Abstract
Mutations in cardiac actin (ACTC) have been associated with different cardiac abnormalities in humans, including dilated cardiomyopathy and septal defects. However, it is still poorly understood how altered ACTC structure affects cardiovascular physiology and results in the development of distinct congenital disorders. A zebrafish mutant (s434 mutation) was identified that displays blood regurgitation in a dilated heart and lacks endocardial cushion (EC) formation. We identified the mutation as a single nucleotide change in the alpha-cardiac actin 1a gene (actc1a), resulting in a Y169S amino acid substitution. This mutation is located at the W-loop of actin, which has been implicated in nucleotide sensing. Consequently, s434 mutants show loss of polymerized cardiac actin. An analogous mutation in yeast actin results in rapid depolymerization of F-actin into fragments that cannot reanneal. This polymerization defect can be partially rescued by phalloidin treatment, which stabilizes F-actin. In addition, actc1a mutants show defects in cardiac contractility and altered blood flow within the heart tube. This leads to downregulation or mislocalization of EC-specific gene expression and results in the absence of EC development. Our study underscores the importance of the W-loop for actin functionality and will help us to understand the structural and physiological consequences of ACTC mutations in human congenital disorders.
INTRODUCTION
Actin is an abundant multifunctional protein involved in many cellular functions, including muscle contraction (18, 19). The highest concentration of actin is found in striated muscles, forming the thin microfilament “rail” of the sarcomere, along which the thick filament, myosin, moves to generate muscle contraction (10, 35). In vertebrates, there are at least six highly conserved types of actin, including two cytoskeletal forms and four tissue-specific isoforms (coded for by the genes shown in parentheses): cardiac (ACTC), skeletal (ACTA1), vascular (ACTA2), and enteric (ACTG2) (19, 40). ACTC is coexpressed with ACTA1 and ACTA2 in the developing heart of zebrafish (4, 37), chickens (28, 32), mice (22, 33), and humans (5). The ACTC isoform of actin has a critical role in the development and function of the heart (19). Mutations in ACTC have been associated with different types of cardiac abnormalities in humans, including idiopathic dilated cardiomyopathy (IDC) and atrial septal defects (24, 27). In studies performed with mice and chickens, the knockout of ACTC caused delayed heart looping, atrial septal defects, and embryonic lethality (19, 24). However, how different ACTC mutations lead to the development of distinct congenital defects is still poorly understood.
In vivo, actin exists in a monomeric form, G-actin, and a polymerized form, F-actin. Under physiological conditions, F-actin is the predominant form; however, a dynamic equilibrium between G- and F-actin is important at the cellular level, such as during cell division, migration, and intracellular transport (18, 34). Actin binds a substantial number of proteins collectively called actin binding proteins (ABPs) (10). A conserved sequence located between subdomains 1 and 3 (the W-loop; residues 165 to 172) is thought to act as an ATP/ADP nucleotide sensor and is critical for interactions with ABPs (18). The physiological importance of the W-loop for actin function in vertebrate development is still not known.
The zebrafish has emerged as an advantageous model system to study early vertebrate development, including cardiac morphogenesis. An optically transparent zebrafish embryo develops a functional circulatory system, including a beating heart, by 24 h postfertilization (hpf). A linear heart tube loops by 48 hpf which is followed by endocardial cushion (EC) formation at the atrioventricular canal (AVC) (2). EC formation is a critical step for subsequent valve morphogenesis. In addition to the genetic control of AVC and EC development, hemodynamics play an important role in restricting expression of EC-specific genes to the AVC region (1, 41). Previous studies have indicated that blood flow is necessary for expression of klf2a, which is needed for EC development (41). An ACTA zebrafish mutant with the cardiofunk gene (cfk) mutation lacks EC development as a result of improper heart function (1). However, the relationships between cardiac function, hemodynamics, and EC development are still poorly understood.
In this study, we have performed positional cloning of a zebrafish mutant (s434 mutation) that displays defects in AVC development and blood circulation and that was previously identified during an N-ethyl-N-nitrosourea (ENU) mutagenesis screen (2). We show here that this phenotype is due to a mutation in the alpha-cardiac actin 1a (actc1a) gene involving a Y169S amino acid substitution within the W-loop region. We show that this mutation results in a decrease of actin polymerization, primarily due to the mutant actin monomer's inability to reanneal after the breakdown of the filament. s434 mutants display reduced cardiac output and altered hemodynamics within the heart, along with misexpression of genes involved in AVC differentiation that lead to the absence of ECs. Our study underscores the importance of the W-loop for actin functionality and adds to our understanding of structural and physiological consequences of ACTC mutations in human congenital disorders.
MATERIALS AND METHODS
Zebrafish strains.
Embryonic and adult zebrafish were raised and maintained under standard laboratory conditions (45). s434 mutants identified in an ENU mutagenesis screen (2) were maintained in the AB wild-type (WT) strain and were crossed into the WIK strain for genetic mapping. The kdrl:EGFPs843 (15) line, expressing enhanced green fluorescent protein (EGFP), was used for analysis of vascular and EC cushion development. Cardiofunk gene (cfks11) carriers were used to breed the s434 mutation into. The etsrp/etv2y11 (30) mutants were used for a startle response analysis.
Mapping of the s434 locus.
The markers used for mapping had the following primer sequences: Z14641 forward, 5′-AAACACATGCACAATGGTAGAAA-3′; Z14641 reverse, 5′-CAGCAAGTTCAGCCAAAACA-3′; fc25c08 forward, 5′-TCTCTTTGGGAAATCCATGC-3′; fc25c08 reverse, 5′-TCTTGTAGAATACCAGCCCTA-3′; Z14263 forward, 5′-CCCATTTAGCCTTGGTATCAG; and Z14263 reverse, 5′-TAGGGTCATACATGCCACTC-3′. A candidate gene approach was used to screen for possible mutations in the s434 locus region. actc1a cDNA from mutant embryos and their wild-type siblings was amplified by PCR and sequenced using the following primers: forward, 5′-AACTCCAGTCTTGCGCTACAA-3′; and reverse, 5′-CAGCACAAGGCACAGTACAAA-3′. Protein alignment of actc1a across species was performed using MacVector and the following accession numbers: zebrafish, NP_001001409; mouse, NP_033738; human, AAB59619; and yeast (Saccharomyces cerevisiae), AAA34391.
In situ hybridization.
Whole-mount in situ hybridization (WISH) was performed on embryos as previously described (16). Antisense RNA probes for the bmp4 and versican protein (42), notch1b (46), nfatc1 (47), amhc (3), vmhc (48), and fli1 (39) genes were synthesized as previously described. klf2a probe was obtained by linearization of a cDNA fragment in pExpress-1 (clone 6997059; Open Biosystems) with SmaI and transcribed using T7 RNA polymerase. actc1a cDNA corresponding to the 3′ untranslated region (3′-UTR) (specific to actc1a) and a 20-bp fragment of the coding sequence were amplified with the following primers: forward, 5′-CCATCGTCCACAGAAAGTGC-3′; and reverse with T7 promoter sequence, 5′-TAATACGACTCACTATAGGTTCGGACACATCAGAACTTTTATTAC-3′. T7 RNA polymerase (Promega) was used to synthesize digoxigenin (DIG)-labeled actc1a probe from the PCR product. Embryos were imaged with an Axio Imager-M2 compound microscope (Zeiss) with a 20×/0.5-numerical aperture (NA) objective.
Morpholino-oligomer knockdown.
Actc1a protein synthesis was inhibited by a translation-blocking morpholino-oligomer (MO) (Gene Tools, LLC) that binds the 5′-UTR: 5′-GATTGCTGTTTTTTAGGATGGTTCA-3′. This MO was targeted to actc1a only, not the other muscle actin isoforms (see Fig. S4 at https://research.cchmc.org/mcb_glenn). Two nanograms of the MO was injected at the 1- to 2-cell stage. Embryos were assessed at 48 to 72 hpf for myocardial function and appearance of heart edema and circulation. Because injection of actc1a MO resulted in minor toxic effects (data not shown), p53 MO (1.2 ng/embryo) was included in the injection mixture to reduce nonspecific p53-mediated apoptosis as reported previously (11). The control MO mixture contained 2 ng of standard control MO (Gene-Tools) and 1.2 ng of p53 MO.
RNA and DNA injection.
Wild-type actc1a was subcloned into the T3TS vector (14), and mRNA was transcribed using the mMessage mMachine T3 kit (Ambion). The RNA was then diluted to 760 pg/nl, and 2 nl was injected into embryos at the 1- to 2-cell stage. Wild-type actc1a genomic DNA containing 2 kb of proximal actc1a promoter and 5 kb of full coding sequence, including introns and 3′-UTR, was amplified from a bacterial artificial chromosome (BAC) containing actc1a (clone HUKGB735B1581Q; Bioscience Life Sciences). The following primers were used for actc1a amplification: forward, 5′-TGGCACAGCAACCCACTGGGC-3′; and reverse, 5′-CACTAGGACACAGTGTGAC-3′. The DNA fragment was injected at a concentration of 25 pg/embryo into the blastomere at the 1-cell stage. In both cases, heart and circulation analyses were performed at 36 to 48 hpf.
Immunohistochemistry and histological analysis.
To detect polymerized actin, embryos were fixed at room temperature for an hour with a mixture of 4% paraformaldehyde, 0.1% Triton, and 0.25% glutaraldehyde in phosphate-buffered saline (PBS). Embryos were rinsed in 0.1% Triton in PBS (PBST) before being stained in a 1:40 solution of rhodamine-phalloidin in PBST. The embryos were stained for 4 h at room temperature or overnight at 4°C, followed by a brief rinse with 0.1% PBST. Myosin was detected using an antibody for sarcomeric myosin (MF20; Iowa Hybridoma Bank), at a dilution of 1:10 in PBST. The C4 monoclonal antibody (Seven Hills Bioreagents) was used to detect both cytoplasmic and muscle actin isoforms, at a dilution of 1:500 in PBST. The Alexa Fluor 568–goat anti-mouse IgG fraction was used to detect the C4 actin antibody, and Alexa Fluor 488- or 594-IgG2B was used to detect MF20, each at a dilution of 1:200 in PBST. To visualize endocardial cushions, kdrl:GFP-positive embryos were fixed for an hour at room temperature with a mixture of 4% paraformaldehyde, 0.1% Triton, and 0.25% gluteraldehyde in PBS. The embryos were stained for 2 h at room temperature with an anti-GFP–rabbit IgG fraction–Alexa Fluor 488 conjugate (Invitrogen) at a dilution of 1:200 in PBST and then rinsed in PBST. Embryos were imaged on a D-Eclipse C1 microscope (Nikon). For histological analysis, embryos were embedded in paraffin and sectioned at 5 μm as previously described (23) and stained with hematoxylin and eosin. Sections were imaged on an Optihop 2 compound microscope (Nikon) with a 10×/0.03-NA objective. For myofibril structural histology, both 72-hpf and 120-hpf embryos were fixed as described above and then embedded in paraffin and sectioned at 4 μm. Tissue was subjected to an antigen retrieval protocol using citrate buffer (0.1 M; pH 6.0) and heat. Antibody staining was performed using MF20 as described earlier. Nuclei were visualized using Vectashield hard-set mounting medium with DAPI (4′,6-diamidino-2-phenylindole) (Vector Labs). Embryos were imaged with an Axio Imager-M2 compound microscope (Zeiss) with a 63×/1.4-NA oil immersion objective. An o-dianisidine stain was used to detect heme in red blood cells, as previously described (21). Stained embryos were imaged on an Achromat S 1.0× dissecting microscope (Zeiss).
Microangiography.
Fluorescein-labeled dextran was injected into the common cardinal vein (just posterior to the atrium) of 48-hpf wild-type and mutant embryos. Injected embryos were visualized for the presence of fluorescein isothiocyanate (FITC) in their vessels and photographed on an Axio Imager-M2 compound microscope (Zeiss) using either the 5× or 10×/0.3-NA objective.
Confocal microscopy and image processing.
Confocal images were acquired using the Nikon D-Eclipse C1, the Nikon Eclipse 90i upright microscope, a Plan Apo 20× 0.75-NA microscope objective, and the 488- and 561-nm laser lines. To obtain high-quality confocal stacks, we collected optical sections at a thickness of 300 nm at a resolution of 512 by 512 or 1,024 by 1,024. Acquired confocal stacks were deconvolved using AutoQuant X.2.2 (Media Cybernetics, Inc.) and saved to generate maximum intensity projected (MIP) tiff files. MIP rendered images were imported into Adobe Photoshop CS2 (San Jose, CA) and adjusted for levels, brightness, contrast, and sharpening.
Protein purification.
F169S actin was purified from F169S actin/Y79S profilin yeast cells (S. cerevisiae) using DNase I (Worthington Biochemistry) affinity chromatography followed by DE52 anion-exchange chromatography as described previously (8). Yeast cells that were mutant for profilin (Y79S) had to be utilized due to the lethality of the F169S mutation, and this profilin mutant previously was found to alleviate actin mutations (44). WT yeast was purified from commercially available yeast cakes by the same procedure. The final G-actin (globular actin) concentration was determined from the A290 (ε290 nm = 25,600 M−1cm−1), and the actin was stored in G buffer (10 mM Tris-HCl [pH 7.5], 0.2 mM CaCl2, 0.1 mM ATP, 0.1 mM dithiothreitol [DTT]) at 4°C for no more than 4 days. Actin purity was determined by 12% SDS-PAGE followed by Coomassie blue staining. A single actin band was observed.
Actin polymerization.
The G-actin was induced to polymerize by the addition of MgCl2 and KCl to final concentrations of 2 and 50 mM, respectively. The polymerization assays were performed in a final volume of 120 μl at 25°C in a microcuvette housed in a thermostatted sample compartment of a FluoroMax-3 spectrofluorometer (Jobin Yvon-Spex). The change in light scattering as an indication of filament formation was recorded using excitation and emission wavelengths of 360 nm. The experiments were repeated at least twice using two different actin preparations with similar results. Aliquots of actin filaments were visualized by negative staining using 1% uranyl acetate, deposited on Formvar-coated copper grids, and visualized with a JEOL JEM-1230 transmission electron microscope in the University of Iowa Electron Microscope Facility. Filament images were recorded with a Gatan UltraScan 1000 2,000-by-2,000 charge-coupled device (CCD) camera.
Phalloidin treatment.
Phalloidin treatment was used in yeast actin polymerization as previously described (18). Phalloidin oleate (Calbiochem, EMD Millipore) was used to treat embryos at a concentration of 2 μM in 1% dimethyl sulfoxide (DMSO). Embryos were dechorionated and treated in 6-well culture dishes starting from 24 hpf and evaluated for cardiac and circulatory function at 48 hpf. To genotype rescued embryos, the following primers were used: forward, GGTCAGTGTCCATCCAATCTG; and reverse, GGTTTTTGAGTACCCACCAG. A 288-bp product was amplified from genomic DNA and subsequently sequenced.
Functional analysis.
Heart rate was determined by counting the heartbeats on live embryos and averaging the number of beats over a minute in wild-type embryos (n = 10) and mutant embryos (n = 10). The average heart rate was subsequently expressed as a frequency value (Hz). A standard two-tailed t test (18 degrees of freedom) was performed, and the P value was calculated assuming a 95% confidence interval.
Systolic, diastolic, contractility, and reverse flow fraction (RFF) measurements were determined by analyzing videos taken on a compound inverted Zeiss microscope with a Hamamatsu high-speed camera at 125 frames per second with a 25× objective. Embryos (48 hpf) were mounted in 1.2% low-melting agarose (Sigma type IX) containing 125 μg/ml of Tricaine.
Contractility was determined by measuring the diameter of the ventricle at systole and diastole using the Image J analysis software. An estimate of ventricular contractility was made using the ventricular shortening fraction (VSF) equation (43): VSF = (ventricular length at diastole − ventricular length at systole)/ventricular length at diastole.
Wild-type (n = 12) and mutant (n = 12) embryos were measured 3 times for VSF values, which were then averaged and used in a standard two-tailed t test (22 degrees of freedom). Significance was calculated assuming a 95% confidence interval. Values are presented as means ± standard errors of the means (SEM).
Wild-type (n = 17) and mutant (n = 13) embryos were measured for RFF values, which were calculated as reported previously (41), by counting the number of frames demonstrating reverse flow and dividing by the total number of frames. Values are presented as means ± standard errors of the means (SEM).
RESULTS
The s434 mutant lacks endocardial cushion development and functional circulation.
The s434 mutant was discovered in a forward genetic screen (2) and displays blood regurgitation at the AVC and defects in EC formation. The s434 mutation is recessively inherited, and a quarter of the progeny (439/1,639) are affected and viable until 7 days postfertilization (dpf).
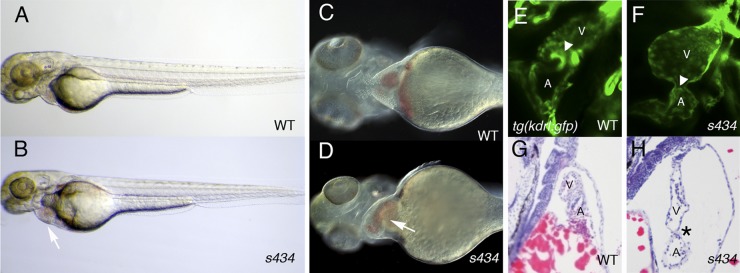
Morphological phenotypic analysis revealed that the s434 mutant embryos develop pericardial edema by 48 hpf (Fig. 1A and B). The heart is dilated and blood regurgitates between the atrium and the ventricle by 48 to 72 hpf (Fig. 1C and D; see Movies 1 and 2 at https://research.cchmc.org/mcb_glenn). Since there was blood regurgitation at 48 hpf, we investigated AVC development and endocardial cushion formation in the mutant embryos. Normally, endocardial cushions are apparent by 96 hpf in zebrafish, formed by endocardial cells clustering at the AVC (Fig. 1E) (2). The s434 mutant embryos demonstrate lack of endocardial cushion formation by 120 hpf, as observed by analyzing kdrl:GFP expression at the AVC (Fig. 1E and F). Transverse sectioning followed by hematoxylin and eosin staining reveals a dilated, elongated heart surrounded by pericardial edema by 72 hpf in the mutant (Fig. 1H) compared to the compact, looped heart in wild-type embryos (Fig. 1G). The heart wall in mutants is also thinner than that of wild-type siblings (Fig. 1G and H).
Fig 1.
s434 mutants possess an enlarged heart and lack endocardial cushions. (A and B) Overall morphology of s434 mutants (B) compared to wild-type siblings (A) at 48 hpf. Pericardial edema is evident by 48 hpf (arrow), although the rest of the body appears morphologically normal. (C and D) Higher magnification of the anterior region of the embryo demonstrates the enlarged heart of s434 mutants at 72 hpf (arrow). (E and F) s434 mutants lack atrioventricular (AVC) endocardial cushions (ECs) by 120 hpf, as shown by lack of kdrl:GFP expression. The arrowhead denotes the AVC. (G and H) Hematoxylin and eosin staining of transverse sections through the heart at 72 hpf. The mutant heart is dilated and surrounded by pericardial edema (asterisk). A, atrium; V, ventricle.
s434 mutants lack functional circulation, but have an intact vascular system.
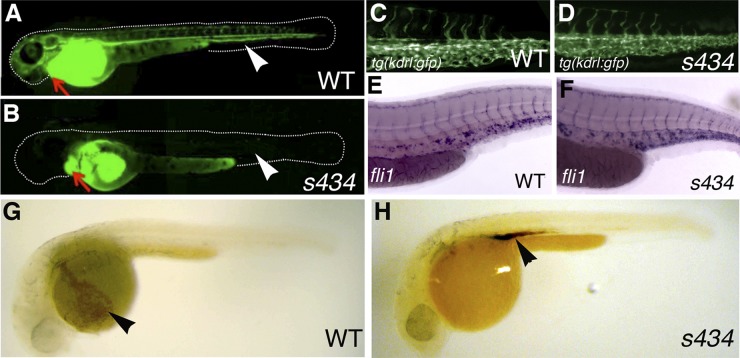
s434 mutant embryos do not develop functional circulation; therefore, a number of assays were performed to assess the development of the vascular system. Microangiography analysis was performed by injecting FITC-labeled dextran (molecular mass of 2 MDa) into the sinus venosus at 48 hpf. In the mutant embryos, dextran remained in the atrium and ventricle when viewed after an hour postinjection, compared to the wild-type embryos, which display FITC fluorescence throughout the vasculature (Fig. 2A and B). To determine if the s434 mutant in fact had a complete vascular system, we outcrossed the s434 line to the vascular endothelium-specific kdrl:GFP reporter line. We observed no difference in GFP expression between the wild-type and mutant embryos (Fig. 2C and D). Whole-mount in situ hybridization (WISH) was performed on 48-hpf embryos for expression of an endothelial cell marker, fli1. There was no difference in fli1 expression, indicating that vascular morphogenesis is not affected in s434 mutants (Fig. 2E and F). To visualize localization of red blood cells shortly after circulation begins, we performed a heme staining with o-dianisidine at 35 hpf. In the wild-type embryos, erythroid cells are distributed throughout the body, although they are most apparent within the broad region of the common cardinal vein, which is located over the yolk (Fig. 2G, arrowhead). In the s434 mutant, red blood cells are not circulating, but rather remain in the intermediate cell mass (ICM) region near the yolk extension, at their formation site (Fig. 2H, arrowhead). These experiments provide evidence for the existence of a complete vascular system in s434 mutants, although blood circulation is not observed.
Fig 2.
s434 mutants do not demonstrate blood circulation; however, they do possess intact vasculature. (A and B) Microangiography was performed at 48 hpf with fluorescein-dextran, injected into the sinus venosus, just posterior to the atrium. An arrow is pointing to the heart in both images. In the WT (A), blood flow carries the labeled dextran out into the distal vessels of the embryo, visualized in green. Arrowheads denote the locations of the dorsal aorta and posterior cardinal vein, which can be used to assess circulation. In the s434 mutants (B), however, the dextran does not circulate throughout the embryo. (C and D) kdrl:GFP expression at 48 hpf in both WT (C) and mutant (D) embryos in the trunk, displaying intact endothelial cells in the vessels. (E and F) In situ hybridization for endothelial marker fli1 expression in the trunk region of WT (E) and mutant (F) embryos at 48 hpf. (G and H) o-Dianisidine heme staining at 33 hpf. The wild-type (G) embryo displays circulation, and broad heme staining is observed in the blood cells within the common cardinal vein (ccv) (arrowhead). The mutant (H), however, does not demonstrate circulation (lack of staining in the ccv), and blood cells are present in the intermediate cell mass region above the yolk extension (arrowhead), at the site of their formation.
s434 encodes a substitution mutation in alpha-cardiac actin.
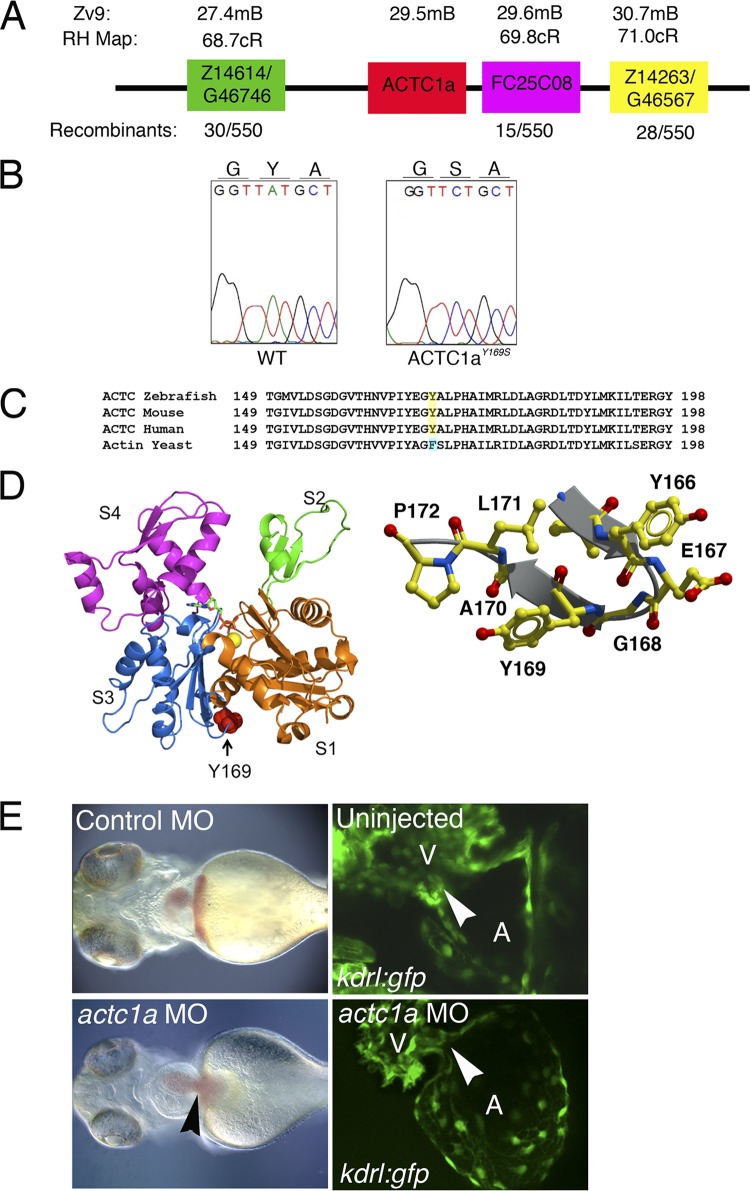
To identify the molecular basis of the s434 phenotype, positional cloning techniques were used to define the genetic locus, which is localized to linkage group 20 (2). Mutant embryos (n = 723) were used for fine mapping of the critical interval of 8.5 Mb (from ZK183N20 to ZC223011). A set of single-strand conformation polymorphism (SSCP) markers were used to map the s434 mutation to the final 3-Mb interval contained by the Z1464 and Z14263 markers (Fig. 3A). We undertook a candidate gene approach and sequenced the coding sequence of alpha-cardiac actin 1a (actc1a), present in this region, using cDNA isolated from the mutant and wild-type sibling embryos (Fig. 3A). After sequence analysis, we detected a single nucleotide change at base pair 572 from A to C (Fig. 3B), resulting in a tyrosine-to-serine substitution at amino acid 171. The first two residues (methionine and cysteine) of the actin protein are cleaved to produce the mature peptide; therefore, this position is known as residue 169 in the mature protein sequence (9, 31). This mutation is located in the third subdomain of G-actin, which is characterized as the W-loop. This region of the actin monomer participates in binding to other monomers while polymerizing into F-actin, as well as interacting with ABPs, such as profilin (12, 13, 18, 26, 29, 34). Crystallization renderings of the actin monomer, highlighting residue 169, are shown in Fig. 3D. The W-loop, including the Y169 residue is 100% conserved among human, mouse, and zebrafish species (Fig. 3C).
Fig 3.
s434 mutants display a point mutation in zebrafish alpha cardiac actin 1a gene (actc1a). (A) The actc1a gene is located on chromosome 20, at 29.5 Mb, between the zebrafish markers Z14614 and FC25C08, based on the Zv9 genome assembly. All markers correspond to the radiation hybrid (RH) map. Recombination is measured in centimorgans (cM). (B) Chromatogram showing that the s434 mutation is an A-to-C substitution at nucleotide 571 of actc1a, resulting in a tyrosine-to-serine amino acid change at amino acid 169 within the final proteolytically processed ACTC1a protein. (C) Protein alignment demonstrating that this tyrosine residue is evolutionarily conserved. Yeast cells have a phenylalanine at residue 169, an analogous aromatic residue. (D) Crystallization renderings of the actin monomer, highlighting the location of residue 169 in the W-loop (right side). Subunits S1 to S4 are marked. This part of the monomer is a nucleotide sensor, participating in ATP-ADP exchange. The image on the left shows the conformation of actin when ATP is bound. (E) Left panels, bright-field images indicate that the actc1a MO injection results in a heart-specific phenotype, with heart enlargement and edema (black arrowhead) observed at 48 hpf. (Right panel) Endocardial cushions do not form in the morphant, as observed in kdrl:GFP embryos (white arrowheads indicate the locations of the AVC). All images are oriented with the anterior portion to the left; the ventral view is shown.
In order to confirm that this mutation was indeed responsible for the phenotype of s434 mutants, morpholino-oligomer knockdown against actc1a was performed. A morpholino oligomer was designed against the 5′ UTR of actc1a and is not expected to target other actin isoforms (see Fig. S1 at https://research.cchmc.org/mcb_glenn). At the one- to two-cell stage, we injected a translation-blocking MO against actc1a and analyzed early heart development. actc1a MO knockdown embryos were morphologically normal, except for a heart-specific phenotype apparent at 48 hpf (Fig. 3E). The actc1a morphants demonstrate heart edema and an enlarged heart, as well as lack of endocardial cushion formation by 120 hpf that is apparent in kdrl:GFP transgenic embryos (Fig. 3E). Injection of a control MO mixture caused no apparent morphological defects and did not affect cardiac morphogenesis (Fig. 3E).
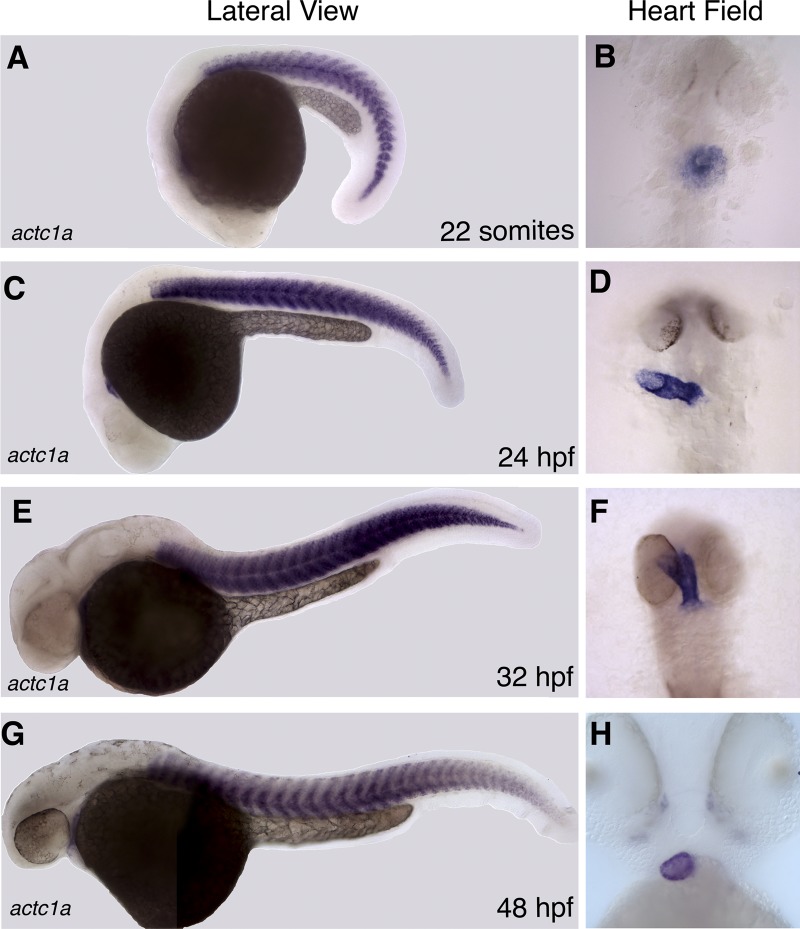
Because there was no comprehensive previous analysis of the zebrafish actc1a temporal expression pattern, WISH analysis was performed for actc1a expression during embryogenesis. At the 22-somite stage, actc1a is expressed in the somitic muscle and the heart field, within the cardiac cone (Fig. 4A and B). Expression in somitic and cardiac muscle is also observed at the 24-hpf and 32-hpf stages (Fig. 4C to F). At 48 hpf, actc1a cardiac expression is preferentially enriched in the ventricle (Fig. 4H). These observations confirm that actc1a is present in the cardiac cells during the stages when s434 mutants display heart developmental defects.
Fig 4.
The expression domain of actc1a is restricted to cardiac and somitic muscle. (A to H) As analyzed by in situ hybridization, actc1a transcript is expressed within the somitic muscle and in the heart tube at the 22-somite (A and B), 24-hpf (C and D), 32-hpf (E and F), and 48-hpf (G and H) stages. At 48 hpf, actc1a cardiac expression becomes enriched in the ventricle (H). (B, D, F, and H) Ventral view of the heart field; the anterior portion is toward the top.
The actc1as434 mutation results in a decreased ability to polymerize F-actin in the heart.
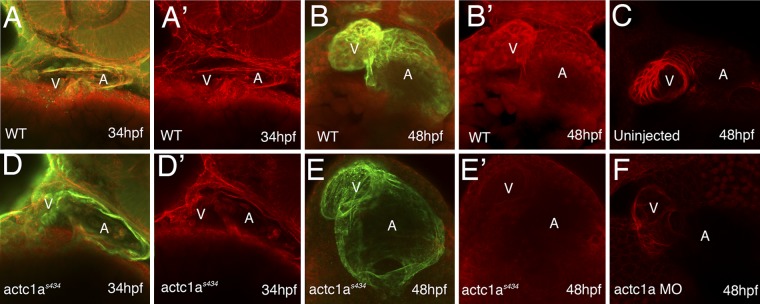
To determine the molecular consequences of the actc1as434 mutation, we assayed actin polymerization in the mutant embryos. Whole-mount immunohistochemical staining was utilized with rhodamine-labeled phalloidin to detect polymerized F-actin in the heart. Embryos were counterstained with an antibody against sarcomeric myosin (MF20) to visualize the myocardium. Early in heart morphogenesis, the staining for polymerized actin (red channel) in mutant embryos at 34 hpf is significantly decreased compared to that in the wild-type siblings (Fig. 5A, A′, D, and D′). Interestingly, actc1as434 mutants do possess an intact myocardium, as shown by MF20 expression (green channel in Fig. 5A and D), indicating that the myocardium forms even when F-actin is dramatically decreased. As development progresses, the loss of F-actin in the heart is very dramatic. In the wild-type embryos at 48 hpf, phalloidin staining reveals a large amount of F-actin in the heart, especially in the ventricle (Fig. 5B and B′). However, in the mutant, phalloidin staining is dramatically decreased, and MF20 staining reveals an enlarged heart (Fig. 5E and E′). Phalloidin staining in the somitic muscle was unchanged between the wild-type and mutant embryos (data not shown). This illustrates the severe reduction in polymerized actin in the heart resulting from the s434 mutation. Similar to s434 mutants, actc1a MO-injected morphants display decreased phalloidin staining, compared to the uninjected control embryos (Fig. 5C and F). These results demonstrate that the actc1as434 phenotype is caused by a severe decrease in polymerized cardiac actin in the heart.
Fig 5.
The actc1as434 mutation results in a dramatic decrease in the amount of polymerized F-actin present in the heart. F-actin was stained with rhodamine-phalloidin (red channel; panels A, A′, B, B′, D, D′, E, and E′), and an MF20 antibody stained for a sarcomeric myosin (green channel). (The green channel is overlaid with the red channel in panels A, B, D, and E.) In the ventral view, the anterior portion is up; maximum-intensity projections of confocal images are shown. At 34 hpf, the F-actin staining in the mutant (D and D′) is reduced in the heart compared to that in the wild type (A and A′). At 48 hpf, the wild-type heart shows strong staining for both MF20 and F-actin staining, which is particularly pronounced in the ventricle (B and B′). In contrast, the mutant shows a dilated heart and unchanged MF20 but vastly decreased actin staining (E and E′). (C and F) Actc1a-morpholino-oligomer-injected embryos (F) demonstrate a dilated heart tube and decreased F-actin staining compared to the uninjected embryo (C). A, atrium; V, ventricle.
Myofibril structure is disrupted in actc1as434 mutant cardiac muscle.
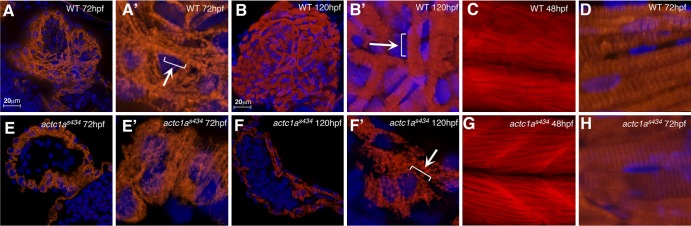
We further investigated the myofibril structure in both the heart and somitic muscle of embryos. We used immunohistochemical staining with MF20 antibody to visualize the myofibrils, as this antibody stains the A bands of the sarcomere (36). At 72 hpf, staining of myofibril structures can be seen in the wild-type heart (Fig. 6A and A′). However, myofibrils are severely disorganized in the hearts of actc1as434 mutants at 72 hpf, and only sparsely distributed myofibril structures can be seen around the edge of the myocardium (Fig. 6E and E′). At 120 hpf, myofibril structures can be clearly visualized with MF20 in the wild-type myocardium (Fig. 6B and B′). In contrast, the mutant heart contains shorter, abnormal myofibril structures compared to the nicely organized striations observed in the wild-type embryos (Fig. 6F and F′). In addition, the heart of a 120-hpf mutant embryo is very distended and fragile, as evident in the magnified image (Fig. 6F′). These data show that myofibrils are highly disorganized in the mutant embryos compared to the wild-type siblings.
Fig 6.
Myofibril structure is altered in the heart of actc1as434 mutants. Myofibril structure was revealed by using the MF20 antibody (red channel; panels A and B, D to F, and H) and by phalloidin staining (red channel; panels C and G). Nuclei were visualized by DAPI staining (blue channel; panels A and B, D to F, and H). (A, A′, B, and B′) The wild-type heart at 72 hpf shows striations in the muscle fibers, revealing myofibrils, which are more evident upon higher magnification in panel A′. (An arrow points to sarcomeres comprising the myofibril, which is outlined in the bracket.) The branching morphology of the muscle is characteristic of cardiac muscle. By 120 hpf, the myofibrils are nicely visualized by MF20, showing a very organized and mature myocardium. (An arrow points to the individual sarcomeres making up the myofibril, which is outlined in the bracket.) Panel B′ is a magnification of a portion of panel B. (E, E′, F, and F′) Myofibril structure in the mutant is not very clear at 72 hpf, although striations are apparent by 120 hpf. Panel E′ is a magnification of a portion of panel E, and panel F′ is a magnification of panel F. However, the mutant myofibrils are very short and not as substantial as the wild-type myofibrils (arrow). (Sarcomeres making up the myofibril are outlined by the bracket.) All of these images were taken with a 63× objective under oil immersion. (C and G) The somitic muscle myofibril organization of both the wild type and mutant at 48 hpf with phalloidin staining revealed no major differences. These images were taken under a 20× objective and then magnified to show detail of the trunk muscle. (D and H) Upon closer inspection, the somitic muscle myofibril organizations were similar in both the wild type and mutants at 72 hpf with MF20 antibody staining. These panels are both magnifications of an image taken from a section on a 63× objective under oil immersion. In panels A and B and panels E and F, the ventral portion is up and the anterior portion is to the left. In panels C and D and panels G and H, the dorsal portion is up and the anterior portion is to the left.
The structure of actc1as434 mutant trunk muscle within the somites did not appear to be disturbed compared to their wild-type siblings (Fig. 6C and D and G and H). We also tested the startle response in wild-type siblings and mutants to confirm that actc1a is not required for skeletal muscle function. When progeny from an actc1as434+/− cross were tested for their startle response at 24 hpf, there was no difference in the reactions of any of the embryos (data not shown). However, actc1as434 mutants did show a slower swimming response to touch stimulus at 72 hpf (see Movie 5 at https://research.cchmc.org/mcb_glenn), and this appears to be an indirect consequence of pericardial edema and absent blood circulation because similar defects are also observed in etsrp/etv2y11−/− mutants, defective in vascular development (30) (data not shown). This is likely due to the fact that other isoforms, including two skeletal actin isoforms (acta1a and acta1b) are also coexpressed strongly in the somitic trunk muscle (37), therefore, actc1a does not appear to have a major requirement in the trunk muscle development.
actc1a and cfk/acta1b have a partially redundant function in zebrafish heart.
Another sarcomeric actin zebrafish mutant, with the cardiofunk (cfk/acta1b) mutation, has been shown to display regurgitant blood flow and EC formation defects (1). To determine if the cfk mutation could complement the actc1as434 mutation, we bred heterozygous carriers of each mutation. Interestingly, acta1b (cfk) fails to complement the actc1as434 phenotype, and cfk+/− s434+/− double heterozygous embryos display a phenotype similar to that of homozygous actc1as434−/− mutants (see Fig. S2 at https://research.cchmc.org/mcb_glenn). WISH analysis was also performed for cfk expression during embryogenesis, which revealed almost an identical expression pattern compared to actc1a. From the 22-somite stage until the 48-hpf stage, cfk is expressed in the somitic muscle and the heart field; however, the expression does not appear as strong as actc1a expression at the same stages (see Fig. S3A to H at https://research.cchmc.org/mcb_glenn). At 48 hpf, cfk cardiac expression is preferentially enriched in the ventricle (see Fig. S3H at at the above URL). These observations confirm that cfk is present in the cardiac cells during the stages when s434 mutants display heart developmental defects. It is likely that acta1b and actc1a can partially substitute for each other; therefore, heterozygous embryos for each mutation do not show a phenotype unless both genes are mutated.
F169S mutation in yeast actin forms unstable filaments that are unable to reanneal.
In order to study the biochemical mechanics of actin polymerization in actc1as434 mutants, yeast (S. cerevisiae) cells were used to create the Y169S mutation in an in vivo culture system. Instead of tyrosine, yeast contains a similar aromatic amino acid, phenylalanine, at position 169 (Fig. 3C). To gain greater insight into the functional consequences of the F169S mutation, we introduced it into the W-loop of yeast actin, which normally has a loop sequence of A167, G168, F169, and S170. We were unsuccessful in recovering cells with this mutation (F169S), indicating it was lethal in an otherwise WT background. The same result was obtained when the Y169S mutation was introduced into yeast cells containing the mammalian skeletal muscle actin loop sequence E167, G168, Y169, and A170 to generate an E-G-S-A sequence. This is an important point, as the E-G-Y-A sequence is viable in yeast, but the E-G-S-A (mutated) sequence is not; therefore, these results demonstrate the biochemical significance of the Y169S mutation in zebrafish.
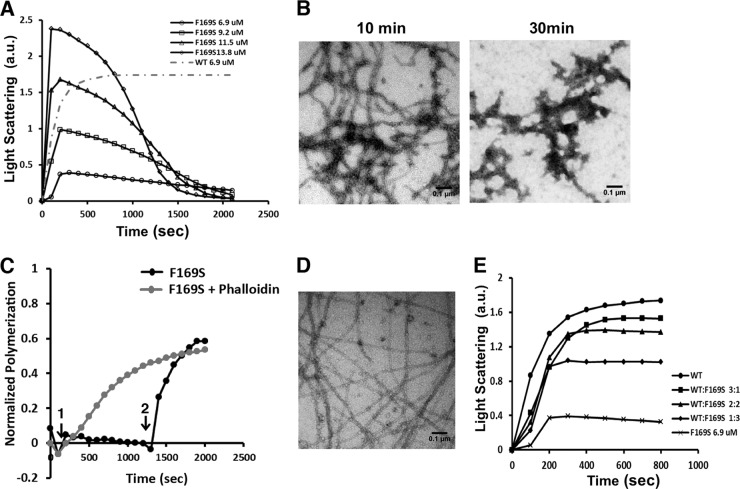
In order to purify mutant actin for biochemical studies, we cultured yeast cells that were double mutant for F169S actin and Y79S profilin. We had previously found that mutations in the actin W-loop could be at least partially suppressed with a profilin Y79S mutation (44). This site lies across from the bottom of the W-loop in the actin-profilin interface, and the mutation presumably allows room for accommodation of steric changes. Indeed, the double mutant cells grew, although at a lower rate than wild-type cells (data not shown). We then purified the actin from the double mutant cells and assessed the actin's ability to polymerize in comparison with WT actin. Figure 7A shows that following induction of polymerization by the addition of salts, light scattering increased as WT actin filaments formed until attainment of a final steady state. In comparison, the mutant actin began to polymerize by showing a rapid increase of light-scattering signal, as shown in Fig. 7A, but after a short time, light scattering returned to baseline. Increasing the amount of actin in the tube resulted in an initial greater extent of light scattering, but in all cases there was a decrease back to baseline. For all concentrations of actin tested, decay to baseline occurred at about the same time. However, failure to fit the decay part of a curve to a first-order equation indicated that multiple processes were occurring at the same time. We examined the reaction solution by electron microscopy at a time just past the peak light-scattering point and following return of the light scattering to near baseline (Fig. 7B). At 10 min, the field is replete with normal-looking filaments, although they appear shorter overall than does WT actin (data not shown). At 30 min, most of the field is empty and what remains appears to be a collection of filament remnants.
Fig 7.
Yeast F169S mutant actin polymerization analysis reveals destabilized filaments and rescue using phalloidin treatment. (A) F169S actin polymerization and visualization of filaments. F169S actin at the concentration indicated was induced to polymerize by the addition of salt as described in Materials and Methods. The increase in light scattering was recorded with time. The gray dotted line shows the extent of polymerization of a concentration of WT actin equal to the lowest concentration of mutant actin examined. a.u., arbitrary units. (B) A 3-μl aliquot was removed from the 13.8 μM F169S actin polymerization reaction mixture in panel A at the time indicated and visualized by electron microscopy as described in Materials and Methods. Each experiment was repeated at least twice with different actin preparations with essentially identical results. (C) Phalloidin rescues F169S actin filament formation. A 5 μM F169S actin sample was induced to polymerize at time point 1, as in panel A. At time point 2, following filament disassembly, 5 μM phalloidin was added. The gray line indicates polymerization of the mutant actin when phalloidin is added at the beginning of the polymerization assay, which can rescue the polymerization defect. The change in light scattering was recorded with time and normalized to the net change in light scattering caused by the polymerization of 5 μM WT actin. (D) Following attainment of the polymerization steady state, a 3-μl mutant actin sample was removed and visualized by electron microscopy as described in Materials and Methods. (E) Copolymerization of F169S actin with WT actin at different WT/mutant ratios. Solutions of 6.9 mM actin at different F169S/WT actin ratios were induced to polymerize by the addition of salt as described in Materials and Methods. The increase of light scattering was recorded with time. The experiments were repeated twice with similar results.
The biphasic polymerization behavior of F169S actin suggests that the second-phase light-scattering decrease is caused by conversion of the F-actin to a nonpolymerizable state. To address the possibility that this represented formation of ADP-G-actin due to ATP depletion, we added back ATP and saw only a small blip in light scattering followed again by a decrease to baseline (data not shown). This result is inconsistent with the nucleotide depletion theory. An alternative possibility is that newly formed ADP-F-actin, following ATP hydrolysis, was unstable and collapsed into very small actin oligomers incapable of reannealing. To test this hypothesis further, we assessed the effects of the filament-stabilizing drug phalloidin (18, 20) on mutant actin behavior. Figure 7C shows the addition of phalloidin to the actin sample following the decrease in light scattering. Phalloidin addition caused a rapid rise in light scattering to near the value achieved with pure WT actin following polymerization. When phalloidin is added at the beginning of the actin polymerization assay, it can also rescue the mutant actin polymerization defect (Fig. 7C, gray line). However, phalloidin-induced polymerization is much faster if phalloidin is added at a later point (a steeper slope for the black line in Fig. 7C) than if phalloidin is added at the beginning (red line). This argues that when phalloidin is added later, it induces repolymerization of small oligomers of actin that resulted from the filament instability due to mutation rather than polymerization of monomeric actin, which is present at the beginning. Furthermore, Fig. 7D demonstrates the existence of fully formed long straight filaments following the repolymerization, consistent with our reannealing hypothesis.
In the zebrafish, a combination of wild-type and mutant actins exists in the same cell. To determine the extent to which one of the two actins might be dominant over the other, we assessed polymerizability of different WT-mutant mixtures at constant total actin concentration. Figure 7E shows that in a mixed filament, the presence of WT actin exerts a dominant filament-stabilizing effect. This assertion is based on the fact that inclusion of 25% WT actin produces a final polymerization extent halfway between that of pure mutant and pure WT actin, whereas a 50:50 mixture results in about 80% polymerization of the entire actin pool.
Phalloidin treatment partially rescues heart functionality in actc1as434 mutants.
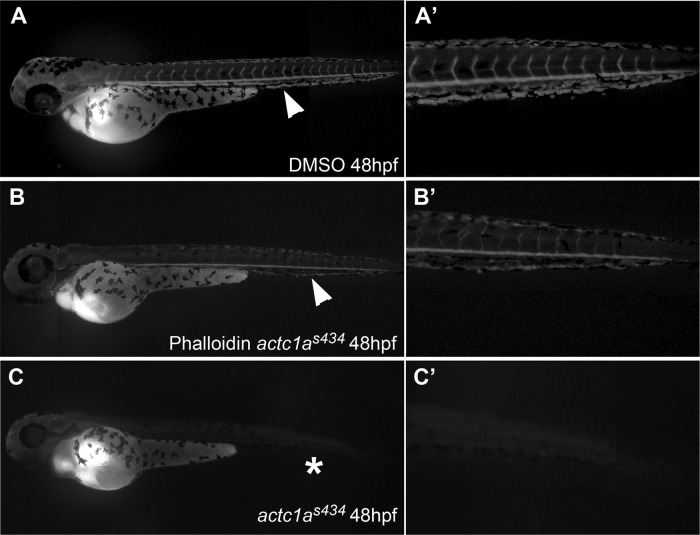
To test if stabilization of actin polymerization would alleviate the zebrafish mutant phenotype, we treated progeny of actc1as434+/− carriers with membrane-permeable phalloidin oleate from 24 to 48 hpf. Following the treatment, all embryos were analyzed for the presence of pericardial edema and the absence of blood circulation, typically observed in actc1as434 mutants. As expected, close to a quarter of either control 1% DMSO-treated embryos (23% [46/203 embryos]), or 2 μM phalloidin treated embryos (27% [93/343 embryos]) displayed pericardial edema (Table 1). However, none of the embryos with pericardial edema in the control group displayed any apparent blood circulation, while 29% (27 out of 93) of the embryos with pericardial edema displayed circulating blood flow (Table 1). Selected embryos from this group of phalloidin-treated embryos that displayed blood circulation were genotyped, which confirmed the presence of homozygous mutant genotype. The presence of circulation was confirmed by microangiography analysis (Fig. 8A to C). These results argue that phalloidin treatment stabilizes actin filaments in the mutant embryos, resulting in the partial alleviation of the observed cardiac defects.
Table 1.
Phalloidin treatment partially rescues circulation defects in s434 mutantsa
| Treatment | No. of embryos with: |
Total no. of embryos treated | |
|---|---|---|---|
| Pericardial edema and no circulation | Pericardial edema and circulation | ||
| DMSO | 46 | 0 | 203 |
| Phalloidin (2 μM) | 66 | 27 | 343 |
Progeny from a cross of actc1as434+/− carriers were treated with either 2 μM DMSO (control) or phalloidin-oleate from 24 hpf to 48 hpf and evaluated for pericardial edema and circulation. Our results show that none of the DMSO-treated embryos with pericardial edema had circulation at 48 hpf. However, of the phalloidin-treated embryos, 66 embryos showed pericardial edema with no circulation, and 27 embryos showed pericardial edema and circulation. The P value for a chi-square test was <0.0001, indicating statistical significance.
Fig 8.
Phalloidin treatment partially rescues circulation in actc1as434 mutants. Microangiography was performed at 48 hpf on wild-type sibling embryos (A), phalloidin-treated embryos that displayed pericardial edema, characteristic of s434 mutants (B), and untreated actc1as434−/− mutants (C). (A) In the wild-type embryos, injection of fluorescein-labeled dextran into the common cardinal vein (CCV) was circulated through the blood vessels of the body, as indicated by the arrowhead. Fluorescence is present in the dorsal aorta, the posterior cardinal vein, and intersegmental vessels (A′), shown in a magnification of panel A. (B) Phenotypically mutant embryos that displayed pericardial edema, alongside their wild-type siblings, were treated 24 to 48 hpf with a 2 μM dose of phalloidin oleate. Injection of dextran into treated mutant embryos showed a partial rescue in circulation, as the dye is present in the dorsal aorta (arrowhead). A magnified view of the trunk is shown in panel B′. (C) Injection of dextran into untreated mutant embryos demonstrated that these embryos do not have circulation, as magnification of their trunk shows there is no dye circulating in their vessels (C′). The images were taken using a 5× objective, with the anterior portion to the left and the dorsal side up.
We also performed injections at the one-cell stage of both actc1a mRNA and DNA constructs in an attempt to rescue the actc1as434 phenotype. However, these injections did not significantly rescue the mutant phenotype. We hypothesize that because actc1a normally is expressed at very high levels, the amount of actin protein that is generated by RNA or DNA injection is insufficient for effective rescue. In addition, it is possible that the mutant phenotype is too latent to be rescued by an injection at the one-cell stage because of the limited half-life of the injected mRNA, while transient DNA expression is typically highly mosaic and is only present in a fraction of cells.
The actc1as434 mutation disrupts proper AVC development.
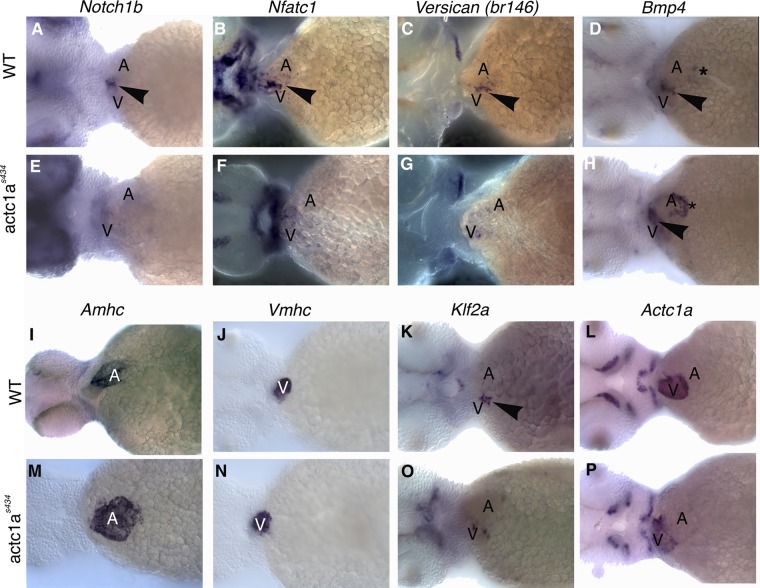
Proper differentiation of the AVC tissues, both endocardium and myocardium, is fundamental to valvulogenesis later in development. To assess AVC development in actc1as434 mutants, WISH analysis for selected endocardial and myocardial AVC markers was performed at 55 hpf. Endocardial notch1b and nfatc1 (nuclear factor of activated T-cell c1) expression is localized to the AVC in wild-type embryos (17, 25, 42), while in the actc1as434 mutants, notch1b expression is absent from the AVC and nfatc1 expression is diffuse and not restricted to the AVC (Fig. 9A, B, E, and F). In wild-type embryos, myocardium-specific vcana (versican a) expression is localized to the AVC (38, 42), while bmp4 (bone morphogenetic protein 4) is expressed at the AVC and partially within the atrium (6, 7) (Fig. 9C and D). In mutant embryos, vcana expression is decreased and not localized to the AVC (Fig. 9G), while bmp4 expression is largely unaffected (Fig. 9H). amhc (atrial myosin heavy chain) expression demonstrates that the atrium is dilated and appears broader in the actc1as434 mutants, while the size of the ventricles as demonstrated by vmhc (ventricular myosin heavy chain) expression is not affected, compared to that in the wild-type embryos at 55 hpf (Fig. 9I, J, M, and N). In addition, endocardial klf2a (kruppel-like factor 2a) expression is mislocalized and not restricted to the AVC in actc1as434 mutants (Fig. 9O). actc1a mRNA expression is not significantly affected, and it remains localized primarily in the ventricle in both wild-type and actc1as434 mutant embryos (Fig. 9L and P). These results indicate that the actc1as434 mutation disrupts proper AVC differentiation.
Fig 9.
The actc1as434 mutation disrupts atrioventricular specification events that precede endocardial cushion formation. In situ hybridization was performed for the following markers in both wild-type (A to D and I to L) and mutant (E to H and M to P) embryos at 55 hpf: notch1b, nfatc1, the versican gene, bmp4, amhc, vmhc, klf2a, and actc1a. (A and E) The notch1b transcript is localized to the endocardial AVC by 55 hpf in the wild-type embryos (arrowhead); however, it appears disorganized and downregulated in the mutants. (B and F) nfatc1 is also localized to the endocardial AVC in wild-type embryos (arrowhead), while it appears decreased and not localized in the mutants. (C and G) The versican gene transcript is restricted to the myocardial AVC in the wild-type embryos by 55 hpf (arrowhead), while it is disorganized and decreased in the mutants. (D and H) bmp4 is expressed in the myocardial AVC (arrowhead) and partially within the atrium (asterisk). In the mutant, bmp4 is still restricted to the AVC (arrowhead), but appears upregulated in the atrium (asterisk). (I and M) Wild-type amhc expression is restricted to the atrium at 55 hpf, while the mutant amhc expression is expanded, indicative of dilated atrium. (J and N) vmhc expression is not affected in the mutant embryos. (K and O) Wild-type expression of klf2a is restricted to the endocardial AVC (arrowhead). klf2a expression is mislocalized in the mutant embryos. (L and P) actc1a expression is restricted mostly to the ventricle in both the wild-type and mutant embryos. No significant change in actc1a expression is apparent.
Similar to actc1as434 mutants, notch1b, klf2a, and vcana expression in actc1a MO-injected embryos was decreased and not localized to the AVC by 48 hpf (see Fig. S4 at https://research.cchmc.org/mcb_glenn). This confirms our earlier data showing that actc1a mutation results in a loss of polymerized cardiac actin, and therefore, actc1a MO knockdown results in a similar phenotype.
Heart function is altered in actc1as434 mutants.
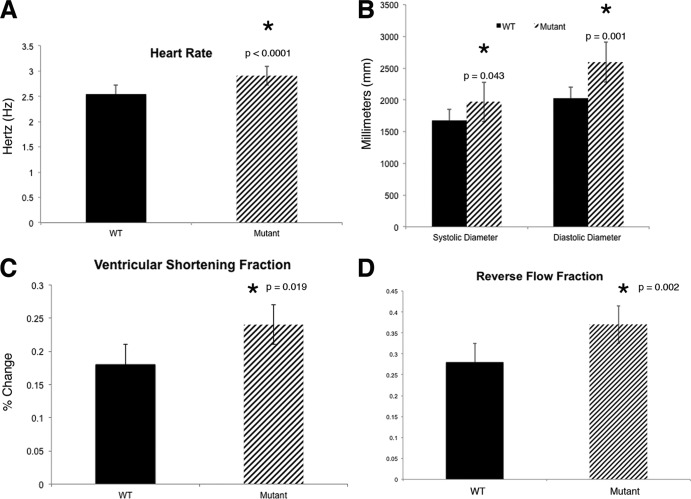
The structural defects in actc1as434 mutants lead to altered heart function. Therefore, functional studies were conducted at 48 hpf to quantify the difference from wild-type heart function. To determine functional parameters, heart function was imaged in live embryos using a high-speed camera (see Movies 3 and 4 at https://research.cchmc.org/mcb_glenn). A statistically significant difference (P < 0.0001) between the mutant heart rate (2.91 Hz), and the wild-type embryo heart rate (2.54 Hz) was observed (Fig. 10A). We measured the diastolic and systolic diameters of the ventricle to determine if there was a dilation of the diameter of the ventricle at diastole, referred to as end-diastolic diameter (Fig. 10B). Indeed, we observed significantly higher systolic (P = 0.043) and diastolic (P = 0.001) diameters in the actc1as434 mutants compared to those in the wild-type embryos. This difference indicates that the mutant ventricle is dilated. In order to determine if the mutant heart has deficiencies in contracting, we measured the ventricular shortening fraction (VSF), which is an estimate of contractility (Fig. 10C) (43). We hypothesize that the heart is still able to contract due to the other actin isoforms present in the zebrafish heart (acta1a, acta1b, and acta2). The mutant VSF was significantly larger than the wild type, indicating that the mutant heart undergoes larger contractions than the wild type. This is an interesting finding, since the mutant heart cannot overcome the resistance of the vascular system even with increased contractility. These results suggest that the mutant hearts are dilated, but fail to produce sufficient force to eject blood into the circulatory system.
Fig 10.
The actc1as434 mutation leads to abnormal heart function. (A) Heart rate was measured in hertz (Hz), as the number of heartbeats divided by 60. The mutant embryos have a faster heart rate than their wild-type siblings (P < 0.0001). (B) Both systolic and diastolic diameters are larger in the mutants than in the wild-type siblings (P = 0.043 and 0.001, respectively). (C) Contractility was measured by calculating the ventricular shortening fraction (VSF), which was found to be higher in the mutants than that in their wild-type siblings (P = 0.019). (D) Reverse flow fraction (RFF) values were measured in the wild-type and mutant embryos, and analysis reveals that mutant embryos have a statistically significant increase in RFF value (P = 0.002). All values are denoted as means ± SEM.
Previous studies have shown that the blood flow pattern within the heart is a critical factor for EC development. It was been suggested that the reverse flow fraction (RFF) is important to restrict klf2a expression to the AVC (41). Either an increase or decrease in the RFF value resulted in mislocalization of klf2a expression and disruption of valvulogenesis. Since expression of klf2a in the mutant embryos is not restricted to the AVC (Fig. 9K and O), we measured RFF values. In mutant embryos, the RFF value at 48 hpf was increased significantly (P = 0.002) to 35% ± 8%, compared to that in wild-type siblings (28% ± 6%) (Fig. 10D). These findings suggest that actc1a mutants fail to initiate EC morphogenesis due to the altered cardiac output. This results in a disrupted blood flow pattern within the heart leading to mislocalized klf2a expression and subsequent failure to initiate EC morphogenesis. These results support the hypothesis that the blood flow pattern within the heart is important for klf2a localization and EC morphogenesis.
DISCUSSION
In this study, we have positionally cloned a zebrafish mutant (s434 mutation) that displays defects in EC morphogenesis and blood circulation. Our results show that actc1as434 mutants have a Y169S amino acid substitution in the W-loop of alpha cardiac actin (actc1a), which results in fragile actin filaments. Yeast cultures demonstrated that F169S mutant actin forms filaments, which then spontaneously disintegrate and are unable to reanneal and form F-actin. The phenotype in both zebrafish and yeast can be partially or fully rescued by treatment with the actin-stabilizing drug phalloidin, respectively. Mutant embryos display altered blood flow within the heart tube, which results in misexpression of genes associated with AVC morphogenesis and leads to the absence of EC formation.
In addition to ACTC, other actins, including skeletal actin (acta1a and acta1b) are expressed in mouse cardiac tissue during embryogenesis and can provide redundant function (32, 33). While expression patterns for all of the zebrafish actin isoforms are not currently known, we have shown that acta1b is present in the heart, as previous studies have shown expression of one zebrafish sarcomeric actin homolog (acta1a) as well as smooth muscle aorta actin (acta2) in the zebrafish heart (1, 37). However, these other isoforms of actin present in the heart (acta1a, acta1b, and acta2) apparently cannot compensate for the lack of actc1a. It is likely that there is partial redundancy between actc1a and acta1b because embryos heterozygous for both actc1as434 and cfk/acta1b show similar defects in cardiac function. A likely explanation is that the overall amount of actin in these double heterozygous embryos is reduced below the critical threshold, similar to actc1as434−/− homozygous embryos.
Our in vitro and in vivo studies suggest that the zebrafish Y169S and analogous yeast F169S mutations have an adverse effect on actin polymerization per se. Yeast in vitro data show that the mutation causes a severe inherent filament destabilization leading to disassembly of the F-actin into small annealing-incompetent filament fragments. This behavior was not due to ATP depletion. Further, it was not due to postpolymerization denaturation of the actin since addition of phalloidin allowed the fragments to reanneal and form normal-appearing actin filaments. Yeast viability requires that stable actin filaments can form, and the phalloidin results suggest that in the cell, an actin-stabilizing protein such as tropomyosin or fimbrin, both present in yeast, might play this role. We also showed that substoichiometric WT actin levels could provide enough normalization of monomer-monomer contacts to restore filament stability. This property, coupled with the presence of filament-stabilizing proteins, could explain why a zebrafish heart producing both mutant and WT actins survives for an extended time and can still partially function. Similar to yeast, the zebrafish mutant phenotype could be partially rescued by treatment with phalloidin, which helped to improve cardiac functionality and restored circulation to a significant fraction of actc1as434 mutants. These results argue that Y169S mutation in s434 mutants affects actin polymerization, which is the primary reason for the observed cardiac defects.
Mutant ACTC alleles that have been previously associated with IDC and atrial septal defects in humans are thought to have a dominant phenotype, since heterozygous carriers are affected (24, 27). However, the actc1as434 mutant phenotype is inherited in a Mendelian recessive manner. We have not observed any developmental defects in heterozygous embryos; therefore, this mutation is unlikely to have a dominant-negative effect. It is possible that heterozygous zebrafish adults may have reduced cardiac performance, which is difficult to detect in the absence of appropriate assays. Because MO knockdown of actc1a phenocopies the actc1as434 mutant phenotype and polymerized actin is greatly reduced in these mutants, this mutation likely renders actin protein function either null or close to null.
Interestingly, a structural mutation in the cardiac actin results in defective EC formation, which is the earliest stage of valvulogenesis. Hemodynamics and oscillatory blood flow have been previously implicated in restricting expression of genes associated with EC morphogenesis, including klf2a, to the AVC (41). Our results show that the blood flow pattern in the heart tube is altered in actc1as434 mutants, and this leads to mislocalization of klf2a and other AVC marker expression as well as an increased RFF and VSF values. Previous studies have also demonstrated that skeletal actin mutants such as the cardiofunk mutant have defective valve formation (1). Similar EC defects are also observed in other mutants that display regurgitant blood flow such as jekyll- and klf2a-MO-injected embryos (41, 42). These studies argue that both proper oscillatory blood flow at the AVC and myocardial function are required for EC formation and valve morphogenesis.
In summary, the actc1as434 mutant provides a tool for understanding the critical role for actin in early heart development. This mutant also demonstrates the essential function of the W-loop of ACTC in a vertebrate model. Understanding the physiological and developmental consequences of different types of actin mutations will provide better insights into and help to develop novel treatments for cardiovascular congenital diseases.
ACKNOWLEDGMENTS
This research was supported by the Cincinnati Children's Hospital Research Foundation, AHA and MOD grants to T.B., and NIH grant DC8803 to P.A.R.
We thank Matt Kofron for aiding in developing a protocol for zebrafish phalloidin staining. The actin crystallization renderings were kindly provided by Rashmi Hegde. We also recognize Michael Craig and Jay Hove for help with the high-speed microscopy and for use of their equipment.
Footnotes
Published ahead of print 2 July 2012
REFERENCES
- 1. Bartman T, et al. 2004. Early myocardial function affects endocardial cushion development in zebrafish. PLoS Biol. 2:E129 doi:10.1371/journal.pbio.0020129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beis D, et al. 2005. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development 132:4193–4204 [DOI] [PubMed] [Google Scholar]
- 3. Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. 2003. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development 130:6121–6129 [DOI] [PubMed] [Google Scholar]
- 4. Bertola LD, Ott EB, Griepsma S, Vonk FJ, Bagowski CP. 2008. Developmental expression of the alpha-skeletal actin gene. BMC Evol. Biol. 8:166 doi:10.1186/1471-2148-8-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boheler KR, et al. 1991. Skeletal actin mRNA increases in the human heart during ontogenic development and is the major isoform of control and failing adult hearts. J. Clin. Invest. 88:323–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boyer AS, et al. 1999. TGFbeta2 and TGFbeta3 have separate and sequential activities during epithelial-mesenchymal cell transformation in the embryonic heart. Dev. Biol. 208:530–545 [DOI] [PubMed] [Google Scholar]
- 7. Chen JN, et al. 1997. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development 124:4373–4382 [DOI] [PubMed] [Google Scholar]
- 8. Cook RK, Blake WT, Rubenstein PA. 1992. Removal of the amino-terminal acidic residues of yeast actin. Studies in vitro and in vivo. J. Biol. Chem. 267:9430–9436 [PubMed] [Google Scholar]
- 9. Cook RK, Sheff DR, Rubenstein PA. 1991. Unusual metabolism of the yeast actin amino terminus. J. Biol. Chem. 266:16825–16833 [PubMed] [Google Scholar]
- 10. dos Remedios CG, et al. 2003. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 83:433–473 [DOI] [PubMed] [Google Scholar]
- 11. Feng L, et al. 1997. Fluorescence probing of yeast actin subdomain 3/4 hydrophobic loop 262–274. Actin-actin and actin-myosin interactions in actin filaments. J. Biol. Chem. 272:16829–16837 [DOI] [PubMed] [Google Scholar]
- 12. Holmes KC, Angert I, Kull FJ, Jahn W, Schroder RR. 2003. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature 425:423–427 [DOI] [PubMed] [Google Scholar]
- 13. Holmes KC, Popp D, Gebhard W, Kabsch W. 1990. Atomic model of the actin filament. Nature 347:44–49 [DOI] [PubMed] [Google Scholar]
- 14. Hyatt TM, ESC. 1999. Vectors and techniques for ectopic gene expression in zebrafish. Methods Cell Biol. 59:117–126 [DOI] [PubMed] [Google Scholar]
- 15. Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY. 2005. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132:5199–5209 [DOI] [PubMed] [Google Scholar]
- 16. Jowett T. 1999. Transgenic zebrafish. Methods Mol. Biol. 97:461–486 [DOI] [PubMed] [Google Scholar]
- 17. Kortschak RD, Tamme R, Lardelli M. 2001. Evolutionary analysis of vertebrate Notch genes. Dev. Genes Evol. 211:350–354 [DOI] [PubMed] [Google Scholar]
- 18. Kudryashov DS, Grintsevich EE, Rubenstein PA, Reisler E. 2010. A nucleotide state-sensing region on actin. J. Biol. Chem. 285:25591–25601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kumar A, et al. 1997. Rescue of cardiac alpha-actin-deficient mice by enteric smooth muscle gamma-actin. Proc. Natl. Acad. Sci. U. S. A. 94:4406–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laudadio RE, et al. 2005. Rat airway smooth muscle cell during actin modulation: rheology and glassy dynamics. Am. J. Physiol. Cell Physiol. 289:C1388–C1395 [DOI] [PubMed] [Google Scholar]
- 21. Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. 2001. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood 98:3087–3096 [DOI] [PubMed] [Google Scholar]
- 22. Lyons GE, Buckingham ME, Mannherz HG. 1991. Alpha-actin proteins and gene transcripts are colocalized in embryonic mouse muscle. Development 111:451–454 [DOI] [PubMed] [Google Scholar]
- 23. Martin RT, Bartman T. 2009. Analysis of heart valve development in larval zebrafish. Dev. Dyn. 238:1796–1802 [DOI] [PubMed] [Google Scholar]
- 24. Matsson H, et al. 2008. Alpha-cardiac actin mutations produce atrial septal defects. Hum. Mol. Genet. 17:256–265 [DOI] [PubMed] [Google Scholar]
- 25. Milan DJ, Giokas AC, Serluca FC, Peterson RT, MacRae CA. 2006. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development 133:1125–1132 [DOI] [PubMed] [Google Scholar]
- 26. Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. 2009. The nature of the globular- to fibrous-actin transition. Nature 457:441–445 [DOI] [PubMed] [Google Scholar]
- 27. Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT. 1998. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science 280:750–752 [DOI] [PubMed] [Google Scholar]
- 28. Ordahl CP. 1986. The skeletal and cardiac alpha-actin genes are coexpressed in early embryonic striated muscle. Dev. Biol. 117:488–492 [DOI] [PubMed] [Google Scholar]
- 29. Paavilainen VO, Oksanen E, Goldman A, Lappalainen P. 2008. Structure of the actin-depolymerizing factor homology domain in complex with actin. J. Cell Biol. 182:51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pham VN, et al. 2007. Combinatorial function of ETS transcription factors in the developing vasculature. Dev. Biol. 303:772–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rubenstein PA. 1990. The functional importance of multiple actin isoforms. Bioessays 12:309–315 [DOI] [PubMed] [Google Scholar]
- 32. Ruzicka DL, Schwartz RJ. 1988. Sequential activation of alpha-actin genes during avian cardiogenesis: vascular smooth muscle alpha-actin gene transcripts mark the onset of cardiomyocyte differentiation. J. Cell Biol. 107:2575–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sassoon DA, Garner I, Buckingham M. 1988. Transcripts of alpha-cardiac and alpha-skeletal actins are early markers for myogenesis in the mouse embryo. Development 104:155–164 [DOI] [PubMed] [Google Scholar]
- 34. Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. 1993. The structure of crystalline profilin-beta-actin. Nature 365:810–816 [DOI] [PubMed] [Google Scholar]
- 35. Sellers J. 1999. Myosins: protein profile, 2nd ed Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 36. Shimizu T, Dennis JE, Masaki T, Fischman DA. 1985. Axial arrangement of the myosin rod in vertebrate thick filaments: immunoelectron microscopy with a monoclonal antibody to light meromyosin. J. Cell Biol. 101:1115–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thisse B, Thisse C. 2004. Fast release clones: a high throughput expression analysis. ZFIN, University of Oregon, Eugene, OR [Google Scholar]
- 38. Thisse C, Thisse B. 1999. Antivin, a novel and divergent member of the TGFbeta superfamily, negatively regulates mesoderm induction. Development 126:229–240 [DOI] [PubMed] [Google Scholar]
- 39. Thompson MA, et al. 1998. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev. Biol. 197:248–269 [DOI] [PubMed] [Google Scholar]
- 40. Vandekerckhove J, Weber K. 1979. The complete amino acid sequence of actins from bovine aorta, bovine heart, bovine fast skeletal muscle, and rabbit slow skeletal muscle. A protein-chemical analysis of muscle actin differentiation. Differentiation 14:123–133 [DOI] [PubMed] [Google Scholar]
- 41. Vermot J, et al. 2009. Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLoS Biol. 7:e1000246 doi:10.1371/journal.pbio.1000246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Walsh EC, Stainier DY. 2001. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science 293:1670–1673 [DOI] [PubMed] [Google Scholar]
- 43. Wang YX, et al. 2005. Requirements of myocyte-specific enhancer factor 2A in zebrafish cardiac contractility. FEBS Lett. 579:4843–4850 [DOI] [PubMed] [Google Scholar]
- 44. Wen KK, McKane M, Stokasimov E, Rubenstein PA. 2011. Mutant profilin suppresses mutant actin-dependent mitochondrial phenotype in Saccharomyces cerevisiae. J. Biol. Chem. 286:41745–41757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Westerfield M. 2007. The zebrafish book, 5th ed Monte Westerfield, Eugene, OR [Google Scholar]
- 46. Westin J, Lardelli M. 1997. Three novel Notch genes in zebrafish: implications for vertebrate Notch gene evolution and function. Dev. Genes Evol. 207:51–63 [DOI] [PubMed] [Google Scholar]
- 47. Wong KS, et al. 2012. Hedgehog signaling is required for differentiation of endocardial progenitors in zebrafish. Dev. Biol. 361:377–391 [DOI] [PubMed] [Google Scholar]
- 48. Yelon D, Horne SA, Stainier DY. 1999. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 214:23–37 [DOI] [PubMed] [Google Scholar]