit had violaxanthin as the major compound both in the skin and in the flesh, accounting for 30% of total carotenoids in flesh. Ripe ‘Granny Smith’ showed a very different carotenoid profile, accumulating more lutein in the skin [4.1 μg (g fresh weight)–1] and flesh [0.28 μg (g fresh weight)–1] than all the other cultivars examined. Lycopene was not found in any of the apples.
Changes in carotenoid accumulation over fruit development
A fruit development series of four maturity dates was sampled for the six apple genotypes (Fig. 4). Carotenoid accumulation patterns during fruit development differed significantly by genotype (Table 1, Supplementary Table S2). ‘Aotea’ had the greatest total carotenoid concentrations at all the fruit developmental stages examined, followed by YAM9. Carotenoid concentration in fruit flesh generally decreased as fruit matured (20–90 days after full bloom, dafb), then increased during ripening (tree ripe, Supplementary Table S2). This trend was observed in all the genotypes studied except for WAM9 and ‘Granny Smith’, in which total carotenoid did not increase during ripening. Total carotenoid concentration in ‘Aotea’ fruit at ripeness increased to the same concentration as 20 dafb (the earliest fruit stage examined). However, in the other cultivars, the carotenoid concentration at ripe stage was lower than at 20 dafb; approximately 60% of 20 dafb concentration in YAM9 ripe fruit, 10% in ‘Royal Gala’, 8.5% in ‘M. 9’, 6% in ‘Granny Smith’, and 5% in WAM9.
Fig. 4.
. Fruit development series of apples used in carotenoid analysis. Fruit were harvested from ‘Aotea’, ‘M. 9’, YAM9, WAM9, ‘Granny Smith’, and ‘Royal Gala’ apple trees, growing under the same environmental conditions, at 20, 50 and 90 days after full bloom (dafb) and ripe fruit stages.
Carotenoid profile at maturity is the end point of the processes that occur during fruit growth. All the apples examined begin development with high concentrations of flesh chlorophyll (Fig. 4, Supplementary Table S2). In fruit flesh, lutein was lost completely at ripeness in all cultivars except ‘Granny Smith’ and ‘M. 9’, with the greatest concentration observed at the earliest fruit stage. In contrast, ripening was characterized by the appearance of xanthophylls; these compounds were not detected at earlier stages of development.
Identification of carotenoid biosynthetic pathway genes in apple
To ascertain the molecular basis of the variation in carotenoid concentration in apple, the apple genes of the carotenoid pathway were identified (Fig. 1; Table 2) by best BLAST match to published apple expressed sequence tag databases and genome sequence (Newcomb et al., 2006; Velasco et al., 2010). Overall, the apple carotenoid genes belong to small gene families, as found in other species such as Arabidopsis and rice (Table 2). There is a single copy of the apple GGPPS identified in the published genome sequence data, while four copies of PSY are present on different linkage groups (LG): PSY1 is on LG 17; PSY2 on LG 9, PSY3 on LG3, and PSY4 on LG11. This is similar to the multiple copies reported for maize (Li et al., 2008). There are two copies of ZDS (LG12 and 4), while a single copy of the recently identified ZISO (Chen et al., 2010) is present on LG15. CRTISO has a single copy located on LG14. Three copies of LCY-β were identified (LG14, LG3, and LG15), while a single copy of LCY-ε was found on LG2. These low gene copy numbers, ranging from one to four, contrasts with the high number of genes in the apple genome, which originated by genome-wide duplication of an ancestor (Velasco et al., 2010).
Table 2.
. Apple carotenoid biosynthetic genes identified from expressed sequence tag (EST) libraries and the apple genome sequence using homology to known gene sequences
| Gene | EST count | Linkage group | mRNA libraries | Arabidopsis | Predicted function | GenBank number | Genome ID |
| GGPPS | 28 | 2, 14.5M | Skin, leaf, buds | AT4G368101 | Farnesyl transtransferase | CN908300 | MDP0000576390 |
| PSY1 | 9 | 17 12M | Leaves, buds, cortex, skin | AT5G17230 | Phytoene synthase, chloroplastic | EB110766 | MDP0000177623 |
| PSY2 | 22 | 9, 11.8M | Fruit skin, leaves, | AT5G17230 | Phytoene synthase, chloroplastic | EB144737 | MDP0000237124 |
| PSY3 | 1 | 3, 1.1M | Pooled library | AT5G17230 | Phytoene synthase, chloroplastic | none | MDP0000151924 |
| PSY4 | 1 | 11, 0.59M | Pooled library | AT5G17230 | Phytoene synthase, chloroplastic | none | MDP0000288336 |
| PDS | 17 | 15 13.3M | Leaves, buds | At4g14210 | Desaturation of phytoene to ζ-carotene | EG631227 | MDP0000148978 |
| ZISO | 3 | 15 13.8M | Roots and various | AT1G10830 | 15-cis-ζ-Carotene isomerase (ZISO) | none | MDP0000139362 |
| ZDS1 | 13 | 4, 25.3M | Senescing leaf, floral bud, fruitlet, shoot, flowers | AT3G04870 | ζ-Carotene desaturase, chloroplast/chromoplast | AF429983 | MDP0000308095 |
| ZDS2 | 8 | 12, 34.5M | Buds, fruit | AT3G04870 | ζ-Carotene desaturase | GO546818 | MDP0000255025 |
| CRTISO | 3 | 14, 6.8M | Shoot internodes | AT1G06820 | Carotenoid isomerase, chloroplastic (CrtISO) | CV880740 | MDP0000180064 |
| LCY-β1 | 17 | 14, 15.8M | Peel, buds, flowers | AT3G10230 | Lycopene β-cyclase, chloroplastic | EB130590 | MDP0000194622 |
| LCY-β2 | 18 | 3, 16.7M | Mature and stored fruit, buds, flowers, leaves | AT3G10230 | Lycopene β-cyclase | CN906791 | MDP0000145663 |
| LCY-β3 | 1 | 15, 27.7M | Pooled cDNA | AT3G10230 | Lycopene β-cyclase | none | MDP0000258205 |
| LCY-ε | 3 | 2, 6.3M | Leaves | AT5G57030 | LUTEIN DEFICIENT 2 | DR999618 | MDP0000158790 |
| CRH-β1 | 20 | 15, 5.2M | Peel, young and mature fruit, buds, etc. | AT1G31800 | CYP97A3, β-ring carotenoid hydroxylase | EB141652 | MDP0000221455 |
| CRH-β2 | 114 | 4, 24.8M | Leaves, skin | AT1G31800 | LUTEIN DEFICIENT 5 | EB141652 | MDP0000278141 |
| CRH-ε | 5 | 14, 20M | Shoots, buds | AT3G53130 | Lutein-deficient 1 | CV091827 | MDP0000253705 |
| CCD1 | 53 | 7, 1.12 | Fruit, seeds, leaves | AT3G63520 | ATNCED1 | EB152793 | MDP0000164529 |
| CCD4 | 45 | 16, 4.2M | Flowers, fruit, buds | AT4G19170 | ATNCED4 | GO511300 | MDP0000774924 |
| ZEP1 | 107 | 2, 17.3M | Skin, leaf, fruit | AT5G67030 | Zeaxanthin epoxidase | CN848844 | MDP0000273958 |
| ZEP2 | 35 | 15, 26.9M | Peel | AT5G67030 | Zeaxanthin epoxidase | FE969463 | MDP0000319667 |
Changes in transcripts of carotenoid genes during fruit development
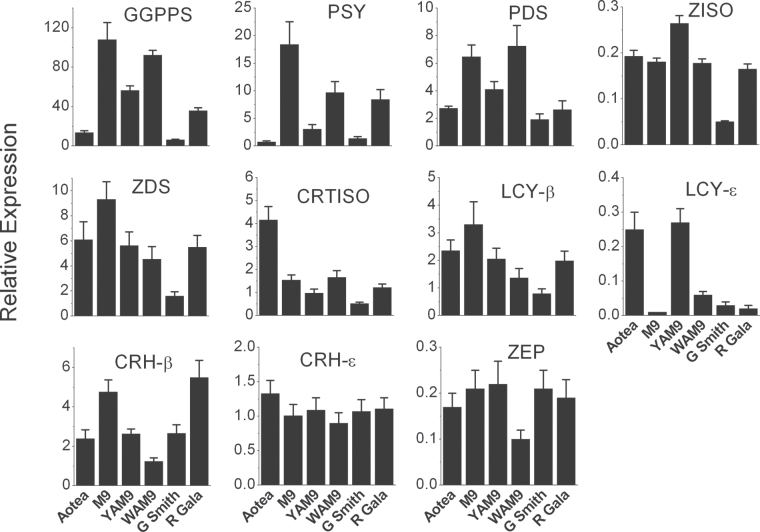
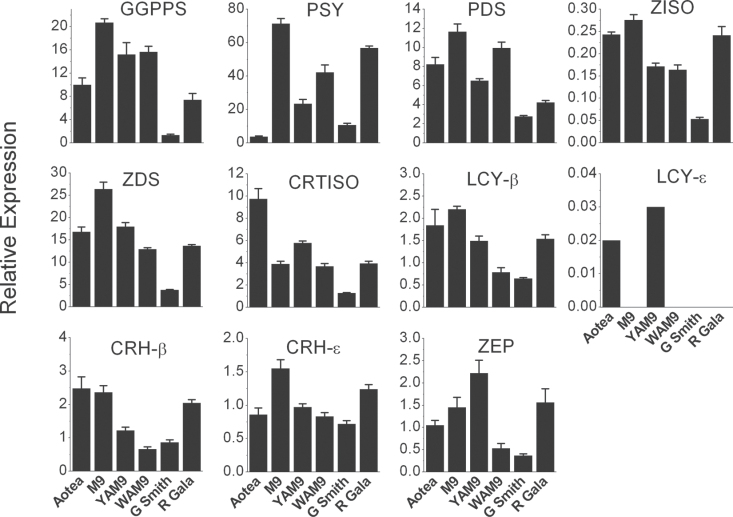
Having established the carotenoid gene number and linkage group location, qRT-PCR primers (Supplementary Table S1) were designed for the genes of the biosynthetic pathway. As total carotenoid concentration increased during fruit ripening (Table 1), gene expression was analysed at this fruit stage to further understand the mechanisms controlling the pathway. GGPPS expression in ripe fruit skin was highest in ‘M. 9’ followed by that in WAM9 and YAM9, with ‘Granny Smith’ showing the lowest level of expression next to ‘Aotea’ (Fig. 5). A similar pattern of reduced expression for ripe ‘Aotea’ for PSY and PDS suggests that, at this stage, expression of these genes may not be the limiting factor for carotenoid biosynthesis. The expression of CRTISO and LCY-ε in the skin at the ripe stage (Fig. 5) showed that ‘Aotea’ had the highest expression. These two genes showed an association between expression level and carotenoid accumulation in the apple genotypes.
Fig. 5.
. Expression of carotenoid biosynthetic pathway genes in ripe fruit skin of the different apple fruit cultivars, as determined by qRT-PCR relative to actin. Error bars are standard errors of the mean from four technical replicates.
In the fruit flesh, a similar pattern was observed at this fruit developmental stage, with CRTISO and LCY-ε showing greatest correlation between relative expression and carotenoid concentration. LCY-ε expression was higher for ‘Aotea’ and YAM9 but its expression was lacking in all other genotypes at the ripe fruit stage (Fig. 6). ‘Aotea’ had the highest expression of CRTISO, followed by YAM9, with ‘M. 9’, WAM9, and ‘Royal Gala’ showing similar transcript levels. ‘Granny Smith’ had the lowest expression level in flesh for all the genes examined except for PSY, where ‘Aotea’ had the lowest transcript level. The expression of PDS, ZDS, and CRH-ε was highest for ‘M. 9’, followed by YAM9 and ‘Aotea’.
Fig. 6.
. Expression of carotenoid biosynthetic pathway genes in ripe fruit flesh of the different apple fruit genotypes, as determined by qRT-PCR relative to actin. Error bars are standard errors of the mean from four technical replicates.
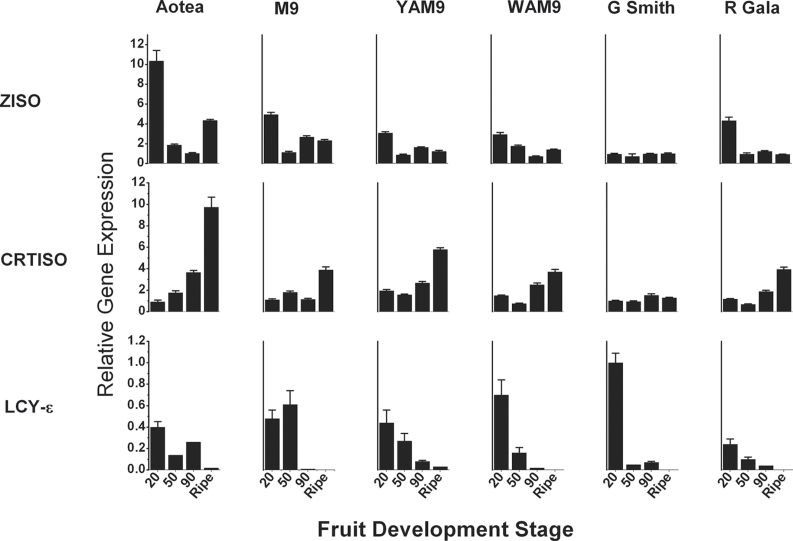
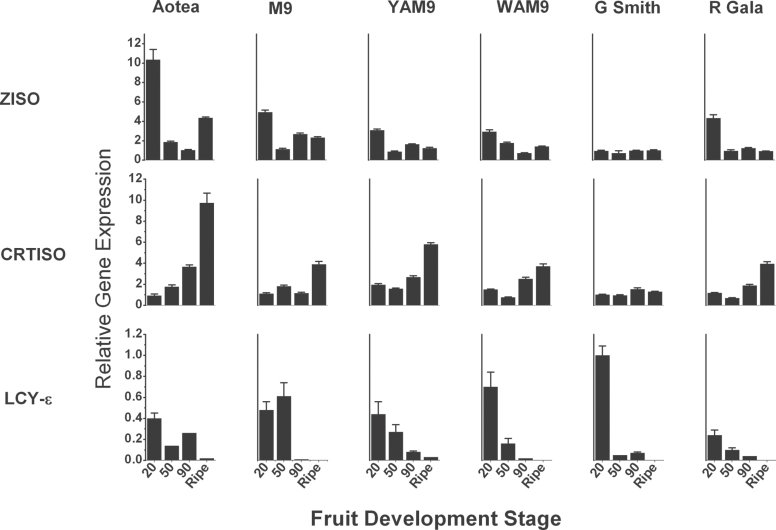
Over the fruit development, in both skin and flesh, increased levels of gene expression was observed for the early pathway genes GGPPS, PSY, PDS, ZDS, CRTISO, and LCY-β (Supplementary Figs. S1 and S2). CRTISO expression in skin was generally high in ‘Aotea’ but was reduced in all the other genotypes compared with LCY-ε expression, which was high in young tissues but was reduced at fruit maturity (Fig. 7). In flesh tissues, CRTISO expression increased as the fruit developed for all the fruit types except ‘Granny Smith’, reaching highest level in the ripe tissue. In a similar pattern to that observed in skin, LCY-ε expression in flesh was down-regulated during the late stages of fruit development, with lowest expression at the ripe fruit stage (Fig. 8). CRH-β expression was down-regulated in the skin at all the stages examined, in contrast to its expression in the flesh where CRH-β expression was high up to 90 dafb before reducing at the ripe fruit stage. CRH-ε was differentially expressed in skin and flesh; expression was reduced in fruit skin during fruit development, but was up-regulated in the flesh (Supplementary Figs. S1 and S2).
Fig. 7.
. Expression patterns of ZISO, CRTISO, LCY-ε and CRH-ε, which showed significant correlation with ripe fruit carotenoid concentrations, in fruit skin during fruit development. Gene expression was measured by qRT-PCR relative to actin. Error bars are standard errors of the mean from four technical replicates.
Fig. 8.
. Expression patterns of ZISO, CRTISO, and LCY-ε, which showed significant correlation with ripe fruit carotenoid concentrations, in fruit flesh during fruit development. Gene expression was measured by qRT-PCR relative to actin. Error bars are standard errors of the mean from four technical replicates.
As PSY, ZDS, LCY-β,and CRH-β are members of gene families (Table 2) gene-specific primers were used to assess the contribution of each family member to the expression profile. For PSY, gene-specific primers could not distinguish between PSY3 and PSY4 transcripts due to the high similarity. PSY1 and PSY2 showed similar pattern and were expressed at higher levels than PSY3 (transcripts were only detectable in WAM9 skin; Supplementary Fig. S3). In ripe apple flesh, PSY1 and PSY2 had similar patterns, with PSY2 having the highest expression (Supplementary Fig. S4). ZDS1 expression was highest in ‘Aotea’ skin and flesh compared to the other tissues, while ZDS2 was higher in ‘M. 9’ and ‘Royal Gala’.
Of the three apple lycopene β-cyclases, LCY-β2 showed higher levels of expression than LCY-β1 and LCY-β3. LCY-β2 was more highly expressed in ‘M. 9’, WAM9, and ‘Royal Gala’ flesh. All three CRH-β genes were more highly expressed in skin than in flesh (Supplementary Figs. S3 and S4) with CRH-β1 and CRH-β3 having higher expression in ‘Aotea’ skin than all other apples. In apple flesh, CRH-β3 showed higher relative expression in ‘Aotea’ than other apples.
Gene expression of apple carotenoid-depleting enzymes
Gene expression profiles of potential carotenoid-depleting enzymes, zeaxanthin epoxidase (ZEP), CCD1, and CCD4, were examined during fruit development (Supplementary Figs. S1 and S2). ZEP expression in fruit skin was typically high at 20 dafb and down-regulated during fruit development for all the genotypes. The expression level was highest in ‘Aotea’ skin, followed by ‘Granny Smith’ and ‘Royal Gala’, with WAM9 having the lowest expression at this stage. In fruit flesh, ZEP expression was highest in YAM9 and appeared stable through fruit development. In ‘Royal Gala’ ZEP expression increased during fruit development, while in ‘Aotea’, WAM9, and ‘Granny Smith’ its expression was down-regulated over fruit development. Gene-specific primers determined that ZEP2 expression was higher than ZEP1 in all tissues, with flesh showing higher expression than skin (Supplementary Figs. S3 and S4). There was reduced expression of CCD1 and CCD4 in fruit skin during fruit development. However, at ripe fruit stage, WAM9 had the highest CCD1 expression, followed by ‘Aotea’ and ‘M. 9’, with ‘Granny Smith’ showing the lowest expression. In contrast, CCD4 expression in skin was down-regulated in all the genotypes and it was barely detectable at the ripe fruit stage. In fruit flesh, CCD1 expression was generally increased in the course of fruit development, while CCD4 expression was down-regulated. CCD1 was highly expressed in WAM9 as well as in ‘Aotea’ and ‘M. 9’ fruit at the ripe stage, while ‘Granny Smith’ had the lowest expression.
Changes in chlorophyll pigment profile during fruit development
Chlorophyll (chl) a and b pigment concentrations generally decreased during fruit development in all the apple genotypes except in ‘Aotea’ flesh, where between 20 and 50 dafb stages chl-a increased (~2 fold) from 10.9 to 20.5 μg (g fresh weight)–1 and chl-b increased more than 2-fold, from 12.8 to 26.3 μg (g fresh weight)–1. However, by 90 dafb, chl-a and chl-b concentrations were 0.3 and 0.2 μg (g fresh weight)–1, respectively. Similarly, chl-a and chl-b concentration in YAM9 reduced from 14.3 μg (g fresh weight)–1 to 4.9 and 5.9 μg (g fresh weight)–1, respectively, while at the same stage, concentrations of chl-a and chl-b in ‘M. 9’ were reduced from 5.2 and 6.5 μg (g fresh weight)–1 to 0.8 and 1.0 μg (g fresh weight)–1, respectively. Chl-b concentrations were greater than or similar to chl-a in all the genotypes examined. In the fruit flesh, concentrations of both pigments were barely detected at the ripe fruit stage.
Comparisons between gene expression and carotenoid concentration
To ascertain how gene expression may be predictive of carotenoid accumulation in mature apple fruit, a Pearson correlation analysis was performed between total carotenoid concentration in ripe fruit and relative gene expression at the individual stages of development or expressed as a total transcript index (Table 3).
Table 3.
. Pearson’s correlation (r) comparing relative gene expression in fruit at different fruit stages (20, 50, 90 dafb and ripe stages), and total transcript index with carotenoid concentration in apple fruit
| Gene | Fruit skin | Fruit flesh | ||||||||
| 20 | 50 | 90 | Ripe | Total transcript index | 20 | 50 | 90 | Ripe | Total transcript index | |
| GGPPS | 0.94** | –0.31 | –0.02 | –0.52 | –0.02 | –0.31 | –0.22 | 0.06 | –0.05 | –0.23 |
| PSY | 0.38 | –0.26 | –0.58 | –0.56 | –0.57 | –0.70 | –0.21 | –0.54 | –0.62 | –0.71 |
| PDS | 0.97** | –0.39 | –0.77 | –0.39 | –0.41 | –0.22 | 0.54 | –0.23 | 0.12 | –0.03 |
| ZISO | 0.94** | 0.81* | 0.53 | 0.13 | 0.86* | 0.97*** | 0.48 | –0.10 | 0.45 | 0.84* |
| ZDS | 0.72 | –0.26 | –0.42 | 0.04 | –0.04 | 0.35 | 0.50 | –0.25 | 0.17 | –0.02 |
| CRTISO | 0.85* | 0.85* | –0.48 | 0.89* | 0.90* | –0.24 | 0.58 | 0.84* | 0.94** | 0.82* |
| LCY-β | 0.90* | –0.18 | 0.40 | 0.15 | 0.36 | –0.19 | –0.45 | 0.50 | 0.39 | 0.11 |
| LCY-ε | 0.66 | 0.52 | 0.99*** | 0.67 | 0.98*** | –0.35 | –0.15 | 0.97*** | 0.66 | 0.02 |
| CRH-β | 0.77 | –0.15 | 0.54 | –0.32 | 0.24 | –0.04 | 0.64 | 0.71 | 0.51 | 0.40 |
| CRH-ε | 0.95** | 0.07 | 0.51 | 0.87* | 0.58 | –0.57 | –0.03 | –0.04 | –0.27 | –0.68 |
| ZEP | 0.64 | 0.22 | –0.31 | –0.05 | 0.47 | 0.54 | 0.07 | 0.04 | 0.12 | 0.07 |
Highlighted values show statistically significant correlation. *P < 0.05; **P < 0.01; ***P < 0.001.
Significant correlation was observed between GGPPS, PDS, and ZISO expression at 20 dafb and carotenoid accumulation in fruit skin. CRTISO expression at various developmental stages, as well as total transcript index, showed significant correlation with carotenoid concentration in both fruit skin and flesh. In the skin significant correlation was found at 20 and 50 dafb and ripe stages (P < 0.05), while in the flesh correlation coefficients of 0.84 (P < 0.05) and 0.94 (P < 0.01) were obtained at 90 dafb and ripe fruit stages. Total transcript indexes in both skin and flesh were significant (r = 0.90, P < 0.05; r = 0.82, P < 0.05, respectively). LCY-ε expression was significantly reduced for all genotypes examined compared with other genes examined. Correlation analysis showed highly significant comparisons (r = 0.99, P < 0.001; r = 0.97, P < 0.001, respectively), with carotenoid accumulation at 90 dafb in fruit skin and flesh. Total transcript index was highly correlated (P < 0.001) with carotenoid accumulation in fruit skin. CRH-ε expression also showed a significant relationship with carotenoid accumulation in fruit skin at 20 dafb and ripe fruit stages (r = 0.95, P < 0.01; r = 0.87, P < 0.05, respectively). There was no such association observed between expression of this gene and carotenoid accumulation in the fruit flesh.
Discussion
Carotenoid accumulation in apple was studied using fruit with extreme phenotypes selected from the germplasm. Gene expression of enzymes acting in the carotenoid metabolic pathway was examined over fruit development and using correlation analyses and pathway steps that potentially control the flux and accumulation of metabolites in apple tissues were identified.
Carotenoid accumulation in apple show genotypic effect
HPLC analysis of extracts indicated a genotypic effect in carotenoid accumulation in apple. Overall, carotenoid concentration varied according to fruit development stage, reducing from the early stage of development until ripening. However, the different genotypes displayed significant variability in accumulating these compounds. Whilst ‘Aotea’ had high carotenoid content, the commercial apple cultivars had low carotenoid contents, similar to observations made in other studies (Yano et al., 2005; Felicetti and Schrader, 2009). This suggests that biosynthetic ability of apple to accumulate carotenoids may have been negatively selected during apple breeding.
Genotypic variation was evident in the carotenoid profiles for each of the apple types examined. The predominant compound at the earliest fruit stage was lutein but this changed as the fruit matured. As lutein made a significant contribution to total carotenoid concentration, its reduction over development had a strong influence on total carotenoid concentrations until the ripening stage, when other compounds increased. Similar variability in carotenoid accumulation is observed in other plant species such as orange and carrots (Alquezar et al., 2008; Clotault et al., 2008; Guzman et al., 2010). The predominant compounds in ‘Aotea’ flesh at ripening were β-cryptoxanthin and β-carotene, whilst ‘Royal Gala’ accumulated violaxanthin and neoxanthin, with lutein still the major compound in ‘Granny Smith’. The preference for one compound over the other in tissues can often be attributed to transcriptional or post-transcriptional factors, which could be occurring in these genotypes (Ronen et al., 1999; Xiangjun et al., 2011).
Carotenoid content in apple influenced by different molecular events
Carotenoid accumulation can be controlled by the biosynthetic or degradative enzyme action on the compounds, so timing and degree of gene expression of these steps is pivotal to the plant (Kato et al., 2004, 2007). In apple, correlations were observed between gene expression and final carotenoid concentration. In fruit skin, GGPPS, PDS, ZISO, CRTISO, LCY-β, and CRH-ε expression at 20 dafb correlated with carotenoid concentration. In fruit flesh, such correlation was not strong, except for ZISO, an indication that different regulatory mechanisms control carotenoid accumulation in these tissues. GGPPS mediates the synthesis of GGPP, which is a substrate for other competing biosynthetic pathways such as chlorophyll, gibberellins, and tocopherol pathways (Botella-Pavia et al., 2004). GGPPS expression will thus be significant to these other pathways compared with PSY, which is the first committed step in the carotenoid pathway. GGPPS and PSY are both early enzymes controlling flux of metabolites into the carotenoid pathway and have been shown to be the critical steps in some species (Shewmaker et al., 1999; Lindgren et al., 2003; Maass et al., 2009; Welsch et al., 2010). Interestingly, PSY expression showed no significant correlation with carotenoid content in apple. The importance of gene expression at early stages of development was shown in maize, where expression of some pathway genes 20–25 days after pollination was predictive of final carotenoid concentration (Vallabhaneni and Wurtzel, 2009). Such expression at an early developmental stage may be sufficient to generate enzyme activity throughout fruit development. The observation that PSY is not a limiting factor in apple, in contrast to other plant species (Isaacson et al., 2004; Ampomah-Dwamena et al., 2009; Vallabhaneni and Wurtzel, 2009), indicates early pathway genes such as PDS and CRTISO may play a more important role in determining high- and low-carotenoid apple cultivars.
ZISO, CRTISO, and LCY-ε have positive influence on apple carotenoid content
The recent discovery of a previously unknown isomerase step in the carotenoid pathway, ZISO, which mediates isomerization of tri-cis-ζ-carotene to a di-cis-ζ-carotene, highlights the important role isomerase enzymes have in the pathway (Li et al., 2007; Chen et al., 2010). In maize, the absence of the ZISO enzyme activity in the y9 mutant results in a block in the biosynthetic pathway, leading to accumulation of the tri-cis-ζ-carotene isomer. The null ZISO mutation can only be partially compensated by light; therefore, its role is critical in tissues that are not exposed to light (Chen et al., 2010). In apple, the significant correlations observed both in fruit skin and flesh suggests its expression early in fruit development is important to carotenoid accumulation. In apple, reduced expression of ZISO will be expected to result in low carotenoid biosynthesis or the accumulation of the intermediate ζ-carotene isomer. However, the detection of downstream carotenoid compounds in these apple fruits suggests ZISO activity is present, although a reduced ZISO expression may limit the amount of flux through these tissues.
CRTISO is required to mediate the conversion of ζ-carotene to trans-lycopene by acting on 7,9,9'-cis-neurosporene and cis-lycopene converting them to 7,9'-cis-neurosporene and all-trans lycopene, respectively (Isaacson et al., 2004). The strong correlation of CRTISO expression with apple carotenoid concentration indicates a significant role for this enzyme. However, an outcome of limiting CRTISO activity would be accumulation of cis-lycopene, as found in tomato (Isaacson et al., 2002, 2004), which does not occur in apple. Reduced activity of ZISO and CRTISO may lead to a general reduction in carotenoid pathway flux due to a feedback mechanism. Feedback regulation has been reported in Arabidopsis, where PSY up-regulation led to increase in upstream MEP pathway enzymes (Rodriguez-Villalon et al., 2009). Also, accumulation of β-carotene in tomato as a result of increased gene expression led to reduction in total carotenoid in fruit due to negative feedback regulation (Romer et al., 2000; Bramley, 2002).
In apple, expression of LCY-ε was highly correlated with carotenoid content in fruit skin but not as strongly in flesh. This differential expression is reflected in the observation that lutein was a major component in the skin but not the flesh. Down-regulation of LCY-ε expression as fruit matured is consistent with a reduction in lutein concentration, as LCY-ε controls the flux towards α-carotene, which is subsequently converted to lutein (Harjes et al., 2008). The observation that lutein content is reduced dramatically during fruit development when LCY-ε is down-regulated indicates that LCY-ε expression is also pivotal to total carotenoid content in apple fruit.
Turnover and sink effects on carotenoid content
The low amounts of carotenoid in apple could suggest a rapid turnover of metabolites from the pathway. CCD1 expression at ripe fruit stage was high in low-lutein genotypes such as ‘Aotea’ and low in lutein-rich genotypes like ‘Granny Smith’ and ‘M. 9’, suggesting an involvement in removing this compound by this enzyme. In strawberry, there is an increase in CCD1 expression during ripening correlated with lutein content, suggesting that lutein may be the main substrate of CCD1 (Garcia-Limones et al., 2008). However, the best BLAST match of apple CCD1 in Arabidopsis is CCD1/NCED1 (At3g63520), which is suggested to act on C27 apocarotenoids exported to the cytosol (Floss et al., 2008), after another CCD cleaves C40 compounds in the plastid (Floss and Walter, 2009).
The current observation that availability of plastid organelles was not a predominant factor in carotenoid accumulation among the apple genotypes strengthens the argument for enzymes controlling carotenoid accumulation in this species. This contrasts with loquat, another fruit of the Rosaceae family. Carotenoid accumulation in loquat fruit follows a similar pattern to that in apple, peaking at the ripe stage. However, loquat mainly accumulates β-carotene (Fu et al., 2011). The difference between the high- and low-carotenoid loquat cultivars was better explained by the presence or absence of plastids, in addition to differential expression of PSY, LCY-β, and CRH-β (Fu et al., 2011). This highlights the complexity associated with the regulation of this pathway in plants and how it may operate differently in closely related species.
Significant differences in carotenoid accumulation between apple fruit genotypes is related to the expression of the genes of the carotenoid biosynthetic pathway. Correlation analyses between gene expression and carotenoid content in fruit identified genes whose expression in general or during certain fruit developmental stages are important determinants of total carotenoids in fruit. These findings are significant to the breeding of apple cultivars for higher levels of health-related compounds.
Supplementary Material
Acknowledgements
This work was funded by the New Zealand Foundation for Research Science and Technology (C06X0812 ‘Exploiting Opportunities from Horticultural Genomics’), the Ministry of Science and Innovation, and PREVAR. The authors thank Tim Holmes for pictures, Natalie How for collecting fruit samples for analyses, and William Laing, Robert Schaffer, Kevin Davies, and Anne Gunson for helpful comments on the manuscript.
Footnotes
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Expression of apple carotenoid biosynthetic genes in fruit skin of different apple genotypes measured at 20, 50, and 90 dafb and ripe fruit stages
Supplementary Fig. S2. Expression of apple carotenoid biosynthetic genes in fruit flesh of different apple genotypes measured at 20, 50, and 90 dafb and ripe fruit stages
Supplementary Fig. S3. Expression of apple carotenoid gene homologues, using gene-specific primers, in ripe fruit skin of different apple genotypes
Supplementary Fig. S4. Expression of apple carotenoid gene homologues, using gene-specific primers, in ripe fruit flesh of different apple genotypes
Supplementary Table S1. Primers for qRT-PCR designed using PRIMER3 software
Supplementary Table S2. Carotenoid concentrations [μg (g fresh weight)–1] in apple fruit flesh measured by HPLC analysis at 20, 50, and 90 dafb and ripe fruit stages
References
- Alquezar B, Rodrigo MJ, Zacarias L. Regulation of carotenoid biosynthesis during fruit maturation in the red-fleshed orange mutant Cara Cara. Phytochemistry. 2008;69:1997–2007. doi: 10.1016/j.phytochem.2008.04.020. [DOI] [PubMed] [Google Scholar]
- Ampomah-Dwamena C, McGhie T, Wibisono R, Montefiori M, Hellens RP, Allan AC. The kiwifruit lycopene beta-cyclase plays a significant role in carotenoid accumulation in fruit. Journal of Experimental Botany. 2009;60:3765–3779. doi: 10.1093/jxb/erp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Kim EH, DellaPenna D, Brutnell TP. Novel lycopene epsilon cyclase activities in maize revealed through perturbation of carotenoid biosynthesis. The Plant Journal. 2009;59:588–599. doi: 10.1111/j.1365-313X.2009.03899.x. [DOI] [PubMed] [Google Scholar]
- Botella-Pavia P, Besumbes O, Phillips MA, Carretero-Paulet L, Boronat A, Rodriguez-Concepcion M. Regulation of carotenoid biosynthesis in plants: evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. The Plant Journal. 2004;40,:188–199. doi: 10.1111/j.1365-313X.2004.02198.x. [DOI] [PubMed] [Google Scholar]
- Bramley PM. Regulation of carotenoid formation during tomato fruit ripening and development. Journal of Experimental Botany. 2002;53:2107–2113. doi: 10.1093/jxb/erf059. [DOI] [PubMed] [Google Scholar]
- Brandi F, Bar E, Mourgues F, Horvath G, Turcsi E, Giuliano G, Liverani A, Tartarini S, Lewinsohn E, Rosati C. Study of ‘Redhaven’ peach and its white-fleshed mutant suggests a key role of CCD4 carotenoid dioxygenase in carotenoid and norisoprenoid volatile metabolism. BMC Plant Biology. 2011:11. doi: 10.1186/1471-2229-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J, Fraser PD, Bramley PM. Identification and quantification of carotenoids, tocopherols and chlorophylls in commonly consumed fruits and vegetables. Phytochemistry. 2003;62:939–947. doi: 10.1016/s0031-9422(02)00710-0. [DOI] [PubMed] [Google Scholar]
- Bus VGM, Chagne D, Bassett HCM, et al. Genome mapping of three major resistance genes to woolly apple aphid (Eriosoma lanigerum Hausm.) Tree Genetics and Genomes. 2008;4:223–236. [Google Scholar]
- Campbell R, Ducreux LJM, Morris WL, Morris JA, Suttle JC, Ramsay G, Bryan GJ, Hedley PE, Taylor MA. The metabolic and developmental roles of carotenoid cleavage dioxygenase4 from potato. Plant Physiology. 2010;154:656–664. doi: 10.1104/pp.110.158733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli CI, Cuttriss AJ, Cossetto SB, Pye W, Crisp P, Whelan J, Finnegan EJ, Turnbull C, Pogson BJ. Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. The Plant Cell. 2009;21:39–53. doi: 10.1105/tpc.108.063131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli CI, Pogson BJ. Source to sink: regulation of carotenoid biosynthesis in plants. Trends in Plant Science. 2010;15:266–274. doi: 10.1016/j.tplants.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plan Molecular Biology Reporter. 1993;116:113–116. [Google Scholar]
- Chen Y, Li FQ, Wurtzel ET. Isolation and characterization of the Z-ISO gene encoding a missing component of carotenoid biosynthesis in plants. Plant Physiology. 2010;153:66–79. doi: 10.1104/pp.110.153916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotault J, Peltier D, Berruyer R, Thomas M, Briard M, Geoffriau E. Expression of carotenoid biosynthesis genes during carrot root development. Journal of Experimental Botany. 2008;59:3563–3573. doi: 10.1093/jxb/ern210. [DOI] [PubMed] [Google Scholar]
- Diretto G, Al-Babili S, Tavazza R, Papacchioli V, Beyer P, Giuliano G. Metabolic engineering of potato carotenoid content through tuber-specific overexpression of a bacterial mini-pathway. PLoS One. 2007;2:e350. doi: 10.1371/journal.pone.0000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducreux LJM, Morris WL, Hedley PE, Shepherd T, Davies HV, Millam S, Taylor MA. Metabolic engineering of high carotenoid potato tubers containing enhanced levels of beta-carotene and lutein. Journal of Experimental Botany. 2005;56:81–89. doi: 10.1093/jxb/eri016. [DOI] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. The Plant Journal. 2007;49,:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez JM, Cantero A, Reindl A, Reichler S, Leon P. 1-Deoxy-d-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. Journal of Biological Chemistry. 2001;276,:22901–22909. doi: 10.1074/jbc.M100854200. [DOI] [PubMed] [Google Scholar]
- Felicetti DA, Schrader LE. Changes in pigment concentrations associated with sunburn browning of five apple cultivars. I. Chlorophylls and carotenoids. Plant Science. 2009;176:78–83. [Google Scholar]
- Floss DS, Schliemann W, Schmidt J, Strack D, Walter MH. RNA interference-mediated repression of MtCCD1 in mycorrhizal roots of Medicago truncatula causes accumulation of C-27 apocarotenoids, shedding light on the functional role of CCD1. Plant Physiology. 2008;148:1267–1282. doi: 10.1104/pp.108.125062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss DS, Walter MH. Role of carotenoid cleavage dioxygenase 1 (CCD1) in apocarotenoid biogenesis revisited. Plant Signaling and Behavior. 2009;4,:172–175. doi: 10.4161/psb.4.3.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Enfissi EMA, Halket JM, Truesdale MR, Yu D, Gerrish C, Bramley PM. Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. The Plant Cell. 2007;19:3194–3211. doi: 10.1105/tpc.106.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Pinto MES, Holloway DE, Bramley PM. Application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. The Plant Journal. 2000;24:551–558. doi: 10.1046/j.1365-313x.2000.00896.x. [DOI] [PubMed] [Google Scholar]
- Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM. Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression) Plant Physiology. 1994;105:405–413. doi: 10.1104/pp.105.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Kong W, Peng G, Zhou J, Azam M, Xu C, Grierson D, Chen K. Plastid structure and carotenogenic gene expression in red- and white-fleshed loquat (Eriobotrya japonica) fruits. Journal of Experimental Botany. 2011;11:11. doi: 10.1093/jxb/err284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Limones C, Schnaebele K, Blanco-Portales R, Luz Bellido M, Luis Caballero J, Schwab W, Munoz-Blanco J. Functional characterization of FaCCD1: a carotenoid cleavage dioxygenase from strawberry involved in lutein degradation during fruit ripening. Journal of Agricultural and Food Chemistry. 2008;56,:9277–9285. doi: 10.1021/jf801096t. [DOI] [PubMed] [Google Scholar]
- Guzman I, Hamby S, Romero J, Bosland PW, O’Connell MA. Variability of carotenoid biosynthesis in orange colored Capsicum spp. Plant Science. 2010;179,:49–59. doi: 10.1016/j.plantsci.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond BR, Wooten BR, Curran-Celentano J. Carotenoids in the retina and lens: possible acute and chronic effects on human visual performance. Archives of Biochemistry and Biophysics. 2001;385:41–46. doi: 10.1006/abbi.2000.2184. [DOI] [PubMed] [Google Scholar]
- Harjes CE, Rocheford TR, Bai L, et al. Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science. 2008;319:330–333. doi: 10.1126/science.1150255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffler JF, Hemmerlin A, Grosdemange-Billiard C, Bach TJ, Rohmer M. Isoprenoid biosynthesis in higher plants and in Escherichia coli: on the branching in the methylerythritol phosphate pathway and the independent biosynthesis of isopentenyl diphosphate and dimethylallyl diphosphate. The Biochemistry Journal. 2002;366:573–583. doi: 10.1042/BJ20020337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T, Ohad I, Beyer P, Hirschberg J. Analysis in vitro of the enzyme CRTISO establishes a poly-cis-carotenoid biosynthesis pathway in plants. Plant Physiology. 2004;136:4246–4255. doi: 10.1104/pp.104.052092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T, Ronen G, Zamir D, Hirschberg J. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. The Plant Cell. 2002;14:333–342. doi: 10.1105/tpc.010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamffer Z, Bindon KA, Oberholster A. Optimization of a method for the extraction and quantification of carotenoids and chlorophylls during ripening in grape berries (Vitis vinifera cv. Merlot) Journal of Agricultural and Food Chemistry. 2010;58:6578–6586. doi: 10.1021/jf1004308. [DOI] [PubMed] [Google Scholar]
- Kato M, Ikoma Y, Matsumoto H, Sugiura M, Hyodo H, Yano M. Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiology. 2004;134,:824–837. doi: 10.1104/pp.103.031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Matsumoto H, Ikoma Y, Kuniga T, Nakajima N, Yoshida T, Yano M. Accumulation of carotenoids and expression of carotenoid biosynthetic genes and carotenoid cleavage dioxygenase genes during fruit maturation in the juice sacs of ‘Tamami,’ ‘Kiyomi’ tangor, and ‘Wilking’ mandarin. Journal of the Japanese Society for Horticultural Science. 2007;76:103–111. [Google Scholar]
- Lee HS, Castle WS, Coates GA. High-performance liquid chromatography for the characterization of carotenoids in the new sweet orange (Earlygold) grown in Florida, USA. Journal of Chromatography A. 2001;913:371–377. doi: 10.1016/s0021-9673(00)01029-3. [DOI] [PubMed] [Google Scholar]
- Li F, Murillo C, Wurtzel ET. Maize Y9 encodes a product essential for 15-cis-ζ-carotene isomerization. Plant Physiology. 2007;144,:1181–1189. doi: 10.1104/pp.107.098996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FQ, Vallabhaneni R, Wurtzel ET. PSY3, a new member of the phytoene synthase gene family conserved in the poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiology. 2008;146:1333–1345. doi: 10.1104/pp.107.111120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Paolillo DJ, Parthasarathy MV, Dimuzio EM, Garvin DF. A novel gene mutation that confers abnormal patterns of beta-carotene accumulation in cauliflower (Brassica oleracea var. botrytis) The Plant Journal. 2001;26:59–67. doi: 10.1046/j.1365-313x.2001.01008.x. [DOI] [PubMed] [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biology. 2010;10:50. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren LO, Stalberg KG, Hoglund AS. Seed-specific overexpression of an endogenous Arabidopsis phytoene synthase gene results in delayed germination and increased levels of carotenoids, chlorophyll, and abscisic acid. Plant Physiology. 2003;132:779–785. doi: 10.1104/pp.102.017053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Van Eck J, Zhou X, et al. The cauliflower or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. The Plant Cell. 2006;18:3594–3605. doi: 10.1105/tpc.106.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass D, Arango J, Wuest F, Beyer P, Welsch R. Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS One. 2009;4:e6373. doi: 10.1371/journal.pone.0006373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb RD, Crowhurst RN, Gleave AP, et al. Analyses of expressed sequence tags from apple. Plant Physiology. 2006;141,:147–166. doi: 10.1104/pp.105.076208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Villalon A, Gas E, Rodriguez-Concepcion M. Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. The Plant Journal. 2009;60,:424–435. doi: 10.1111/j.1365-313X.2009.03966.x. [DOI] [PubMed] [Google Scholar]
- Romer S, Fraser PD, Kiano JW, Shipton CA, Misawa N, Schuch W, Bramley PM. Elevation of the provitamin A content of transgenic tomato plants. Nature Biotechnology. 2000;18:666–669. doi: 10.1038/76523. [DOI] [PubMed] [Google Scholar]
- Ronen G, Cohen M, Zamir D, Hirschberg J. Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. The Plant Journal. 1999;17:341–351. doi: 10.1046/j.1365-313x.1999.00381.x. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sauret-Gueto S, Botella-Pavia P, Flores-Perez U, Martinez-Garcia JF, San Roman C, Leon P, Boronat A, Rodriguez-Concepcion M. Plastid cues posttranscriptionally regulate the accumulation of key enzymes of the methylerythritol phosphate pathway in Arabidopsis . Plant Physiology. 2006;141,:75–84. doi: 10.1104/pp.106.079855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiggert RM, Steingass CB, Heller A, Esquivel P, Carle R. Characterization of chromoplasts and carotenoids of red- and yellow-fleshed papaya (Carica papaya L.) Planta. 2011;234:1031–1044. doi: 10.1007/s00425-011-1457-1. [DOI] [PubMed] [Google Scholar]
- Shewmaker CK, Sheehy JA, Daley M, Colburn S, Ke DY. Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. The Plant Journal. 1999;20,:401–412. doi: 10.1046/j.1365-313x.1999.00611.x. [DOI] [PubMed] [Google Scholar]
- Southon S. Increased fruit and vegetable consumption within the EU: potential health benefits. Food Research International. 2000;33:211–217. [Google Scholar]
- Vallabhaneni R, Wurtzel ET. Timing and biosynthetic potential for carotenoid accumulation in genetically diverse germplasm of maize. Plant Physiology. 2009;150,:562–572. doi: 10.1104/pp.109.137042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, et al. The genome of the domesticated apple (Malus × domestica Borkh.) Nature Genetics. 2010;42:833. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- Wellburn AR. The spectral determination of chlorophyll-a and chlorophyll-b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. Journal of Plant Physiology. 1994;144:307–313. [Google Scholar]
- Welsch R, Arango J, Baer C, Salazar B, Al-Babili S, Beltran J, Chavarriaga P, Ceballos H, Tohme J, Beyer P. Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. The Plant Cell. 2010;22:3348–3356. doi: 10.1105/tpc.110.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiangjun Z, McQuinn R, Zhangjun F, Wolters AMA, Van Eck J, Brown C, Giovannoni JJ, Li L. Regulatory control of high levels of carotenoid accumulation in potato tubers. Plant, Cell and Environment. 2011;34,:1020–1030. doi: 10.1111/j.1365-3040.2011.02301.x. [DOI] [PubMed] [Google Scholar]
- Xu C-J, Fraser PD, Wang W-J, Bramley PM. Differences in the carotenoid content of ordinary citrus and lycopene-accumulating mutants. Journal of Agricultural and Food Chemistry. 2006;54:5474–5481. doi: 10.1021/jf060702t. [DOI] [PubMed] [Google Scholar]
- Yano M, Kato M, Ikoma Y, Kawasaki A, Fukazawa Y, Sugiura M, Matsumoto H, Oohara Y, Nagao A, Ogawa K. Quantitation of carotenoids in raw and processed fruits in Japan. Food Science and Technology Research. 2005;11:13–18. [Google Scholar]
- Zhou C, Zhao D, Sheng Y, Tao J, Yang Y. Carotenoids in fruits of different persimmon cultivars. Molecules (Basel, Switzerland) 2011;16:624–636. doi: 10.3390/molecules16010624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.