Abstract
Fcp1 dephosphorylates the C-terminal domain of the largest subunit of RNA polymerase II (Pol II) to recycle it into a form that can initiate a new round of transcription. Previously, we identified Drosophila Fcp1 as an important factor in optimal Hsp70 mRNA accumulation after heat shock. Here, we examine the role of Fcp1 in transcription of heat shock genes in vivo. We demonstrate that Fcp1 localizes to active sites of transcription including the induced Hsp70 gene. The reduced Hsp70 mRNA accumulation seen by RNA interference (RNAi) depletion of Fcp1 in S2 cells is a result of a loss of Pol II in the coding region of highly transcribed heat shock-induced genes: Hsp70, Hsp26, and Hsp83. Moreover, Fcp1 depletion dramatically increases phosphorylation of the non-chromatin-bound Pol II. Reexpression of either wild-type or catalytically dead versions of Fcp1 demonstrates that both the reduced Pol II levels on heat shock genes and the increased levels of phosphorylated free Pol II are dependent on the catalytic activity of Fcp1. Our results indicate that Fcp1 is required to maintain the pool of initiation-competent unphosphorylated Pol II, and this function is particularly important for the highly transcribed heat shock genes.
INTRODUCTION
Proper temporal and spatial expression of RNA transcripts is vital to the development and health of all organisms. At the heart of eukaryotic transcription is RNA polymerase II (Pol II), the enzyme that catalyzes the synthesis of RNA from a DNA template for protein-coding genes. Transcription is a cyclic process that can be divided into three distinct phases: initiation, elongation, and termination (41). During initiation, the Pol II complex assembles around the DNA at promoters and catalyzes the synthesis of the first phosphodiester bond in the gene's RNA transcript. Elongation involves the processive synthesis of the RNA transcript. Termination of the transcription cycle results in the release of both the nascent transcript and Pol II from the DNA template, and terminated Pol II can then be recycled for subsequent rounds of transcription.
The C-terminal domain (CTD) of the largest subunit of Pol II contains a series of heptad repeats (YSPTSPS) that are differentially modified during distinct phases of the transcription cycle. CTD residues are targets of various modifications, including methylation, phosphorylation, glycosylation, and proline isomerization (13). The best-studied of these CTD modifications is phosphorylation. In particular, phosphorylation on serine 5, serine 7, and serine 2 of the CTD repeats is readily apparent during Pol II's progression though the transcription cycle (8, 13, 39). Phosphorylation of serine 5 occurs early in the cycle, between initiation and elongation, and is predominantly catalyzed by the Cdk7 kinase associated with the general transcription factor (GTF) TFIIH (1, 46). Serine 7 is also phosphorylated early in the transcription cycle by Cdk7, but phosphorylation of this residue further increases toward the 3′ end of genes (8), mediated by the kinase Cdk9 (1, 44). Serine 2 phosphorylation occurs at the transition into productive elongation and can be catalyzed by two kinases: Cdk9 of P-TEFb and Cdk12 (BUR1 and CTDK-I, respectively, in Saccharomyces cerevisiae) (4, 32).
As a result of these modifications, the unphosphorylated Pol II (Pol IIa) that initiates transcription is radically transformed to the hyperphosphorylated Pol II (Pol IIo) that transcribes through the gene body during productive elongation (26, 39). Importantly, these marks serve as a platform for the recruitment of factors with functions relevant to particular stages in the transcription cycle (13). For example, early in the transcription cycle, the serine 5-phosphorylated CTD is bound by the mRNA capping enzymes (14, 15), and during elongation, the serine 2-phosphorylated CTD is bound by several factors, including elongation factors (30), RNA processing factors (35), and termination factors (27, 33). Thus, the phosphorylated CTD serves as a scaffold for the timely recruitment of factors during the transcription cycle to ensure proper mRNA biogenesis.
Since unphosphorylated Pol II forms the preinitiation complex, dephosphorylation of the CTD is critical for the recycling of terminated Pol II into a form that can initiate transcription (11). The mechanistic details of how termination interfaces with Pol II dephosphorylation are unknown (5), but the conversion of Pol IIo back to Pol IIa is catalyzed by CTD phosphatases. These phosphatases target different phosphorylated residues of the CTD repeat (34). The CTD phosphatases Rtr1, SCP1, and Ssu72 all target serine 5 phosphorylation, and abrogation of Ssu72 leads to defects in transcription in yeast (25, 36, 40, 47). Fcp1 is an essential CTD phosphatase in yeast and Drosophila, and although there is detailed information about how it binds Pol II (7, 20, 22, 43), the target of Fcp1 is less clear. In vitro assays have implicated both serine 2 and serine 5 as possible targets (18, 28), and serine 2 has been shown to be the in vivo target in yeast (10).
Several studies have indicated that Fcp1 has a direct role in transcription. Both in vitro biochemical studies and in vivo studies in yeast have shown that Fcp1 dephosphorylation increases transcription (10, 11, 23), and expression of Drosophila Fcp1 affected luciferase expression from reporter genes (45). Moreover, a role for Fcp1 in metazoan gene transcription in vivo is supported by a study showing FLAG-tagged Fcp1 colocalizes with bulk Rpb1 on Drosophila polytene chromosomes (45), and an RNA interference (RNAi) screen identified Drosophila Fcp1 as an important factor in optimal Hsp70 mRNA accumulation after heat shock (HS) (2). However, another study could not observe localization of the FLAG-tagged Fcp1 on the induced Hsp70 gene by chromatin immunoprecipitation (ChIP) (45). To reconcile and extend these studies, we examine the role of Fcp1 in Hsp70 gene regulation in vivo. Using immunostaining and ChIP, we show that Fcp1 colocalizes with phosphorylated Pol II at active sites of transcription, including the induced Hsp70 gene, in Drosophila polytene chromosomes and S2 cell culture. Moreover, RNAi depletion of Fcp1 in S2 cells (Fcp1-RNAi cells) results in the loss of Pol II in the coding region of heat shock-induced Hsp70. Intriguingly, this loss of Pol II signal correlates with a dramatic increase in phosphorylation of the non-chromatin-bound Pol II, and both of these effects are dependent on the catalytic activity of Fcp1. These findings indicate that the decrease in Pol II levels at Hsp70 in Fcp1-depleted cells are a consequence of free phosphorylated Pol II that cannot be recycled for additional rounds of transcription.
MATERIALS AND METHODS
Polytene immunostaining.
Immunofluorescence of proteins on fixed polytene chromosomes was performed as described by Schwartz et al. (42), utilizing 1:50 H14 antibody (Covance), 1:25 Fcp1 antibody, 1:25 Scp1 antibody, and 1:20 Ssu72 antibody.
Antibody generation.
Fcp1 and SCP1 were amplified from the Drosophila cDNA (Open Biosystems) and cloned into pET30a. The His-tagged protein was expressed in Escherichia coli BL21 and purified using Ni-nitrilotriacetic acid (NTA) magnetic agarose beads (Qiagen) according to the manufacturer's recommendation. The purified recombinant protein was used as the antigen for the generation of rabbit antibodies, according to published protocols (17).
Ssu72 was amplified from S2 cDNA and cloned into pET28a. The His-tagged protein was expressed in E. coli BL21 and purified using Ni-NTA-agarose (Qiagen), and the purified recombinant protein was sent to Pocono Rabbit Farm and Laboratory for the generation of guinea pig antibodies.
Generation of transgenic cell lines.
The Fcp1 coding region was cloned into the pDONR221 vector from S2 cDNA using the Gateway system (Invitrogen). The catalytically dead D215N version was created using QuikChange mutagenesis (Stratagene). The clones were transferred to a Cu-inducible Drosophila expression vector with an N-terminal FLAG tag based on the DEST48 vector (Invitrogen). The vectors were stably transfected into S2 cells by cotransfection with pCoBlast vector (Invitrogen) using Effectene transfection reagent (Qiagen). The primers used are listed in the supplemental material.
RNAi depletion.
The template DNA was amplified from Drosophila genomic DNA using the primers indicated in the supplemental material, and 1 μg of template DNA was used per 25 μl of the T7 polymerase reaction mixture (40 mM Tris-Cl, pH 8.0, 10 mM dithiothreitol [DTT], 2 mM spermidine-HCl, 20 mM MgCl2, 0.1 μl of T7 polymerase [lab stock]). The reaction mixture was incubated at 37°C for 4 h, and then the DNA was digested with DNase I. After extraction with phenol-chloroform, the RNAs were precipitated with 500 mM ammonium acetate (NH4Ac) and 2 volumes of ethanol. The RNAs were resuspended in diethyl pyrocarbonate (DEPC)-treated water, denatured at 80°C for 3 min, and annealed on ice. Drosophila S2 cells were grown in M3-Bacto peptone-yeast extract (BPYE) supplemented with 10% serum to a density between 3 × 106 and 5 × 106 cells/ml. After cultures were split to 1 × 106 cells/ml in serum-free M3 medium (at least a 1:3 split), the desired volume of cells was mixed with 10 μg/ml double-stranded RNA (dsRNA) and incubated at 25°C for 45 min, and an equal volume of M3-BPYE medium supplemented with 20% serum was added. After 5 days, the cells were harvested for the experiments.
Chromatin immunoprecipitation.
Drosophila S2 cells were grown in M3-BPYE–10% serum to approximately 6 × 106 cells/ml. To prepare the heat shock-induced chromatin, an equal volume of M3-BPYE medium (no serum) at 48°C was added to the cells, and the culture was incubated at 36.5°C for the desired time. Then, the same volume of M3-BPYE medium (no serum) at 4°C was added to bring the cells to room temperature, and formaldehyde was immediately added to a 1% final concentration. After samples were mixed for 2 min at room temperature, the cross-linking was quenched with the addition of glycine to a 125 mM final concentration. After samples were mixed for 2 min room temperature, the cells were cooled on ice for 2 min and centrifuged for 5 min at 4°C. After the medium was thoroughly removed, the cells were resuspended to 1 × 108 cells/ml in sonication buffer (20 mM Tris-Cl, pH 8.0, 2 mM EDTA, 0.5 mM EGTA, 0.5% SDS, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitor cocktail [Roche]). The cells were sonicated 12 times for 20 s each time with a 1-min rest in between at 4°C using a Bioruptor sonicator (Diagenode) on the highest setting. The sonicated material was centrifuged at 20,000 × g for 10 min at 4°C, and the supernatant was saved for the immunoprecipitation (IP). The non-heat shock (NHS)-induced chromatin was prepared in the same manner, except that 2 volumes of room temperature M3-BPYE medium (no serum) were added to the cells before cross-linking with formaldehyde. For each IP, 25 μl of material was mixed with 1 ml of IP buffer (20 mM Tris-Cl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 10% glycerol, 0.5% Triton X-100) and 30 μl of 50% protein A-agarose (EMD Millipore) at 4°C for 1 to 2 h. The cleared material was mixed with the antiserum at 4°C overnight; proteins were immunoprecipitated with 4 μl of rabbit anti-Rpb3 antiserum (Lis laboratory stock), 2 μl of rabbit anti-heat shock transcription factor (anti-HSF) antiserum (Lis laboratory stock), 10 μl of rabbit anti-Fcp1 (Reinberg lab stock), 10 μl of mouse monoclonal IgM H5 antibody (Covance) for phosphorylated serine 2 CTD, and 10 μl of mouse monoclonal IgM H14 antibody (Covance) for phosphorylated serine 5 CTD. Rabbit anti-mouse IgM antibody (20 μl) was added to the mouse monoclonal IP mixtures (to allow pulldown with protein A-agarose). Each IP mixture was incubated with 60 μl of protein A-agarose for 2 h at 4°C and washed once with low-salt buffer (20 mM Tris-Cl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), three times with high-salt buffer (20 mM Tris-Cl, pH 8.0, 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), once with LiCl buffer (10 mM Tris-Cl, pH 8.0, 250 mM LiCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate), and twice with Tris-EDTA (TE) buffer, pH 8.0. The bound protein/DNA was eluted twice with 250 μl of elution buffer (1% SDS, 100 mM NaHCO3). The cross-links were reversed by the addition of 20μl of 5 M NaCl and incubation at 65°C for 4 h. After extraction with phenol-chloroform, the immunoprecipitated DNA was precipitated and resuspended in double-distilled H2O (ddH2O), and each immunoprecipitated DNA was quantified using a Roche LightCycler 480 along with a standard curve of 10%, 1%, 0.1%, and 0.01% of input DNA (the standard curve was used to determine the amount of DNA immunoprecipitated). The primers used are listed in the supplemental material.
RT-qPCR.
After heat shock treatment, cells were pelleted, and RNA was isolated using an Omega E.Z.N.A. Total RNA kit I (R6834) and quantified using a NanoDrop 1000 spectrophotometer. Duplicate reverse transcription (RT) reactions were performed with 200 ng of total RNA and an oligo(dT) primer using Superscript III reverse transcriptase (18080; Invitrogen). Upon completion, the reaction mixtures were diluted 10-fold with 10 mM Tris-Cl (pH 8.0), and 2 μl was used in 10-μl quantitative PCRs (qPCRs) using the following primer sets (the number indicates the midpoint of the primer set relative to the TSS): Hsp70Ab+2211 primer set, Hsp26+624 primer set, Hsp83+3686 primer set, and RpL32+563 primer set (for primer sets, see the supplemental material). The qPCR was run on a Roche LightCycler 480, and the level of each mRNA was calculated relative to that of RpL32 using the 2−ΔCT method (where CT is threshold cycle).
Cellular fractionation.
The protocol for fractionation of proteins (free versus chromatin bound) was adapted from Aygun et al. (3). Briefly, cells were centrifuged at 1,000 rpm for 5 min at 4°C and resuspended in nucleus lysis buffer (20 mM Tris-Cl, pH 7.5, 3 mM EDTA, 10% glycerol, 150 mM potassium acetate [KAc], 1.5 mM MgCl2, 1 mM DTT, 0.1% NP-40, 1 mM PMSF, and protease inhibitors [Roche]) to 1 × 108 cells/ml. The cells were immediately homogenized with 60 strokes in a 2-ml Teflon Dounce homogenizer and centrifuged at 15,000 × g for 5 min at 4°C; the supernatant (free fraction) was transferred to a new tube, and the pellet was resuspended in nucleus lysis buffer to the equivalent of 1 × 108 cells/ml (chromatin fraction).
Microarray data accession number.
Genomic data described in this work have been deposited in the Gene Expression Omnibus (GEO) database under accession no. GSE38748.
RESULTS
Fcp1 localizes to sites of active transcription.
To investigate the role of Fcp1 in transcription, we generated an antibody to the previously identified Drosophila Fcp1 homolog, CG12252 (45). Immunoblotting using the antibody detected one major band at the predicted size of 97 kDa (see Fig. 2A). Additionally, this protein is depleted in Fcp1-RNAi cells, demonstrating that the antibody recognizes Fcp1 (see Fig. 2A). In order to assess the global distribution of Fcp1 at gene loci in vivo, Drosophila polytene chromosomes were immunostained for Fcp1 and phosphorylated Pol II (H14 monoclonal antibody). Fcp1 colocalized with phosphorylated Pol II at many interband loci, including developmental puffs at 2B, 23E, 74E, and 75B (Fig. 1A), although not always with the same intensity. This agrees with previous results indicating that Fcp1 localizes to most sites of active transcription (45).
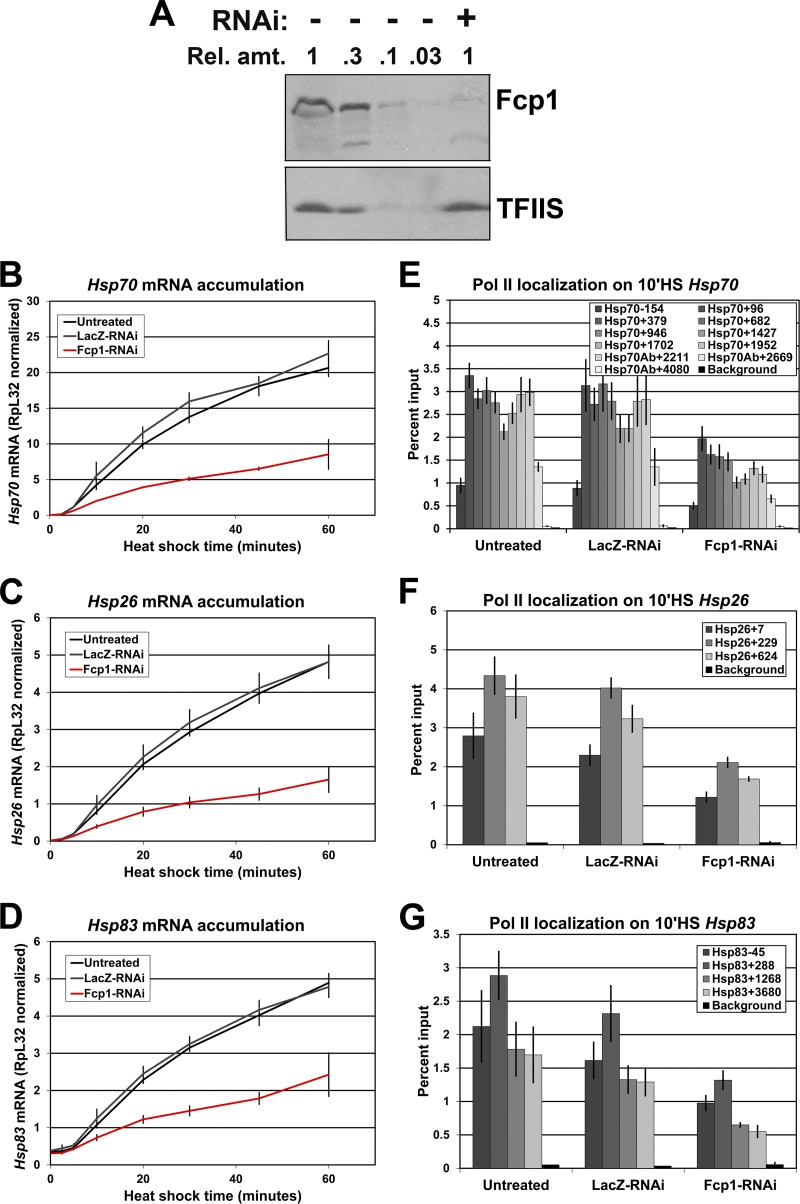
Fig 2.
Fcp1 depletion by RNAi diminishes the levels of Pol II on Hsp genes. (A) Western blots of whole-cell extracts from control (−) and Fcp1-RNAi (+) cells probed with antibodies for Fcp1 (1:1,000; lab stock) and TFIIS (1:3,000; lab stock loading control). The relative amount loaded is indicated (where 1 = 1 × 106 cells). (B to D) RT-qPCR results for heat shock time course in untreated, LacZ-RNAi, and Fcp1-RNAi cells. Total RNA was reverse transcribed with oligo(dT) and amplified with primer sets to the Hsp70, Hsp26, and Hsp83 genes. (E to G) ChIP results for the Pol II subunit Rpb3 in untreated, LacZ-RNAi, and Fcp1-RNAi S2 cells at 10 min of HS on the Hsp70, Hsp26, and Hsp83 genes. The legend indicates the midpoint of each PCR fragment. The y axis shows the percentage of input DNA immunoprecipitated (error bars indicate standard error of the mean of at least three biological replicates).
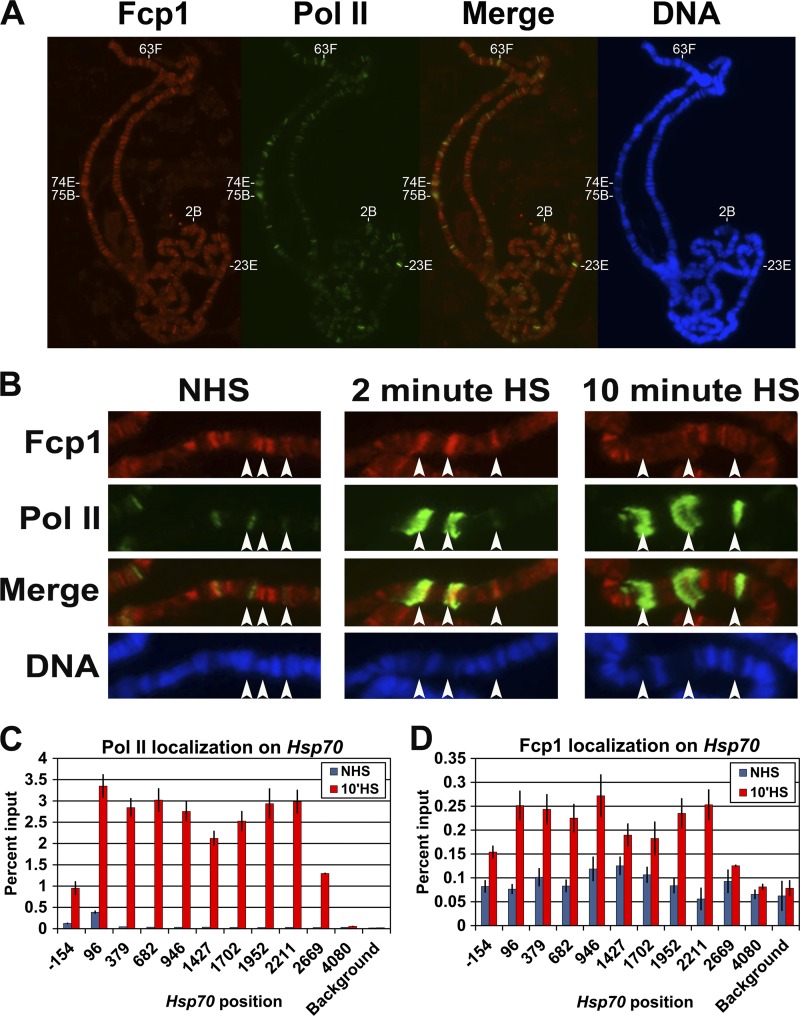
Fig 1.
Fcp1 localizes to transcriptionally active loci. (A and B) Drosophila spread polytene chromosomes immunostained with antibodies to Fcp1 (red) and serine 5-phosphorylated Pol II CTD (H14 antibody, green). The DNA is stained with 4′,6′-diamidino-2-phenylindole (blue). Merge is an overly of Fcp1 and serine 5-phosphorylated Pol II CTD. Panel A shows chromosomes from salivary glands under NHS conditions. In panel B, Hsp70 loci (87A and 87C [endogenous] and a single Hsp70 transgene at 87E) are marked by arrows in salivary glands under NHS and HS conditions. (C) ChIP results showing the enrichment of Pol II (Rpb3) at the Hsp70 gene in Drosophila S2 cells under NHS and HS conditions. (D) ChIP results of the Fcp1 enrichment on the Hsp70 gene in Drosophila S2 cells under NHS and HS conditions. The x axis shows the midpoint of each PCR fragment along the Hsp70 gene, and the y axis shows the percentage of input DNA immunoprecipitated (error bars indicate the standard error of the mean of at least four biological replicates).
Our previous work found that Fcp1 depletion decreases Hsp70 transcript levels by ∼50% compared to RNAi control cells treated with β-galactosidase dsRNA (LacZ-RNAi cells) (2). To further study the role of Fcp1 in HS gene regulation, Fcp1 and phosphorylated Pol II localization was examined on polytene chromosomes derived from salivary glands heat shocked at 37°C for various times. The fixed polytene chromosomes show that Fcp1 was recruited to the endogenous Hsp70 genes at the 87A and 87C loci after heat shock, as well as to a transgenic Hsp70 gene inserted at the 87E locus (arrows in Fig. 1B). Fcp1 immunostaining can be observed after 2 and 10 min of heat shock, albeit with reduced signal at 10 min (Fig. 1B). The weaker Fcp1 signal at 10 min of HS is likely due to decondensation of the loci as 60 min of recovery after heat shock results in the return of a strong immunofluorescence signal (see Fig. S1 in the supplemental material). A similar pattern of recruitment was observed for green fluorescent protein (GFP)-Fcp1 in living cells (see Fig. S2 in the supplemental material). Interestingly, despite the fact that our previous work showed that RNAi depletion of SCP1 and Ssu72 results in a modest effect on Hsp70 gene expression (2), both factors are also recruited to active Hsp70 loci (see Fig. S3 in the supplemental material). Taken together, these immunostaining and live-cell imaging experiments indicate that Fcp1 localizes to transcribing Hsp70 loci.
In order to assess the distribution of Fcp1 at higher resolution, we performed ChIP for Fcp1 at the uninduced and induced Hsp70 gene in Drosophila S2 cells. Under NHS conditions, Fcp1 is not enriched on the Hsp70 gene in comparison to the signal in a background region 30 kb away from the Hsp70 gene. In contrast, at 10 min of HS, Fcp1 is enriched on the transcribed region of Hsp70 compared to either region downstream of the transcribing Pol II (Fig. 1D, position 4080) or the background region (Fig. 1D). Fcp1 localizes evenly across the Hsp70 gene at 10 min of heat shock, and the pattern of Fcp1 enrichment is similar to that of Pol II (Fig. 1C). These findings suggest that Fcp1 associates with the elongating Pol II complex, similar to results from Saccharomyces cerevisiae (10).
Fcp1 depletion affects transcription of Hsp70 during heat shock.
Previously, we showed that RNAi knockdown of Drosophila Fcp1 resulted in a 2-fold reduction in Hsp70 mRNA accumulation after 20 min of heat shock (2). To further characterize this effect, we performed a heat shock time course in control and Fcp1 knockdown cells and examined the level of mRNA from three heat shock genes: Hsp70, Hsp26, and Hsp83. RNAi depletion of Fcp1 was performed using a dsRNA targeting the fifth exon of Fcp1 (see region A in Fig. S4A in the supplemental material). RNAi knockdown reduced Fcp1 protein levels by at least 90% as assayed by Western blotting (Fig. 2A). In agreement with the previous work, Fcp1 knockdown reduces Hsp70 mRNA levels 2- to 3-fold at heat shock time points of 5 min or longer (Fig. 2B). Hsp26 mRNA accumulation is similarly affected by Fcp1 knockdown (Fig. 2C), and Hsp83 mRNA accumulation is reduced, but less so (Fig. 2D).
The localization of Fcp1 on Hsp70 during heat shock suggests that these Fcp1 knockdown effects on Hsp70 mRNA levels may be due to direct effects on transcription. To investigate this, we used ChIP to assay Pol II localization at the active Hsp70 gene in control and Fcp1-depleted cells. Compared to untreated or LacZ-RNAi control cells, Fcp1 knockdown results in a reduction of Pol II throughout the Hsp70 transcription unit at 10 min of heat shock (Fig. 2E). The Pol II ChIP signal is slightly more reduced toward the 3′ end of the gene (from about 40% at 5′ end to about 60% at the 3′ end). Additionally, ChIP for Fcp1 showed that the knockdown reduced Fcp1 levels on the Hsp70 gene close to levels at the background region (see Fig. S4D in the supplemental material). Fcp1 knockdown also results in a reduction of Pol II levels on the transcription unit of induced Hsp26 and Hsp83 (Fig. 2F and G). Notably, the loss of Pol II signal is similar to the decrease in Hsp70 mRNA levels observed in Fcp1-RNAi cells (2). The comparable decrease in Hsp70 mRNA accumulation and Pol II levels indicates that Fcp1 knockdown affects transcription directly.
To ensure that the effects seen are a result of Fcp1 knockdown and not due to depletion of an unintended target, we depleted Fcp1 using a different dsRNA targeting a nonoverlapping region of the gene (see region B in Fig. S4A in the supplemental material). The new dsRNA showed comparable knockdown of Fcp1 (see Fig. S4B, region A compared to B), and a similar reduction in Pol II levels (see Fig. S4C). Given the highly unlikely overlap of any possible unintended targets for these two dsRNAs, this indicates that the effects seen are due to Fcp1 depletion.
We next investigated whether Fcp1 depletion also perturbs levels of the promoter-proximally paused Pol II. To test this, we used ChIP to examine the distribution of Pol II on Hsp70 in Fcp1-depleted cells under non-heat shock (NHS) conditions. We did not observe any effect of Fcp1 knockdown on the level of paused Pol II at Hsp70 in uninduced cells (see Fig. S5A in the supplemental material).
Constitutively expressed genes are not detectably affected by Fcp1 depletion.
Given the effect of Fcp1 depletion on transcription of Hsp70 during heat shock, we investigated whether Fcp1 depletion also affects transcription of constitutively expressed genes under NHS conditions. We performed ChIP for Pol II in NHS control and Fcp1-RNAi cells. Surprisingly, we failed to see significant changes in Pol II levels on any genes in Fcp1-depleted cells, even at highly expressed genes (Hsp83 and Thor) or moderately expressed genes (RpL32, β-1-tubulin, pnr, Hsp26, and Hsp70) (see Fig. S5B to G in the supplemental material). To exhaustively investigate constitutively expressed genes, we also performed global run-on sequencing (GRO-seq) in control LacZ-RNAi and Fcp1-RNAi cells to comprehensively quantify the transcriptionally engaged polymerases genome-wide. Comparison of biological replicates for the LacZ-RNAi control and Fcp1-RNAi cells failed to identify any genes with significantly reduced polymerase levels in Fcp1-depleted cells, and only seven genes (T48, Appl, mfas, GlcAT-P, amon, corn, and Rgk1) had increased polymerase levels (see Fig. S6). Moreover, it was also surprising to find that under NHS conditions Fcp1 depletion did not influence the expression of highly transcribed genes (according to GRO-seq gene body read density). The observed effects on heat shock-induced genes could be due to a requirement for Fcp1 under heat shock conditions. To investigate this, we examined Pol II on the constitutively expressed genes at 10 min of heat shock. Although the levels of Pol II on these genes is lower due to a general shutdown of transcription during heat shock, the Pol II levels are comparable for control and Fcp1-depleted cells (see Fig. S7 in the supplemental material). Thus, at this level of Fcp1 depletion, transcription is impaired on only the extremely highly expressed heat shock-induced genes.
Fcp1 depletion results in an increase in CTD phosphorylation of unengaged Pol II.
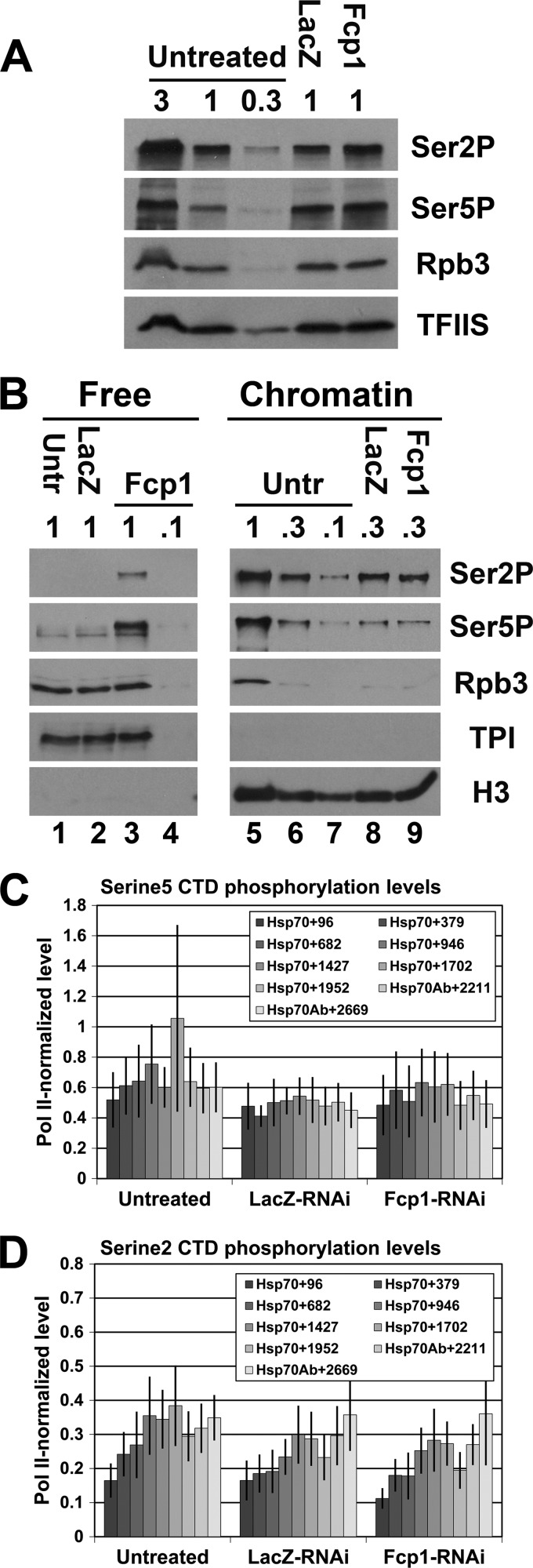
Next, we used Western blotting to examine the CTD phosphorylation level in Fcp1-RNAi cells for serine 5 and serine 2 phosphorylation (using the 3E10 and 3E8 monoclonal antibodies, respectively). Surprisingly, there were no dramatic changes in the overall levels of either epitope in whole-cell extracts (Fig. 3A). Previous studies have shown that hypophosphorylated Pol IIa initiates transcription (9, 21, 26, 31). Therefore, we next investigated whether the level of terminated non-chromatin-bound (free) unphosphorylated Pol II is reduced when Fcp1 is depleted. To do this, we examined free and chromatin-bound fractions of Pol II for changes in CTD phosphorylation in Fcp1-RNAi-treated cells. The free (cytoplasmic and nucleoplasmic) proteins were separated from chromatin-bound proteins with a modified version of a previously developed procedure (3). Histone H3 is enriched on the chromatin fraction, and triose phosphate isomerase (TPI) is enriched in the free fraction as expected (Fig. 3B). In addition, chromatin-bound Pol II in control cells had high levels of both serine 5 and serine 2 phosphorylation, and free Pol II had extremely low levels of phosphorylation (Fig. 3B, lanes 5 and 1, respectively). Although Fcp1 knockdown did not dramatically change levels of chromatin-bound phosphorylated Pol II (Fig. 3B, lane 9 compared to lanes 6 and 8), knockdown did increase the levels of free phosphorylated Pol II (Fig. 3B, lanes 3 and 4 compared to lanes 1 and 2). Similar levels of the Pol II subunit, Rpb3, show that the increase in phosphorylated CTD was not due to an increase in overall free Pol II in the Fcp1-RNAi cells (Fig. 3B, lanes 1 through 3). Interestingly, both serine 5 and serine 2 phosphorylation increased, indicating that Fcp1 is important for dephosphorylation of serine 2 and serine 5 in vivo (Fig. 3B). These results indicate that changes in the phosphorylation of free Pol II constitute a small fraction of the total phosphorylated Pol II in the cell.
Fig 3.
Fcp1 knockdown does not significantly change phosphorylation level of Pol II on the Hsp70 gene. (A) Western blots of whole-cell extracts from untreated, LacZ-RNAi, and Fcp1-RNAi cells probed with antibodies for phosphorylated CTD serine 2 (3E10 at 1:250; EMD Millipore), phosphorylated CTD serine 5 (3E8 at 1:250; EMD Millipore), Rpb3 (1:1,000; lab stock loading control), and TFIIS (1:3,000; lab stock loading control). The relative amount loaded is indicated (where 1 = 6 × 105 cells). (B) Western blots of free and chromatin-bound protein fractions from untreated, LacZ-RNAi, and Fcp1-RNAi cells probed with antibodies for phosphorylated CTD serine 5 (3E8 at 1:250; EMD Millipore), phosphorylated CTD serine 2 (3E10 at 1:250; EMD Millipore), Rpb3 (1:1,000; lab stock loading control), triose phosphate isomerase (1:1,000; lab stock loading control) and histone H3 (ab1791 at 1:500; Abcam). The relative amount loaded is indicated (where 1 = 1 × 106 cells). (C and D) ChIP results of the serine 5- and serine 2-phosphorylated Pol II CTD enrichment relative to Pol II enrichment on the Hsp70 gene in untreated, LacZ-RNAi, and Fcp1-RNAi S2 cells at 10 minutes of HS. The legend indicates the midpoint of each PCR fragment. The y axis shows the ratio of the percentages of input DNA immunoprecipitated (error bars indicate standard error of the mean of at least three biological replicates).
Although fractionation showed that the level of chromatin-bound Pol II was unaffected under NHS conditions, we next investigated whether the Pol II reduction on the Hsp70 gene body in Fcp1-depleted cells might be associated with abnormal Pol II phosphorylation levels on the gene during heat shock. ChIP using antibodies to serine 5- and serine 2-phosphorylated CTD (using the H14 and H5 monoclonal antibodies, respectively) showed reduced levels of phosphorylated Pol II across Hsp70 at 10 min of heat shock, comparable to the Pol II reduction. We also saw a similar reduction in serine 5-phosphorylated CTD and serine 2-phosphorylated CTD using the 3E8 and 3E10 antibodies, respectively (data not shown). Therefore, Pol II-normalized phosphorylation levels of both serine 5 and serine 2 showed no significant change in any region of Hsp70 (Fig. 3C and D). The relatively uniform reduction of all forms of Pol II across Hsp70 in Fcp1 knockdown cells indicates that Pol II modifications during elongation occurred normally and suggests that it is the Pol II initiation rate that is affected in induced cells by Fcp1 knockdown (Fig. 2B).
Transcription defects of Fcp1 depletion are dependent on Fcp1 phosphatase activity.
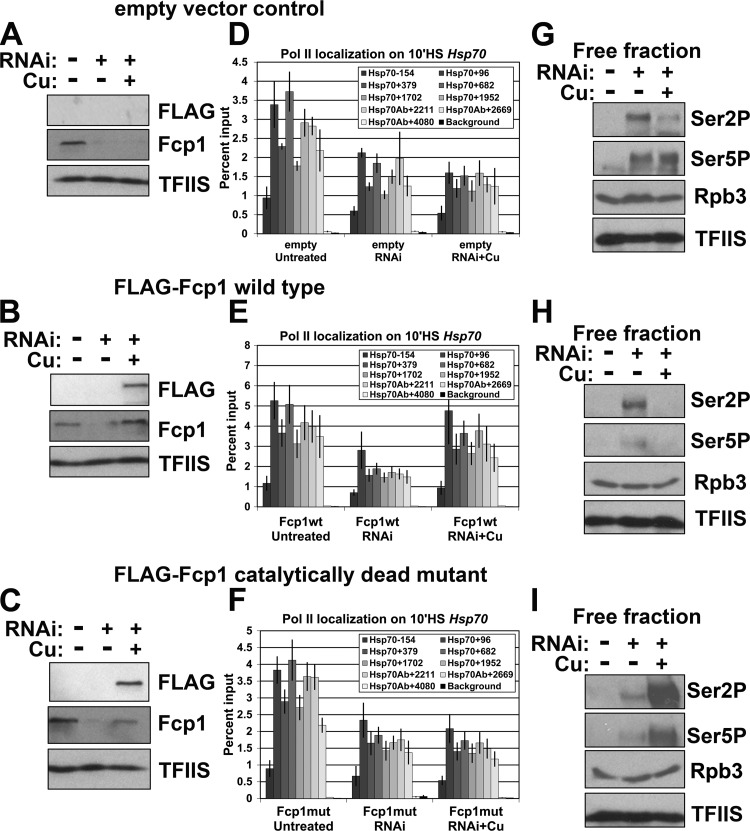
Previous work has shown that some functions of yeast Fcp1 can occur independently of its catalytic activity (11). Therefore, the effects seen in our various assays could be due to the loss of the Fcp1 phosphatase activity or loss of the protein itself, independent of its catalytic function. Fcp1 is the founding member of the Fcp1 homology domain (FCPH) family of phosphatases, which contain a highly conserved DXDX(T/V) active site. Mutation of either aspartate residue abolishes Fcp1 phosphatase activity (19). To test if the effects we saw were dependent on the phosphatase activity of Fcp1, we stably transfected a copper-inducible FLAG-tagged transgene with either a wild-type Fcp1 or a catalytically dead version (in which the second catalytic aspartate was mutated to asparagine) into S2 cells. A dsRNA targeting the Fcp1 3′ untranslated region (UTR) (see Fig. S4A, region C, in the supplemental material) was used to knock down endogenous Fcp1 to similar levels as the other dsRNAs (see Fig. S4B). RNAi-resistant wild-type or mutant versions of Fcp1 were then reexpressed by addition of CuSO4 to the cell culture medium (Fig. 4A to C). We examined Pol II distribution in untreated cultures and in RNAi cultures with or without CuSO4. In all cell lines, 3′ UTR RNAi depletion reduced Pol II levels on heat shock-induced Hsp70 to levels similar to those of other dsRNAs (Fig. 4D to F). Reexpression of the wild-type Fcp1 partially restored the Fcp1 knockdown in Hsp70 Pol II after 10 min of HS to untreated levels (Fig. 4E). In contrast, neither an empty vector control nor the catalytically dead version restored Pol II levels (Fig. 4D and F). Both Hsp26 and Hsp83 showed some rescue upon reexpression of the wild-type Fcp1 but not the catalytically dead mutant (see Fig. S8 in the supplemental material). Similar to the other dsRNAs, 3′ UTR RNAi depletion also increased the phosphorylated free Pol II (Fig. 4G to I, middle lanes). Cells reexpressing the wild type had levels of phosphorylated free Pol II similar to levels of untreated cells (Fig. 4H, right lane), but, interestingly, cells reexpressing the catalytically dead Fcp1 further increased the amount of phosphorylated free Pol II above Fcp1 knockdown alone (Fig. 4I, right lane). To determine if this additional increase in phosphorylation of free Pol II has an effect on the transcription of constitutively expressed genes under NHS conditions, we performed ChIP for Pol II under NHS conditions in cells reexpressing the mutant Fcp1, but we did not see any effect (see Fig. S9 in the supplemental material). The rescue of Pol II levels on heat shock-induced genes by wild-type Fcp1, but not the catalytic mutant, demonstrates that the effects of Fcp1 knockdown are due to loss of the Fcp1 phosphatase activity.
Fig 4.
Reexpression of wild-type Fcp1 rescues Pol II levels on heat shock-induced Hsp70. (A to C) Western blots of whole-cell extracts with and without Fcp1-RNAi and with and without Cu induction of the transgenic Fcp1 from control (empty vector) (A), FLAG-tagged wild-type Fcp1 transgene cells (B), and FLAG-tagged catalytically dead mutant Fcp1 transgene cells (C) probed with antibodies for FLAG (1:5,000; Stratagene), Fcp1 (1:1,000; lab stock), and TFIIS (1:3,000; lab stock loading control). (D to F) ChIP results for the Pol II subunit Rpb3 enrichment on the Hsp70 gene at 10 minutes of HS for control (empty vector) (D), FLAG-tagged wild-type Fcp1 transgene (Fcp1wt) cells (E), and FLAG-tagged catalytically dead mutant Fcp1 transgene (Fcp1mut) cells (F). The legend indicates the midpoint of each PCR fragment. The y axis shows the percentages of input DNA immunoprecipitated (error bars indicate standard error of the mean of three biological replicates). (G to I) Western blots of serine 2-phosphorylated CTD (3E10 at 1:250; EMD Millipore), serine 5 phosphorylated CTD (3E8 at 1:250; EMD Millipore), Rpb3 (1:1,000; lab stock loading control), and TFIIS (1:3,000; lab stock loading control) on the free fraction with and without Fcp1-RNAi and with and without Cu induction of the transgene from control (empty vector) (G), FLAG-tagged wild-type Fcp1 transgene cells (H), and FLAG-tagged catalytically dead mutant Fcp1 transgene cells (I). The relative amount loaded is indicated (where 1 = 1 × 106 cells).
Codepletion of Fcp1 and P-TEFb restores Pol II levels at the 5′ end of Hsp70.
Levels of Pol II on a gene are controlled at multiple steps during the transcription cycle. For example, the level of Pol II on the 5′ ends of genes depends upon both the rate of initiation and the rate of pause escape (12). This is exemplified at the Hsp70 gene, where under uninduced conditions, the Pol II initiation rate is higher than the pause escape rate, and thus the 5′ end is highly occupied by a transcriptionally engaged Pol II. In contrast, during an optimal heat shock, Pol II is efficiently released into productive elongation, and the Hsp70 genes are fully occupied with a transcribing Pol II complex every 80 bp (29). Thus, if our hypothesis is that Fcp1 knockdown diminishes levels of Pol II on Hsp70 by reducing initiation, we predict that the level of Pol II on the 5′ end of Hsp70 during Fcp1 knockdown will increase back to its fully occupied, induced levels by reducing the pause escape rate.
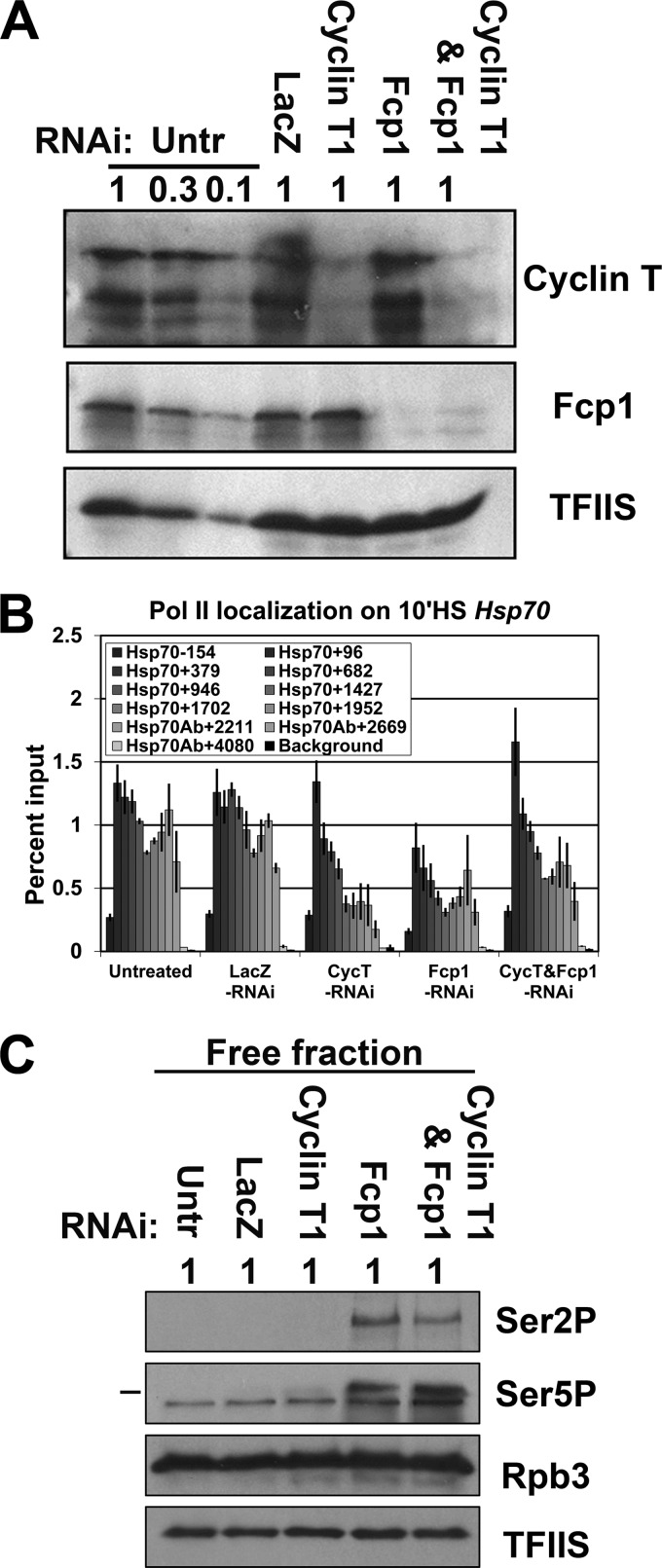
Since P-TEFb activity is required for pause escape (37), we reasoned that the pause escape rate could be reduced by depleting the P-TEFb subunit, cyclin T1. We therefore used RNAi to deplete the cyclin T1 (cyclin T1-RNAi cells) alone or in combination with Fcp1 (cyclin T1-Fcp1-RNAi cells), and performed Pol II ChIP at 10 min of heat shock. Cyclin T1 is reduced by about 90% when knocked down alone or in combination with Fcp1, and Fcp1 depletion levels are similar when Fcp1 is knocked down alone or in combination with cyclin T1 (Fig. 5A). As we expected, ChIP for Pol II showed that the rate of pause escape was reduced in cells depleted of cyclin T1. Levels of Pol II in the pause region at the 5′ end of the gene were unaffected, remaining fully occupied with paused Pol II, but levels of Pol II in the downstream gene body region were reduced, indicative of the lower rate of pause escape (Fig. 5B). Consistent with previous experiments, Fcp1 knockdown reduced Pol II levels in the pause region (Hsp70+96 primer set) to approximately half the control level. As we hypothesized, depletion of cyclin T1 in conjunction with Fcp1 increased the Pol II level in the paused region compared to Fcp1 depletion alone, restoring full Pol II occupancy on the 5′ end of the gene in cells depleted of both Fcp1 and cyclin T1, similar to control or cyclin T1 knockdown alone (Fig. 5B). Similar results were seen on both Hsp26 and Hsp83 (see Fig. S10 in the supplemental material).
Fig 5.
Cyclin T1 and Fcp1 double knockdown increases Pol II levels at the 5′ end of Hsp70 during heat shock. (A) Western blots of whole-cell extracts from untreated, LacZ-RNAi, cyclin T1-RNAi, Fcp1-RNAi, and double knockdown cyclin T1-Fcp1-RNAi cells probed with antibodies for cyclin T1 (1:1,000; lab stock), Fcp1 (1:1,000; lab stock), and TFIIS (1:3,000; lab stock loading control). The relative amount loaded is indicated (where 1 = 1.5 × 106 cells). (B) ChIP results for the Pol II subunit Rpb3 enrichment on the Hsp70 gene in untreated, LacZ-RNAi, cyclin T1-RNAi, Fcp1-RNAi, and double knockdown cyclin T1-Fcp1-RNAi cells at 10 min of heat shock. The x axis shows the midpoint of each PCR fragment along the Hsp70 gene, and the y axis shows the percentage of input DNA immunoprecipitated (error bars indicate the standard error of the mean of at least four biological replicates). (C) Phosphorylated CTD serine 5 (3E8 at 1:250; EMD Millipore), serine 2 (3E10 at 1:250; EMD Millipore), and TFIIS (1:3,000; lab stock loading control) Western blots of nonchromatin (free) fractions from untreated, LacZ-RNAi, cyclin T1-RNAi, Fcp1-RNAi, and double knockdown cyclin T1-Fcp1-RNAi cells. The relative amount loaded is indicated (where 1 = 1 × 106 cells).
P-TEFb phosphorylates the CTD on serine 2, the presumed target of Fcp1 in Drosophila; therefore, we investigated if the codepletion's rescue of the 5′ levels of Pol II on induced HS genes could be caused by the codepletion reducing the high level of phosphorylated free Pol II seen in the Fcp1 knockdown. Western blotting showed that the levels of phosphorylated free Pol II (both serine 5 and serine 2) remained high in the codepleted cells, similar to levels with Fcp1 knockdown alone (Fig. 5C). Taken together, our results support the model that the most highly expressed genes depend on Fcp1 phosphatase activity to provide sufficient levels of unphosphorylated Pol II to support correspondingly high initiation rates.
DISCUSSION
Our previous studies showed that Fcp1 depletion in Drosophila S2 cells results in reduced Hsp70 mRNA accumulation after heat shock (2). In this study, we set out to further investigate the role of Fcp1 in transcription in vivo. Consistent with a direct role in transcription, we have demonstrated that Drosophila Fcp1 localizes to actively elongating Pol II complexes. In particular, Fcp1 colocalizes with Pol II at many loci on polytene chromosomes under NHS conditions. Although the ratio between Pol II and Fcp1 signals varied at different loci, the variation in relative signal may represent differences in the transcription level at each locus. Immunostaining at 2 min of heat shock, when Pol II is being recruited to the 87A and 87C loci, showed strong Fcp1 signal, but the Fcp1 signal was more diffuse at 10 min of heat shock when the loci are saturated with Pol II and maximally decondensed (48). At higher resolution, our ChIP experiments showed that Fcp1 localization on heat shock-induced Hsp70 was evenly distributed across the gene in the same pattern as Pol II. These findings are consistent with previous in vitro and ChIP experiments in yeast showing that Fcp1 colocalizes with elongating Pol II (6, 10, 24).
Fcp1 temperature-sensitive mutants in yeast have increased serine 2 phosphorylation on genes at the restrictive temperature (10). Therefore, it was surprising to find that Pol II-normalized CTD phosphorylation levels on heat shock-induced Hsp70 did not change in Fcp1 depleted cells. There are several possible explanations. First, RNAi-treated cells may still contain enough Fcp1 to transiently associate with the elongation complex and prevent abnormal phosphorylation levels. Second, CTD phosphorylation may be maximal on heat shock-induced Hsp70 and therefore cannot increase further in Fcp1-RNAi cells. Finally, Fcp1 may not catalyze CTD dephosphorylation of the elongating complex. Although in vivo experiments in S. cerevisiae found evidence for Fcp1 catalytic activity during transcription and posttermination, an in vitro study indicated that free Pol II is the preferred substrate of Fcp1 (24). Our ChIP results are consistent with dephosphorylation occurring after elongation. In addition, Fcp1 depletion does not change the amount of phosphorylation or total Pol II in the chromatin fraction but dramatically increased the amount of phosphorylated Pol II in the free fraction.
Strikingly, Fcp1 depletion resulted in a reduction of Pol II levels across all regions of the induced Hsp70, Hsp26, and Hsp83 genes, similar in magnitude to the decrease in the corresponding mRNAs (2). However, we failed to see significant changes in Pol II levels on any genes in Fcp1-depleted cells under non-heat shock conditions by ChIP or GRO-seq. Although we cannot eliminate the possibility that Fcp1 depletion affects heat shock signaling, we believe it is unlikely because recruitment of the activator HSF to Hsp70 is unaffected (data not shown). The detection of a 2-fold reduction in Hsp70 transcription with no detectable changes in transcription of constitutively expressed genes may be explained by the extremely high levels of transcription on induced heat shock genes compared to NHS genes. It has been estimated that an optimally induced Hsp70 gene has Pol II complexes every 80 bp (29). This translates into a high turnover of Pol II, with a new Pol II initiating about every 4 s, corresponding to greater than a 100-fold increase in transcription (16). Thus, a decrease in unphosphorylated free Pol II, the form which is required for initiation (26), may slow initiation on induced Hsp70 sufficiently to cause an increase in the spacing between elongating Pol II complexes on heat shock-induced Hsp70. This increased spacing would cause a corresponding decrease in Pol II ChIP along the Hsp70 transcription unit (see Fig. S11, panel B versus panel A, in the supplemental material). Based on GRO-seq gene body reads, both Hsp83 and Thor were among the highest expressed in uninduced cells, but, notably, Hsp83 is known to be transcribed at a 11-fold higher level in induced cells based on pulse labeling measurements in vivo (38). Thus, no constitutively expressed gene in S2 cells has a density of Pol II approaching that of induced Hsp70, Hsp26, or Hsp83. The fact that only super highly expressed HS genes are affected indicates that the concentration of unphosphorylated Pol II, which is required for initiation, is not limiting for the vast majority of genes expressed (see Fig. S11, panel B versus panel A). In agreement with this model, slowing the rate of pause escape by codepletion of the P-TEFb subunit cyclin T1 with Fcp1 depletion restored Pol II levels on the 5′ end of induced Hsp70 to control levels by making pause escape sufficiently slow that the reduced initiation rate could still fill the pause site to its normal level.
Overall, our study demonstrates that Fcp1 depletion causes reduced HS gene expression and a corresponding reduction of Pol II on induced Hsp70, and it also causes a dramatic increase in phosphorylation of both serine 2 and serine 5 on free Pol II. Although these results suggest that Fcp1 dephosphorylates both serine 2 and serine 5, we cannot rule out that its activity is coupled to a second phosphatase. Further studies are required to determine if both residues are direct targets of Drosophila Fcp1 or if dephosphorylation of these different residues is indeed coupled. Taken together, our results are consistent with Fcp1 depletion impairing the ability to recycle Pol II, reducing the pool of initiation-competent polymerase, and leading to reduced levels of transcribing Pol II on the highly transcribed heat shock genes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Janis Werner for performing all of the polytene staining experiments. We also thank Ross MacIntyre for generously providing the triose phosphate isomerase antibody.
This work was supported by NIH grant GM25232 to J.T.L., NRSA NIH grant GM087003 to M.S.B., and by the Howard Hughes Medical Institute and NIH grant GM37120 to D.R.
Footnotes
Published ahead of print 25 June 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Akhtar MS, et al. 2009. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell 34:387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ardehali MB, et al. 2009. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 28:1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aygün O, Svejstrup J, Liu Y. 2008. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl. Acad. Sci. U. S. A. 105:8580–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartkowiak B, et al. 2010. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 24:2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buratowski S. 2005. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 17:257–261 [DOI] [PubMed] [Google Scholar]
- 6. Calvo O, Manley JL. 2005. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 24:1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chambers RS, Wang BQ, Burton ZF, Dahmus ME. 1995. The activity of COOH-terminal domain phosphatase is regulated by a docking site on RNA polymerase II and by the general transcription factors IIF and IIB. J. Biol. Chem. 270:14962–14969 [DOI] [PubMed] [Google Scholar]
- 8. Chapman RD, et al. 2007. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318:1780–1782 [DOI] [PubMed] [Google Scholar]
- 9. Chesnut JD, Stephens JH, Dahmus ME. 1992. The interaction of RNA polymerase I1 with the adenovirus-2 major of the C-terminal late promoter is precluded by phosphorylation domain of subunit IIa*. J. Biol. Chem. 267:10500–10506 [PubMed] [Google Scholar]
- 10. Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cho H, et al. 1999. A protein phosphatase functions to recycle RNA polymerase II. Genes Dev. 13:1540–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Core LJ, Lis JT. 2008. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319:1791–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Egloff S, Murphy S. 2008. Cracking the RNA polymerase II CTD code. Trends Genet. 24:280–288 [DOI] [PubMed] [Google Scholar]
- 14. Fabrega C, Shen V, Shuman S, Lima CD. 2003. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol. Cell 11:1549–1561 [DOI] [PubMed] [Google Scholar]
- 15. Ghosh A, Shuman S, Lima CD. 2011. Structural insights to how mammalian capping enzyme reads the CTD code. Mol. Cell 43:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gilmour DS, Lis JT. 1985. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol. Cell. Biol. 5:2009–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harlow E, Lane D. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 18. Hausmann S, Koiwa H, Krishnamurthy S, Hampsey M, Shuman S. 2005. Different strategies for carboxyl-terminal domain (CTD) recognition by serine 5-specific CTD phosphatases. J. Biol. Chem. 280:37681–37688 [DOI] [PubMed] [Google Scholar]
- 19. Hausmann S, Shuman S. 2002. Characterization of the CTD phosphatase Fcp1 from fission yeast. Preferential dephosphorylation of serine 2 versus serine 5. J. Biol. Chem. 277:21213–21220 [DOI] [PubMed] [Google Scholar]
- 20. Kamenski T, Heilmeier S, Meinhart A, Cramer P. 2004. Structure and mechanism of RNA polymerase II CTD phosphatases. Mol. Cell 15:399–407 [DOI] [PubMed] [Google Scholar]
- 21. Kang ME, Dahmus ME. 1993. RNA polymerases IIA and IIO have distinct roles during transcription from the TATA-less murine dihydrofolate reductase promoter. J. Biol. Chem. 268:25033–25040 [PubMed] [Google Scholar]
- 22. Kimura M, Suzuki H, Ishihama A. 2002. Formation of a carboxy-terminal domain phosphatase (Fcp1)/TFIIF/RNA polymerase II (pol II) complex in Schizosaccharomyces pombe involves direct interaction between Fcp1 and the Rpb4 Subunit of pol II. Mol. Cell. Biol. 22:1577–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kobor MS, et al. 1999. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol. Cell 4:55–62 [DOI] [PubMed] [Google Scholar]
- 24. Kong SE, et al. 2005. Interaction of Fcp1 phosphatase with elongating RNA polymerase II holoenzyme, enzymatic mechanism of action, and genetic interaction with elongator. J. Biol. Chem. 280:4299–4306 [DOI] [PubMed] [Google Scholar]
- 25. Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. 2004. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol. Cell 14:387–394 [DOI] [PubMed] [Google Scholar]
- 26. Laybourn PJ, Dahmus ME. 1990. Phosphorylation of RNA polymerase IIA occurs subsequent to interaction with the promoter and before the initiation of transcription. J. Biol. Chem. 265:13165–13173 [PubMed] [Google Scholar]
- 27. Licatalosi DD, et al. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9:1101–1111 [DOI] [PubMed] [Google Scholar]
- 28. Lin PS, Dubois M-F, Dahmus ME. 2002. TFIIF-associating carboxyl-terminal domain phosphatase dephosphorylates phosphoserines 2 and 5 of RNA polymerase II. J. Biol. Chem. 277:45949–45956 [DOI] [PubMed] [Google Scholar]
- 29. Lis J. 1998. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harbor Symp. Quant. Biol. 63:347–356 [DOI] [PubMed] [Google Scholar]
- 30. Liu J, et al. 2011. Solution structure of tandem SH2 domains from Spt6 protein and their binding to the phosphorylated RNA polymerase II C-terminal domain. J. Biol. Chem. 286:29218–29226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu H, Flores O, Weinmann R, Reinberg D. 1991. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc. Natl. Acad. Sci. U. S. A. 88:10004–10008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marshall NF, Peng J, Xie Z, Price DH, Chem JB. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal Domain kinase. J. Biol. Chem. 271:27176–27183 [DOI] [PubMed] [Google Scholar]
- 33. Meinhart A, Cramer P. 2004. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430:223–226 [DOI] [PubMed] [Google Scholar]
- 34. Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. 2005. A structural perspective of CTD function. Genes Dev. 19:1401–1415 [DOI] [PubMed] [Google Scholar]
- 35. Morris DP, a Greenleaf L. 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 275:39935–39943 [DOI] [PubMed] [Google Scholar]
- 36. Mosley AL, et al. 2009. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell 34:168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ni Z, et al. 2008. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol. Cell. Biol. 28:1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Connor D, Lis JT. 1981. Two closely linked transcription units within the 63B heat shock puff locus of D. melanogaster display strikingly different regulation. Nucleic Acids Res. 9:5075–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Payne JM, Laybourn PJ, Dahmus ME. 1989. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J. Biol. Chem. 264:19621–19629 [PubMed] [Google Scholar]
- 40. Reyes-Reyes M, Hampsey M. 2007. Role for the Ssu72 C-terminal domain phosphatase in RNA polymerase II transcription elongation. Mol. Cell. Biol. 27:926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saunders A, Core LJ, Lis JT. 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7:557–567 [DOI] [PubMed] [Google Scholar]
- 42. Schwartz BE, Werner JK, Lis JT. 2004. Indirect immunofluorescent labeling of Drosophila polytene chromosomes: visualizing protein interactions with chromatin in vivo. Methods Enzymol. 376:393–404 [DOI] [PubMed] [Google Scholar]
- 43. Suh M-H, et al. 2005. Fcp1 directly recognizes the C-terminal domain (CTD) and interacts with a site on RNA polymerase II distinct from the CTD. Proc. Natl. Acad. Sci. U. S. A. 102:17314–17319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tietjen JR, et al. 2010. Chemical-genomic dissection of the CTD code. Nat. Struct. Mol. Biol. 17:1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tombácz I, Schauer T, Juhász I, Komonyi O, Boros I. 2009. The RNA Pol II CTD phosphatase Fcp1 is essential for normal development in Drosophila melanogaster. Gene 446:58–67 [DOI] [PubMed] [Google Scholar]
- 46. Watanabe Y, et al. 2000. Modulation of TFIIH-associated kinase activity by complex formation and its relationship with CTD phosphorylation of RNA polymerase II. Genes Cells 5:407–423 [DOI] [PubMed] [Google Scholar]
- 47. Yeo M, Lin PS, Dahmus ME, Gill GN. 2003. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J. Biol. Chem. 278:26078–26085 [DOI] [PubMed] [Google Scholar]
- 48. Zobeck KL, Buckley MS, Zipfel WR, Lis JT. 2010. Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol. Cell 40:965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.