Abstract
Cell migration is a fundamental biological function, critical during development and regeneration, whereas deregulated migration underlies neurological birth defects and cancer metastasis. MARCKS-like protein 1 (MARCKSL1) is widely expressed in nervous tissue, where, like Jun N-terminal protein kinase (JNK), it is required for neural tube formation, though the mechanism is unknown. Here we show that MARCKSL1 is directly phosphorylated by JNK on C-terminal residues (S120, T148, and T183). This phosphorylation enables MARCKSL1 to bundle and stabilize F-actin, increase filopodium numbers and dynamics, and retard migration in neurons. Conversely, when MARCKSL1 phosphorylation is inhibited, actin mobility increases and filopodium formation is compromised whereas lamellipodium formation is enhanced, as is cell migration. We find that MARCKSL1 mRNA is upregulated in a broad range of cancer types and that MARCKSL1 protein is strongly induced in primary prostate carcinomas. Gene knockdown in prostate cancer cells or in neurons reveals a critical role for MARCKSL1 in migration that is dependent on the phosphorylation state; phosphomimetic MARCKSL1 (MARCKSL1S120D,T148D,T183D) inhibits whereas dephospho-MARCKSL1S120A,T148A,T183A induces migration. In summary, these data show that JNK phosphorylation of MARCKSL1 regulates actin homeostasis, filopodium and lamellipodium formation, and neuronal migration under physiological conditions and that, when ectopically expressed in prostate cancer cells, MARCKSL1 again determines cell movement.
INTRODUCTION
MARCKS-like protein 1 (MARCKSL1) is an actin binding protein that is predominantly expressed in immature brain (1, 27). The MARCKSL1 homologue MARCKS has been more extensively studied and has been shown to bind actin with a stoichiometry of 1:2, thereby facilitating cross-linking (56; reviewed in reference 39). Binding to actin occurs via an effector domain (ED) that is 87% identical to the corresponding domain of MARCKSL1. Surprisingly, however, full-length MARCKSL1 does not cross-link F-actin (49; reviewed in reference 39), although the MARCKSL1 effector domain alone interacts with actin. This indicates that in a physiological context, another level of regulation is required for MARCKSL1 to regulate actin bundling. The only known critical function of MARCKSL1 is in early development of the nervous system, as genetic disruption of MARCKSL1 results in neural tube closure defects (5, 51), events that depend on coordinated control of actin functions, cell shape, and cell migration. MARCKSL1 is also associated with cell spreading (23); however, the mechanism whereby MARCKSL1 regulates F-actin in a cellular context has remained obscure.
c-Jun N-terminal kinase 1 (JNK1) and JNK2, like MARCKSL1, are required for neural tube closure (20, 36). However, the identity of the JNK effectors mediating this event remains unresolved. JNK activity is highly elevated in neuronal cells (8, 42), and although initially unexpected (7, 8, 53), it is now accepted that a number of important physiological substrates for JNK exist in the brain, many of which are microtubule regulators, including microtubule-associated protein 2 (MAP2), DCX, and SCG10 (3, 4, 13, 22, 42). As a consequence, JNK functions in neurite growth and axonal transport (6, 30, 28, 42). However, a convincing literature also indicates that JNK activity facilitates migration of many cell types, including epithelium-like, fibroblast, endothelial, and glioblastoma cells (15, 17, 25, 35, 40). In contrast to the findings of these studies in nonneuronal cells, JNK activity retards migration of central nervous system (CNS) neurons (48). In an effort to better understand the mechanism of JNK function in the brain, we screened for novel CNS substrates of JNK. Here we identify MARCKSL1 as a JNK substrate. We show that JNK phosphorylates MARCKSL1 in intact cells on sites that are not conserved in MARCKS. When phosphorylated on these sites, MARCKSL1 induces F-actin bundling, reduces actin turnover in cells, and retards cell migration. This reveals that MARCKSL1 is a newly identified effector of JNK that links directly to the actin cytoskeleton, thereby reducing actin plasticity and impeding forward migration of neurons.
MATERIALS AND METHODS
Materials.
Anti-PJNK(Thr183/Tyr185) was from Cell Signaling Technologies, and anti-SAPK1b/SAPKβ/JNK3 was from Upstate Biotechnology (Lake Placid, NY). Rabbit anti-MARCKSL1 (ProteinTech Group Inc.) was used to detect MARCKSL1. Mouse anti-green fluorescent protein (anti-GFP) (clone JL-8) was from Clontech (Mountain View, CA). Antibodies against glutathione S-transferase (GST) and phospho-c-Jun(Ser63) were from Santa Cruz Biotechnology (Santa Cruz, CA). The actin antibody (clone AC-4700) was from Sigma (St. Louis, MO). Anti-mouse β-tubulin (clone KMX-1) was from Chemicon (Temecula, CA). Alexa 568-phalloidin and secondary antibodies for immunofluorescence staining and anti-rabbit antibody–Alexa 546 and anti-mouse antibody–Alexa 488 and 555 as well as Hoechst 33342 and Mowiol mounting media were from Molecular Probes (Eugene, OR). SP600125 was from Calbiochem (San Diego, CA).
Plasmids.
Mouse MARCKSL1, mouse cofilin-1, and human paxillin-β were cloned by PCR from mouse brain and human neuroblastoma cDNA. Genes were ligated into the EcoRI site of pXFP-N1 vectors for MARCKSL1 and pXFP-C1 vectors for paxillin and cofilin (Clontech) as indicated. MARCKSL1-S120A/T148A/T183A (MARCKSL1-AAA) and MARCKSL1-S120D/T148D/T183D (MARCKSL1-DDD) were prepared by insertional overlapping PCR as previously described (42). Small interfering RNA (siRNA)-insensitive mutants mouse MARCKSL1-WTi, MARCKSL1-AAAi, and MARCKSL1-DDDi were prepared by insertional overlapping PCR using mutagenic primers and inserted into p1xCherry-N1. For expression in Escherichia coli, MARCKSL1, MARCKSL1S120A,T148A,T183A, and MARCKSL1S120D,T148D,T183D were ligated into the EcoRI site of pGEX-6P3 (Amersham Biosciences). ECFP-NES-JBD was prepared by replacing the enhanced GFP (EGFP) coding sequence from EGFP-NES-JBD (3) with enhanced cyan fluorescent protein (ECFP) using the NheI and BsrGI cloning sites. Rat β-actin, obtained by PCR as described previously (7), was ligated into the EcoRI site of the pVenus vector. pcDNA3-MKK7-JNK1 and pcDNA3-MKK7-JNK2 were gifts from R. Davis (Howard Hughes Medical Institute [HHMI], Worcester, MA).
Mass spectrometry. (i) In vivo analysis of phosphorylation sites.
The tryptic digests were passed over a TiO2 column slurry packed in an Eppendorf tip and eluted with trifluoroacetic acid (TFA) as described previously (43). Samples were analyzed with an LTQ-Orbitrap instrument (ThermoFisher, Bremen, Germany), coupled to a high-performance liquid chromatography (HPLC) splitless Eksigent two-dimensional (2D) NanoLC system (Eksigent Technologies, Dublin, CA). The HPLC autosampler injected 5 μl of peptide sample, which was trapped on the precolumn (Zorbax 300SB-C18; Agilent Technologies, Santa Clara, CA) (5 mm by 0.3 mm; 5-μm-pore-size membrane) and washed (15 min) with solvent A (0.1% fluorescent antibody [FA]) only. Peptides were separated on a reverse-phase analytical column (Zorbax 300SB-C18; Agilent Technologies) (150 mm by 75 μm; 3.5-μm-pore-size membrane) at a flow rate of 400 nl/min. A gradient of 5% to 40% acrylonitrite (ACN) was run from over 55 min, held for 5 min, and then raised to 80% (over 0.1 min). The latter concentration was kept for 15 min before a return to 5% and reequilibration. The same protocol was used for analysis of the protein standard except that pooled labeled ADH peptides were diluted 10-fold before injection. Mass spectra of all samples were always acquired in the positive-ionization mode, with an m/z scan range of 400 to 2,000 Da. After the three most intense precursor masses in mass spectroscopy (MS) mode were selected, their tandem MS (MS/MS) scans (with an m/z range of 50 to 2,000 Da) were performed with a dynamic exclusion range of 240 s after the second peptide count. The collision-induced dissociation (CID) and high-energy CID (HCD) collision energies were set at 35 and 45 eV, respectively.
(ii) Database analysis.
All LC-MS/MS data were processed using the Proteios2 Software Environment (version 2.10.0; available at http://www.proteios.org) and analyzed with Mascot. The enzyme was set to trypsin with two allowed miscleavages and the instrument to ion trap and peptide and fragment mass tolerances of 5 ppm and 0.5 Da, respectively, with 0.01 as the false-discovery rate. Searches were done against IPI mouse database version 3.71, including a decoy data set. Carbamidomethyl-Cys was set as a fixed modification and oxidation of Met and phosphorylation of Ser/Thr and Tyr as variable modifications.
In vitro analysis of phosphorylation sites.
GST-MARCKSL1 (1 μg) was phosphorylated in vitro with GST-JNK1α1 as described previously (48), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and subjected to in-gel digestion with trypsin (Promega, Madison, WI). Tryptic peptides were dissolved in 0.2 M acetic acid–30% ACN (vol/vol) and phosphopeptides isolated using a PHOS-Select (Sigma) iron affinity matrix. Peptide digest (1 pmol) was loaded slowly onto a nanoscale immobilized metal affinity column (IMAC) prepared in a 10-μl gel loader tip and washed 3 times with 0.2 M acetic acid–30% ACN and 1 time with water before elution with 1.5% NH3 directly into 5% formic acid. Acidified peptides were desalted using a nanoscale column packed with POROS oligoR3 (PerSeptive Biosystems) chromatography material or graphite powder (Sigma). The oligoR3 material was used for desalting the phosphopeptides at positions 111 to 144 and 167 to 200, containing phosphorylation sites S120 and T183, respectively, while the hydrophilic phosphopeptide at positions 145 to 157 containing phosphorylation site T148 was captured on graphite powder. Samples were desalted by repeated washing with 0.1% formic acid and elution with matrix solution (dihydroxybenzoic acid [DHB] 50% ACN–1% phosphoric acid) for matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis (Voyager-DE PRO; Applied Biosystems) or by step elution with 2.5% formic acid–10% to 50% methanol for nano-liquid chromatography–electrospray quadrupole–time of flight mass spectrometer (nESI-qQTOF) analysis (QStar Pulsar; Applied Biosystems). Approximately 20% of the IMAC eluate was removed prior to acidification for phosphatase treatment using 0.05 U per μl alkaline phosphatase (Boehringer Mannheim) in 0.1 M NH4HCO3. Analysis of fragment ion spectra was done using Analyst QS software (Applied Biosystems) and interpreted both manually and using MASCOT software (Matrix Science).
Phosphorylation analysis.
Metabolic labeling with 32P was performed as previously described (48) with minor changes. HEK-293 cells that were nearly confluent were transfected using the calcium precipitation method. At 24 h later, cells were washed with phosphate-free Dulbecco's modified Eagle's medium (DMEM) (Gibco catalog no. 11971-025) containing 0.1% fetal calf serum (FCS) and incubated for 19 h. Cells were incubated in phosphate-free medium containing 0.5 mCi/ml [32P]orthophosphate (MP Biomedicals) for 2 h in 5% CO2 at 37°C. Where indicated, cells were irradiated with UV (254 nm, 80 J/m2) and incubated for 2 h before lysis in immunoprecipitation buffer (20 mM HEPES [pH 7.4], 2 mM EGTA, 50 mM β-glycerophosphate, 1 mM dithiothreitol [DTT], 1 mM Na3VO4, 1% [vol/vol] Triton X-100, 10% [vol/vol] glycerol, 0.5 mM benzamidine, 10 nM microcystin-leucine-argenine [microcystin-LR], phenylmethylsulfonyl fluoride [PMSF; 100 μg/ml], and 1 μg/ml each of aprotonin, leupeptin, and pepstatin). GFP-tagged proteins were immunoprecipitated at 4°C by overnight incubation with a polyclonal anti-rabbit GFP antibody (Clontech) and collected using protein G-Sepharose (Upstate Biotechnology). Following several washes, immunopurified proteins were mixed with Laemmli sample buffer and analyzed by SDS-PAGE. Quantification of 32P-labeled protein was done using a Fuji BAS-1800 phosphorimager. Kinetic analysis of JNK substrate phosphorylation was performed as previously described (3, 48).
Cell culture and transfection.
Cortical neurons were prepared from P0 Sprague-Dawley rats and transfected with LF2000 as previously described (42). For cell migration analysis in Transwells (Corning Inc.), 2.2 × 105 neurons were seeded in 6.5-mm-diameter 5-μm-pore-size Transwells precoated with laminin (10 μg/ml). Mouse embryonal fibroblasts (MEFs) were cultured in minimal essential medium supplemented with 10% (vol/vol) bovine calf serum, 2 mM glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, and nonessential amino acids (Sigma). All cells were cultured in a humidified 5% CO2 atmosphere at 37°C. MEFs were transfected with Lipofectamine 2000, according to the instructions of the manufacturer (Invitrogen, Carlsbad, CA). PC-3 cells were cultured in RPMI 1640 (Sigma) and HEK-293T cells in Dulbecco's modified Eagle's medium (Sigma), both supplemented with 10% (vol/vol) fetal calf serum (FCS), 2 mM glutamine and 50 U/ml penicillin, and 50 μg/ml streptomycin.
Lentiviral generation of stable knockdown PC-3 cell lines.
Two 19-nucleotide sequences (for cell line 5, 5′-GAA CGG AAC AGA UGA GGC A-3′; for cell line 6, 5′-GGC CUG UCC UUC AAG AGA A-3′) targeting human MARCKSL1 (NM_023009) were inserted into GFP-expressing biscistronic pGIPZ lentiviral vector. A total of 5 × 104 cells/well were seeded on day 0 and 24 h later were transduced with lentiviral particles (at a multiplicity of infection [MOI] of 200) in serum-free media. At 48 h postransduction, puromycin (1 μg/ml) was added for 10 days. PC-3 cells expressing GFP, indicative of lentiviral integration, were collected by fluorescence-assisted cell sorting. The extent of knockdown was confirmed by measurement of mRNA (reverse transcription-PCR [RT-PCR]) and protein levels (Western blot analysis).
Transwell migration assay.
For PC-3 cells, 8-μm-pore-size Transwells were used. PC-3 cells stably transduced with human MARCKSL1 small hairpin RNA (shRNA) or nontargeting shRNA were seeded in RPMI 1640 medium with 1% FCS at 130,000 cells per well. The lower chamber was filled with 500 μl RPMI 1640 containing 10% FCS. Cells were allowed to migrate for 8 h, after which the cells that had migrated through the filter were counted (in a blinded manner) using an Olympus IX70 fluorescence microscope. For rescue experiments, stable knockdown PC-3 cells were plated in plastic wells and transfected 24 h later with 0.28 μg of DNA using Lipofectamine LTX (Invitrogen). Following 24 h of expression, transfected cells were resuspended in 1% fetal calf serum–RPMI 1640 medium and plated in Transwells. For neuronal migration analysis, cortical neurons in Transwells (5 μm pore size) were transfected during plating with LTX (Invitrogen). The medium was replaced after 4 h and fixed 48 h postplating. The number of migrated cells was counted using an Olympus IX70 fluorescence microscope.
Wound-healing assays.
MEFs on coverslips, transfected at 24 h postplating, were grown to confluence, after which the cell layer was scored using a sterile 0.5-to-20-μl tip. Cells were fixed and stained with Hoechst 33342 14 h later. Fluorescence images were taken using a 4× objective. GFP-positive cells that migrated into a defined region of the scored area were counted and expressed as a percentage of cell counts from equal-sized neighboring regions flanking the scored area.
Actin polymerization.
Pyrene-labeled G-actin (Cytoskeleton Inc., Denver, CO) (10 μM) was suspended in buffer containing 5 mM Tris-HCl (pH 8.0), 0.2 mM ATP, and 0.2 mM CaCl2 and incubated on ice for 10 min with GST, GST-MARCKSL1, GST-MARCKSL1-AAA, GST-MARCKSL1-DDD, or GST-cofilin (1 or 4 μM). The reaction mixture was placed at 24°C in a quartz cuvette and G-actin fluorescence monitored for 5 min before initiation of polymerization by addition of a 1/10 volume of actin polymerization buffer (500 mM KCl, 20 mM MgCl2, 10 mM ATP, 100 mM Tris-HCl, pH 7.5). Fluorescence was recorded every 20 s using a Cary Eclipse fluorescence spectrophotometer (Varian, Australia) (excitation, 365 nm; emission, 407 nm; bandwidth, 5 nm).
F-actin binding assay.
F-actin, prepared as described above, was incubated with 4 μM recombinant protein for 50 min at room temperature. Actin polymers were isolated by centrifugation at 150,000 × g for 2 h at 20°C. The resulting F-actin pellet was gently washed and suspended in Laemmli buffer for analysis of F-actin binding proteins.
G-actin interaction.
A solution of G-actin (1 μM) in low-salt buffer (5 mM Tris-HCl [pH 8.0], 0.2 mM CaCl2, 1 mM DTT; used to prevent actin filament formation) was incubated with 1 μM GST or GST-tagged MARCKSL1, MARCKSL1-AAA, MARCKSL1-DDD, or cofilin for 30 min. GST fusions were sequestered by incubation for 1 h with glutathione (GSH)-Sepharose (Amersham Pharmacia) at 4°C. The Sepharose was washed 5 times in low-salt buffer containing 0.1% Tween 20 before G-actin binding was assessed by immunoblotting.
Extraction of digitonin-insoluble F-actin.
Transfected MEF cells were washed briefly in permeabilization buffer (20 mM HEPES [pH 7.4], 280 mM sucrose, 200 mM KCl, 2 mM EGTA) to remove media and permeabilized on ice for 10 min with digitonin (0.008%) in permeabilization buffer. Digitonin extracts were collected as described previously (48a) and cells washed with permeabilization buffer to remove residual soluble protein. Digitonin-insoluble material was suspended in Laemmli buffer. Fractions were analyzed by Western blotting.
Fluorescence recovery after photobleaching (FRAP) analysis was carried out as previously described with minor modifications (48). MEFs on 3.5-cm-diameter glass-bottomed dishes (MatTek, Ashland, MA) were transfected with venus-actin together with MARCKSL1-CFP, MARCKSL1-AAA-CFP, MARCKSL1-DDD-CFP, CFP-cofilin, or CFP as indicated. At 24 h later, regions of interest were bleached using 300 iterations of a 488-nm laser (100% transmission) and a Zeiss 510-META confocal microscope and environmental control (5% CO2, 37°C, humidified). Postbleach images were acquired at 6-s intervals using 514-nm (5% transmission) and 488-nm (0.05% transmission) lasers for excitation. Venus-actin fluorescence emission was collected using a META detector at 529 to 593.2 nm. A 63× oil objective and maximum scan speed and resolution (708 by 708) were used to minimize fading. Background and fading correction was performed as previously described (32, 42). Data were normalized to the prebleach baseline. Curve fitting of normalized data was performed using FrapCalc software (Rolf Sara; Turku Centre for Biotechnology, Finland). Mobile fractions and half-maximum values for the fast (0 to 90 s) and total recovery curves were calculated using the following equations: I(t) = A(1− eτt) and t½ = (ln 0.5)/−τ.
Time-lapse analysis of actin loss after neuronal permeabilization.
Primary neurons on glass coverslips were transfected with venus–β-actin together with pECFP-MARCKSL1 constructs or pECFP-NES-JBD as indicated. At 24 h posttransfection, cells were rinsed in Mg2+-free Locke's buffer (154 mM NaCl, 5.6 mM KCl, 3.6 mM NaHCO3, 1.3 mM CaCl2, 5.6 mM d-glucose, 5 mM HEPES [pH 7.4]). Subsequently, cells were incubated in 1 ml buffer {80 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 6.5), 5 mM EGTA, 2 mM MgCl2, 0.1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (AEBSF)}. Cells were imaged using an Olympus IX81/ZDC-CellR fluorescence microscope with a 20× objective and CFP/yellow fluorescent protein (CFP/YFP) signals imaged using appropriate filters. Data were acquired from a region of interest in the neuronal cell soma. Data from approximately 40 cells were averaged and plotted as a percentage of initial fluorescence. Data analysis was carried out using CellR-analySIS software (Olympus).
Time-lapse imaging.
Cortical neurons were cultured for 6 days in vitro in 3.5-cm-diameter glass-bottomed dishes (MatTek Corp., Ashland, MA) and transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. After 24 h of expression of MARCKSL1-GFP or GFP-CAAX, wide-field imaging with sequential acquisition settings was performed using a Zeiss Axiovert 200 M microscope (Carl Zeiss AG, Oberkochen, DE) with a Plan-Neofluar 63×/1.25-numerical-aperture (NA) oil objective, a neutral-density ND4 filter, and an ORCA 1394 ERG camera (Hamamatsu Photonics, Japan). Images were acquired with Wasabi software (Hamamatsu Photonics, Japan) in continuous acquisition mode with a 1-s exposure time for 8 min.
Motility dynamics measurements.
Motility of filopodia in neuronal processes was visualized by arithmetical subtraction of consecutive images acquired at 10-s intervals from an 8-min time-lapse recording using ImageJ. The subtracted images were averaged and inverted to obtain a single image displaying motility changes as a gray scale, with darker pixels representing higher motility (12). To quantify the degree of displacement or motility of filopodia, the area occupied by processes was obtained using a maximal-intensity projection and the integrated density was measured from this area.
Immunostaining.
Immunocytochemical staining was carried out as described previously. Acin was stained with Alexa 568-phalloidin (0.005 U/μl). For immunohistochemistry, 21 samples from cases of primary prostate carcinoma were collected from the pathology archive of Turku University Central Hospital and stained for MARCKSL1 in a Lab Vision Autostainer. The primary antibody MARCKS-like-1 (ProteinTech Group, Chicago, IL) was diluted 1:300, and detection was performed using a BrightVision+ poly-horseradish peroxidase (poly-HRP) IgG kit (Immunologic, Duiven, the Netherlands).
Bioinformatic analysis of MARCKSL1 expression.
To evaluate MARCKSL1 expression in various healthy and malignant human tissues, we used the in silico transcriptomics database (19) containing integrated gene expression data from 9,783 samples covering 175 types of healthy and pathological human tissues. The majority of samples (n = 9,456) contained information on MARCKSL1 expression. A comparison of MARCKSL1 mRNA expression levels in healthy and malignant tissues was done using Student′s t test (fold changes and P values are indicated in Results).
Statistical analysis.
Statistical analysis of variance was done using SPSS for Windows 11.0.1. Testing of homogeneous sample variances was performed using Levene's test prior to one-way analysis of variance (ANOVA) to determine levels of statistical significance. A Fisher least-significant-difference (LSD) post hoc test was used in cases with more than two variable groups.
RESULTS
Proteomics screening revealed that JNK phosphorylates MARCKSL1 on C-terminal residues S120, T148, and T183 in vitro and in brain ex vivo.
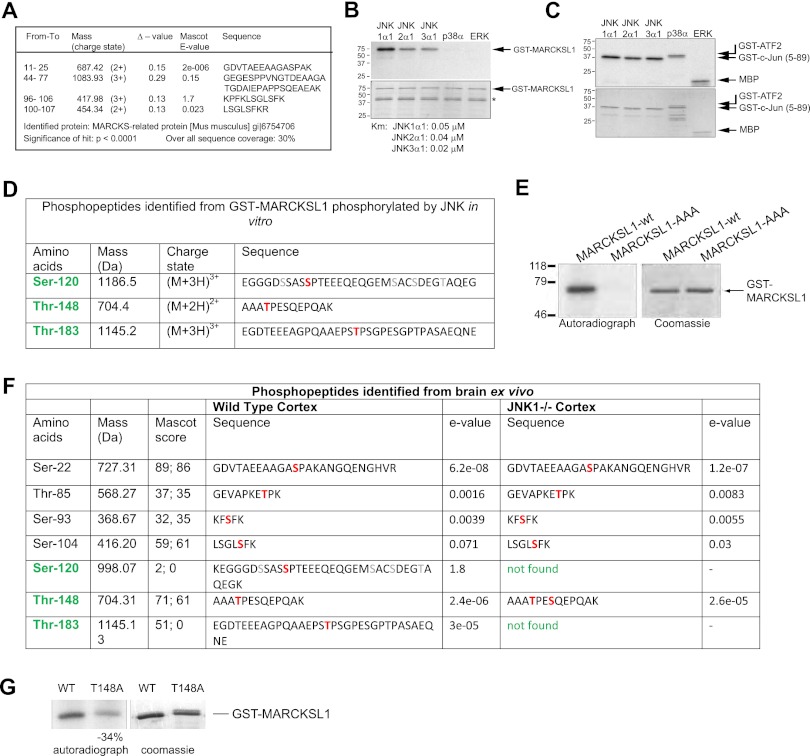
To determine targets for JNK in the central nervous system, we screened brain lysate for proteins that were strongly phosphorylated by recombinant JNK, as previously described (3). We identified a prominently phosphorylated ∼40-kDa protein that migrated as distinct spots with descending pIs consistent with multisite phosphorylation (see Fig. S1a in the supplemental material). Mass spectrometry sequencing identified the protein as MARCKS-like protein 1 (MARCKSL1), also known as MRP, MacMARCKS, and F52 (Fig. 1A). To further analyze MARCKSL1 as a potential JNK target, we prepared recombinant MARCKSL1 and studied its phosphorylation in vitro. All three JNKs (JNK1, JNK2, and JNK3) phosphorylated MARCKSL1 in vitro; however, no phosphorylation was detected by p38 or extracellular signal-regulated kinase (ERK) analysis (Fig. 1B), though ERK is reported to phosphorylate the related MARCKS protein (37). This was not due to variability in kinase activities, as ERK and p38 both showed normal activity when tested against known substrates (Fig. 1C), indicating that all kinases were similarly active. A kinetic analysis of MARCKSL1 phosphorylation by JNK yielded Km values of 0.05, 0.04, and 0.02 μM for JNK1, -2, and -3, indicating high affinity for the substrate.
Fig 1.
Identification of brain-derived MARCKSL1 as a JNK substrate. (A) LC-ESI-qTOF MS/MS sequencing identified this protein as a MARCKS-related protein (MARCKSL1). Identified peptides are shown. (B) Bacterially expressed GST-MARCKSL1 was phosphorylated by active recombinant JNK1α1, JNK2α1, JNK3α1, p38α, and ERK in vitro (top panel). While JNK isoforms strongly phosphorylated MARCKSL1, p38α and ERK did not. (*, degradation product). A kinetic analysis of GST-MARCKSL1 phosphorylation yielded Km values of 0.05, 0.04, and 0.02 μM for JNK1, JNK2, and JNK3, respectively. (C) In vitro kinase assay showing the relative activity of JNKs toward c-Jun and p38α and ERK toward ATF2 and myelin basic protein (MBP). (D) To identify which sites on MARCKSL1 were phosphorylated by JNK, in vitro kinase assays using GST-MARCKSL1 were carried out and phosphopeptides identified by metal affinity chromatography followed by mass spectrometry. S120, T148, and T183 were identified as JNK-phosphorylated sites. (E) S/T-to-A mutations of MARCKSL1 (GST-MARCKSL1S120A/T148A/T183A) eliminated JNK-dependent phosphorylation. (F) The sites of MARCKSL1 phosphorylated in wild-type and jnk1−/− mouse P7 cortex were analyzed. All three JNK-phosphorylated sites contained constitutively bound phosphate in the wild-type cortex, in addition to other phosphorylated residues. Cortex from jnk1−/− mice lacked phosphate on S120 and T183, confirming that these sites were phosphorylated by JNK1 nonredundantly. (G) In vitro phosphorylation of GST-MARCKSL1T148→A148 showed 34% less phosphate incorporation than GST-MARCKSL1WT, indicating that T148 is directly phosphorylated by JNK, but this phosphorylation is compensated in vivo in brain.
To determine which sites on MARCKSL1 were phosphorylated by JNK, we analyzed in vitro-phosphorylated GST-MARCKSL1 using mass spectrometry. MS/MS sequencing of enriched phosphopeptides revealed the presence of phosphate on sites S120, T148, and T183 (Fig. 1D; see also Fig. S1a in the supplemental material). This was validated by a characteristic neutral loss of 80 Da, corresponding to the loss of a HPO3 from the phosphopeptides upon treatment with phosphatase (see Fig. S1b in the supplemental material). Mutation of all three sites to alanine (MARCKSL1-AAA; MARCKSL1S120A,T148A,T183A) resulted in complete loss of JNK-mediated phosphorylation (Fig. 1E), indicating that no JNK phosphorylation sites remained unidentified.
To determine if JNK phosphorylated MARCKSL1 in vivo in brain, we made use of JNK1−/− mice, in which constitutive JNK activity is substantially reduced (42). TiO2-enriched phosphopeptides isolated from wild-type (WT) and JNK1−/− brains were analyzed by MS/MS. This analysis revealed seven sites on MARCKSL1 that were basally phosphorylated in brain (Fig. 1F). Among these were the JNK sites S120, T148, and T183 and protein kinase C (PKC) sites Ser-93 and Ser-104 as well as previously uncharacterized phosphorylation sites (Ser-22 and Thr-85). Notably, in JNK1−/− brains, phosphopeptides encoding S120 and T183 were not detected, while the PKC sites and the other uncharacterized phosphorylation sites remained unchanged compared to those of wild-type brains. This result indicated that S120 and T183 are phosphorylated by JNK1 in vivo in a nonredundant manner. However, the third JNK phosphorylation site (T148) remained phosphorylated in JNK1−/− brains, suggesting that another kinase compensates for JNK in JNK1−/− mice. To determine whether this is the case, we measured the phosphorylation of MARCKSL1-T148A, where T148 was mutated to alanine (Fig. 1G). There was a 34% reduction in phosphate incorporation upon mutation of this site, indicating that T148 was indeed directly phosphorylated by JNK1, though in JNK1−/− brain, other kinases, possibly JNK2 and -3, compensate for the absence of JNK1. These results were further supported by metabolic labeling experiments that indicated that MARCKSL1 was phosphorylated by JNK in intact cells (see Fig. S1b in the supplemental material).
Phosphomimetic MARCKSL1S120D,T148D,T183D induces F-actin bundling but MARCKSL1-WT does not.
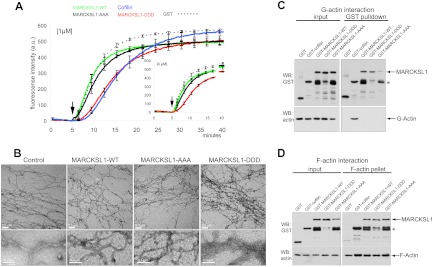
We next asked what could be the functional consequence of MARCKSL1 phosphorylation by JNK. While the effector domain of MARCKSL1 stimulates actin polymerization in vitro, full-length MARCKSL1 does not (49, 50). This raised the possibility that full-length MARCKSL1 must be posttranslationally modified before it can regulate actin dynamics. To test this, we used pyrene-labeled actin, the fluorescence of which increases upon polymerization and is proportional to actin filament length (9). While recombinant full-length MARCKSL1 did not alter the actin polymerization rate compared to the GST control (Fig. 2A), phosphomimetic MARCKSL1-DDD markedly retarded initiation of actin polymerization, as did cofilin.
Fig 2.
Pseudophosphorylated MARCKSL1S120D,T148D,T183D induces F-actin bundling. (A) To test whether MARCKSL1 regulated actin filament formation in vitro, we measured the polymerization rate of pyrene-labeled G-actin in the presence of 1 μM or 4 μM (inset) bacterially purified MARCKSL1s or cofilin as a control. After initiation of polymerization (arrows), fluorescence was measured at 20-s intervals. Like cofilin, MARCKSL1-DDD delayed actin polymerization onset. Mean values ± standard errors of the means (SEM) of the results of 3 to 4 repeat experiments are shown. (B) Electron microscope images of negatively stained actin (10 μM) polymerized in the presence of 1 μM GST (control), GST-MARCKSL1, GST-MARCKSL1-AAA, or GST-MARCKSL1-DDD. Only GST-MARCKSL1-DDD induced F-actin bundling. (C) To assess whether MARCKSL1 or MARCKSL1 phosphorylation mutants bind to G-actin, GST-tagged MARCKSL1s or cofilin was incubated in buffer that was nonpermissive with respect to polymerization. GST-MARCKSL1 proteins were isolated and analyzed by Western blotting (WB). GST-MARCKSL1s did not interact with G-actin, but GST-cofilin did. (D) To test whether MARCKSL1 or MARCKSL1 phosphorylation mutants bound F-actin, MARCKSL1s were incubated in actin polymerization-permissive buffer. F-actin was isolated by ultracentrifugation. MARCKSL1-WT, MARCKSL1-DDD, MARCKSL1-AAA, and cofilin each copelleted with F-actin.
It is known that delayed onset of pyrene-labeled actin filament growth can be a result of F-actin bundling (21), and, indeed, cofilin bundles F-actin in vitro (31). To test whether MARCKSL1-DDD could induce F-actin bundling, we used electron microscopy. F-actin polymerized in the presence of MARCKSL1-WT, MARCKSL1-AAA, or MARCKSL1-DDD was visualized using negative staining, and the results revealed the formation of fine actin filaments in control samples polymerized in the presence of GST or MARCKSL1-WT (Fig. 2B). However, actin polymerized in the presence of MARCKSL1-DDD produced a dense meshwork with significantly thicker actin filaments; the actin caliber was >100 nm in some regions (Fig. 2B). These findings suggest that JNK phosphorylation of MARCKSL1 induces bundling of F-actin, possibly explaining the delayed filament growth.
We next tested if MARCKSL1-DDD, like cofilin (29), bound directly to monomeric G-actin. This was done by incubating GST-MARCKSL1 variants or GST-cofilin with G-actin at a 1:1 stoichiometry in low-salt buffer. As expected, GST-cofilin bound directly to G-actin (29); however, MARCKSL1-WT, MARCKSL1-AAA, and MARCKSL1-DDD did not (Fig. 2C). In sharp contrast, MARCKSL1-WT, MARCKSL1-AAA, and MARCKSL1-DDD all bound equally well to filamentous actin (F-actin; Fig. 2D). In summary, although all MARCKSL1 variants bound to F-actin, only phosphomimetic-MARCKSL1 was able to induce actin bundling.
Expression of MARCKSL1S120D,T148D,T183D or active JNK stabilizes cellular actin.
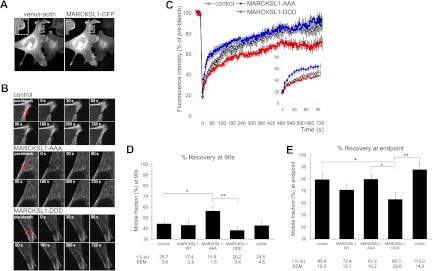
To determine whether JNK phosphorylation of MARCKSL1 influences actin dynamics in intact cells, we expressed venus-actin in MEFs (which do not express endogenous MARCKSL1; data not shown) and measured the effect of CFP-tagged MARCKSL1 and JNK-site mutants on actin mobility. Exogenously expressed venus-actin integrated into the F-actin cytoskeleton (Fig. 3A). Regions rich in venus-actin were photobleached, and fluorescence recovery was monitored in the presence of MARCKSL1 phosphorylation-site mutants as indicated (Fig. 3B). Actin turnover was increased in cells expressing MARCKSL1-AAA at both early and late time points (Fig. 3C and D), while MARCKSL1-DDD expression led to a decrease in actin turnover (Fig. 3E), indicating decreased dynamics. This was likely due to increased actin bundling in the presence of MARCKSL1-DDD (Fig. 2).
Fig 3.
Phosphomimetic MARCKSL1S120D,T148D,T183D inhibits actin dynamics, and MARCKSL1S120A,T148A,T183A increases it. (A) Venus-actin expressed in MEFs incorporates into stress fibers and colocalizes with MARCKSL1-CFP at the lamellipodia. (B) FRAP analysis was carried out to determine whether JNK phosphorylation of MARCKSL1 regulates F-actin turnover in cells. Representative images of MEFs expressing venus-actin and CFP empty vector (control), CFP-tagged MARCKSL1-AAA, or MARCKSL1-DDD are shown. The region to be bleached is marked with a red square. (C) FRAP curves for venus-actin corrected for background/fading and normalized to prebleach intensity. The fast-recovery phase (0 to 90 s) is shown in the inset. (D) Calculated mobile fraction and half-maximum times for the fast-recovery phase (0 to 90 s). (E) Calculated mobile fraction and half-maximum (t½) times for the FRAP curve (0 to 720 s). Mean values ± SEM of the results of 9 or 10 separate experiments are shown. Significance levels: *, P < 0.05; **, P < 0.01.
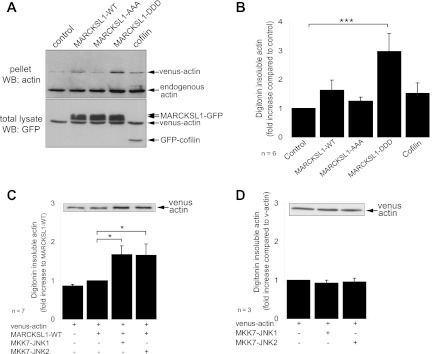
As a complementary approach to the FRAP analysis, we used digitonin permeabilization to quantify the effect of MARCKSL1 on actin stability. GFP-tagged MARCKSL1s were expressed in fibroblasts and the levels of digitonin-soluble and -insoluble actin were measured. Notably, MARCKSL1-DDD increased insoluble actin levels 3-fold (Fig. 4A and B). This is consistent with decreased actin dynamics in cells expressing MARCKSL1-DDD (Fig. 3). To determine if active JNK also stabilized actin, we expressed dominant active JNK chimeras (MKK7-JNK1/2) together with MARCKSL1-WT (Fig. 4C). Active JNK increased the amount of stable actin in cells expressing MARCKSL1 (Fig. 4C). Significantly, however, in cells lacking MARCKSL1, active JNK did not alter actin stability (Fig. 4D). These results indicate that JNK regulation of actin is mediated by MARCKSL1.
Fig 4.
JNK phosphorylation of MARCKSL1 increases actin stability. (A) To determine the effect of JNK phosphorylation on stable actin levels, MEFs expressing venus-actin in the presence or absence of GFP-tagged MARCKSL1s (or GFP-cofilin) were treated with digitonin to extract soluble proteins with minimal loss of cell membrane. Total lysate and pellet following extraction were blotted for actin and GFP as shown. (B) Quantified data from panel A are shown. (C) To determine the effect of JNK activation on actin stability, MEFs were transfected with venus-actin, MARCKSL1-GFP, and active JNK chimera MKK7-JNK1 or MKK7-JNK2 as shown. JNK activators facilitate MARCKSL1-GFP stabilization of actin. A representative blot of lysate following digitonin extraction is shown (inset). (D) Expression of active JNK chimeras in the absence of MARCKSL1 did not alter actin stability. Mean data ± SEM are shown. Significance levels: *, P < 0.05; ***, P < 0.001.
Expression of MARCKSL1 increases migration in MEFs, and active JNK or MARCKSL1S120D,T148D,T183D is inhibitory.
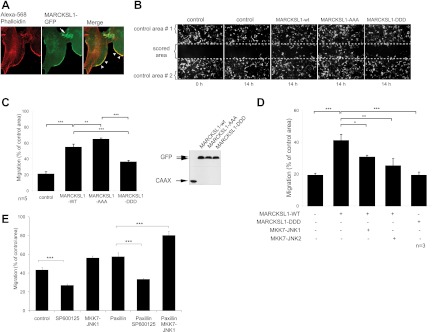
Remodeling of actin at the leading edge of cells is a key driving force for cell migration (47). We observed that MARCKSL1-GFP frequently colocalized with F-actin at the leading edge in MEFs and enriched at points of cell-cell contact (Fig. 5A). This prompted us to analyze cell migration. Wound-healing assays were carried out in cells transfected with GFP-CAAX to label the plasma membrane and GFP-tagged MARCKSL1 variants. Interestingly, expression of MARCKSL1-WT increased migration and expression of MARCKSL1-AAA induced migration even more than expression of MARCKSL1-WT. Conversely, MARCKSL1-DDD significantly reduced migration relative to MARCKSL1-WT (Fig. 5B and C). These data indicate that MARCKSL1 induces migration unless phosphorylated on JNK sites S120, T148, and T183, where it restricts cell movement. To test whether this switch was a JNK-dependent event, we examined cells expressing the dominant active JNK chimeras together with MARCKSL1-WT. The constitutively active JNK chimeras retarded migration to almost the same extent as MARCKSL1-DDD (Fig. 5D).
Fig 5.
Activation of JNK or expression of a JNK site-phosphomimicking MARCKSL1 mutant reduces MEF cell migration. (A) Fluorescent micrographs of MEF cells transfected with MARCKSL1-GFP (green) and stained with phalloidin (red). MARCKSL1 colocalizes with actin in lamellipodia and at cell-cell junctions. (B) To test whether MARCKSL1 regulated cell migration, MEFs were transfected with GFP-CAAX (control) and GFP-tagged MARCKSL1-WT or phosphorylation mutants. Wound-healing assays were carried out and images taken at 0 h or 14 h after scoring. Total cell density was monitored from Alexa 546-phalloidin staining at h 0 (not shown). (C) The number of transfected cells that migrated into the scored region is expressed as a proportion of cells in the adjacent control area. MARCKSL1S120A,S148A,S183A increased migration, while MARCKSL1S120D,S148D,S183D inhibited it. A Western blot shows the integrity of constructs used. (D) Cells were transfected as indicated and migration measured as described for panel C. In cells expressing MARCKSL1, the active MKK7-JNK1 chimera decreased migration, as did MARCKSL1S120D,S148D,S183D. (E) The effect of JNK activation on paxillin-induced migration was examined. Cells were treated with SP600125 (10 μM) to inhibit JNK activity. In cells expressing paxillin, JNK facilitated migration. Significance levels: *, P < 0.05, **, P < 0.01; ***, P < 0.001.
A substantial literature indicates that JNK facilitates cell migration, at least in nonneuronal cells (15, 16, and 18; reviewed in reference 52). Similarly, in our hands, the JNK inhibitor SP600125 reduced migration of fibroblasts, while active JNK increased it, so long as MARCKSL1 was not present (Fig. 5D and E). Likewise, when we expressed GFP-paxillin, an alternative JNK substrate, SP600125, slowed migration and MKK7-JNK1 increased it. Our data show that MARCKSL1, a protein predominantly expressed in brain, switched the migration phenotype from one facilitated by JNK to one inhibited by JNK, possibly contributing to the negative role played by JNK in radial migration during cortical development (48).
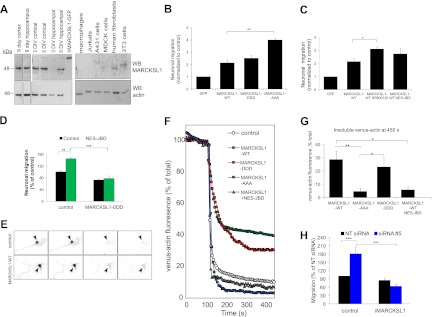
Expression of MARCKSL1S120D,T148D,T183D stabilizes actin and inhibits migration in neurons.
MARCKSL1 mRNA levels are high in brain during early postnatal weeks and low in other organs (24, 27). Moreover, MARCKSL1 is often considered to be widely expressed in different cell types. To determine the status of MARCKSL1 protein in brain, we immunoblotted brain tissues, neuronal lysates, and nonneuronal cell lines using MARCKSL1 antibody that detects both rodent and human MARCKSL1 (Fig. 6A). MARCKSL1 protein levels were relatively high in brain (cortex and hippocampus) and enriched in neurons isolated from these regions (cortical neurons, hippocampal neurons). However, we did not detect MARCKSL1 in unstimulated macrophages, Jurkat cells, fibroblasts, or epithelial cell lines (Fig. 6A). Given that MARCKSL1 is predominant in neuronal cells, we examined the effect of phosphorylation site mutations on migration of neurons in Transwells (Fig. 6B and C). MARCKSL1-DDD reduced migration relative to MARCKSL1-AAA, as observed in fibroblasts (Fig. 5C). In contrast to fibroblasts, however, neurons expressing MARCKSL1-WT mimicked MARCKSL1-DDD. This was likely due to phosphorylation of MARCKSL1-WT by endogenous JNK activity that was elevated in neurons but not in other cell types (7, 8, 42). Consistent with this, inhibition of endogenous JNK with the JNK inhibitor SP600125 or with NES-JBD, a structurally independent inhibitor, increased migration in MARCKSL1-WT-expressing neurons (Fig. 6C). Moreover, we found that pseudophosphorylated MARCKSL1-DDD recovered a normal migration phenotype in cells where JNK activity was inhibited (Fig. 6D), indicating once again that phosphorylation of MARCKSL1 by JNK retards migration, whereas preventing phosphorylation of MARCKSL1 on the JNK sites enhances it.
Fig 6.
C-terminal phosphorylation of MARCKSL1 inhibits neuronal migration. (A) Immunoblotting of brain tissues, neuronal cultures, and human cell lines for MARCKSL1. hMARCKSL1-GFP was added as a positive control to demonstrate antibody species specificity. (B) The effect of MARCKSL1 on neuronal migration in 1-day in vitro neurons using Transwell chambers. Neurons were transfected with MARCKSL1-GFP constructs as indicated. The number of transfected neurons migrating through the membrane was counted 24 h posttransfection. (C) Neurons were transfected with MARCKSL1-GFP with or without JNK inhibitor SP600125 (3 μM) or GFP-NES-JBD. Neuronal migration was measured as described for panel A. (D) Neuronal migration was measured in Transwells from cells expressing dsRed2 in the presence or absence of GFP-NES-JBD or MARCKSL1-DDD as indicated. (E) Actin stability in neurons was measured in neurons transfected with CFP-tagged MARCKSL1s together with venus-actin as described here, and venus-actin fluorescence loss after Triton X-100 permeabilization was measured over time. Representative images and (F) raw traces showing fluorescence loss from the YFP channel over time are shown. (G) Venus-actin fluorescence remaining at 450 s postpermeabilization is shown. (H) MARCKSL1 expression in neurons was knocked down using siRNA 5 or nontargeting (NT) siRNA. Then, neuronal migration was assessed in Transwells in the absence or presence of iMARCKSL1 (siRNA-insensitive mutant). Data represent means ± SEM. Significance levels: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We next tested whether MARCKSL1 regulated actin stability in neurons using time-lapse fluorescence imaging. Neurons transfected with venus-actin and MARCKSL1-CFP or phosphorylation site mutants of MARCKSL1 were permeabilized using Triton X-100. Loss of venus-actin upon plasma membrane permeabilization was visualized as a loss of venus-actin fluorescence (Fig. 6E and F). The steady-state fluorescence that ensued some minutes after permeabilization was taken to be a representation of stable actin, with the lost fluorescence indicating soluble actin. In control neurons expressing venus-actin only, permeabilization led to a rapid and almost total loss of venus-actin (Fig. 6F and G). Expression of MARCKSL1-WT or MARCKSL1-DDD increased the amount of stable actin by 2.5-fold (Fig. 6G). We then inhibited high neuronal JNK activity by expressing the cytosolic JNK inhibitor (GFP-NES-JBD). Under these conditions, MARCKSL1-WT no longer stabilized actin (Fig. 6G). These results indicate that MARCKSL1-WT can stabilize actin in neurons only when phosphorylated by JNK.
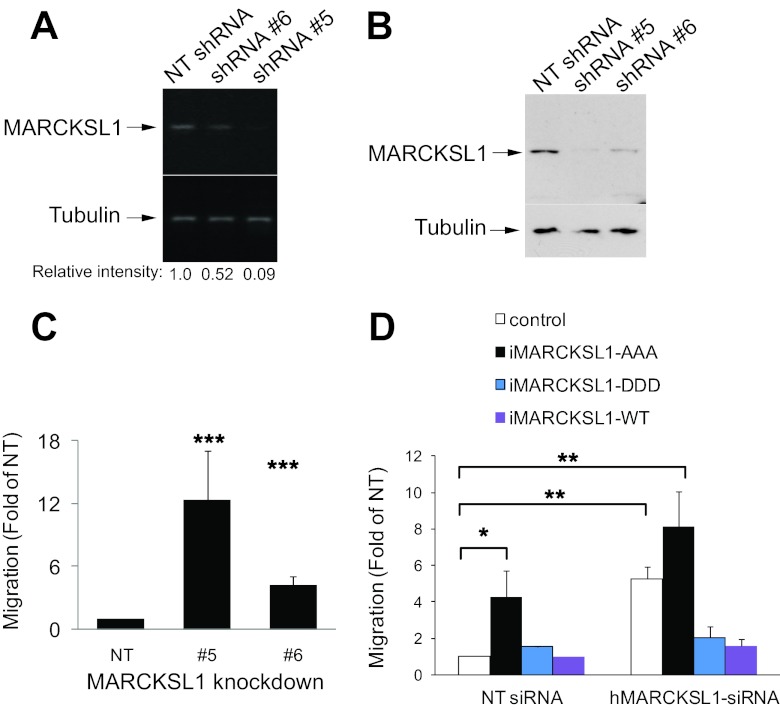
To determine if MARCKSL1 is a critical player in neuronal migration, we knocked down MARCKSL1 expression in cortical neurons and measured migration in Transwells (Fig. 6H; see also Fig. 9B, where the efficiency of knockdown performed using siRNA 5 is shown). Knockdown of neuronal MARCKSL1 increased migration. Notably, a normal migration phenotype was recovered upon expression of iMARCKSL1 (a MARCKSL1 mutant that is insensitive to siRNA 5). Together, the results in Fig. 6 show that MARCKSL1 is a critical regulator of neuronal migration. The repressor function that MARCKSL1 plays during migration is dependent on its phosphorylation by JNK.
Fig 9.
C-terminal phosphorylation of MARCKSL1 inhibits migration in PC-3 cells. (A) Stable knockdown lines of PC-3 cells were generated using nontargeting shRNA (NT) or MARCKSL1 shRNA 5 or 6. The relative silencing of MARCKSL1 is shown. (B) Immunoblots of cell line lysates (NT, cell line 5, and cell line 6), blotted for MARCKSL1. The levels of MARCKSL1 protein in NT and knockdown cell lines were also measured. (C) Migration of NT and knockdown cell lines 5 and 6 was examined in Transwells. Knockdown of MARCKSL1 with shRNA 5 led to increased migration. (D) The influence of shRNA-insensitive MARCKSL1 (iMARCKSL1s) on migration was examined. Control blots showing iMARCKSL1 mutant expression are shown in Fig. S2 in the supplemental material. Readdition of iMARCKSL1-DDD rescued a normal migration phenotype in cells expressing the shRNA 5 phenotype. Data represent means ± SEM. Significance levels: *, P < 0.05; **, P < 0.005; ***, P < 0.001.
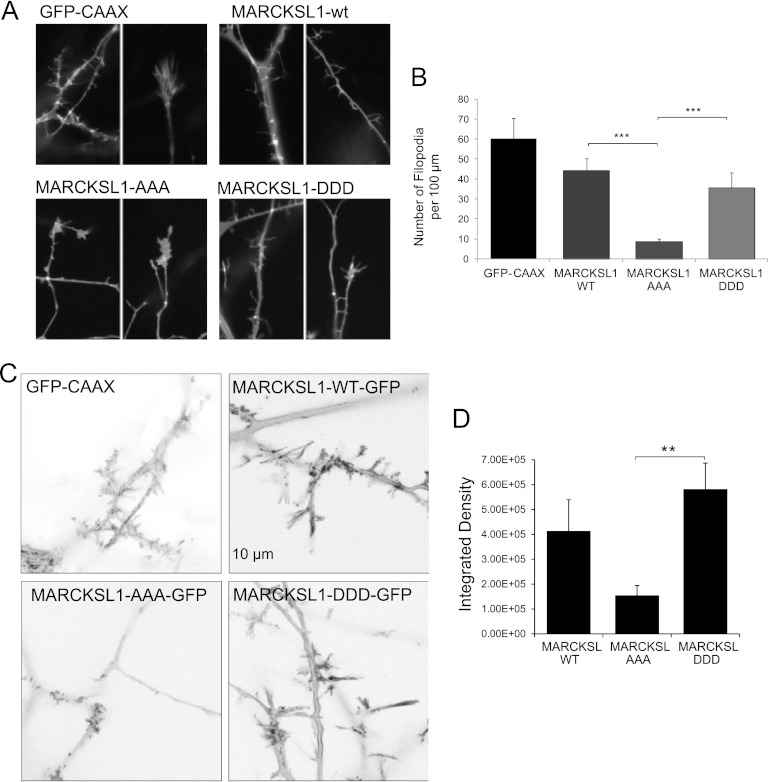
Phosphorylation of MARCKSL1S120,T148,T183 determines dendritic filopodium generation in neurons, and MARCKSL1-AAA promotes formation of lamellae.
Several actin structures govern forward movement of cells, among these filopodia and lamelipodia (54). We therefore examined the effect of MARCKSL1 phosphorylation variants on cell morphology in cortical neurons (Fig. 7). Neurons expressing MARCKSL1-WT or MARCKSL1-DDD generated filopodia along the dendrites that were notably longer and more prominent than those in control cells expressing GFP-CAAX (Fig. 7A and B). This difference can also be observed from the time-lapse recordings (see Fig. 7; see also movies in the supplemental material). The observation of prominent filopodia upon expression of MARCKSL1-DDD is not surprising, given that MARCKSL1-DDD bundles actin (Fig. 2B) and that actin cross-linking is required for filopodium formation (38). Strikingly, however, cells expressing MARCKSL1-AAA largely failed to generate filopodia (Fig. 7A and B; see also movies in the supplemental material). These cells generated tiny protrusions along the neurites, but these protrusions remained transient and failed to develop into stable filopodia. Instead, MARCKSL1S120A,T148A,T183A-expressing cells generated large, flat lamellipodial protrusions. This is consistent with the enhanced migration observed in MARCKSL1S120A,T148A,T183A-expressing cells (Fig. 6).
Fig 7.
JNK phosphorylation sites on MARCKSL1 are important for filopodium formation and motility. (A) Snapshots of cortical neurons expressing MARCKSL1-GFP phosphorylation site variants are shown. Cells expressing MARCKSL1-AAA typically displayed large lamellipodia and very few if any filopodium-enriched regions. Conversely, cells expressing MARCKSL1-DDD displayed prominent and long filopodia. Time-lapse movies of these fields of view are available in the supplemental material. (B) Quantitative data on filopodial numbers from neurons expressing MARCKSL1-WT, MARCKSL1-AAA, or MARCKSL1-DDD are shown. (C) To quantify filopodial dynamics, maximum-intensity projections representing sequentially subtracted images were used to provide an index of motility. Increased intensity reflects increased movements. Cells expressing MARCKSL1-DDD or the WT showed greater filopodial dynamics. (D) Quantitative data depicting integrated densities from multiple maximum-intensity projections, as depicted in panel C, are shown. Data represent means ± SEM. Significance levels: **, P < 0.01; ***, P < 0.001.
To quantify the extent of filopodial dynamics, we used a method described by Fischer and Kaech wherein a sequence of time-lapse images are sequentially subtracted, with the remaining pixels providing a measure of global dynamics. Averaged projections of global dynamics are shown in Fig. 7C for cells expressing GFP-CAAX and MARCKSL1 variants. MARCKSL1-WT and MARCKSL1S120D,T148D,T183D projections reveal high pixel intensity (representing high motility) in the filopodial regions, while MARCKSL1S120A,T148A,T183A intensity projections show little filopodial movement. The integrated density quantified in Fig. 7D illustrates the substantial difference in global dynamics measured in the processes of MARCKSL1S120D,T148D,T183D- and MARCKSL1S120A,T148A,T183A-expressing cells. Quantitation of integrated density from GFP-CAAX-expressing cells was precluded by the large level of vesicular movements (GFP-CAAX labels cytosolic vesicles in addition to the plasma membrane).
MARCKSL1 gene expression is significantly deregulated in a wide range of cancers.
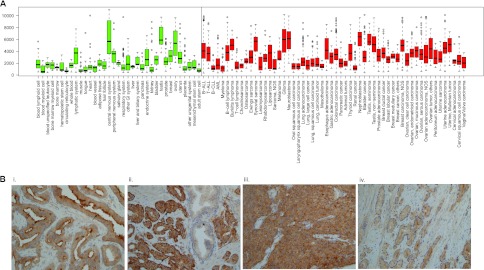
MARCKSL1 was recently reported to be anomalously upregulated in breast cancer (45). To determine whether irregular MARCKSL1 expression is a feature of other cancers and, indeed, to determine the extent of MARCKSL1 deregulation in breast cancer, we searched the In Silico Transcriptomics (IST) database. The data indicated that MARCKSL1 expression is deregulated in a wider range of cancers than previously appreciated (Fig. 8A). In normal tissues, MARCKSL1 mRNA was highly expressed in the central nervous system, testis, ovary, and lymphatic organs. In addition to breast cancer (fold change, 1.44; P < 0.0279), there was significant upregulation of MARCKSL1 in lung cancer (fold change, 1.729; P < 0.0001), muscle-derived cancer (fold change, 3.312; P < 0.0001, including rhabdomyosarcoma and leiomyosarcoma), and uterine cancer (fold change, 1.426; P < 0.0011). In Wilms's tumor samples, MARCKSL1 was also expressed at high levels and in prostate adenocarcinoma (fold change, 1.444; P < 0.0001). This is consistent with reports from others of MARCKSL1 upregulation in prostate cancer (11, 34).
Fig 8.
MARCKSL1 expression is deregulated in a broad range of cancers, including prostate cancer, where it shows prominent apical and basal plasma membrane staining. (A) Box-plot presentation of normalized median expression levels for MARCKSL1 mRNA across major normal tissues (green) and cancers (red). The box extends to the third quartile of the data, and the median is indicated. Whiskers extend to the extreme values unless there are outliers (data that lie >1.5× the interquartile range lower than the first quartile or 1.5× the interquartile range higher than the third quartile), which are indicated as circles. MARCKSL1 is highly expressed in the central nervous system, testis, and ovary among normal tissues, whereas in cancers, MARCKSL1 mRNA is more widely expressed. The minimum number of samples for each group is set to 8. Abbreviations: other GI, other gastrointestinal tissues; B-ALL, precursor B acute lymphoblastic leukemia; T-ALL, adult T-cell leukemia; B-CLL, B-cell chronic lymphocytic leukemia; AML, acute myeloid leukemia; NOS, not otherwise specified. (B) Primary prostate carcinomas were stained for MARCKSL1. MARCKSL1 displayed clear basal and apical immunoreactivity in Gleason G3 carcinomas. Noncancerous regions were negative for MARCKSL1. (i) Gleason G3 prostate carcinoma, cytoplasmic and strong apical membrane staining; (ii) Gleason G3 prostate carcinoma, cytoplasmic and strong basal membrane staining; (iii) Gleason G4 prostate carcinoma, cytoplasmic staining; (iv) Gleason G3 prostate carcinoma, cytoplasmic staining. *, normal glands (MARCKSL1 negative).
MARCKSL1 protein expression is high and distinctly localized in prostate carcinomas.
Given that MARCKS1 transcript is upregulated in prostate cancer (Fig. 8), we examined whether MARCKSL1 protein expression is induced in primary prostate carcinomas (Fig. 8B). Gleason grade 3 pattern carcinomas displayed cytoplasmic and strongly positive apical membrane staining (Fig. 8B, panel i; arrows, large lumens) or strong basal membrane immunoreactivity for MARCKSL1 (Fig. 8B, panel ii; arrows, small lumens). Notably, the normal glands stained negative for MARCKSL1 (Fig. 8B, panel ii; asterisk). High-grade, Gleason grade 4 prostate carcinomas with a solid growth pattern displayed prominent MARCKSL1 immunoreactivity in the cytoplasm (Fig. 8B, panel iii).
MARCKSL1 is a critical determinant of PC3 cell migration, and C-terminal phosphorylation is inhibitory.
Having demonstrated that MARCKSL1 protein is upregulated in primary prostate carcinomas, we investigated whether MARCKSL1 functioned in prostate cancer cell migration and, in particular, whether C-terminal phosphorylation would influence this event. We therefore generated two stable knockdown lines of PC-3 cells (lines 5 and 6), showing strong and moderate knockdown, respectively, of MARCKSL1 mRNA and protein (Fig. 9A and B). With both knockdown lines, the migration rate was significantly increased (Fig. 9C). To determine the influence of MARCKSL1 phosphorylation on migration in PC-3 cells, we used shRNA-insensitive MARCKSL1 mutants where JNK phosphorylation sites were mutated to aspartate (iMARCKSL1-DDD) or alanine (iMARCKSL1-AAA), mimicking phospho- and dephospho-MARCKSL1, respectively (Fig. 9D). Readdition of iMARCKSL1-DDD reduced migration, thereby recovering a normal migration phenotype. These results suggest that MARCKSL1 is a critical regulator of migration in PC3 cells. Dephosphorylated MARCKSL1 (MARCKSL1-AAA) enhances migration, while phosphorylation on S120, T148, and T183 switches this function to a repressor role.
DISCUSSION
This report identifies MARCKSL1 as a novel JNK substrate that regulates the dynamics of the actin cytoskeleton in mammalian cells. We classify three JNK phosphorylation sites in the C-terminal region of MARCKSL1. Phosphomimetics of C-terminal-phosphorylated MARCKSL1 stabilize and increase bundling of F-actin. The functional consequences of MARCKSL1 phosphorylation for these sites are a reduction of neuronal migration and alterations in filopodium morphology and dynamics. We also show that MARCKSL1 mRNA is ectopically expressed in a wide range of cancers and that C-terminal phosphorylation of MARCKSL1 regulates the migration rate in prostate cancer cells. These data provide a novel mechanism whereby phosphorylation of MARCKSL1 regulates actin bundling, leading to altered cell migration in contexts as divergent as neuronal and cancer cell migration.
MARCKSL1 as a substrate for PKC has been well studied (44; reviewed in reference 2). PKC phosphorylation sites are conserved in MARCKSL1 and MARCKS and reside within the effector domain (ED). While PKC phosphorylation of MARCKSL1 inhibits actin cross-linking (14), we show that JNK phosphorylates MARCKSL1 on unique C-terminal sites and facilitates actin bundling. This bundling (Fig. 2B) resembles that induced by MARCKS (14) and cofilin (31). Phosphorylation of MARCKSL1 on S120, T148, and T183 may be necessary for bundling, as MARCKSL1S120A,T148A,T183A binds but fails to bundle F-actin. These results may resolve the paradoxical finding that the MARCKSL1 ED alone facilitates actin bundling whereas full-length MARCKSL1 does not (49, 50).
Precisely how MARCKSL1 facilitates actin bundling is not clear. However, in the absence of crystallographic data, we carried out molecular modeling, which indicated that the JNK phosphorylation sites on MARCKSL1 are surface accessible and surround the ED (see Fig. S3 in the supplemental material). Upon phosphorylation, the negative charges introduced to these sites likely induce a conformational change in MARCKSL1 which may expose the polycationic ED. Unshielding of the ED would in turn enable lateral alignment of actin filaments (i.e., bundling) due to an electrostatic mechanism described by Tang and Janmey (41). In support of this model (see Fig. S3 in the supplemental material), the positively charged lysines and arginines in the N-terminal, central, and C-terminal regions of the MARCKSL1 ED are known to be essential for bundling (50). The functional consequences of JNK phosphorylation of MARCKSL1 are increased rigidity in the actin cytoskeleton and retarded migration, while MARCKSL1S120A,T148A,T183A increases actin dynamics and advances migration (Fig. 2 to 6). The mechanism of MARCKSL1S120A,T148A,T183A action appears to involve binding to actin filaments, thereby causing dispersal of the actin network in a way that promotes actin plasticity and forward movement of cells.
How might these changes in actin stability result in altered cell migration? Cell motility universally requires protrusion at the leading edge. We show that MARCKSL1S120A,T148A,T183A-expressing cortical neurons generate unusually flat lamellipodia that are highly motile. Lamellipodia are characterized by an actin network consisting of short, branched actin, and protrusion force is generated by actin treatmilling at the plasma membrane (33). The reduced rigidity and increased actin turnover in MARCKSL1S120A,T148A,T183A-expressing cells (Fig. 3) may facilitate their enhanced migration. Alternatively, it may be that the increase in numbers of dendritic filopodia, in neurons expressing MARCKSL1S120D,T148D,T183D, hinders forward movement by amplifying the number of adhesion sites. Moreover, neurons expressing MARCKSL1-DDD generate long filopodial extensions along the length of the dendrites. This kind of dendritic extension of filopodia was not observed in cells expressing GFP-CAAX or MARCKSL1-AAA. Thus, C-terminal-phosphorylated MARCKSL may be necessary for initiation of new dendritic arbors. Consistent with this, we have previously shown that JNK1 regulates the dendritic architecture in brain. On the other hand, these protrusions may indicate a role for C-terminal-phosphorylated MARCKSL1 in the formation of immature dendritic spines.
We show that neurons provide the appropriate cellular environment for MARCKSL1 regulation of actin and migration. Neurons express high levels of MARCKSL1 relative to other cells (Fig. 6A), and JNK activity is elevated there (8), providing a mechanism for homeostatic regulation of actin remodeling. Interestingly, during early brain development, both MARCKSL1 and JNK play critical roles during neurulation and mice lacking either MARCKSL1 or JNK1 and -2 fail to complete neural tube closure, exhibiting neural tube defects that include exencephaly (5, 20, 36, 51). It is notable that among the JNK substrates reported to date, MARCKSL1 is the only example where targeted deletion of the kinase substrate phenocopies targeted deletion of JNK1 and -2. Cell spreading and filopodial protrusion are conserved mechanisms that are known to be necessary for fusion of epithelial sheets during dorsal closure (26) and expected to be essential during neural tube closure. Here, we show that MARCKSL1 induces migration of fibroblasts (Fig. 5A) and alters filopodium formation and motility when expressed in neurons (Fig. 7). The common neural tube defects exhibited by MARCKSL1−/− and JNK1−/−2−/− mice suggest that these molecules share a pathway during early development of the central nervous system. The neuronal migration phenotype observed upon knockdown of MARCKSL1 (Fig. 6G) may signify a role for MARCKSL1 also at later stages of brain development, when waves of neurons migrate to final destinations. Certainly, JNK1 negatively regulates radial migration and multipolar stage transition, the tubulin destabilizing factor SCG10 being an important JNK1 effector (48). MARCKSL1 could also play a role. The MARCKSL1 homolog MARCKS has already been implicated in regulation of cortical lamination (46).
While the physiological function of MARCKSL1 has begun to be revealed, the significance of ectopically expressed MARCKSL1 in cancers is not yet clear but may play a role in cancer cell invasion, given the MARCKSL1 regulation of migration (Fig. 5 and 6) and adhesion (11). Induction of MARCKSL1 in breast cancer has been reported previously (45). Here we demonstrate that MARCKSL1 is more widely expressed in cancers than previously appreciated (Fig. 8) and that MARCKSL1 protein is ectopically expressed in 100% of the screened high-grade primary prostate carcinomas. We show that MARCKSL1 increases migration in PC3 cells when in the dephosphorylated form, while C-terminal phosphorylation of MARCKSL1 is inhibitory (Fig. 9). Our data indicate that the resultant effect of MARCKSL1 on cell migration, be it facilitatory or inhibitory, is dependent on the C-terminal phosphorylation of MARCKSL1. This phosphorylation may be pivotal in cancers, where MARCKSL1 could theoretically enhance or retard cell motility. Indeed, we find that increased MARCKSL1 expression in breast cancer is correlated with an increased risk for distant metastasis (16a). Understanding the significance of MARCKSL1 upregulation in cancers naturally requires more study; however, our data suggest that the phosphorylation status is critical to determining the outcome in terms of migration.
An interesting outcome of this study is the demonstration that JNK activity can either promote or hinder migration in a single cell type, merely by utilizing different effector proteins. Accordingly, when paxillin is expressed in fibroblasts, JNK facilitates migration; however, when MARCKSL1 is expressed, JNK inhibits migration (Fig. 6). It is worth emphasizing that MARCKSL1 protein is enriched in the brain and in neurons (Fig. 7A), extending indications that this was the case from in situ hybridization studies (27). We recently showed that JNK1 retards migration in neurons via the microtubule regulator SCG10 (48). Here we describe MARCKSL1 as an additional substrate that negatively regulates migration when phosphorylated. Indeed, we find that expression of MARCKSL1 in nonneuronal cells switches the mode of migration from a nonneuronal cell phenotype (where JNK inhibits migration) to a neuronal cell phenotype (where JNK facilitates migration). We propose that the mechanism involves the acquired capacity of MARCKSL1 to fix the actin cytoskeleton once phosphorylated by JNK, thereby impeding the forward movement of cells.
Supplementary Material
ACKNOWLEDGMENTS
The work was funded by the Academy of Finland (grants 206497, 125860, and 218125 to E.T.C.); Åbo Akademi University, Åbo Akademi University Foundation, Magnus Ehrnrooths Foundation, the Sigrid Juselius Foundation, the National Graduate School in Information and Structural Biology, the Turku Graduate School of Biomedical Sciences, the Sigrid Juselius Foundation, the Molecular Medicine graduate school, K. Albin Johanssons Stiftelse, and CIMO provided financial support to B.B., A.P., N.W., L.P., and H.M.
We thank Jukka Westermarck for performing pilot experiments with cancer cell lines and Olli Carpen for guidance on cancer tissue analysis. We thank J. P. Slotte for use of the Cary Eclipse spectrophotometer and J. Meriluoto for Microcystin-LR. We acknowledge the Turku Centre for Biotechnology Cell Imaging Core and Proteomics unit, the Multimodal Imaging Centre, and the BioMater Centre at the University of Eastern Finland for TEM analysis.
Footnotes
Published ahead of print 2 July 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Aderem A. 1992. The MARCKS brothers: a family of protein kinase C substrates. Cell 71: 713– 716 [DOI] [PubMed] [Google Scholar]
- 2. Arbuzova A, Schmitz AA, Vergères G. 2002. Cross-talk unfolded: MARCKS proteins. Biochem. J. 362: 1– 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Björkblom B, et al. 2005. Constitutively active cytoplasmic c-Jun N-terminal kinase 1 is a dominant regulator of dendritic architecture: role of microtubule-associated protein 2 as an effector. J. Neurosci. 25: 6350– 6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang L, Jones Y, Ellisman MH, Goldstein LS, Karin M. 2003. JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev. Cell 4: 521– 533 [DOI] [PubMed] [Google Scholar]
- 5. Chen J, et al. 1996. Disruption of the MacMARCKS gene prevents cranial neural tube closure and results in anencephaly. Proc. Natl. Acad. Sci. U. S. A. 93: 6275– 6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ciani L, Salinas PC. 2007. c-Jun N-terminal kinase (JNK) cooperates with Gsk3beta to regulate Dishevelled-mediated microtubule stability. BMC Cell Biol. 8: 27 doi:10.1186/1471-2121-8-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coffey ET, Courtney MJ. 1997. Regulation of SAPKs in CNS neurons. Biochem. Soc. Trans. 25: S568. [DOI] [PubMed] [Google Scholar]
- 8. Coffey ET, Hongisto V, Dickens M, Davis RJ, Courtney MJ. 2000. Dual roles for c-Jun N-terminal kinase in developmental and stress responses in cerebellar granule neurons. J. Neurosci. 20: 7602– 7613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cooper JA, Walker SB, Pollard TD. 1983. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 4: 253– 262 [DOI] [PubMed] [Google Scholar]
- 10. Reference deleted. [Google Scholar]
- 11. Finlayson AE, Freeman KW. 2009. A cell motility screen reveals role for MARCKS-related protein in adherens junction formation and tumorigenesis. PLoS One 4: e7833 doi:10.1371/journal.pone.0007833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fischer M, Kaech S, Knutti D, Matus A. 1998. Rapid actin-based plasticity in dendritic spines. Neuron 20: 847– 854 [DOI] [PubMed] [Google Scholar]
- 13. Gdalyahu A, et al. 2004. DCX, a new mediator of the JNK pathway. EMBO J. 23: 823– 832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hartwig JH, et al. 1992. MARCKS is an actin filament crosslinking protein regulated by protein kinase C and calcium-calmodulin. Nature 356: 618– 622 [DOI] [PubMed] [Google Scholar]
- 15. Huang C, Rajfur Z, Borchers C, Schaller MD, Jacobson K. 2003. JNK phosphorylates paxillin and regulates cell migration. Nature 424: 219– 223 [DOI] [PubMed] [Google Scholar]
- 16. Javelaud D, Laboureau J, Gabison E, Verrecchia F, Mauviel A. 2003. Disruption of basal JNK activity differentially affects key fibroblast functions important for wound healing. J. Biol. Chem. 278: 24624– 24628 [DOI] [PubMed] [Google Scholar]
- 16a. Jonsdottir K., et al. The prognostic value of MARCKS-like1 in lymph node negative breast cancer. Breast Cancer Res. Treat., in press. [DOI] [PubMed] [Google Scholar]
- 17. Karin M, Gallagher E. 2005. From JNK to pay dirt: jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life 57: 283– 295 [DOI] [PubMed] [Google Scholar]
- 18. Kavurma MM, Khachigian LM. 2003. ERK, JNK, and p38 MAP kinases differentially regulate proliferation and migration of phenotypically distinct smooth muscle cell subtypes. J. Cell Biochem. 89: 289– 300 [DOI] [PubMed] [Google Scholar]
- 19. Kilpinen S, et al. 2008. Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol. 9: R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuan CY, et al. 1999. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron 22: 667– 676 [DOI] [PubMed] [Google Scholar]
- 21. Kwon HJ, et al. 2006. Anisotropic nucleation growth of actin bundle: a model for determining the well-defined thickness of bundles. Biochemistry 45: 10313– 10318 [DOI] [PubMed] [Google Scholar]
- 22. Kyriakis JM, Avruch J. 1990. pp54 microtubule-associated protein 2 kinase. A novel serine/threonine protein kinase regulated by phosphorylation and stimulated by poly-L-lysine. J. Biol. Chem. 265: 17355– 17363 [PubMed] [Google Scholar]
- 23. Li J, Zhu Z, Bao Z. 1996. Role of MacMARCKS in integrin-dependent macrophage spreading and tyrosine phosphorylation of paxillin. J. Biol. Chem. 271: 12985– 12990 [DOI] [PubMed] [Google Scholar]
- 24. Lobach DF, Rochelle JM, Watson ML, Seldin MF, Blackshear PJ. 1993. Nucleotide sequence, expression, and chromosomal mapping of Mrp and mapping of five related sequences. Genomics 17: 194– 204 [DOI] [PubMed] [Google Scholar]
- 25. Malchinkhuu E, et al. 2005. Role of p38 mitogen-activated kinase and c-Jun terminal kinase in migration response to lysophosphatidic acid and sphingosine-1-phosphate in glioma cells. Oncogene 24: 6676– 6688 [DOI] [PubMed] [Google Scholar]
- 26. Martín-Blanco E, Knust E. 2001. Epithelial morphogenesis: filopodia at work. Curr. Biol. 11: R28– R31 [DOI] [PubMed] [Google Scholar]
- 27. McNamara RK, Lenox RH. 1998. Distribution of the protein kinase C substrates MARCKS and MRP in the postnatal developing rat brain. J. Comp. Neurol. 397: 337– 356 [PubMed] [Google Scholar]
- 28. Morfini GA, et al. 2009. Pathogenic huntingtin inhibits fast axonal transport by activating JNK3 and phosphorylating kinesin. Nat. Neurosci. 12: 864– 871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nishida E, Maekawa S, Sakai H. 1984. Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry 23: 5307– 5313 [DOI] [PubMed] [Google Scholar]
- 30. Oliva AA, Atkins CM, Copenagle L, Banker GA. 2006. Activated c-Jun N-terminal kinase is required for axon formation. J. Neurosci. 26: 9462– 9470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pfannstiel J, et al. 2001. Human cofilin forms oligomers exhibiting actin bundling activity. J. Biol. Chem. 276: 49476– 49484 [DOI] [PubMed] [Google Scholar]
- 32. Phair RD, Gorski SA, Misteli T. 2004. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375: 393– 414 [DOI] [PubMed] [Google Scholar]
- 33. Pollard TD, Borisy GG. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112: 453– 465 [DOI] [PubMed] [Google Scholar]
- 34. Romanuik TL, et al. 2009. Novel biomarkers for prostate cancer including noncoding transcripts. Am. J. Pathol. 175: 2264– 2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rush S, et al. 2007. c-jun amino-terminal kinase and mitogen activated protein kinase 1/2 mediate hepatocyte growth factor-induced migration of brain endothelial cells. Exp. Cell Res. 313: 121– 132 [DOI] [PubMed] [Google Scholar]
- 36. Sabapathy K, et al. 1999. Defective neural tube morphogenesis and altered apoptosis in the absence of both JNK1 and JNK2. Mech. Dev. 89: 115– 124 [DOI] [PubMed] [Google Scholar]
- 37. Schönwasser DC, Palmer RH, Herget T, Parker PJ. 1996. p42 MAPK phosphorylates 80 kDa MARCKS at Ser-113. FEBS Lett. 395: 1– 5 [DOI] [PubMed] [Google Scholar]
- 38. Small JV, Stradal T, Vignal E, Rottner K. 2002. The lamellipodium: where motility begins. Trends Cell Biol. 12: 112– 120 [DOI] [PubMed] [Google Scholar]
- 39. Sundaram M, Cook HW, Byers DM. 2004. The MARCKS family of phospholipid binding proteins: regulation of phospholipase D and other cellular components. Biochem. Cell Biol. 82: 191– 200 [DOI] [PubMed] [Google Scholar]
- 40. Takino T, et al. 2005. JSAP1/JIP3 cooperates with focal adhesion kinase to regulate c-Jun N-terminal kinase and cell migration. J. Biol. Chem. 280: 37772– 37781 [DOI] [PubMed] [Google Scholar]
- 41. Tang JX, Janmey PA. 1998. Two distinct mechanisms of actin bundle formation. Biol. Bull. 194: 406– 408 [DOI] [PubMed] [Google Scholar]
- 42. Tararuk T, et al. 2006. JNK1 phosphorylation of SCG10 determines microtubule dynamics and axodendritic length. J. Cell Biol. 173: 265– 277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thingholm TE, Jensen ON, Robinson PJ, Larsen MR. 2008. SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol. Cell. Proteomics 7: 661– 671 [DOI] [PubMed] [Google Scholar]
- 44. Verghese GM, et al. 1994. Protein kinase C-mediated phosphorylation and calmodulin binding of recombinant myristoylated alanine-rich C kinase substrate (MARCKS) and MARCKS-related protein. J. Biol. Chem. 269: 9361– 9367 [PubMed] [Google Scholar]
- 45. Wang J, Jarrett J, Huang CC, Satcher RL, Levenson AS. 2007. Identification of estrogen-responsive genes involved in breast cancer metastases to the bone. Clin. Exp. Metastasis 24: 411– 422 [DOI] [PubMed] [Google Scholar]
- 46. Weimer JM, et al. 2009. MARCKS modulates radial progenitor placement, proliferation and organization in the developing cerebral cortex. Development 136: 2965– 2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Welch MD, Mallavarapu A, Rosenblatt J, Mitchison TJ. 1997. Actin dynamics in vivo. Curr. Opin. Cell Biol. 9: 54– 61 [DOI] [PubMed] [Google Scholar]
- 48. Westerlund N, et al. 2011. Phosphorylation of SCG10/stathmin-2 determines multipolar stage exit and neuronal migration rate. Nat. Neurosci. 14: 305– 313 [DOI] [PubMed] [Google Scholar]
- 48a. White P, Doctor RB, Dahl RH, Chen J. 2000. Coincident microvillar actin bundle disruption and perinuclear actin sequestration in anoxic proximal tubule. Am. J. Physiol. Renal Physiol. 278: F886– F893 [DOI] [PubMed] [Google Scholar]
- 49. Wohnsland F, Schmitz AA, Steinmetz MO, Aebi U, Vergéres G. 2000. Interaction between actin and the effector peptide of MARCKS-related protein. Identification of functional amino acid segments. J. Biol. Chem. 275: 20873– 20879 [DOI] [PubMed] [Google Scholar]
- 50. Wohnsland F, Steinmetz MO, Aebi U, Vergères G. 2000. MARCKS-related protein binds to actin without significantly affecting actin polymerization or network structure. Myristoylated alanine-rich C kinase substrate. J. Struct. Biol. 131: 217– 224 [DOI] [PubMed] [Google Scholar]
- 51. Wu M, Chen DF, Sasaoka T, Tonegawa S. 1996. Neural tube defects and abnormal brain development in F52-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 93: 2110– 2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xia Y, Karin M. 2004. The control of cell motility and epithelial morphogenesis by Jun kinases. Trends Cell Biol. 14: 94– 101 [DOI] [PubMed] [Google Scholar]
- 53. Xu X, Raber J, Yang D, Su B, Mucke L. 1997. Dynamic regulation of c-Jun N-terminal kinase activity in mouse brain by environmental stimuli. Proc. Natl. Acad. Sci. U. S. A. 94: 12655– 12660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xue F, Janzen DM, Knecht DA. 2010. Contribution of filopodia to cell migration: a mechanical link between protrusion and contraction. Int. J. Cell Biol. 2010: 507821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamauchi E, Nakatsu T, Matsubara M, Kato H, Taniguchi H. 2003. Crystal structure of a MARCKS peptide containing the calmodulin-binding domain in complex with Ca2+-calmodulin. Nat. Struct. Biol. 10: 226– 231 [DOI] [PubMed] [Google Scholar]
- 56. Yarmola EG, Edison AS, Lenox RH, Bubb MR. 2001. Actin filament cross-linking by MARCKS: characterization of two actin-binding sites within the phosphorylation site domain. J. Biol. Chem. 276: 22351– 22358 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.