Abstract
Peroxisome proliferator-activated receptor γ (PPARγ) is a master regulator of adipocyte differentiation and function. We and others have previously mapped PPARγ binding at a genome-wide level in murine and human adipocyte cell lines and in primary human adipocytes. However, little is known about how binding patterns of PPARγ differ between brown and white adipocytes and among different types of white adipocytes. Here we have employed chromatin immunoprecipitation combined with deep sequencing to map and compare PPARγ binding in in vitro differentiated primary mouse adipocytes isolated from epididymal, inguinal, and brown adipose tissues. While these PPARγ binding profiles are overall similar, there are clear depot-selective binding sites. Most PPARγ binding sites previously mapped in 3T3-L1 adipocytes can also be detected in primary adipocytes, but there are a large number of PPARγ binding sites that are specific to the primary cells, and these tend to be located in closed chromatin regions in 3T3-L1 adipocytes. The depot-selective binding of PPARγ is associated with highly depot-specific gene expression. This indicates that PPARγ plays a role in the induction of genes characteristic of different adipocyte lineages and that preadipocytes from different depots are differentially preprogrammed to permit PPARγ lineage-specific recruitment even when differentiated in vitro.
INTRODUCTION
Mammals have fat depots at various locations in the body. Classically, these tissues have been characterized as either white adipose tissue (WAT) or brown adipose tissue (BAT). Both tissues store energy in the form of triglycerides; however, whereas WAT releases the energy as fatty acids that can be converted to metabolic energy in other tissues, BAT metabolizes the fatty acids in the adipocytes and releases the energy as heat. This unique energy-dispersing function of BAT relies on the high number of mitochondria in the adipocytes and the expression and activation of uncoupling protein 1 (UCP1), residing in the inner mitochondrial membrane (9). Previously, BAT was thought to be present only in newborn and hibernating mammals; however, recently it was demonstrated that also human adults have discrete BAT depots (13, 38, 46, 58, 61). It has been suggested that activation of these BAT depots could increase energy expenditure and lead to weight loss. Consistent with this, there seems to be a negative correlation between body mass index and the presence of BAT (13, 68, 69)
Notably, it has also been indicated that different WAT depots are not identical but have different properties (57, 62). It is currently unclear to what extent differences between the distinct WAT depots reflect differences at the cellular level between adipocytes in the depots or reflect context-dependent differences (e.g., microenvironment); however, recent evidence indicates that different subtypes of white adipocytes do exist. Thus, several studies have demonstrated different gene expression profiles between visceral and subcutaneous WAT and preadipocytes and adipocytes from these tissues (18, 47, 54, 56, 63, 65).
Interestingly, a third type of adipocyte, named brite (brown-in-white) (42) or beige (27), has been identified in WAT and in cultures of human (6, 7, 16, 26) and rodent (3, 12, 17, 19, 20, 25, 34, 42, 59, 67) white adipocytes exposed to sustained treatment with high-affinity peroxisome proliferator-activated receptor γ (PPARγ) agonists (e.g., the thiazolidinedione rosiglitazone) and/or β-adrenergic agonists. These brite/beige cells (here referred to as brite cells) express many of the genes specifically expressed in brown adipocytes, but they also express genes that are characteristic for brite cells (e.g., the homeobox c9 [Hoxc9] and short stature homeobox 2 [Shox2] genes) (42, 65). Interestingly, some WAT depots are more prone to induction of brite adipocytes than others. Thus, inguinal WAT (iWAT), representing subcutaneous WAT, expresses significantly higher levels of brite characteristic markers compared to, e.g., epididymal WAT (eWAT), representing visceral WAT, upon induction by, e.g., cold (49, 65). This further supports the notion that fundamentally different types of white adipocytes exist.
Both brown and white adipocytes arise by commitment and differentiation of mesenchymal stem cells (MSCs) residing in the adipose tissues. However, interestingly, the MSCs that give rise to these two different types of adipocytes have been shown to be fundamentally different. Thus, brown adipocyte precursors have been shown to share many characteristics with the myoblast lineage that are absent in the white adipocyte precursors (28, 55). The recent discoveries of differences among the white adipocytes from different depots suggest the existence of disparate types of white adipocyte precursors. It remains to be shown to what extent the adipocyte precursors in a particular depot are a homogenous or a mixed pool of precursors; e.g., it is presently unclear whether all preadipocytes in the iWAT have the potential to induce UCP1 or whether it is restricted to a subpopulation of precursors (3, 42).
In vitro as well as in vivo studies have demonstrated that peroxisome proliferator-activated receptor γ (PPARγ) acts as a master regulator of both white and brown adipocyte differentiation (2, 16, 42, 45, 60). PPARγ binds to DNA as a heterodimer with retinoid X receptor (RXR) and is activated by polyunsaturated fatty acids and fatty acid derivatives such as prostaglandins. In addition, insulin-sensitizing, antidiabetic thiazolidinediones, such as rosiglitazone, are potent and specific activators of PPARγ (1). We and others have recently profiled the genome-wide binding of PPARγ in 3T3-L1 adipocytes using chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) (36, 40, 48, 52), ChIP combined with microarray (ChIP-chip) (33, 64), or ChIP-paired-end tagging (ChIP-PET) (21). Results from these studies have revealed that PPARγ binding is highly enriched in the vicinity of genes upregulated during adipogenesis (33, 40). Specifically, we found that ∼74% of genes that are highly induced during adipogenesis have PPARγ:RXR binding sites within 50 kb of their transcriptional start site (TSS) (40, 52), indicating that PPARγ is directly involved in the activation of most genes of the adipocyte gene program. These profiles provide important insight into the basic mechanism of PPARγ function in adipocytes. However, while 3T3-L1 adipocytes have been shown to recapitulate many of the features of primary adipocytes, there are some notable differences. For example, 3T3-L1 adipocytes express much lower levels of some adipocyte-specific genes, e.g., leptin (35), compared with primary adipocytes. Thus, it remains to be determined to what extent the specific positions of the binding sites in 3T3-L1 adipocytes reflect those found in primary adipocytes. In addition, it is unknown whether the PPARγ binding profiles are similar between white and brown adipocytes and between different types of white adipocytes.
In this study, we have used ChIP-seq to map all PPARγ binding sites in primary mouse adipocytes differentiated in vitro from the stromal-vascular fraction (SVF) isolated from epididymal, inguinal, or brown adipose tissues. This allows us for the first time to report PPARγ binding profiles in primary mouse white and brown adipocytes and to compare binding profiles from adipocytes from different depots. More than half of the binding sites are common between the different in vitro differentiated primary adipocytes, but there are also clear depot-selective binding sites, several of which represent depot-selective PPARγ binding at the same sites in adipocytes isolated directly from the tissue. Intriguingly, the depot-selective binding sites correlate with depot-specific expression of nearby genes.
MATERIALS AND METHODS
Animals and isolation and differentiation of preadipocytes.
Outbred, male, Naval Medical Research Institute (NMRI) mice, purchased from Taconic, were used for preparation of primary cultures of white and brown adipocytes. Mice were kept at room temperature (∼20°C) for 7 days after arrival. At the age of 4 to 5 weeks, mice were deprived of food for ∼2 h and killed between 9 and 11 a.m. by cervical dislocation. BAT was isolated from the interscapular, cervical, and axillary depots and pooled, and WAT was isolated from the epididymal (eWAT) and inguinal/dorsolumbar (iWAT) depots. Samples from 15 mice were pooled in order to obtain enough material for ChIP-seq. Samples for validation of selected ChIP-seq sites and measuring of corresponding mRNA levels were pooled from 10 to 15 mice in at least two independent experiments. The pooled adipose tissues were carefully minced and treated with collagenase (type II; Sigma), essentially as described in reference 42, to separate the SVF from mature adipocytes. Mature adipocytes were harvested for analyses of mRNA expression, and the SVFs were induced to undergo in vitro differentiation to adipocytes. Pellets containing the SVFs were suspended in ∼4 ml DMEM/animal for all tissues and seeded in six-well plates (2 ml cell suspension/10 cm2/well; Corning). The cells were cultured under differentiation-inducing conditions in a medium consisting of Dulbecco modified Eagle medium (DMEM) (Sigma), 10% newborn calf serum (NCS) (HyClone), 30 nM insulin, 25 μg/ml sodium ascorbate, 10 mM HEPES, pH 7.4, at 37°C, 4 mM l-glutamine (Invitrogen), 100 μg/ml streptomycin, 62.5 μg/ml penicillin, and 1 μM rosiglitazone (Novo Nordisk). Cells of SVF were cultured at 37°C and 8% CO2. Medium was changed 1 day after seeding and subsequently every second day. The cultures were harvested at the days indicated in the figure legends. For comparison between the in vitro differentiated adipocytes, only cultures with a >50% degree of differentiation were used.
The 3T3-L1 preadipocytes were differentiated to adipocytes by stimulation with 3-isobutyl-1-methylxanthine, dexamethasone, and insulin as described previously (24). Oil Red O staining to assess lipid accumulation was performed as previously described (22).
RNA extraction, cDNA synthesis, and quantitative real-time PCR.
RNA extraction, cDNA synthesis, and quantitative real-time PCR (qPCR) were performed as previously described (8). Sequences of primers used for real-time PCR are available upon request.
Western blotting and ECL detection.
Whole-cell extracts for Western blotting were prepared as previously described (22). Protein concentration was determined using the Bio-Rad RCDC protein assay. Samples in SDS-containing buffer were subjected to Western blotting as previously described (39). The membrane was probed with following primary antibodies: anti-PPARγ (sc-7273; Santa Cruz Biotechnologies, Santa Cruz, CA), anti-TFIIB (sc-225; Santa Cruz), and anti-UCP1 (42). The secondary antibodies for enhanced chemiluminescence detection (ECL) were horseradish peroxidase-conjugated swine anti-rabbit IgG (P0399; Dako, Vancouver, Canada) and goat anti-mouse IgG (P0447; Dako).
ChIP and ChIP-seq.
eWAT-, iWAT-, and BAT-derived adipocytes were cross-linked using 0.5 M disuccinimidyl glutarate (DSG) (Proteochem, Denver, CO) for 45 min followed by cross-linking using 1% formaldehyde for 10 min. Cross-linking was stopped by the addition of glycine to a final concentration of 0.125 M for 10 min, followed by addition of ChIP lysis buffer (1% Triton X-100, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, and 20 mM Tris, pH 8.0). Samples were sonicated using the Diagenode Bioruptor twin (3 × 14 cycles, 30 s on/off, maximum level). Samples were centrifuged for 1 min at 10,000 × g, the fat layer was removed, and the supernatant was used for subsequent chromatin IP using anti-PPARγ (sc-7196; Santa Cruz).
Freshly isolated eWAT, iWAT, and BAT were carefully minced and cross-linked for 30 min with 0.5 M DSG followed by cross-linking with 1% formaldehyde for 30 min. Cross-linking was stopped by addition of glycine as described above. Subsequently, nuclei were isolated by addition of cell lysis buffer (0.02 M Tris, pH 8, 0.5% NP-40, 0.085 M KCl) followed by repeated rounds of manual Dounce homogenization. Pooled nuclei were resuspended in cell lysis buffer, run through a mesh filter, and sonicated (2 × 12 cycles, 30 s on/off, maximum level).
Chromatin from eWAT-, iWAT-, and BAT-derived adipocytes and mature adipocytes isolated directly from tissue was diluted in ChIP lysis buffer, and chromatin IP was performed using anti-PPARγ (sc-7196; Santa Cruz). For single IPs, chromatin from ∼1,000,000 cells was used, and for ChIP-seq experiments, chromatin from ∼15,000,000 cells was used. Following 3 h rotation at 4°C, 50 μl protein A beads (Amersham Pharmacia Biotech) was added and samples were incubated overnight at 4°C with rotation. Beads were washed at 4°C once with IP wash buffer 1 (1% Triton, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, 20 mM Tris, pH 8.0, and 0.1% sodium deoxycholate [NaDOC]), twice with IP wash buffer 2 (1% Triton, 0.1% SDS, 500 mM NaCl, 1 mM EDTA, 20 mM Tris, pH 8.0, and 0.1% NaDOC), once with IP wash buffer 3 (0.25 M LiCl, 0.5% NP-40, 1 mM EDTA, 20 mM Tris, pH 8.0, and 0.5% NaDOC), and finally twice with IP wash buffer 4 (10 mM EDTA and 200 mM Tris, pH 8), all at 4°C. DNA-protein complexes were eluted with 400 μl of elution buffer (1% SDS and 0.1 M NaHCO3) and de-cross-linked by adding NaCl to a final concentration of 0.2 M and shaking for ∼3 h at 65°C. ChIPed DNA was purified using phenol-chloroform and analyzed by qPCR or prepared for sequencing according to the manufacturer's instructions (Illumina).
ChIP-seq data analysis.
Sequence reads from each ChIP-seq library were trimmed to 40 bases, and each library was collapsed using FastX (http://hannonlab.cshl.edu/fastx_toolkit/) (i.e., only one copy of each unique read was kept for downstream alignment). Each collapsed library was aligned to the genome (mm9) using Bowtie (31) with the following parameters: -m 3 –best –strata, with all other parameters set at default. For PPARγ tracks from eWAT-, iWAT-, and BAT-derived adipocytes, the numbers of aligned reads were comparable, ranging from 5.8 to 7.5 million mapped reads. Likewise, analyses of previously published PPARγ data sets from 3T3-L1 (40, 52) and hASC (36) produced 5.2 million and 10.0 million mapped reads, respectively. The resulting bowtie alignment files were used as input for the ChIP-seq software suite HOMER (23) used for various downstream analyses.
Peak calling and analyses.
Regions enriched for PPARγ binding were identified using HOMER with the “-factor” setting and a false-discovery rate (FDR) cutoff of <0.1%. Moreover, only peaks with ≥4-fold tag count enrichment compared to both input control and local tag density (10-kb region around putative peak) were used for downstream analyses. To allow quantitative analysis of PPARγ binding across different samples, we used a tag count-based approach. First, the identified peaks in eWAT-, iWAT-, and BAT-derived adipocytes were merged into a single peak file, and tag count information for all three primary data sets was obtained for the merged peak set using HOMER (i.e., the number of tags was counted for the three primary data sets in a 220-bp window around merged peak centers). In all cases, tag counts were normalized to 10 million reads in all subsequent analyses, unless stated otherwise. Identification of different subsets of peaks was subsequently performed using a normalized tag count cutoff corresponding to the FDR < 0.1% setting. Moreover, a fold enrichment cutoff (≥2.5-fold difference in normalized tag count) was introduced to identify subsets of peaks with different PPARγ binding patterns across ChIP-seq libraries. Clusters were defined by several tag count-based filters. For instance, cluster I sites (i.e., “eWAT-enriched” sites) were defined by the following quantitative filters. (i) Sites must have a minimum normalized tag count fulfilling the initial criterion FDR < 0.1% (i.e., a normalized tag count cutoff of ≥12). (ii) Sites must have a normalized tag count of ≤6 for data sets from iWAT- and BAT-derived adipocytes, or sites in eWAT-derived adipocytes must have a ≥2.5-fold enrichment over iWAT- and BAT-derived adipocytes. As another example, cluster VII sites (i.e., “primary shared”) were defined by eWAT-, iWAT-, and BAT-derived adipocytes all having a normalized tag count of ≥12. Thus, using different combinations of tag count-based filters, every site was allocated uniquely to a single cluster.
The previously published data sets 3T3-L1 day 6 (PPARγ ChIP-seq and DHS-seq) (40) and hASC (PPARγ ChIP-seq) (36) were reanalyzed using HOMER with the same parameters for peak calling as described above. The only exception was that 3T3-L1 day 6 DHS peaks and hASC PPARγ peaks were called only on the basis of ≥4-fold enrichment versus local background along with the FDR < 0.1% setting.
Genomic distribution of peaks.
HOMER was used to annotate each peak to its nearest transcription start sites (TSS). Three random peak sets with comparable features as the primary adipocyte peak set (∼40,000 sites, 220-bp peak width) were generated using shuffleBed in the BedTools suite (44). The spatial distributions relative to RefSeq TSSs were determined using distance cutoffs as stated. Finally, the mean values for the three random peak sets, along with the corresponding values for the PPARγ peak set, were used to calculate the “fold enrichment versus random” values.
Conservation of PPARγ binding sites.
The UCSC LiftOver tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver) was used to lift the human hASC PPARγ binding sites from the human hg19 to the mm9 mouse assembly, using minMatch = 0.1. To filter out lifted sites spanning either a very short or very long region, a size cutoff was applied, using Q1 − IQR as the lower limit and Q3 + IQR as the upper size limit for the lifted sites, where Q1, Q3, and IQR correspond to the 25th and 75th percentile and the interquartile range, respectively. Using this cutoff, 57% of the total original hASC sites could be lifted to the mm9 assembly. Overlaps between the lifted hASC sites and the primary and/or 3T3-L1 peak sets were calculated using HOMER. A given peak was considered to overlap a lifted hASC PPARγ site if the distance between their peak centers was ≤150 bp, which approximately corresponds to a 30% actual peak overlap.
Functional enrichment analysis.
PPARγ sites enriched in eWAT or BAT were assigned to the nearest gene, and the resulting gene lists were used for functional enrichment analysis using HOMER, KEGG pathways (29, 30), and WikiPathways (http://www.wikipathways.org) databases.
RESULTS
In vitro differentiation of preadipocytes from eWAT, iWAT, and BAT.
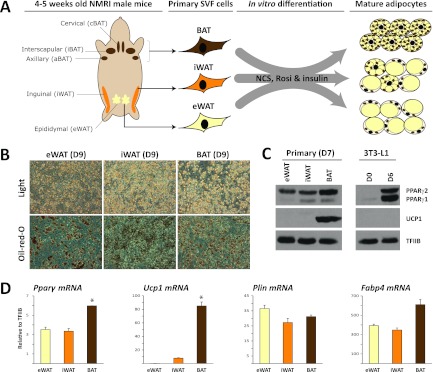
To compare the genome-wide binding pattern of PPARγ in primary in vitro differentiated adipocytes from different origins, we isolated the SVF of eWAT, iWAT, and BAT from 15 NMRI mice. This mouse strain was chosen because it is genetically more competent of developing brite cells than, e.g., strain C57BL/6 (42). Pools of preadipocytes from the 15 different mice were cultured and differentiated in vitro into mature adipocytes (here referred to as eWAT-, iWAT-, and BAT-derived adipocytes), using an identical differentiation protocol for all three SVFs (Fig. 1A). Within 9 days of induction, more than 50% of the cells differentiated into lipid-filled adipocytes (Fig. 1B). Importantly, eWAT-, iWAT, and BAT-derived adipocytes display similar levels of adipocyte marker gene expression, perilipin (PLIN), and fatty acid binding protein 4 (FABP4), confirming that a comparable degree of differentiation was obtained (Fig. 1D). Similarly, comparable mRNA and protein levels of PPARγ are observed for the eWAT-, iWAT-, and BAT-derived adipocytes, although a slightly higher expression of PPARγ is seen in BAT-derived adipocytes (Fig. 1C and D). Compared to the 3T3-L1 adipocytes, the primary adipocyte cultures display a much higher ratio of PPARγ2/1 protein (Fig. 1C). As expected, mRNA and protein levels of the brown adipocyte-specific marker UCP1 are high in BAT-derived adipocytes, moderate in iWAT-derived adipocytes, and insignificant in eWAT-derived adipocytes (Fig. 1D). Thus, the eWAT-, iWAT-, and BAT-derived adipocytes display UCP1 levels characteristic of their respective depots (65).
Fig 1.
In vitro adipocyte differentiation of stromal-vascular cells isolated from eWAT, iWAT, and BAT. (A) Stromal-vascular fractions were isolated from eWAT, iWAT, and BAT of 4- to 5-week-old male NMRI mice and differentiated in vitro to mature adipocytes in a medium containing newborn calf serum (NCS), insulin, and rosiglitazone. (B) Cultures of eWAT-, iWAT-, and BAT-derived preadipocytes were differentiated for 9 days and examined by phase-contrast microscopy. Accumulation of neutral lipids was visualized by Oil Red O staining. (C) Protein expression of PPARγ and UCP1 in day 7 in vitro differentiated adipocytes obtained from eWAT, iWAT, and BAT. Levels of these markers in 3T3-L1 preadipocytes (day 0) and adipocytes (day 6) are shown for comparison. Protein expression was determined by Western blotting and ECL using TFIIB as a loading control. (D) mRNA expression levels of Pparγ, Ucp1, Plin, and Fabp4 in eWAT-, iWAT-, and BAT-derived adipocytes were determined using qPCR and normalized to levels of TFIIB. Data are presented as means of three biological replicates (*P < 0.005 compared to eWAT and iWAT) and are representative of three independent experiments.
Genome-wide profiles of PPARγ binding reveals common as well as depot-selective binding sites.
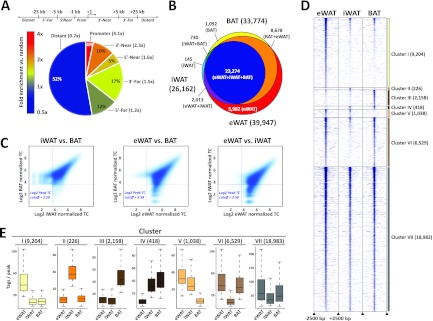
Genome-wide PPARγ binding profiles in eWAT-, iWAT-, and BAT-derived adipocytes were generated by ChIP-seq. For peak calling we used HOMER (23) and then merged peaks called in eWAT-, iWAT-, and BAT-derived adipocytes into a single peak file. This allowed us to count tags in all three primary data sets around merged peak centers and to identify peaks in the three data sets based on a tag count cutoff corresponding to an FDR of <0.1% (i.e., 12 tags). Using this tag count-based strategy, we were able to detect 39,947, 26,162, and 33,774 PPARγ binding sites in eWAT-, iWAT-, and BAT-derived adipocytes, respectively. Consistent with previous analyses of binding sites of PPARγ and other nuclear receptors (5, 10, 40, 66), the majority of binding sites are located distant from genes, although binding sites are most highly enriched (4.1-fold over random genomic positions) in the promoter regions (Fig. 2A). Comparison of the three PPARγ profiles show that there is a high degree of overlap between profiles of PPARγ binding sites of eWAT-, iWAT-, and BAT-derived adipocytes (i.e., primary-shared sites) (Fig. 2B). A total of 41,914 PPARγ binding sites are found in one or more of the primary in vitro differentiated adipocytes, and ∼55% of these are found in all three types of adipocytes (Fig. 2B). The overall similarity in PPARγ binding is further supported by density plots demonstrating that the most intense PPARγ binding sites are shared between the primary adipocytes (Fig. 2C) and by correlation analyses indicating that the relative intensities of PPARγ binding in eWAT-, iWAT-, and BAT-derived adipocytes are similar (see Table S1 in the supplemental material).
Fig 2.
Comparison of PPARγ binding profiles in eWAT-, iWAT-, and BAT-derived adipocytes. (A) Genomic position of primary-shared PPARγ binding sites relative to nearest transcriptional start sites in day 7 eWAT-, iWAT-, and BAT-derived adipocytes. Fold enrichment of PPARγ binding sites compared to random distribution of transcription factor binding sites is indicated by numbers in brackets and by color coding as shown. (B) Overlap between PPARγ binding sites in day 7 eWAT (yellow), iWAT (orange), and BAT (brown)-derived adipocytes detected by ChIP-seq. (C) PPARγ binding intensity in eWAT-, iWAT-, and BAT-derived adipocytes (log2 to the number of normalized reads per peak). Horizontal and vertical lines indicate log2 peak tag count (TC) cutoff at 3.58 (corresponding to a tag count of 12). (D) Heat map showing PPARγ binding site intensity in a ±2.5-kb window around peak centers in eWAT-, iWAT-, and BAT-derived adipocytes. Clusters are defined as follows: clusters I to III, PPARγ binding sites enriched in eWAT-, iWAT-, and BAT-derived adipocytes, respectively; cluster IV, PPARγ binding sites enriched in iWAT- and BAT-derived adipocytes; cluster V, PPARγ binding sites enriched in iWAT- and eWAT-derived adipocytes; cluster VI, PPARγ binding sites enriched in eWAT- and BAT-derived adipocytes; and cluster VII, PPARγ binding sites enriched in eWAT-, iWAT-, and BAT-derived adipocytes (primary shared). (E) Box plots illustrating the distribution of number of tags per peak in the seven clusters.
Despite the overall similarity in PPARγ profile among eWAT-, iWAT, and BAT-derived adipocytes, our analyses also indicate the existence of depot-selective PPARγ binding sites. To focus our further analyses of these depot-selective sites (i.e., peaks with a ≥2.5-fold-higher tag count in one type of adipocytes compared with other types), we grouped PPARγ binding sites into seven clusters based on their relative binding intensity in eWAT-, iWAT-, and BAT-derived adipocytes. Clusters I to III represent PPARγ binding sites enriched in eWAT-, iWAT-, or BAT-derived adipocytes, respectively; clusters IV to VI represent dual PPARγ binding sites (i.e., enriched in iWAT- and BAT-derived, eWAT- and iWAT-derived, or eWAT- and BAT-derived adipocytes, respectively); and cluster VII represents binding sites shared between all types of adipocytes (Fig. 2D and E). These analyses demonstrate that depot-selective binding sites are not just borderline peaks but also include high-intensity peaks.
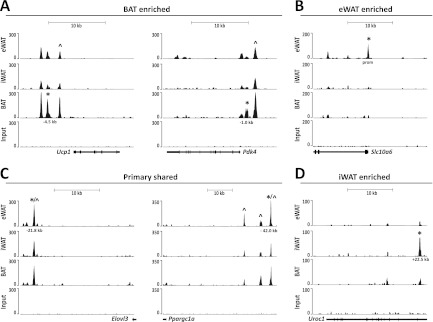
Examples of such depot-selective PPARγ binding sites are shown in Fig. 3. PPARγ binding in the vicinity of the Pdk4 and Ucp1 promoters is enriched in BAT-derived adipocytes, although there is also significant binding in both eWAT- and iWAT-derived adipocytes (Fig. 3A). The kb −2.5 PPARγ binding site in the Ucp1 locus has previously been reported (4, 15, 51, 60); however, the other sites are all novel. Similarly, in the Pdk4 locus the kb −2.7 binding site has been reported at an orthologous position in human cells (14) but to our knowledge sites have not yet been reported in the mouse genome (Fig. 3A). Conversely, PPARγ binding in the vicinity of the Slc10a6 promoter is highly enriched in eWAT-derived adipocytes, whereas intronic binding in the Uroc1 gene is highly enriched in iWAT-derived adipocytes, and binding sites in the vicinity of the Elovl3 and Ppargc1a genes are of similar intensity in all three types of adipocytes (Fig. 3B to D). The cell type-specific patterns were validated by ChIP-qPCR in at least two independent experiments (see Fig. 5A and 6A; also see Fig. S1 in the supplemental material).
Fig 3.
Binding of PPARγ to selected loci in primary mouse eWAT-, iWAT-, and BAT-derived adipocytes. UCSC Genome Browser tracks are shown. (A) PPARγ binding sites enriched in BAT-derived adipocytes at the Ucp1 and Pdk4 loci. (B) PPARγ binding sites enriched in eWAT-derived adipocytes at the Slc10a6 locus. (C) Primary-shared PPARγ binding sites at the Elovl3 and Pgc-1α loci. (D) PPARγ binding sites enriched in iWAT-derived adipocytes at the UroC1 locus. *, PPARγ binding sites validated by ChIP-PCR (see Fig. 5 and 6; also see Fig. S1 in the supplemental material) in this study; ^, previously published PPARγ binding sites.
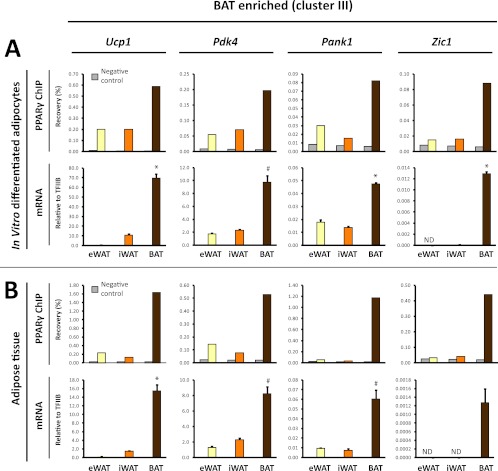
Fig 5.
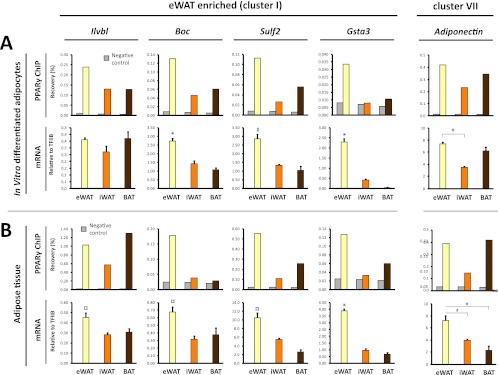
BAT-enriched PPARγ binding is associated with BAT-selective mRNA expression of the corresponding genes. (A) PPARγ binding (upper panel) at selected BAT-enriched sites (cluster III sites: Ucp1 [kb −4.5], Pdk4 [kb −1], Pank1 promoter, Zic1 [kb +13]) (see Fig. 3 and Fig. S3 in the supplemental material) in eWAT-, iWAT-, and BAT-derived adipocytes. Binding was determined by ChIP-qPCR and expressed as % recovery compared to input sample. mRNA expression (lower panel) of the corresponding genes relative to TFIIB in eWAT-, iWAT-, and BAT-derived adipocytes. (B) PPARγ binding (upper panel) in freshly isolated eWAT, iWAT, and BAT to the same sites as in panel A, as investigated by ChIP-qPCR. mRNA expression of the corresponding genes relative to TFIIB in mature adipocytes isolated directly from eWAT, iWAT, and BAT as indicated. mRNA data are presented as means of three biological replicates and representative of at least two independent experiments. ChIP-qPCR data are representative of two independent experiments. *, P < 0.005; #, P < 0.01 compared to eWAT- and iWAT-derived adipocytes or eWAT and iWAT. ND, not detected.
Fig 6.
eWAT-enriched PPARγ binding is associated with eWAT-selective mRNA expression of the corresponding genes. (A) PPARγ binding (upper panel) at selected eWAT-enriched binding sites (cluster I sites: Ilvbl TSS, Boc [kb +2.6], Sulf2 [kb +2], Gsta3 promoter [see Fig. S3 in the supplemental material]) in eWAT-, iWAT-, and BAT-derived adipocytes. The PPARγ binding site at adipoq kb −2 is contained within cluster VII, which represents sites present in all types of adipocytes. Binding was determined by ChIP-qPCR and expressed as percent recovery compared to input sample. mRNA expression (lower panel) of the corresponding genes relative to TFIIB in eWAT-, iWAT-, and BAT-derived adipocytes. (B) PPARγ binding (upper panel) in freshly isolated eWAT, iWAT, and BAT to the same sites as in panel A, as investigated by ChIP-qPCR. mRNA expression (lower panel) of the corresponding genes relative to TFIIB in mature adipocytes isolated directly from eWAT, iWAT, and BAT as indicated. mRNA data are presented as means of three biological replicates and representative of at least two independent experiments. ChIP-qPCR data are representative of two independent experiments. *, P < 0.005; #, P < 0.01; and ¤, P < 0.02 compared to eWAT- and BAT-derived adipocytes or eWAT and BAT.
Thus, the overall PPARγ binding profiles are similar between eWAT-, iWAT-, and BAT-derived adipocytes. However, we also observe distinct depot-selective PPARγ binding sites, which may reflect distinct subsets of genes regulated by PPARγ in eWAT-, iWAT-, and BAT-derived adipocytes.
PPARγ binding sites specific to primary adipocytes are located in regions with a closed chromatin configuration in 3T3-L1 adipocytes.
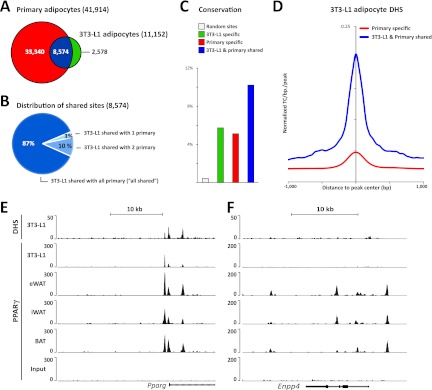
We compared a total of 41,914 PPARγ binding sites identified in eWAT-, iWAT-, and BAT-derived adipocytes with our previously reported PPARγ binding profile in 3T3-L1 adipocytes (40). Of note, 3T3-L1 and primary mouse adipocytes display comparable levels of PPARγ protein expression (Fig. 1C) and the sequencing depths of the PPARγ ChIP-seq were comparable. Our analyses show that the majority (∼77%) of PPARγ binding sites in 3T3-L1 adipocytes are present in eWAT-, iWAT-, and BAT-derived adipocytes, with only a few sites that are specific for 3T3-L1 adipocytes (Fig. 4A). On the other hand, ∼80% of the PPARγ binding sites in eWAT-, iWAT-, and BAT-derived adipocyte cells could not be found in 3T3-L1 adipocytes (Fig. 4A), and PPARγ binding profiles of eWAT-, iWAT-, and BAT-derived adipocytes are more similar to each other than to that of 3T3-L1 adipocytes (see Table S1 and Fig. S2 in the supplemental material). Thus, most PPARγ binding sites in 3T3-L1 adipocytes are also found in primary in vitro differentiated adipocytes; however, 3T3-L1 adipocytes clearly lack most sites present in primary cells. Notably, the majority (87%) of the sites that are shared between 3T3-L1 and eWAT-, iWAT-, and BAT-derived adipocytes are present in all three primary adipocytes (“all shared”), indicating that those sites represent robust PPARγ binding sites important for adipocyte biology (Fig. 4B). This is further supported by conservation analysis showing that PPARγ binding sites shared between eWAT-, iWAT-, and BAT-derived adipocytes and 3T3-L1 adipocytes display a higher degree of conservation (i.e., 10%) in primary human in vitro differentiated adipocytes (36) than PPARγ binding sites enriched only in 3T3-L1 or eWAT-, iWAT-, and BAT-derived adipocytes (i.e., 6% and 5%, respectively) (Fig. 4C). Of the primary-specific PPARγ binding sites, 35% are associated with a total of 6,196 novel putative target genes, whereas 28% are associated with 3,395 genes that also have PPARγ-associated peaks in 3T3-L1 adipocytes, and 37% are not assigned to any genes (within 50 kb). These differences in the number of putative 3T3-L1 target genes might account for some of the functional differences between primary and 3T3-L1 adipocytes.
Fig 4.
Most PPARγ binding sites in 3T3-L1 cells are shared with primary adipocytes. (A) Genome-wide overlap between the PPARγ binding sites in eWAT-, iWAT-, and BAT-derived adipocytes with sites previously detected in 3T3-L1 adipocytes (38). (B) Characterization of PPARγ binding sites shared between 3T3-L1 adipocytes and primary mouse in vitro differentiated adipocytes. “3T3-L1 shared with 1 primary” indicates that the PPARγ binding sites in 3T3-L1 adipocytes overlap PPARγ binding sites in either eWAT-, iWAT-, or BAT-derived adipocytes, and “3T3-L1 shared with 2 primary” denotes that that PPARγ binding sites in 3T3-L1 adipocytes overlap PPARγ binding sites in two out of the three adipose-derived adipocytes. The majority (87%) of shared sites can be detected in all three types of primary adipocytes (“all shared”). (C) Retention of mouse PPARγ binding sites at orthologous positions in the human genome. Retention frequency is compared for sites specific to eWAT-, iWAT-, and BAT-derived adipocytes (red), specific to 3T3-L1 adipocytes (green), and shared between eWAT-, iWAT-, and BAT-derived adipocytes and 3T3-L1 adipocytes (blue). (D) Comparison of the average chromatin openness in the 3T3-L1 genome for sites that are occupied by PPARγ only in primary eWAT-, iWAT-, and BAT-derived adipocytes (red) and sites that are occupied by PPARγ in both 3T3-L1 and primary adipocytes (blue). Chromatin openness is based on previously published DNase I hypersensitivity (DHS) tag counts (41). (E) Alignment of the PPARγ ChIP-seq profile and DHS-seq profile previously reported for 3T3-L1 adipocytes (38, 41) with the PPARγ ChIP-seq profiles from eWAT-, iWAT-, and BAT-derived adipocytes. Examples from the Pparg and Enpp4 loci are shown.
We have previously determined the positions of open chromatin regions in 3T3-L1 adipocytes using DNase I hypersensitivity assays combined with deep sequencing (DHS-seq) (52). Interestingly, aggregate plots showed that PPARγ binding sites that are shared between eWAT-, iWAT-, and BAT-derived adipocytes and 3T3-L1 adipocytes are generally found in open chromatin regions in 3T3-L1 adipocytes, whereas sites specific for eWAT-, iWAT-, and BAT-derived adipocytes are much less DNase I hypersensitive in 3T3-L1 adipocytes (Fig. 4D). Examples of sites shared between 3T3-L1 adipocytes and eWAT-, iWAT-, and BAT-derived adipocytes are found in the Pparg2 promoter region, and these sites overlap DHS sites in 3T3-L1 cells (Fig. 4E). In contrast, the sites in the Enpp4 locus are located in closed chromatin regions in 3T3-L1 adipocytes and are not bound by PPARγ in these cells (Fig. 4E and F). This indicates that PPARγ binding in 3T3-L1 adipocytes is strongly correlated with open chromatin structure.
Thus, these results indicate that PPARγ binding sites observed in 3T3-L1 adipocytes faithfully recapitulate a subfraction of sites observed in eWAT-, iWAT-, and BAT-derived adipocytes. However, ∼80% of the binding sites observed in eWAT-, iWAT-, and BAT-derived adipocytes cannot be found in the 3T3-L1 system. Binding sites that are lost in 3T3-L1 adipocytes appear to be located primarily in regions of closed chromatin structure in the 3T3-L1 cells. Whether PPARγ binding is the cause or the result (or both) of the open chromatin structure remains to be determined.
Depot-selective PPARγ binding correlates with gene expression.
Although the majority of PPARγ binding sites are shared between eWAT-, iWAT-, and BAT-derived adipocytes, our analyses clearly demonstrate the existence of depot-selective binding sites. To investigate the functional relevance of such depot-selective binding, we assigned PPARγ binding sites that were enriched in either eWAT-derived (cluster I) or BAT-derived (cluster III) adipocytes to the nearest gene within 50 kb and performed functional enrichment analyses. The PPARγ binding sites enriched in iWAT-derived adipocytes were not included in these analyses due to a low number of assigned genes. Functional enrichment analyses of PPARγ-associated genes revealed that many GO terms were shared between the different types of adipocytes, which include adipocyte (P < 0.02), PPAR signaling (P < 0.01), and adipocytokine signaling pathways (P < 0.02) (see Table S2 in the supplemental material). PPARγ binding sites enriched in BAT-derived adipocytes are associated with genes designated as being involved in fatty acid beta oxidation, fatty acid elongation, mitochondrial function, and bile acid synthesis (Table 1). In contrast, PPARγ binding sites enriched in eWAT-derived adipocytes are, e.g., associated with genes related to estrogen metabolism and signaling, as well as different growth factor signaling pathways (Table 1).
Table 1.
Functional enrichment analysisa
| Adipocyte type, pathway, and GO term | P value | No. of genes in BAT or eWAT/no. of genes in GO termb | fBAT/eWAT or feWAT/BATc |
|---|---|---|---|
| BAT enriched | |||
| Wiki | |||
| FA beta oxidation | 3.3E-12 | 26/73 | 7.5 |
| FA-, TG-, and ketone body metabolism | 0.0002 | 10/35 | 3.1 |
| Nicotine activity on dopaminergic neurons | 0.0023 | 8/33 | 6.9 |
| Mitochondrial LC-FA beta oxidation | 0.0065 | 5/17 | 2.5 |
| KEGG | |||
| FA elongation | 7.5E-24 | 27/43 | 35 |
| FA metabolism | 2.4E-14 | 34/125 | 3.2 |
| Biosynthesis of unsaturated FAs | 3.7E-07 | 14/47 | 2.5 |
| eWAT enriched | |||
| Wiki | |||
| Estrogen metabolism | 2.5E-12 | 38/53 | 11 |
| Androgen receptor signaling pathway | 0.0004 | 37/87 | 5.4 |
| Leptin signaling pathway | 0.0006 | 34/79 | 3.2 |
| Estrogen signaling pathway | 0.0007 | 16/29 | 4.6 |
| IL-2 signaling pathway | 0.0021 | 23/51 | 6.6 |
| TP53 network | 0.0024 | 15/29 | 4.3 |
| IL-1 signaling pathway | 0.0038 | 27/65 | 7.8 |
| Signaling by Notch | 0.0044 | 13/25 | —d |
| Osteopontin signaling | 0.0068 | 10/18 | —d |
| KEGG | |||
| Prion diseases | 3.6E-15 | 47/69 | 6 |
| Biotin metabolism | 2.2E-06 | 9/9 | —d |
| Nicotinate and nicotinamide metabolism | 0.0015 | 21/48 | 2.7 |
| Fc gamma R-mediated phagocytosis | 0.0020 | 34/91 | 2.9 |
| Epithelial cell signaling in Helicobacter pylori infection | 0.0062 | 25/66 | 3.2 |
PPARγ sites enriched in eWAT- or BAT-derived adipocytes were assigned to the nearest gene (within 50 kb), and the resulting target gene lists were used for functional enrichment analysis using WikiPathways and KEGG pathways databases. The most significantly (P < 0.01) enriched gene ontology (GO) terms meeting the criterion ƒ ≥ 2.5, where ƒ represents the ratio between the fraction of gene lists for eWAT- and BAT-enriched pathways, are shown. Thus, GO terms presented in this table are those that are different between eWAT- and BAT-derived adipocytes. GO terms that are shared between eWAT- and BAT-derived adipocytes are shown in Table S2 in the supplemental material. Abbreviations: FA, fatty acid; TG, triglyceride; LC, long chain; IL, interleukin; TP, tumor protein.
Genes in GO term near BAT-enriched PPARγ sites or genes in GO term near eWAT-enriched PPARγ sites.
fBAT/eWAT for genes enriched near BAT-selective PPARγ binding sites or feWAT/BAT for genes enriched near eWAT-selective PPARγ binding sites.
—, no genes in GO term near BAT-enriched PPARγ sites.
To further examine the relationship between depot-selective PPARγ binding and depot-specific gene expression, we selected a subset of eWAT-derived (cluster I) or BAT-derived (cluster III) PPARγ peaks located within 15 kb of a known TSS. The selected PPARγ binding sites represent prominent peaks in their respective loci but vary in intensity from weak (Zic1 and Gsta3) to moderate (Pdk4 and Boc) and very strong (Ucp1 and Ilvbl) PPARγ binding sites (Fig. 3; see Fig. S3 in the supplemental material). In addition, we included PPARγ peaks associated with the adiponectin gene, since adiponectin has previously been reported to be produced primarily in visceral compared to subcutaneous adipose tissue (11, 37, 53). In our in vitro differentiated adipocytes none of the PPARγ binding sites in the vicinity of the adiponectin gene had an eWAT tag count of >2.5-fold over the tag count in iWAT- and BAT-derived adipocytes (see Fig. S3C in the supplemental material; data not shown). Notably, evaluation of the mRNA expression from the selected loci displayed a high concordance between depot-selective PPARγ binding and depot-selective expression of mRNA (Fig. 5 and 6A). Thus, high levels of PPARγ binding at prominent peaks in the Ucp1, Pdk4, and Pank1 and Zic1 loci are associated with significantly higher levels of mRNA expression from these loci in BAT-derived adipocytes compared with eWAT- and iWAT-derived adipocytes (Fig. 5A). Similarly, high levels of PPARγ binding at prominent peaks in the Boc, Sulf2, and Gsta3 loci are associated with significantly higher levels of expression of these genes in eWAT-derived adipocytes than in iWAT- and BAT-derived adipocytes (Fig. 6A). This indicates that PPARγ may play an important role in the activation of these depot-selective genes.
To investigate to what extent the depot-selective differences in PPARγ recruitment and expression of neighboring genes reflect conditions in mature adipocytes in the respective tissues, we isolated chromatin directly from the tissues and determined the relative binding of PPARγ to these sites. We isolated mature adipocytes in parallel directly from the different tissues and determined the mRNA level by qPCR. Intriguingly, mature adipocytes from the different adipose depots display depot-selective PPARγ binding and mRNA expressions that correspond to those observed in in vitro differentiated adipocytes (Fig. 5B and 6B). These data indicate that the eWAT-, iWAT-, and BAT-derived adipocytes are able to faithfully recapitulate the depot-selective patterns of PPARγ recruitment and gene expression observed in vivo. Moreover, the data further support the notion that depot-specific recruitment of PPARγ plays a role in depot-specific gene expression.
DISCUSSION
PPARγ is a well-accepted master regulator of adipocyte differentiation in human and rodents (2, 16, 42, 45, 60), yet the profiling of binding sites of this transcription factor in primary mouse adipocytes has not been reported. We used ChIP-seq to generate genome-wide profiles of PPARγ binding sites in in vitro differentiated primary adipocytes isolated from eWAT, iWAT, and BAT, which were differentiated to a comparable degree and with comparable PPARγ expression. We identify a total of 41,914 PPARγ binding sites in eWAT-, iWAT-, and BAT-derived adipocytes, 23,274 of which are shared between eWAT-, iWAT-, and BAT-derived adipocytes. The high degree of overlap between PPARγ binding in the different types of adipocytes is remarkable given that brown preadipocytes are derived from the myoblast lineage (50, 55). Interestingly, we also identify robust depot-selective binding sites, where PPARγ occupancy is much higher in one type of adipocytes compared to the others. Importantly, similar depot-selective PPARγ binding is observed in the different adipose tissues, indicating that the eWAT-, iWAT-, and BAT-derived adipocytes faithfully recapitulate many of the specific features of the gene programs in adipocytes in the respective tissues.
Comparison with PPARγ ChIP-seq profiles from 3T3-L1 adipocytes (40) revealed that ∼76% of the binding sites in 3T3-L1 adipocytes are found in eWAT-, iWAT-, and BAT-derived adipocytes. Binding sites that are specific to eWAT-, iWAT-, and BAT-derived adipocytes appear to be located primarily in regions with closed chromatin configuration in 3T3-L1 adipocytes, suggesting that these sites are inaccessible to PPARγ in these cells. Whether this is due to epigenetic factors or missing cooperating or priming factors remains to be determined. As the primary cells were not deprived of rosiglitazone prior to the isolation of chromatin, it cannot be excluded that we identified additional PPARγ binding sites as a result of ligand treatment. However, our unpublished observation from genome-wide studies in 3T3-L1 adipocytes indicates that binding profiles do not change much in response to rosiglitazone. In any case, we predict that sites shared between 3T3-L1 and eWAT-, iWAT-, and BAT-derived adipocytes contain most of the key adipogenic PPARγ binding sites. Consistent with this, the shared sites display a higher degree of conservation in human adipocytes, compared to sites specific to eWAT-, iWAT-, and BAT-derived adipocytes or 3T3-L1 adipocytes.
Intriguingly, we could show that depot-selective binding of PPARγ in eWAT-, iWAT-, and BAT-derived adipocytes and in their respective tissues is correlated with depot-selective expression of the corresponding genes. This indicates that PPARγ plays a role as an activator of these depot-selective genes and that its recruitment to specific sites is lineage dependent. Similar cell type-specific occupancy and association with gene expression have been reported for PPARγ in 3T3-L1 adipocytes and macrophages (32) and for lipid X receptor in macrophages and B cells (23). In both cases, cell type-specific binding of the nuclear receptor appeared to be due at least in part to expression of different cooperating transcription factors. However, in the case of the depot-specific differences observed in the present study, it should be noted that PPARγ binding is much less restrictive than the mRNA expression. Thus, PPARγ binds to a number of binding sites in the vicinity of BAT-specific genes in eWAT- and iWAT-derived adipocytes where these genes are not expressed, demonstrating that PPARγ is not sufficient to activate these genes. Presumably, other transcription factors and/or cofactors are needed to fully activate the gene programs specific to BAT and eWAT. In BAT such program-specific coactivators could include PR domain containing 16 (PRDM16), and PPARγ coactivator 1α (PGC-1α), which have been shown to be of major importance for the ability of PPARγ to activate the BAT-specific genes (41, 43). However, it is also possible that specific combinations of transcription factors at hotspots (52) where PPARγ binds are important for the ability of PPARγ to activate BAT-specific genes.
The finding that in vitro differentiation of preadipocytes from the different adipocyte depots results in adipocytes that reflect the depot-selective nature of the in vivo differentiated adipocytes in the respective tissues is remarkable. First of all, it shows that the preadipocytes in different depots are different and are preprogrammed to become a particular adipocyte lineage(s). This preprogramming appears to be so robust that it is maintained even after exposure to a strong adipogenic cocktail. Second, it demonstrates that eWAT-, iWAT-, and BAT-derived adipocytes may be used as model systems for studying depot-selective differences in the adipogenic gene programs.
In conclusion, we generated genome-wide maps of PPARγ binding sites in primary epididymal, inguinal, and brown mouse adipocytes. While the profiles overall are very similar, our analyses demonstrate the existence of clear depot-selective binding sites that are associated with highly depot-specific gene expression. This indicates that PPARγ plays a role in the induction of genes characteristic of different adipocyte lineages, and that preadipocytes from different depots are differentially preprogrammed to permit PPARγ lineage-specific recruitment. Future investigations will reveal the molecular components of these preprogramming systems.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Mandrup laboratory for fruitful discussions and the Lazar laboratory for input to the development of ChIP protocol for adipose tissue. Rosiglitazone was a kind gift from Per Sauerberg, Novo Nordisk A/S.
This work has been supported in part by grants from the Lundbeck Foundation, the Novo Nordisk Foundation, the Danish Independent Research Council Natural Sciences, and the Swedish Research Council and by grants from NordForsk given to the Nordic Centre of Excellence MitoHealth.
Footnotes
Published ahead of print 25 June 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Aagaard MM, Siersbaek R, Mandrup S. 2011. Molecular basis for gene-specific transactivation by nuclear receptors. Biochim. Biophys. Acta 1812: 824– 835 [DOI] [PubMed] [Google Scholar]
- 2. Barak Y, et al. 1999. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell 4: 585– 595 [DOI] [PubMed] [Google Scholar]
- 3. Barbatelli G, et al. 2010. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am. J. Physiol. Endocrinol. Metab. 298: E1244– E1253 [DOI] [PubMed] [Google Scholar]
- 4. Barbera MJ, et al. 2001. Peroxisome proliferator-activated receptor alpha activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J. Biol. Chem. 276: 1486– 1493 [DOI] [PubMed] [Google Scholar]
- 5. Boergesen M, et al. 2012. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol. Cell. Biol. 32: 852– 867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bogacka I, Ukropcova B, McNeil M, Gimble JM, Smith SR. 2005. Structural and functional consequences of mitochondrial biogenesis in human adipocytes in vitro. J. Clin. Endocrinol. Metab. 90: 6650– 6656 [DOI] [PubMed] [Google Scholar]
- 7. Bogacka I, Xie H, Bray GA, Smith SR. 2005. Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes 54: 1392– 1399 [DOI] [PubMed] [Google Scholar]
- 8. Bugge A, et al. 2010. A novel intronic peroxisome proliferator-activated receptor gamma enhancer in the uncoupling protein (UCP) 3 gene as a regulator of both UCP2 and -3 expression in adipocytes. J. Biol. Chem. 285: 17310– 17317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cannon B, Nedergaard J. 2004. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84: 277– 359 [DOI] [PubMed] [Google Scholar]
- 10. Chong HK, et al. 2010. Genome-wide interrogation of hepatic FXR reveals an asymmetric IR-1 motif and synergy with LRH-1. Nucleic Acids Res. 38: 6007– 6017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cnop M, et al. 2003. Relationship of adiponectin to body fat distribution, insulin sensitivity and plasma lipoproteins: evidence for independent roles of age and sex. Diabetologia 46: 459– 469 [DOI] [PubMed] [Google Scholar]
- 12. Collins S, Daniel KW, Petro AE, Surwit RS. 1997. Strain-specific response to beta 3-adrenergic receptor agonist treatment of diet-induced obesity in mice. Endocrinology 138: 405– 413 [DOI] [PubMed] [Google Scholar]
- 13. Cypess AM, et al. 2009. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360: 1509– 1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Degenhardt T, et al. 2007. Three members of the human pyruvate dehydrogenase kinase gene family are direct targets of the peroxisome proliferator-activated receptor beta/delta. J. Mol. Biol. 372: 341– 355 [DOI] [PubMed] [Google Scholar]
- 15. del Mar Gonzalez-Barroso M, et al. 2000. Transcriptional activation of the human ucp1 gene in a rodent cell line. Synergism of retinoids, isoproterenol, and thiazolidinedione is mediated by a multipartite response element. J. Biol. Chem. 275: 31722– 31732 [DOI] [PubMed] [Google Scholar]
- 16. Elabd C, et al. 2009. Human multipotent adipose-derived stem cells differentiate into functional brown adipocytes. Stem Cells 27: 2753– 2760 [DOI] [PubMed] [Google Scholar]
- 17. Festuccia WT, Blanchard PG, Richard D, Deshaies Y. 2010. Basal adrenergic tone is required for maximal stimulation of rat brown adipose tissue UCP1 expression by chronic PPAR-gamma activation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 299: R159– R167 [DOI] [PubMed] [Google Scholar]
- 18. Gesta S, et al. 2006. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc. Natl. Acad. Sci. U. S. A. 103: 6676– 6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghorbani M, Himms-Hagen J. 1997. Appearance of brown adipocytes in white adipose tissue during CL 316,243-induced reversal of obesity and diabetes in Zucker fa/fa rats. Int. J. Obes. Relat. Metab. Disord. 21: 465– 475 [DOI] [PubMed] [Google Scholar]
- 20. Guerra C, Koza RA, Yamashita H, Walsh K, Kozak LP. 1998. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. J. Clin. Invest. 102: 412– 420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hamza MS, et al. 2009. De-novo identification of PPARgamma/RXR binding sites and direct targets during adipogenesis. PLoS One 4: e4907 doi:10.1371/journal.pone.0004907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hansen JB, et al. 1999. Activation of peroxisome proliferator-activated receptor gamma bypasses the function of the retinoblastoma protein in adipocyte differentiation. J. Biol. Chem. 274: 2386– 2393 [DOI] [PubMed] [Google Scholar]
- 23. Heinz S, et al. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576– 589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Helledie T, et al. 2002. The gene encoding the Acyl-CoA-binding protein is activated by peroxisome proliferator-activated receptor gamma through an intronic response element functionally conserved between humans and rodents. J. Biol. Chem. 277: 26821– 26830 [DOI] [PubMed] [Google Scholar]
- 25. Himms-Hagen J, et al. 2000. Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. Am. J. Physiol. Cell Physiol. 279: C670– C681 [DOI] [PubMed] [Google Scholar]
- 26. Huttunen P, Hirvonen J, Kinnula V. 1981. The occurrence of brown adipose tissue in outdoor workers. Eur. J. Appl. Physiol. Occup. Physiol. 46: 339– 345 [DOI] [PubMed] [Google Scholar]
- 27. Ishibashi J, Seale P. 2010. Medicine. Beige can be slimming. Science 328: 1113– 1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kajimura S, et al. 2009. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature 460: 1154– 1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28: 27– 30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular datasets. Nucleic Acids Res. 40(Database issue): D109– D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 doi:10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lefterova MI, et al. 2010. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol. Cell. Biol. 30:2078– 2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lefterova MI, et al. 2008. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 22: 2941– 2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Madsen L, et al. 2010. UCP1 induction during recruitment of brown adipocytes in white adipose tissue is dependent on cyclooxygenase activity. PLoS One 5: e11391 doi:10.1371/journal.pone.0011391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mandrup S, Loftus TM, MacDougald OA, Kuhajda FP, Lane MD. 1997. Obese gene expression at in vivo levels by fat pads derived from s.c. implanted 3T3-F442A preadipocytes. Proc. Natl. Acad. Sci. U. S. A. 94:4300– 4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mikkelsen TS, et al. 2010. Comparative epigenomic analysis of murine and human adipogenesis. Cell 143: 156– 169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Motoshima H, et al. 2002. Differential regulation of adiponectin secretion from cultured human omental and subcutaneous adipocytes: effects of insulin and rosiglitazone. J. Clin. Endocrinol. Metab. 87: 5662– 5667 [DOI] [PubMed] [Google Scholar]
- 38. Nedergaard J, Bengtsson T, Cannon B. 2007. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 293: E444– E452 [DOI] [PubMed] [Google Scholar]
- 39. Nielsen R, Grontved L, Stunnenberg HG, Mandrup S. 2006. Peroxisome proliferator-activated receptor subtype- and cell-type-specific activation of genomic target genes upon adenoviral transgene delivery. Mol. Cell. Biol. 26: 5698– 5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nielsen R, et al. 2008. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 22: 2953– 2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ohno H, Shinoda K, Spiegelman BM, Kajimura S. 2012. PPARg agonists induce a white-to-brown fat conversion through the stabilization of PRDM16 protein. Cell Metab. 15: 395– 404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petrovic N, et al. 2010. Chronic peroxisome proliferator-activated receptor (PPAR) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classical brown adipocytes. J. Biol. Chem. 285: 7153– 7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Puigserver P, et al. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92: 829– 839 [DOI] [PubMed] [Google Scholar]
- 44. Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841– 842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rosen ED, et al. 1999. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 4: 611– 617 [DOI] [PubMed] [Google Scholar]
- 46. Saito M, et al. 2009. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58: 1526– 1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Samaras K, Botelho NK, Chisholm DJ, Lord RV. 2010. Subcutaneous and visceral adipose tissue gene expression of serum adipokines that predict type 2 diabetes. Obesity (Silver Spring) 18: 884– 889 [DOI] [PubMed] [Google Scholar]
- 48. Schmidt SF, et al. 2011. Cross species comparison of C/EBPalpha and PPARgamma profiles in mouse and human adipocytes reveals interdependent retention of binding sites. BMC Genomics 12: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schulz TJ, et al. 2011. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc. Natl. Acad. Sci. U. S. A. 108: 143– 148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Seale P, et al. 2008. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454: 961– 967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sears IB, MacGinnitie MA, Kovacs LG, Graves RA. 1996. Differentiation-dependent expression of the brown adipocyte uncoupling protein gene: regulation by peroxisome proliferator-activated receptor gamma. Mol. Cell. Biol. 16: 3410– 3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Siersbaek R, et al. 2011. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J. 30: 1459– 1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Staiger H, et al. 2003. Relationship of serum adiponectin and leptin concentrations with body fat distribution in humans. Obes. Res. 11: 368– 372 [DOI] [PubMed] [Google Scholar]
- 54. Tchkonia T, et al. 2007. Identification of depot-specific human fat cell progenitors through distinct expression profiles and developmental gene patterns. Am. J. Physiol. Endocrinol. Metab. 292: E298– E307 doi:10.1152/ajpendo.00202.2006 [DOI] [PubMed] [Google Scholar]
- 55. Timmons JA, et al. 2007. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc. Natl. Acad. Sci. U. S. A. 104: 4401– 4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tinahones FJ, et al. 2010. Obesity and insulin resistance-related changes in the expression of lipogenic and lipolytic genes in morbidly obese subjects. Obes. Surg. 20: 1559– 1567 [DOI] [PubMed] [Google Scholar]
- 57. Tran TT, Yamamoto Y, Gesta S, Kahn CR. 2008. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metab. 7: 410– 420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. van Marken Lichtenbelt WD, et al. 2009. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360: 1500– 1508 [DOI] [PubMed] [Google Scholar]
- 59. Vegiopoulos A, et al. 2010. Cyclooxygenase-2 controls energy homeostasis in mice by de novo recruitment of brown adipocytes. Science 328: 1158– 1161 [DOI] [PubMed] [Google Scholar]
- 60. Villarroya F, Iglesias R, Giralt M. 2007. PPARs in the control of uncoupling proteins gene expression. PPAR Res. 2007: 74364 doi:10.1155/2007/74364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Virtanen KA, et al. 2009. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 360: 1518– 1525 [DOI] [PubMed] [Google Scholar]
- 62. Virtanen KA, et al. 2002. Glucose uptake and perfusion in subcutaneous and visceral adipose tissue during insulin stimulation in nonobese and obese humans. J. Clin. Endocrinol. Metab. 87: 3902– 3910 [DOI] [PubMed] [Google Scholar]
- 63. Vohl MC, et al. 2004. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes. Res. 12: 1217– 1222 [DOI] [PubMed] [Google Scholar]
- 64. Wakabayashi K, et al. 2009. The peroxisome proliferator-activated receptor gamma/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol. Cell. Biol. 29: 3544– 3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Walden TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. 2012. Recruited versus nonrecruited molecular signatures of brown, “brite” and white adipose tissues. Am. J. Physiol. Endocrinol. Metab. 302: E19– E31 doi:10.1152/ajpendo.00249.2011 [DOI] [PubMed] [Google Scholar]
- 66. Welboren WJ, et al. 2009. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 28: 1418– 1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xue B, Coulter A, Rim JS, Koza RA, Kozak LP. 2005. Transcriptional synergy and the regulation of Ucp1 during brown adipocyte induction in white fat depots. Mol. Cell. Biol. 25: 8311– 8322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yoneshiro T, et al. 2011. Age-related decrease in cold-activated brown adipose tissue and accumulation of body fat in healthy humans. Obesity (Silver Spring) 19: 1755– 1760 [DOI] [PubMed] [Google Scholar]
- 69. Zingaretti MC, et al. 2009. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J. 23: 3113– 3120 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.