It has been well over a century since the responses of bacteria to chemical stimuli were first documented by Engelman (1) and Pfeffer (2). In the 1960s, Adler reinitiated the study of bacterial chemotaxis (3), and during the past few decades, numerous laboratories have investigated the molecular basis of stimulus-response coupling in this system. The ability to study behavior in a unicellular organism with facile genetics has made bacterial chemotaxis an attractive model system for a diverse group of researchers, including geneticists, biochemists, biophysicists, and molecular, structural, and theoretical biologists. The biochemistry of the signaling pathway has been elucidated, and atomic resolution structures are available for most components of the signaling circuitry. This raises the possibility of eventually achieving a complete understanding of the mechanisms through which protein modifications and protein–protein interactions control the flow of information in this model sensory system. However, with the opportunity to pursue investigations at an ever-increasing level of molecular detail, there is a tendency to lose sight of the bigger picture. There is still a lot to be learned from the swimming responses of bacterial cells. In fact, it is the behavioral responses that molecular investigations seek to explain.
In this issue of the Proceedings, Jasuja et al. (4) report an elegant physiological study of chemotactic responses in Escherichia coli and Salmonella typhimurium by using photolabile caged chemoeffectors and computer-assisted motion analysis to precisely measure the stimulus-response relation. Response amplitudes were found to increase logarithmically with stimulus strength, indicating involvement of cooperative interactions in the excitation pathway. This feature, which facilitates responses to broad ranges of stimuli concentrations, underscores the sophistication of the bacterial chemotaxis system and suggests the exquisite complexity of interactions among the relatively small number of protein components that control swimming behavior.
Bacteria migrate in chemical gradients, toward attractant chemoeffectors or away from repellents, by using a temporal sensing mechanism that involves a rudimentary memory. When the concentration of attractant increases over time, cells suppress tumbling behavior and increase the length of swimming runs, producing a biased random walk that results in net migration up an attractant gradient. More than 20 years ago, Koshland proposed a basic scheme for the biochemical system controlling this behavior, invoking a “response regulator” molecule (5). He proposed that the level of the response regulator relative to a threshold value determined the flagellar response, and that the level of the response regulator was itself controlled by environmental stimuli. This scheme has been validated; the response regulator as well as the components that control its activity have been identified, and the biochemical pathways involved in chemotaxis have been established (for reviews, see refs. 6 and 7).
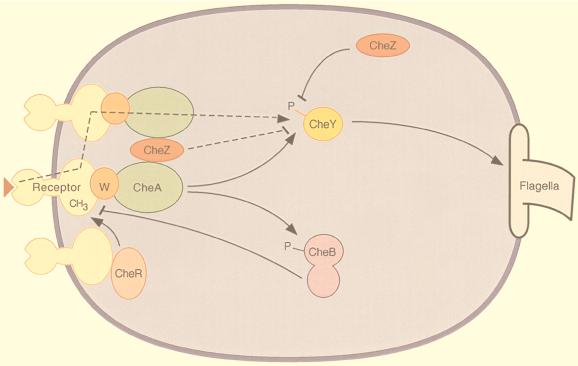
The response regulator is phosphorylated CheY, which binds to a component of the flagellar motor, effecting clockwise flagellar rotation and a tumbling response. The rest of the signal transduction proteins function either directly or indirectly to modulate the level of CheY phosphorylation (Fig. 1). A family of transmembrane chemoreceptors sense chemical stimuli, and through the adapter protein CheW, modulate the activity of the histidine protein kinase CheA. The signaling activity of the receptors is influenced not only by ligand occupancy, but also by the state of methylation of specific glutamate residues within their cytoplasmic domains. The reversible modifications of the receptors are catalyzed by methyltransferase CheR and methylesterase CheB, an enzyme that is also activated by phosphorylation. CheA provides phosphoryl groups to both CheY and CheB. Autophosphatase activity of CheY results in a phosphorylated half-life of less than a minute, and the lifetime is reduced even further by CheZ, a protein that accelerates the dephosphorylation of CheY.
Figure 1.
Schematic diagram of bacterial chemotaxis signal transduction. Transmembrane receptor proteins bind chemoeffectors and, through an adapter protein, CheW, control the activity of histidine protein kinase CheA. The cytoplasmic domains of the receptors are methylated by methyltransferase CheR and demethylated by methylesterase CheB. Attractant binding decreases kinase activity, while receptor methylation increases kinase activity. CheA provides phosphoryl groups to CheY and CheB, producing active forms of these proteins. Phosphorylated CheB demethylates receptors, providing a feedback loop that contributes to adaptation. The response regulator, phosphorylated CheY, binds to the flagellar motor, inducing clockwise flagellar rotation and a tumbling response. CheZ accelerates the dephosphorylation of CheY. The dashed lines indicate two postulated mechanisms for amplification of the excitation signal. Arrowheads and bars represent positive and negative regulation of phosphorylation, methylation, and clockwise flagellar rotation.
This relatively simple biochemical scheme controls an extremely fine-tuned behavior. The system is exquisitely sensitive over a very broad range, allowing chemotactic responses to changes in attractant concentration of less than 1% over a concentration range spanning 5 orders of magnitude (8). The system also exhibits adaptation. Cells show transient responses to changes in chemoeffector concentrations, returning to prestimulus steady state behavior even though the alteration in chemoeffector concentration is maintained. Our current understanding of the biochemical pathways can qualitatively explain these behavioral phenomena, but to date, computer models have not been able to recapitulate all essential features of chemotactic behavior (9).
One critical parameter missing from the description of chemotactic behavior is the relation between stimulus and response. This relation, referred to as chemotactic gain, is defined as the fractional increase in counterclockwise motor bias divided by the fractional change in receptor occupancy. Our current understanding of gain derives from early studies that, while technically elegant, lacked the necessary time resolution and/or population statistics. These studies showed the change in behavior to be proportional to the rate of change in chemoeffector concentration when examined under conditions where the rate of change in chemoeffector concentration was slow relative to chemotactic excitation time, thus allowing contributions from both excitation and adaptation pathways (10, 11). A subsequent analysis with greater temporal resolution was limited to a single attractant concentration jump at one prestimulus chemoeffector concentration (12). In the absence of additional data, it has been generally assumed that chemotactic gain is invariant over different ranges of stimulus.
Jasuja and colleagues (4) have now examined the stimulus-response relation using rapid and defined concentration jumps over a broad range of chemoeffector concentrations. Their extensive study was facilitated by two technologies. Computer-assisted motion analysis was used to examine relatively large populations of bacteria with 33-msec time-frame resolution (13) and rapid, defined jumps in chemoeffector concentration were generated by photolysis of caged chemoeffectors (14). Both the motility response and the change in attractant concentration could be rigorously quantified. However, it should be noted that calculation of gain requires values for receptor occupancy that are calculated using dissociation constants for receptor-attractant interaction determined under conditions that are not identical to the present study.
In contrast to prevailing assumptions, Jasuja et al. find that gain is not invariant with changes in receptor occupancy. Rather, responses increase logarithmically with stimulus strength. Thus gain increases with decreasing changes in receptor occupancy, effectively extending responses over a greater range of stimulus. The logarithmic relation is maintained over a broad range of receptor occupancy, with little change in sensitivity at the extremes of the response range, where receptors are expected to have either very low or very high occupancy.
The nonlinear stimulus-response relation is intriguing for several reasons. The observed relation is not consistent with stoichiometric signaling between the chemoreceptors and the response regulator; the excitation signal is amplified. Increased attractant binding to the chemoreceptors results in a decrease in the concentration of phosphorylated CheY, producing counterclockwise flagellar rotation and suppression of tumbling. Thus the amplified signal is inhibition of a phosphorylation cascade rather than stimulation of an enzymatic pathway as is commonly observed in other sensory transduction systems (15–17). Additionally, the nonlinear stimulus-response relation contrasts with that of adaptation which varies linearly with changes in receptor occupancy (18, 19). Therefore, adaptation times are not proportional to the excitation signal. This suggests that excitation and adaptation, which are both triggered by changes in receptor occupancy, are mediated by divergent pathways.
The molecular basis of both the amplified excitation response and the proportional adaptive response to changes in ligand binding to the chemoreceptors remains to be determined. Jasuja et al. discuss several possible mechanisms that have been previously proposed to explain amplification of the excitation signal (see Fig. 1). One possible mechanism involves amplification through receptor clusters, such that ligand binding to a single receptor can inactivate more than one histidine kinase CheA (20). The clustering of receptors (21) and the nonstoichiometric interaction of components within receptor–CheW–CheA complexes (22) support such a model. An alternative mechanism involves amplification through release of receptor-sequestered CheZ, which catalytically accelerates the dephosphorylation of CheY (23). Sorting out the contributions of each component to excitation and adaptation is not an easy task. Although the data of Jasuja et al. suggest that the adaptation pathway must diverge before amplification of the excitatory signal, many protein components appear to function within both pathways. For instance, inactivation of histidine kinase CheA not only affects CheY, but also lowers phosphorylation of methylesterase CheB, one of the two receptor modification enzymes involved in adaptation. CheZ also potentially contributes to adaptation, as its activity is regulated by oligomerization with phosphorylated CheY (24). Indeed, signaling schemes can be envisaged that circumvent these overlaps, albeit by involving increased complexity of interactions between the components of the signaling pathways.
The assay developed by Jasuja et al. is extremely powerful in that it provides temporal resolution sufficient to assess excitation responses independent from influences of adaptation. Examination of the stimulus-response relation in a variety of chemotaxis mutants should readily provide data that limit plausible mechanisms for signal amplification. Once again, we are reminded that observations of the behavior of the intact organism are essential to understanding this sensory system in molecular detail.
Footnotes
The companion to this Commentary begins on page 11346.
References
- 1.Engelmann W. Bot Ztg. 1882;40:419–426. [Google Scholar]
- 2.Pfeffer W. Untersuch Bot Inst Tübingen. 1884;1:363–482. [Google Scholar]
- 3.Adler J. Science. 1966;153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 4.Jasuja R, Yu-Lin, Trentham D R, Khan S. Proc Natl Acad Sci USA. 1999;96:11346–11351. doi: 10.1073/pnas.96.20.11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koshland D E., Jr Science. 1977;196:1055–1063. doi: 10.1126/science.870969. [DOI] [PubMed] [Google Scholar]
- 6.Bourret R B, Borkovich K A, Simon M I. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- 7.Falke J J, Bass R B, Butler S L, Chervitz S A, Danielson M A. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mesibov R, Ordal G W, Adler J. J Gen Physiol. 1973;62:203–223. doi: 10.1085/jgp.62.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morton-Firth C J, Shimizu T S, Bray D. J Mol Biol. 1999;286:1059–1074. doi: 10.1006/jmbi.1999.2535. [DOI] [PubMed] [Google Scholar]
- 10.Brown D A, Berg H C. Proc Natl Acad Sci USA. 1974;71:1388–1392. doi: 10.1073/pnas.71.4.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Block S M, Segall J E, Berg H C. J Bacteriol. 1983;154:312–323. doi: 10.1128/jb.154.1.312-323.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segall J E, Block S M, Berg H C. Proc Natl Acad Sci USA. 1986;83:8987–8991. doi: 10.1073/pnas.83.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan S, Castellano F, Spudich J L, McCray J A, Goody R S, Reid G P, Trentham D R. Biophys J. 1993;65:2368–2382. doi: 10.1016/S0006-3495(93)81317-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jasuja R, Keyoung J, Reid G P, Trentham D R, Khan S. Biophys J. 1999;76:1706–1719. doi: 10.1016/S0006-3495(99)77329-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pugh E N, Jr, Lamb T D. Biochim Biophys Acta. 1993;1141:111–149. doi: 10.1016/0005-2728(93)90038-h. [DOI] [PubMed] [Google Scholar]
- 16.Mombaerts P, Wang F, Dulac C, Vassar R, Chao S K, Nemes A, Mendelsohn M, Edmondson J, Axel R. Cold Spring Harbor Symp Quant Biol. 1996;61:135–145. [PubMed] [Google Scholar]
- 17.Herness M S, Gilbertson T A. Annu Rev Physiol. 1999;61:873–900. doi: 10.1146/annurev.physiol.61.1.873. [DOI] [PubMed] [Google Scholar]
- 18.Berg H C, Tedesco P M. Proc Natl Acad Sci USA. 1975;72:3235–3239. doi: 10.1073/pnas.72.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spudich J L, Koshland D E., Jr Proc Natl Acad Sci USA. 1975;72:710–713. doi: 10.1073/pnas.72.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borkovich K A, Alex L A, Simon M I. Proc Natl Acad Sci USA. 1992;89:6756–6760. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maddock J, Shapiro L. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Levit M, Lurz R, Surette M G, Stock J B. EMBO J. 1997;16:7231–7240. doi: 10.1093/emboj/16.24.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardina P J, Manson M D. Science. 1996;274:425–426. doi: 10.1126/science.274.5286.425. [DOI] [PubMed] [Google Scholar]
- 24.Blat Y, Gillespie B, Bren A, Dahlquist F W, Eisenbach M. J Mol Biol. 1998;284:1191–1199. doi: 10.1006/jmbi.1998.2224. [DOI] [PubMed] [Google Scholar]