Abstract
Controlled renewal of the epithelium with precise cell distribution and gene expression patterns is essential for colonic function. GATA6 is expressed in the colonic epithelium, but its function in the colon is currently unknown. To define GATA6 function in the colon, we conditionally deleted Gata6 throughout the epithelium of small and large intestines of adult mice. In the colon, Gata6 deletion resulted in shorter, wider crypts, a decrease in proliferation, and a delayed crypt-to-surface epithelial migration rate. Staining techniques and electron microscopy indicated deficient maturation of goblet cells, and coimmunofluorescence demonstrated alterations in specific hormones produced by the endocrine L cells and serotonin-producing cells. Specific colonocyte genes were significantly downregulated. In LS174T, the colonic adenocarcinoma cell line, Gata6 knockdown resulted in a significant downregulation of a similar subset of goblet cell and colonocyte genes, and GATA6 was found to occupy active loci in enhancers and promoters of some of these genes, suggesting that they are direct targets of GATA6. These data demonstrate that GATA6 is necessary for proliferation, migration, lineage maturation, and gene expression in the mature colonic epithelium.
INTRODUCTION
The mammalian colon is lined by a highly differentiated epithelium comprised of specialized absorptive and secretory cells, with cell distribution and gene expression patterns evolved to facilitate the absorption of large quantities of water. The colonic epithelium is maintained through a process of continuous cellular renewal in which stem cells, located at the base of the crypts (4), produce 14 to 21 transient amplifying cells per hour (19) that give rise to all differentiated cell types. The three main types of differentiated cells, colonocytes, goblet cells, and enteroendocrine cells, differentiate while migrating up to the surface epithelium, with a turnover time of 4 to 6 days. Colonocytes have both absorptive and secretory functions, absorbing sodium, water, and short-chain fatty acids while secreting potassium and bicarbonate. Goblet cells produce mucins that help protect the mucosa from injury, and enteroendocrine cells secrete hormones that regulate gastrointestinal function. Controlled renewal of the epithelium with precise cell distribution and gene expression patterns is essential for colonic function and is controlled by molecular mechanisms that are only beginning to be elucidated.
GATA factors are evolutionarily conserved zinc finger transcription factors that play key roles in proliferation, differentiation, and gene regulation in multiple organs (24). GATA6 is coexpressed with GATA4 in the proximal small intestine and is expressed independently of GATA4 in the ileum (7, 9) and colon (11, 12, 14, 18, 33). In the ileum, GATA6 promotes crypt cell proliferation, Paneth cell differentiation, and enteroendocrine cell commitment and regulates the expression of specific absorptive enterocyte genes (7). In the proximal small intestine, GATA4 and GATA6 are both capable of mediating these functions (7). In cell culture models, GATA6 activates the promoters of genes expressed in the colon (1, 10, 29). Gata6 expression is upregulated in colon cancer epithelial cells (5, 14, 29), as well as in nonmalignant cells along the stromal margins in humans (14), suggesting a role in proliferation. Collectively, these reports delineate complex functions for GATA6 in the colon. Using genetic loss-of-function experiments, we provide the first in vivo evidence that GATA6 is necessary for colonic epithelial differentiation and regeneration. We find that GATA6 is expressed in proliferating crypt cells, differentiated goblet cells, enteroendocrine cells, and surface colonocytes. We demonstrate that GATA6 is required for crypt cell proliferation and migration, secretory cell differentiation, and the expression of specific goblet cell and colonocyte genes.
MATERIALS AND METHODS
Mice.
Previously established and confirmed Gata6loxP/loxP, VillinCreERT2-positive (G6del) mice were used in this study to produce conditional, inducible deletion of Gata6 in the colonic epithelium (7). Gata6loxP/loxP, VillinCreERT2-negative mice served as controls (Ctl). DNA was obtained from tail biopsy specimens, and genotypes were determined by semiquantitative PCR using previously validated primers (9, 30). Male and female mice 6 to 8 weeks of age were treated with five daily intraperitoneal injections of tamoxifen (Sigma Chemical Company, St. Louis, MO) (100 μl, 10 mg/ml; dissolved in ethanol-sunflower oil = 1:9 [vol/vol]) as described previously (8, 9), and tissue was collected 28 days after the last injection (see Fig. S1A in the supplemental material). Selected mice were treated with bromodeoxyuridine (BrdU) (0.1 ml of 10 mg/ml) 28 days after the last tamoxifen treatment, and tissue was collected 1 h or 3 days after the BrdU injection (see Fig. S1B). For a specific experiment, mice 6 to 8 weeks of age were treated with tamoxifen twice a day for 5 days and tissue was collected the day of the last injection (see Fig. S1C). Approval was obtained from the Institutional Animal Care and Use Committee.
Tissue isolation and processing.
Mice were anesthetized and dissected, 0.2- to 0.4-cm-long segments ∼2 cm distal to the ileocecal junction were used for RNA isolation, and the next 1 cm was fixed in 4% paraformaldehyde for 4 h and dehydrated overnight, as previously described (8, 9). In selected mice, a 4-cm-long segment of midcolon was dissected for the isolation of epithelial cell nuclear extracts. Fixed tissue was paraffin embedded and sectioned (5 μm2), and selected samples were stained with hematoxylin and eosin (H&E) or with the periodic acid-Schiff (PAS) reaction in the Imaging Core of the Harvard Digestive Disease Center at Beth Israel-Deaconess Medical Center (Boston, MA). In selected mice, well-oriented sections were fixed in 1.25% glutaraldehyde–4% formaldehyde–0.1 M cacodylic acid buffer (pH 7.4) and processed by the Imaging Core for transmission electron microscopy, as previously described (7).
Immunohistochemistry and immunofluorescence.
Immunohistochemistry and immunofluorescence analyses were conducted as previously described (8, 9). Primary antibodies used included goat anti-GATA6 (R&D Systems; catalog no. AF1700) (1:50), rabbit anti-chromogranin A (anti-CHGA) (Leica Microsystems, Inc., Buffalo Grove, IL; catalog no. CHROM 430 CE) (1:2,000), goat anti-chromogranin A (anti-CHGA) (Santa Cruz Biotechnology, Santa Cruz, CA; catalog no. sc-1488) (1:100), rabbit anti-Ki67 (Santa Cruz; catalog no. sc-15402) (1:200), mouse anti-BrdU (Thermo Fisher Scientific, Inc., Fremont, CA; catalog no. MS-1058-PO) (1:250), rabbit anti-cleaved caspase-3 (anti-CASP3) (Cell Signaling Technology, Inc., Danvers, MA; catalog no. 9664) (1:100), rabbit anti-mucin 2 (anti-MUC2) (Santa Cruz; catalog no. sc-15334) (1:200), rabbit anti-trefoil factor 3 (anti-TFF3) (gift from D. Podolsky, University of Texas, Southwestern) (1:2,000), goat anti-glucagon-like peptide 1 (anti-GLP1) (Santa Cruz; catalog no. sc-7782) (1:100), goat anti-peptide YY (anti-PYY) (Santa Cruz; catalog no. sc-4318) (1:50), rabbit antiserotonin (5-HT) (Immunostar, Inc., Hudson, WI; catalog no. 20080) (1:10,000), and rabbit anti-SLC9A2 (gift from M. Donowitz, Johns Hopkins University School of Medicine, Baltimore, MD [17]) (1:400). Secondary antibodies included biotinylated donkey anti-rabbit IgG, donkey anti-goat IgG, and donkey anti-mouse IgG (all from Vector Labs, Burlingame, CA), Alexa Fluor 488–anti-rabbit IgG, and Alexa Fluor 594–anti-goat IgG (both from Life Technologies, Carlsbad, CA). For immunohistochemistry, biotinylated secondary antibodies were linked to avidin-horseradish peroxidase conjugates (Vector Labs), visualized using 3,3′-diamino benzidine (DAB; Sigma) for 2 to 5 min, and lightly counterstained with methyl green. For immunofluorescence, sections were mounted in SlowFade Gold (Life Technologies) containing 4′,6-diamino-2-phenylindol dihydrochloride (DAPI).
RNA isolation and gene expression analysis.
RNA was isolated using an RNeasy minikit (Qiagen, Inc., Valencia, CA). mRNA abundances were determined by quantitative reverse transcriptase-PCR (qRT-PCR) as described previously (8, 9) using validated primer pairs (see Fig. S2 and S3 in the supplemental material). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA abundance was measured for each sample and used to normalize the data. All data were expressed relative to the mean value of Ctl colon.
Western blotting.
Nuclear extracts were isolated from 4-cm-long segments of midcolon using a protocol originally established for mouse small intestinal epithelium (9, 35). Western analysis for GATA6 was carried out as previously described (9) using rabbit anti-GATA6 (1:1,000; Cell Signaling, A403).
Tissue measurements and cell counting.
Crypt depth and width were determined from H&E-stained slides using ImageJ software (available at http://rsb.info.nih.gov/ij/). Ten well-oriented crypts per slide from 5 different animals per group were measured. The level of Ki67-positive cells in mouse colon was determined by immunostaining, and the number was defined as a percentage of total epithelial cells or as the total number per well-oriented crypt. CHGA-positive cells were identified by immunofluorescence, and the number was defined as a percentage of total epithelial cells, identified by DAPI-positive (blue) nuclei in the epithelial layer. For both calculations, a minimum of 250 cells per slide were counted from a minimum of 5 different animals per group. To quantify specific enteroendocrine subpopulations, coimmunofluorescence analysis was performed for CHGA and GLP1, CHGA and PYY, or CHGA and 5-HT. In multiple microscopic fields, the number of CHGA-positive cells was determined (green or red filter). In the same field, the numbers of GLP1-positive cells (red filter), PYY-positive cells (red filter), or 5-HT-positive cells (green filter) were then determined. A minimum of 100 CHGA-positive cells were counted per animal, and 5 animals in each group were included.
Cell culture and lentiviral infection.
LS174T cells were cultured in RPMI 1640 (Mediatech, Inc., Mooresville, NC), and 293T and Caco-2 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Mediatech) containing 4.5 g/liter glucose, l-glutamine, and sodium pyruvate. Both media were supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and penicillin (100 U/ml)-streptomycin (1 mg/ml) (Sigma), and both lines were maintained in 5% CO2 at 95% relative humidity and 37°C. Media were replaced every 2 to 3 days. To produce lentivirus for the knockdown of Gata6, 293T cells were plated at 105 cells/ml on 6-cm plates and transfected after 24 h with 1 μg of vesicular stomatitis virus glycoprotein G (VSV-G) envelope-expressing plasmid pMD.G, 1 μg of pCMV-dR8.91 (Delta 8.9) plasmid containing gag, pol, and rev genes, and 1 μg of shRNA-pLKO.1 plasmids expressing a knockdown short hairpin RNA (shRNA; Sigma) for green fluorescent protein (GFP) (Ctl) (SHC005) or human Gata6 (G6kd) (TRCN0000005392) using 6 μl of FuGENE 6 reagent (Roche). Media from the transfected 293T cells were changed to RPMI media (recipient cell media) 16 h after transfection. Culture media containing lentiviral particles were collected and filtered (0.45 μm pore size) 48 h after transfection and transferred immediately to LS174T cells plated at 30% confluence to infect for 2 h using 0.5 μl of Polybrene/ml. Infection was repeated the next day followed by selection of infected cells using 4 μg/ml puromycin. Infected cells were kept under conditions of selection until the day of harvesting. Trypsinized cells were homogenized using a QIA shredder (Qiagen), and RNA was isolated as described above for mouse tissue. Nuclear extracts were isolated as described previously (21). Gata6 knockdown was determined by qRT-PCR for human Gata6 and by Western blot analysis as previously described (9) using anti-rabbit GATA6 (Cell Signaling).
ChIP.
To determine direct association of GATA6 with putative target genes, we identified possible targets using publicly available databases and confirmed enhancer identity and GATA6 occupancy using chromatin immunoprecipitation (ChIP) assays. To identify putative targets, we downloaded the H3K4me2 and GATA6 ChIP-seq data for proliferating Caco-2 cells and the Caco-2 cell input from GEO data sets (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23436) (37) and uploaded these two files into Cistrome (22). We identified positive sites of H3K4me2 enrichment, a characteristic of active loci (6, 15, 16), and GATA6 occupancy using the Model-based Analysis for ChIP-Seq (MACS) peak-calling tool (38) and a P value cutoff of 10−5. We then mapped the peaks to the closest transcription start site (TSS) using the Peak2Gene tool in Cistrome. Using the Integrated Genome Browser (Affymetrix, Santa Clara, CA) (23), we mapped the positions and heights of the peaks. This enabled identification of putative GATA6 occupancy sites in human chromatin in an intestinal cell line. It also enabled the identification of loci to be used as putative positive (enhancer of Usp-12) and negative (α-amylase TSS) controls.
For ChIP assays, LS174T cells were incubated in RPMI medium containing 1% formaldehyde (Fisher Scientific, Pittsburgh, PA) for 10 min at 37°C. The cells were washed 2 times with phosphate-buffered saline (PBS), scraped, and resuspended in lysis buffer (50 mM Tris-Cl [pH 8.1], 10 mM EDTA, 1% sodium dodecyl sulfate [SDS]) containing protease inhibitor cocktail and PMSF) (10 μl/ml). The samples were sonicated to obtain chromatin fragments of between 400 and 1,000 bp. Sonicated samples were resuspended in ChIP dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1) and incubated overnight at 4°C with Dynabeads beads (Life Technologies) conjugated with one of the following antibodies: rabbit GATA6 (Cell Signaling; catalog no. 4253S), rabbit H3K27ac (Abcam, Cambridge, MA; catalog no. Ab4729), or rabbit IgG (negative control) (Millipore, Temecula, CA; catalog no. PP64B).
The IP sample was washed 6 times with radioimmunoprecipitation (RIPA) buffer (50 mM HEPES [pH 7.6], 0.5 M LiCl, 1 mM EDTA, 1% Nonidet P-40, 0.7% sodium deoxycholate), and the DNA was recovered by reverse cross-linking in 1% SDS–0.1 M NaHCO3 for 7 h at 65°C. DNA was purified using a QIAquick PCR purification kit (Qiagen) and quantified by Picogreen (Life Technologies). One nanogram of DNA was used per qPCR using primers displayed in Fig. S3 in the supplemental material.
Statistical analyses.
Data are expressed as means ± standard deviations (SD). Statistically significant differences were determined by the two-tailed Student's t test. Differences were considered statistically significant at P < 0.05.
RESULTS
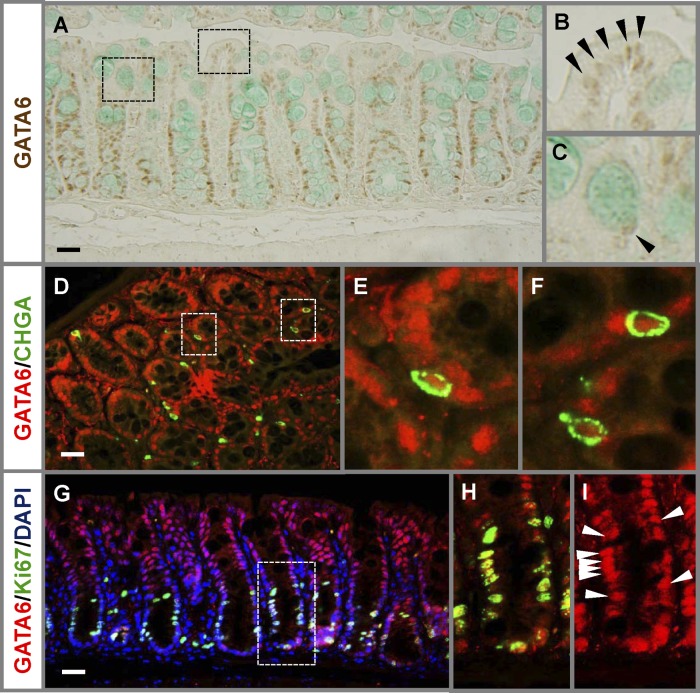
To determine which mouse colon cells could be under direct regulation by GATA6, we examined the expression of GATA6 in mice in situ. GATA6 was readily detected in the nuclei of colonocytes and goblet cells (Fig. 1A to C), as previously shown in humans (14). As shown by coimmunofluorescence, GATA6 was present in CHGA-positive enteroendocrine cells (Fig. 1D to F). Specifically, red GATA6 immunofluorescence was found in 93% of all CHGA-positive cells (>200 cells counted) for which nuclear DAPI blue fluorescence was also detected, indicating that GATA6 is generally expressed in the enteroendocrine cell population. GATA6 was also coexpressed with Ki67 in the nuclei of cells in the proliferative compartment (Fig. 1G to I). Together, these data show that GATA6 is expressed in all differentiated and proliferating cells of the mature mouse colonic epithelium.
Fig 1.
GATA6 is expressed in all differentiated and proliferating cells in the mature mouse colonic epithelium. (A to C) Immunostaining reveals that GATA6 is expressed in nuclei of surface colonocytes (B) and goblet cells (C) (arrowheads). (D to I) Coimmunofluorescence with GATA6 and CHGA (D to F) or GATA6 and Ki67 (G to I) reveals that GATA6 is also expressed in CHGA-positive enteroendocrine cells and Ki67-positive cells in the proliferative compartment. Arrowheads indicate examples of GATA6-positive cells. Bars: 100 μm.
To determine GATA6 function in these cells, we specifically ablated Gata6 with tamoxifen-induced, VillinCreERT2-mediated recombination in mice homozygous for the Gata6loxP allele (30), which results in deletion of Gata6 specifically in the small and large intestines (7). Quantitative RT-PCR indicated that Gata6 mRNA abundance, measured 28 days after the completion of tamoxifen treatment, was reduced >90% in G6del colon compared to Ctl colon (see Fig. S4A in the supplemental material), consistent with a reduced expression of GATA6 protein determined by Western blot analysis (see Fig. S4B) and by immunostaining (see Fig. S4C and D). These data verify knockout of Gata6 in adult mouse colon after tamoxifen treatment. Phenotypically, the overall health status, judged by activity and fur quality, of the G6del mice was indistinguishable from that of the Ctl mice. However, we observed that softer and lighter-in-color stools were produced by the G6del mice compared to the Ctl mice. To define potential compensation by other GATA factors, we tested the levels of expression of Gata4 and Gata5 after Gata6 deletion and found a 3-fold increase in Gata4 mRNA but no difference in Gata5 mRNA in G6del colon compared to Ctl colon (see Fig. S4E). However, it should be noted that the levels of Gata4 mRNA in Ctl colon are nearly undetectable (<0.5% of wild-type jejunum) and that the biological influence of a 3-fold increase of such a low level is unknown.
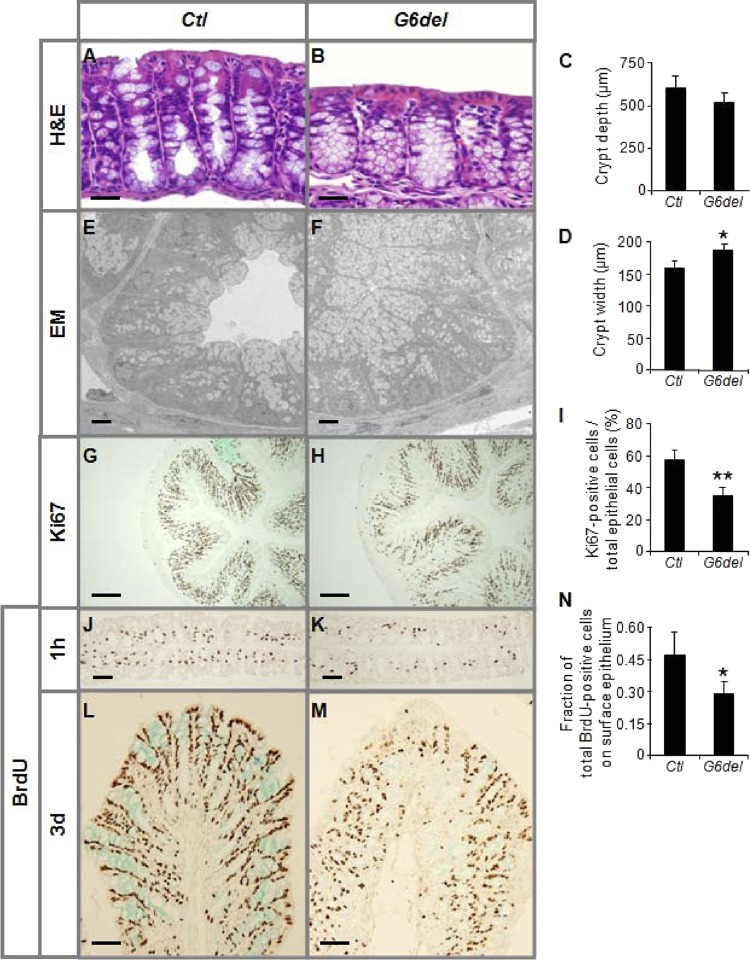
As shown by standard H&E staining (Fig. 2A and B), G6del colon showed shorter, wider crypts with an accumulation of goblet-like cells and fewer mature goblet cells on or near the surface epithelium. The crypt depth of G6del colon was modestly (12%) but not significantly reduced (Fig. 2C), but the crypt width was significantly increased 18% (P < 0.05) (Fig. 2D). The increase in goblet-like structures in crypts of G6del colon was confirmed by transmission electron microscopy (EM) (Fig. 2E and F). The number of Ki67-positive cells per total epithelial cells was decreased 39% (P < 0.01) (Fig. 2G to I), indicating a reduction in proliferating cell numbers. Immunostaining for BrdU on tissue samples harvested 1 h after BrdU injection demonstrated fewer cells containing BrdU labeling in G6del colon compared to Ctl colon (Fig. 2J and K), revealing a reduction in numbers of cells undergoing DNA synthesis and confirming a decrease in proliferating cells. We did not observe a decrease in the number of cleaved caspase 3-positive cells (see Fig. S5 in the supplemental material), indicating that apoptosis was unchanged in the G6del mice. To determine whether cell migration was affected, we applied BrdU pulse-chase analysis. After a 3-day chase, we found a lower percentage of BrdU-positive cells in surface colonocytes in G6del compared to Ctl mice (Fig. 2L to N), suggesting a reduced rate of migration when Gata6 is conditionally deleted. The reduced migration rate may have been caused by a diminished underlying drive resulting from a decreased proliferation rate, which could, in turn, account for the wider crypt morphology.
Fig 2.
Intestinal Gata6 deletion results in a reduction in the proliferation and migration rates in the mature mouse colonic epithelium. (A to D) H&E staining reveals an increase in goblet-like structures in crypts of G6del colon (A and B), a modest (nonsignificant) reduction in crypt depth (C), and significant increase in crypt width (D). (E and F) Transmission EM confirmed an accumulation of goblet-like cells in crypts. (G to K) The number of Ki67-positive cells as a percentage of total epithelial cells is decreased in G6del colon compared to Ctl colon (G to I) and is supported by a decrease in the number of BrdU-positive cells in G6del colon harvested 1 h after BrdU injection (J and K). (L to N) BrdU immunostaining 3 days after BrdU injection revealed a significantly lower percentage of cells at the surface epithelium (n = 4 in each group). *, P < 0.05; **, P < 0.01. Bars: A and B, 100 μm; E and F, 10 μm; G and H, 400 μm; J, K, L, and M, 200 μm.
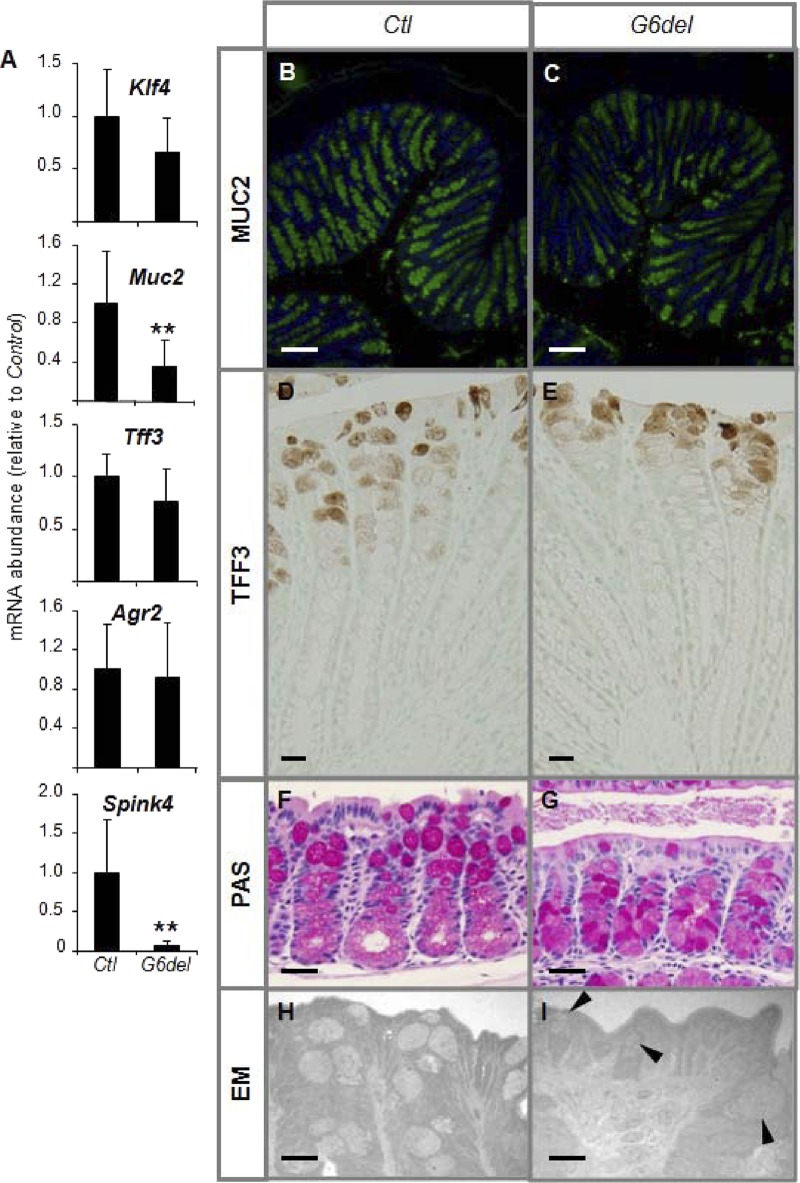
To determine the effect of intestinal Gata6 deletion on goblet cell differentiation in the colon, we measured the mRNA abundances of Krüppel-like factor 4 (Klf4), which is necessary for goblet lineage differentiation in developing colon (20), and the goblet cell markers Muc2, Tff3, anterior gradient homolog 2 (Agr2), and serine protease inhibitor, Kazal type 4 (Spink4). We failed to find differences in the mRNA abundances of Klf4, Tff3, and Agr2 (Fig. 3A), suggesting that goblet cell commitment remained intact. However, the mRNA abundance of Muc2 was reduced 64% (P < 0.01) and of Spink4 93% (P < 0.01) (Fig. 3A), suggesting that, although goblet cells are committed, they do not exhibit the endogenous levels of expression of all goblet genes. The intensity of MUC2 immunofluorescence was generally decreased in G6del compared to Ctl colon (Fig. 3B and C). Further, TFF3-positive goblet cells localize higher in the crypts, mainly near the surface epithelium, in G6del colon compared to Ctl colon (Fig. 3D and E). PAS staining indicated that the intense pink color of mature goblet cells that normally occurs in the upper half of the crypt (Fig. 3F) was more dispersed throughout the crypt in the G6del colon (Fig. 3G). Transmission EM of surface epithelium revealed a reduction of mature goblet cell numbers and the presence of poorly defined goblet cells with irregular shapes, granular cytoplasm, and fewer secretory granules in the upper half of the crypts of G6del colon (Fig. 3H and I, arrowheads). Together, these data suggest a delayed or deficient maturation process of committed goblet cells. The significant downregulation of Spink4, and possibly Muc2, further suggests that GATA6 is specifically required for the expression of these genes within goblet cells.
Fig 3.
Intestinal Gata6 deletion results in impaired terminal differentiation of goblet cells in the mature mouse colon. (A) Quantitative RT-PCR shows a decrease in Muc2 and Spink4 mRNA abundance in G6del colon compared to Ctl colon (n = 7 in each group). (B and C) Immunostaining reveals a reduction in MUC2-positive goblet cells, mainly in the lower half of the crypts, in G6del colon compared to Ctl colon. (D and E) Immunostraining reveals that TFF3-positive goblet cells localize higher in the crypts in G6del compared to Ctl colon. (F and G) PAS staining demonstrates an absence of intense staining in the upper half of the crypt of G6del colon. (H and I) Transmission EM reveals disorganized Golgi structures (arrowheads) in crypt goblet cells of G6del colon. **, P < 0.01. Bars: B and C, 400 μm; D and E, 50 μm; F and G, 100 μm; H and I, 40 μm.
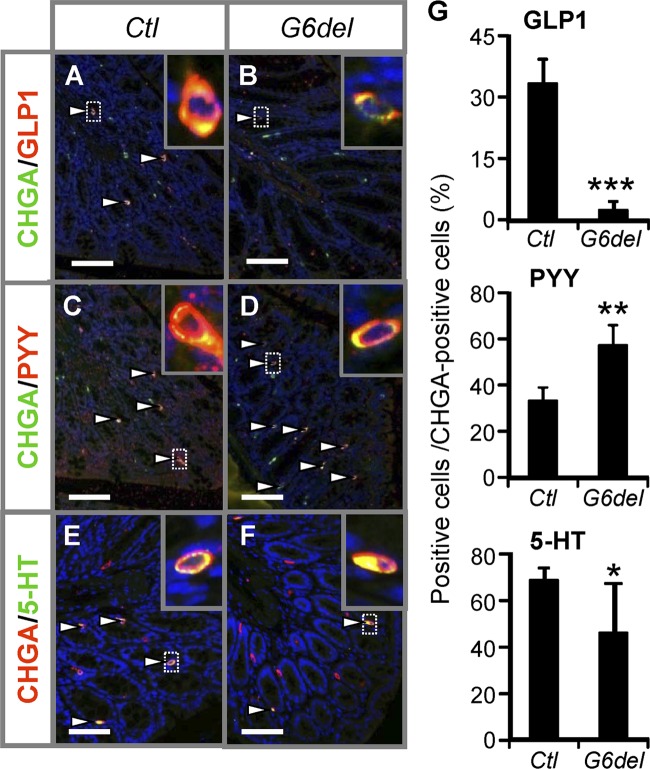
To determine the effect of intestinal Gata6 deletion on enteroendocrine cells in the colon, we measured the mRNA abundances of different hormones produced in the colon. The mRNA abundances for the pan-endocrine cell marker Chga (see Fig. S6A in the supplemental material) and the numbers of CHGA-positive cells (see Fig. S6B) were indistinguishable between G6del colon and Ctl colon, suggesting that enteroendocrine cell commitment remains intact after Gata6 deletion. However, the mRNA abundances of the hormones produced by endocrine L cells (26) such as secretin (Sct), cholecystokinin (Cck), and glucagon (Gcg) were reduced 81% (P < 0.05), 91% (P < 0.001), and 94% (P < 0.001), respectively, whereas that of Pyy was increased 79% (P < 0.05) in G6del colon compared to Ctl colon (see Fig. S6C). Coimmunofluorescence analysis performed with GLP1 and CHGA (Fig. 4A and B), PYY and CHGA (Fig. 4C and D), or serotonin (5-HT) and CHGA (Fig. 4E and F) demonstrated a 93% reduction in the number of GLP1-expressing cells (P < 0.001), a 72% increase in the number of PYY-expressing cells (P < 0.01), and a 33% reduction in the number of 5-HT-expressing cells (P < 0.05) per total number of CHGA-expressing cells (Fig. 4G) in G6del colon compared to Ctl colon. This indicates that the total number of enteroendocrine cells does not change but that the production of specific hormones within endocrine cells in the colon is altered by Gata6 deletion. These data are consistent with the hypothesis that GATA6 specifically regulates the hormone production within different enteroendocrine sublineages (L cells and serotonin-producing cells) in the mature mouse colon.
Fig 4.
Intestinal Gata6 deletion results in alterations in enteroendocrine subpopulations in the colon. (A to G) Cell counts on sections stained with GLP1 and CHGA (A and B), PYY and CHGA (C and D), or 5-HT and CHGA (E and F) reveal decreases in the numbers of GLP1-positive and 5-HT-positive and an increase in the number of PYY-positive cells per total number of CHGA-positive cells in G6del compared to Ctl colon (G) (n = 4 in each group). Arrowheads indicate positive cells. Bars: 200 μm.
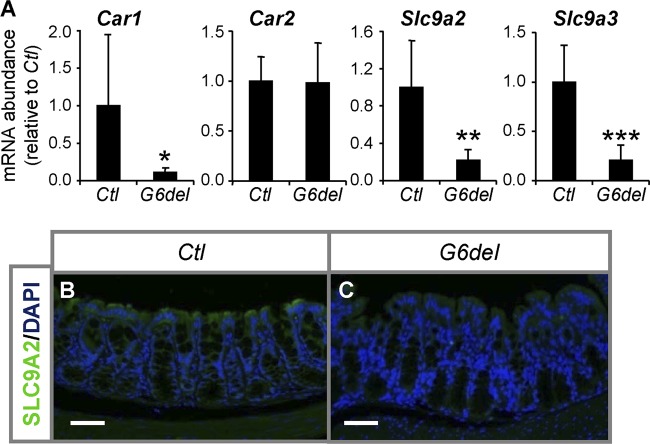
To determine the consequences of Gata6 deletion with respect to colonocytes, we measured the mRNA abundances of terminal differentiation genes specific to this lineage. We did not find differences in the mRNA abundances of the colonocyte markers anion exchanger 1 (Slc4a1, also known as Ae1) and 2 (Slc4a2, also known as Ae2); ATPase, Na+/K+ transporting, polypeptide beta 1 (Atp1b1) and 3 (Atp1b3); solute carrier family 2 (facilitated glucose transporter), member 1 (Slc2a1); and carbonic anhydrase 2 (Car2) between G6del and Ctl colon (Fig. 5A; see Fig. S7 in the supplemental material), indicating that colonocyte commitment is not affected by conditional Gata6 deletion. However, we found 89% (P < 0.05), 78% (P < 0.01), and 79% (P < 0.001) reductions in the mRNA abundances of the colonocyte markers carbonic anhydrase 1 (Car1) and solute carrier family 9 (sodium/hydrogen exchanger), members 2 and 3 (Slc9a2 and Slc9a3, also known as Nhe2 and Nhe3), respectively, in G6del colon compared to Ctl colon (Fig. 5A). The protein levels of SLC9A2, as determined by the intensity of the immunofluoresence signal, followed its mRNA pattern (Fig. 5B and C), confirming decreased expression. Together, these data suggest that GATA6 activates the expression of specific colonocyte genes within the mature mouse colon.
Fig 5.
GATA6 is required for the expression of specific colonocyte genes in the mature mouse colon. (A) Quantitative RT-PCR shows decreases in Car1, Slc9a2, and Slc9a3 mRNA abundance in G6del colon compared to Ctl colon (n = 7 in each group). *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B and C) Immunofluorescence analysis performed with anti-SLC9A2 antibody shows a reduction in intensity in the fluorescence signal (green) for SLC9A2 protein in surface colonocytes of G6del colon compared to Ctl colon. Bars: 100 μm.
To determine if the observed altered phenotype after Gata6 deletion represents a short-term effect on proliferation and differentiation rather than a reestablishment of a new homeostatic balance over time, we analyzed mouse colon samples 5 days after initiation of tamoxifen treatment. We found a 68% decrease of Gata6 mRNA abundance (see Fig. S8A in the supplemental material) and a reduction in GATA6 protein (see Fig. S8B and C) in G6del colon compared to Ctl colon. We were unable to detect a difference in the numbers of Ki67-positive cells per crypt (Fig. 6A). This could have been due to a latent effect on homeostasis or to incomplete Gata6 knockout. In spite of the incomplete Gata6 knockout, we observed a significant decrease in a goblet target (Spink4) and a colonocyte target (Car1) and a trend toward a reduction in an enteroendocrine target (Gcg) (Fig. 6B). These data indicate that the changes in cellular differentiation after Gata6 deletion are a short-term effect rather than due to a reestablishment of a new homeostatic balance over time.
Fig 6.
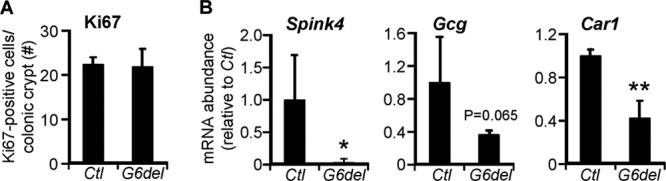
Short-term Gata6 deletion. (A) The number of Ki67-positive cells as a percentage of total epithelial cells is unchanged in G6del colon compared to Ctl colon. (B) Quantitative RT-PCR shows decreases in Car1, Spink4, and Gcg mRNA abundance in G6del colon compared to Ctl colon (n = 3 or 4 in each group). *, P < 0.05; **, P < 0.01.
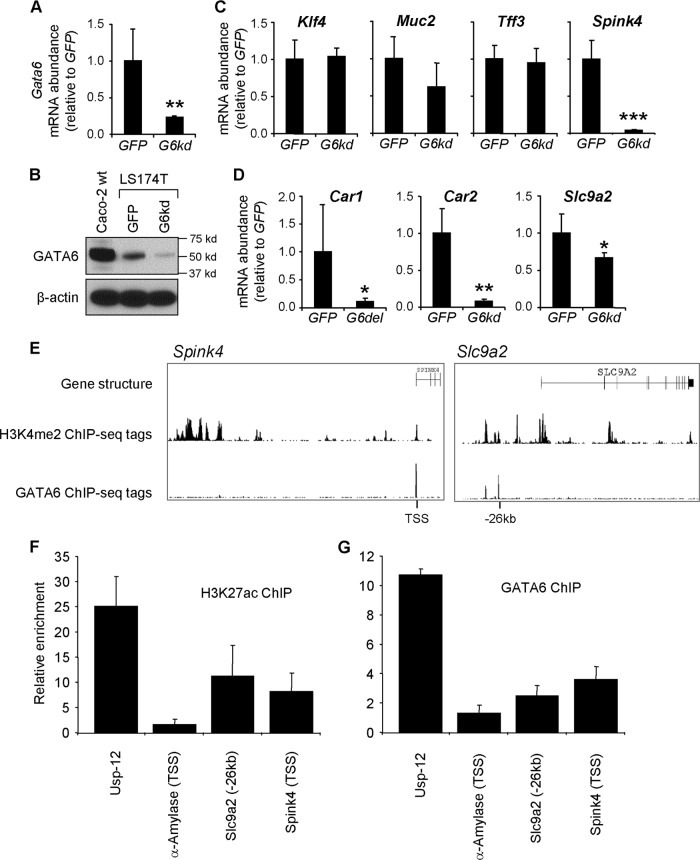
To test the hypothesis that GATA6 directly regulates specific genes in the colon, we characterized the regulation of colonic genes in the LS174T human colonic adenocarcinoma cell line (31), which express multiple colonocyte and goblet cell genes and abundant GATA6. Using a lentiviral RNA interference (RNAi) knockdown approach in which a short hairpin RNA (shRNA) for the Gata6 transcript is expressed, we established a 74% knockdown of Gata6 mRNA (Fig. 7A) that was reflected by a reduced expression of GATA6 in Western blots (Fig. 7B). Like the goblet cell markers in the colon of G6del mice (see Fig. 3A), Klf4, Tff3, and Agr2 were not affected by Gata6 knockdown, but expression of Muc2 was slightly reduced (nonsignificant) and expression of Spink4 was greatly reduced (P < 0.001) (Fig. 7C). Further, expression of the colonocyte genes Car1, Car2, and Slc9a2 was significantly reduced (Slc9a3 was below levels of detectability in these cells) (Fig. 7D). Taken together, these results suggest remarkably high concordance of colonic GATA6 function in mouse colon and a human colonic cell line. However, whereas Car1 but not Car2 transcript abundance was reduced in the mouse colon, Car2 was also reduced in the LS174T cells, suggesting a possible species difference in GATA6 regulation. Overall, these data further implicate GATA6 in the regulation of goblet and colonocyte genes, and yet whether GATA6 functions to directly or indirectly activate these targets is unclear.
Fig 7.
GATA6 occupies active chromatin sites in the Spink4 and Slc9a2 genes. (A and B) LS174T cells infected with a lentivirus expressing an shRNA for Gata6 (G6kd) demonstrate a decrease in Gata6 mRNA abundance (qRT-PCR; n = 4 independent infections) (A) and GATA6 protein abundance (Western blotting) (B) compared to control LS174T cells infected with a lentivirus expressing an shRNA for GFP. (C and D) Among the goblet (C) and colonocyte (D) markers, Spink4, Car1, Car2, and Slc9a2 were downregulated in the Gata6 knockdown LS174T cells. (E) Using publicly available ChIP-seq databases of the human intestinal cell line Caco-2 (37), GATA6 enrichment was found in enhancers, identified by H3K4me2 analysis (6, 15, 16), that were assigned to Spink4 and Slc9a2. (F) ChIP analysis using LS174T cells and an antibody for the H3K27ac enhancer mark, demonstrating enrichment at these sites. (G) ChIP analysis using LS174T cells and a GATA6 antibody, showing GATA6 enrichment at these sites.
We next sought to characterize direct occupancy of GATA6 at loci of genes modulated by Gata6 knockdown. Using publicly available ChIP-seq data from the human intestinal cell line Caco-2 (37) (such data are not available for LS174T cells), we identified GATA6 enrichment in enhancers or promoters, identified by the presence of H3K4me2 (6, 15, 16), that were assigned to Spink4 and Slc9a2 (Fig. 7E). Within these loci, GATA6 enrichment was found at the TSS of Spink4 and 26 kb upstream of the TSS in Slc9a2. To confirm that these loci were in an open or active state in LS174T cells, we performed ChIP analysis using an antibody for H3K27ac and found enrichment at these sites (Fig. 7F). To determine whether GATA6 also occupies these sites in LS174T cells, we performed ChIP analysis using a GATA6 antibody and found GATA6 enrichment at both sites (Fig. 7G). In contrast, GATA6 ChIP showed no enrichment at the α-amylase promoter, which served as a negative control. An active locus in the Usp-12 gene shown previously to bind GATA6 in Caco-2 cells (37) was used as a positive control. Taken together, these data suggest that GATA6 directly regulates specific colon genes, including Spink4 and Slc9a2.
DISCUSSION
The mammalian colonic epithelium is comprised of specialized cells with specific cell distribution and gene expression patterns designed to facilitate the absorption of large quantities of water. The regeneration and differentiation of the colonic epithelium are controlled by evolutionarily conserved mechanisms that are only beginning to be elucidated. Here, we show that GATA6 is expressed in differentiated and proliferating cells of the mature mouse colon (Fig. 1) and that conditional deletion of Gata6 (see Fig. S4 in the supplemental material) results in alterations in crypt structure, a decrease in cellular proliferation and migration (Fig. 2), a delayed maturation of goblet cells (Fig. 3), alterations in hormone production within enteroendocrine subpopulations (Fig. 4), and decreases in the expression of specific goblet cell genes (Fig. 3 and 7) and colonocyte genes (Fig. 5 and 7). GATA6 was found to occupy active chromatin loci within some of these genes, suggesting that they are direct GATA6 targets (Fig. 7). These data demonstrate that GATA6 is necessary for the maintenance of colonic epithelial regeneration and differentiation.
Cellular proliferation is required for the continuous renewal of the small intestinal and colonic epithelium. Under physiological conditions, crypt cell production and surface epithelial cell loss in the epithelium of the mature colon are in equilibrium. However, situations exist in which increased proliferation leads to an expansion of intestinal epithelium cell numbers. A beneficial increase in proliferation occurs during the adaptive response after loss of functional epithelial surface (13). When increased proliferation becomes uncontrolled, this can lead to undesirable polyp and tumor formation. GATA6 has been reported to be oncogenic in multiple cancers and precancerous lesions, including Barrett's esophagus, gastric cancer, pancreatic cancer, and colon cancer (2). Gata6 expression is upregulated in colon cancer epithelial cells (5, 14, 29), as well as in nonmalignant cells along the stromal margins in humans (14). Although it remains to be determined whether the increase in Gata6 expression is a correlative, causative, or protective event, our data determined here in the colon (Fig. 2), and previously in small intestine (7), that show that GATA6 promotes cellular proliferation are consistent with the hypothesis that GATA6 is oncogenic in the gastrointestinal tract (2). Further work remains to be done, perhaps through forced overexpression experiments, to confirm whether or not GATA6 is truly oncogenic.
The expression of multiple goblet cell markers in G6del colon indicates that the goblet lineage is specified, but their patterns of expression suggest that goblet maturation is delayed and/or defective (Fig. 3). It has been previously shown that deletion of Muc2 results in impaired goblet cell differentiation (36). It has also been shown that GATA factors are capable of activating the Muc2 promoter (25, 34). Thus, although further work is needed for confirmation, it is possible that the decrease in Muc2 mRNA in G6del colon results in a delayed or defective maturation of colonic goblet cells. It is also possible that the delayed migration rate in G6del colon somehow disrupts the normal maturation process of goblet cells, perhaps through defects in receipt of exogenous signaling, as they migrate to the surface epithelium.
In the colon, GATA6 is required for the normal maturation of goblet cells, whereas in the small intestine, GATA6 does not play a role in commitment, differentiation, or migration of goblet cells on villi (7). Deletion of Klf4 results in impaired goblet cell differentiation in the colon but has no effect on goblet cell differentiation in the small intestine (20). Together, these data demonstrate that goblet cell differentiation is regulated differently by both GATA6 and KLF4 in the colon compared to the small intestine.
We show here that GATA6 is expressed in enteroendocrine cells in the colonic mouse epithelium (Fig. 1), in contrast to a reported absence of GATA6 expression in human colonic enteroendocrine cells (14). We also show that, although conditional deletion of Gata6 does not alter the overall population of enteroendocrine cells, conditional deletion of Gata6 results in alterations in the production of hormones in the L-cell and serotonin-producing enteroendocrine subpopulations (Fig. 4). L-cell enteroendocrine cells in the colon are thought to originate from the PYY-producing enteroendocrine progenitor cells (32) from which all L-cell subpopulations arise, whereas serotonin-producing cells are thought to originate from progenitors that never express PYY (26). Our data suggest that GATA6 is not required for enteroendocrine cell commitment in the mature mouse colon but defines the terminal differentiation of specific enteroendocrine sublineages, including L cells and serotonin-producing cells. It is possible that the observed alterations represent a defective process of maturation of enteroendocrine cells or a defective regulation of specific genes within the mature enteroendocrine cells. Further work is required to define the underlying mechanism by which GATA6 regulates enteroendocrine cells in the mature colon.
The colonic surface epithelium is mainly responsible for the concentration of fecal effluent through water and electrolyte absorption. This absorption is facilitated by a large osmotic gradient established by enzymes and transporters that are expressed within the colonocytes. Here, we show that, although conditional Gata6 deletion has no general effect on colonocyte differentiation, specific colonocyte genes such as Car1, Slc9a2, and Slc9a3 are significantly downregulated (Fig. 5). Carbonic anhydrases catalyze the conversion of carbonic acids to bicarbonate and protons, whereas the sodium-hydrogen exchangers utilize the protons generated by carbonic anhydrases to drive the internalization of Na+ and the subsequent absorption of water across the apical surface. CAR2, which is not affected by conditional Gata6 deletion in mice (Fig. 5), is likely able to compensate for the decrease in CAR1. Deletion of Slc9a2 exhibits no overt disease phenotype (3, 27). However, deletion of Slc9a3 results in slight diarrhea and a blood pH that is mildly acidotic (28). Taken together, these data indicate that SLC9A3 is the major absorptive sodium-hydrogen exchanger in the colon. The significant downregulation of both Slc9a2 and Slc9a3 by conditional Gata6 deletion (Fig. 5) could therefore result in a decrease in the efficiency of water absorption by colonocytes, possibly explaining the observed softer and lighter-in-color stools produced by the G6del mice.
Interestingly, our data indicate that GATA6 regulates the same genes differently in the ileum and colon. For example, expression of Car1 and Slc9a2 are downregulated in the colon (Fig. 6) but upregulated in the ileum (7); expression of Car2 is not changed in the colon (Fig. 5) but is upregulated in the ileum (7); and expression of Slc9a3 is downregulated in the colon (Fig. 5) but not changed in the ileum (7) after Gata6 deletion. These data indicate that the same genes are regulated by the same transcription factor (GATA6) differently in different organs within the gastrointestinal tract. Our data also suggests that GATA6 controls both proliferation and differentiation. GATA6, like GATA factors in other systems, might insert its regulation in multiple pathways where the specific regulation is dependent on the cell state (i.e., proliferating versus differentiated). This could result from differences in the chromatin state in which GATA6 has differential access to specific loci that is dependent on the cell state. It could also be due to differential expression of GATA6 partner proteins that enable specific regulation at distinct sites.
Using an RNAi knockdown approach in the LS174T human colonic cell line, we found that a panel of goblet and colonocyte genes are downregulated (Fig. 7C and D) in a manner similar to that found in G6del mouse colon, although Car2 was also downregulated in the LS174T cells (Fig. 7D), suggesting a species difference. We further verified direct occupancy of GATA6 in active loci in enhancers or promoters of these targets, namely, Spink4 and Slc9a2 (Fig. 7E to G). These data provide strong evidence for the direct regulation of individual target genes within specific lineages by GATA6.
The pleiotropic effects we observed upon Gata6 deletion in the colon highlight the integral role this factor plays in colonic differentiation and regeneration, and going forward we anticipate that our genetic model will be useful in studies of colonic disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Hagan and A. Calhoun for technical assistance and insight with electron microscopy, D. K. Podolsky for the TFF3 antibody, M. Donowitz for the SLC9A2 antibody, and J. C. Fleet, R. J. Grand, and R. K. Montgomery for critical feedback on the manuscript.
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases grants RO1-DK-061382 (S.D.K.) and K01-DK-088868 (M.P.V.), fellowship no. 1987 from the Crohn's and Colitis Foundation of America (M.P.V.), the Harvard Digestive Disease Center (grant 5P30-DK-34854), the Nutricia Research Foundation (E.B. and B.E.A.), the Foundation De Drie Lichten (E.B.), the Foundation Doctor Catharine van Tussenbroek (E.B.), KWF Kankerbestrijding (B.E.A.), Prins Bernhard Cultuurfonds (B.E.A.) in The Netherlands, and the European Society for Pediatric Research (B.E.A.) in Switzerland.
Footnotes
Published ahead of print 25 June 2012
E.B. and B.E.A. contributed equally to the article.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Al-azzeh ED, Fegert P, Blin N, Gott P. 2000. Transcription factor GATA-6 activates expression of gastroprotective trefoil genes TFF1 and TFF2. Biochim. Biophys. Acta 1490: 324–332 [DOI] [PubMed] [Google Scholar]
- 2. Ayanbule F, Belaguli NS, Berger DH. 2011. GATA factors in gastrointestinal malignancy. World J. Surg. 35: 1757–1765 [DOI] [PubMed] [Google Scholar]
- 3. Bachmann O, et al. 2004. The Na+/H+ exchanger isoform 2 is the predominant NHE isoform in murine colonic crypts and its lack causes NHE3 upregulation. Am. J. Physiol. Gastrointest. Liver Physiol. 287: G125–G133 [DOI] [PubMed] [Google Scholar]
- 4. Barker N, et al. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 [DOI] [PubMed] [Google Scholar]
- 5. Belaguli NS, et al. 2010. GATA6 promotes colon cancer cell invasion by regulating urokinase plasminogen activator gene expression. Neoplasia 12: 856–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bernstein BE, et al. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- 7. Beuling E, et al. 2011. GATA factors regulate proliferation, differentiation, and gene expression in small intestine of mature mice. Gastroenterology 140: 1219–1229.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beuling E, et al. 2008. GATA4 mediates gene repression in the mature mouse small intestine through interactions with friend of GATA (FOG) cofactors. Dev. Biol. 322: 179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bosse T, et al. 2006. Gata4 is essential for the maintenance of jejunal-ileal identities in the adult mouse small intestine. Mol. Cell. Biol. 26: 9060–9070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brewer AC, Sparks EC, Shah AM. 2006. Transcriptional regulation of the NADPH oxidase isoform, Nox1, in colon epithelial cells: role of GATA-binding factor(s). Free Radic. Biol. Med. 40: 260–274 [DOI] [PubMed] [Google Scholar]
- 11. Divine JK, et al. 2004. GATA-4, GATA-5, and GATA-6 activate the rat liver fatty acid binding protein gene in concert with HNF-1alpha. Am. J. Physiol. Gastrointest. Liver Physiol. 287: G1086–G1099 [DOI] [PubMed] [Google Scholar]
- 12. Dusing MR, Florence EA, Wiginton DA. 2003. High-level activation by a duodenum-specific enhancer requires functional GATA binding sites. Am. J. Physiol. Gastrointest. Liver Physiol. 284: G1053–G1065 [DOI] [PubMed] [Google Scholar]
- 13. Fabrikant JI. 1987. Adaptation of cell renewal systems under continuous irradiation. Health Phys. 52: 561–570 [DOI] [PubMed] [Google Scholar]
- 14. Haveri H, et al. 2008. Transcription factors GATA-4 and GATA-6 in normal and neoplastic human gastrointestinal mucosa. BMC Gastroenterol. 8: 9 doi: 10.1186/1471-230X-8-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He HH, et al. 2010. Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 42: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heintzman ND, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoogerwerf WA, et al. 1996. NHE2 and NHE3 are human and rabbit intestinal brush-border proteins. Am. J. Physiol. 270: G29–G41 [DOI] [PubMed] [Google Scholar]
- 18. Huggon IC, et al. 1997. Molecular cloning of human GATA-6 DNA binding protein: high levels of expression in heart and gut. Biochim. Biophys. Acta 1353: 98–102 [DOI] [PubMed] [Google Scholar]
- 19. Karam SM. 1999. Lineage commitment and maturation of epithelial cells in the gut. Front. Biosci. 4: D286–D298 [DOI] [PubMed] [Google Scholar]
- 20. Katz JP, et al. 2002. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129: 2619–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krasinski SD, Van Wering HM, Tannemaat MR, Grand RJ. 2001. Differential activation of intestinal gene promoters: functional interactions between GATA-5 and HNF-1 alpha. Am. J. Physiol. Gastrointest. Liver Physiol. 281: G69–G84 [DOI] [PubMed] [Google Scholar]
- 22. Liu T, et al. 2011. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 12: R83 doi: 10.1186/gb-2011-12-8-r83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patient RK, McGhee JD. 2002. The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 12: 416–422 [DOI] [PubMed] [Google Scholar]
- 25. Ren CY, Akiyama Y, Miyake S, Yuasa Y. 2004. Transcription factor GATA-5 selectively up-regulates mucin gene expression. J. Cancer Res. Clin. Oncol. 130: 245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schonhoff S, et al. 2005. Energy homeostasis and gastrointestinal endocrine differentiation do not require the anorectic hormone peptide YY. Mol. Cell. Biol. 25: 4189–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schultheis PJ, et al. 1998. Targeted disruption of the murine Na+/H+ exchanger isoform 2 gene causes reduced viability of gastric parietal cells and loss of net acid secretion. J. Clin. Invest. 101: 1243–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schultheis PJ, et al. 1998. Renal and intestinal absorptive defects in mice lacking the NHE3 Na+/H+ exchanger. Nat. Genet. 19: 282–285 [DOI] [PubMed] [Google Scholar]
- 29. Shureiqi I, et al. 2007. The transcription factor GATA-6 is overexpressed in vivo and contributes to silencing 15-LOX-1 in vitro in human colon cancer. FASEB J. 21: 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sodhi CP, Li J, Duncan SA. 2006. Generation of mice harbouring a conditional loss-of-function allele of Gata6. BMC Dev. Biol. 6: 19 doi: 10.1186/1471-213X-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tom BH, et al. 1976. Human colonic adenocarcinoma cells. I. Establishment and description of a new line. In Vitro 12: 180–191 [DOI] [PubMed] [Google Scholar]
- 32. Upchurch BH, Fung BP, Rindi G, Ronco A, Leiter AB. 1996. Peptide YY expression is an early event in colonic endocrine cell differentiation: evidence from normal and transgenic mice. Development 122: 1157–1163 [DOI] [PubMed] [Google Scholar]
- 33. Valente AJ, et al. 2008. Regulation of NOX1 expression by GATA, HNF-1alpha, and Cdx transcription factors. Free Radic. Biol. Med. 44: 430–443 [DOI] [PubMed] [Google Scholar]
- 34. van der Sluis M, et al. 2004. The murine Muc2 mucin gene is transcriptionally regulated by the zinc-finger GATA-4 transcription factor in intestinal cells. Biochem. Biophys. Res. Commun. 325: 952–960 [DOI] [PubMed] [Google Scholar]
- 35. van Wering HM, et al. 2004. Complex regulation of the lactase-phlorizin hydrolase promoter by GATA-4. Am. J. Physiol. Gastrointest. Liver Physiol. 287: G899–G909 [DOI] [PubMed] [Google Scholar]
- 36. Velcich A, et al. 2002. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 295: 1726–1729 [DOI] [PubMed] [Google Scholar]
- 37. Verzi MP, et al. 2010. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell 19: 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.