Abstract
Junctional adhesion molecule A (JAM-A) is a unique tight junction (TJ) transmembrane protein that under basal conditions maintains endothelial cell-cell interactions but under inflammatory conditions acts as a leukocyte adhesion molecule. This study investigates the fate of JAM-A during inflammatory TJ complex remodeling and paracellular route formation in brain endothelial cells. The chemokine (C-C motif) ligand 2 (CCL2) induced JAM-A redistribution from the interendothelial cell area to the apical surface, where JAM-A played a role as a leukocyte adhesion molecule participating in transendothelial cell migration of neutrophils and monocytes. JAM-A redistribution was associated with internalization via macropinocytosis during paracellular route opening. A tracer study with dextran-Texas Red indicated that internalization occurred within a short time period (∼10 min) by dextran-positive vesicles and then became sorted to dextran-positive/Rab34-positive/Rab5-positive vesicles and then Rab4-positive endosomes. By ∼20 min, most internalized JAM-A moved to the brain endothelial cell apical membrane. Treatment with a macropinocytosis inhibitor, 5-(N-ethyl-N-isopropyl)amiloride, or Rab5/Rab4 depletion with small interfering RNA oligonucleotides prevented JAM-A relocalization, suggesting that macropinocytosis and recycling to the membrane surface occur during JAM-A redistribution. Analysis of the signaling pathways indicated involvement of RhoA and Rho kinase in JAM-A relocalization. These data provide new insights into the molecular and cellular mechanisms involved in blood-brain barrier remodeling during inflammation.
INTRODUCTION
The central event in central nervous system (CNS) inflammation is the recruitment of leukocytes. This multifactorial process involves chemotactic signals that promote directed migration, adhesion receptor-ligand interaction at the microvascular endothelial cell surface, and production of the matrix metalloproteinase needed for extracellular matrix breakdown and leukocyte extravasation (1, 10, 20, 38, 39). The presence of destructive leukocytes in the brain parenchyma can cause a cascade of events promoting injury and further amplifying the inflammatory response (1, 38, 39). Regulation of leukocyte entry is, therefore, a critical event for controlling inflammation. Remodeling of the brain endothelial cell surface and the tight junction (TJ) complexes between brain endothelial cells plays a pivotal role in regulating leukocyte recruitment (1, 39, 53). In recent years, some components of the TJ complex, such as junctional adhesion molecules (JAMs), have emerged as being critical in leukocyte-endothelial cell interactions and promoting leukocyte transmigration (53, 54).
Junctional adhesion molecule A (JAM-A) is a member of the immunoglobulin superfamily (IgSF) (3, 25). Structurally, this molecule is composed of a single membrane-spanning domain, two IgG-like extracellular domains, an extracellular N-terminal loop with the dimerization domain, and a short C-terminus cytoplasmic tail with a PDZ-binding domain. The PDZ-binding domain facilitates interactions with TJ-associated scaffold proteins such as zonula occludens 1 (ZO-1), afadin 6 (AF-6), ASIP/Par3, cingulin, and domain substrates for protein kinase C (PKC) and protein kinase A (PKA) (4, 5, 15, 17, 25, 34, 42, 55). In general, JAM-A is expressed at cell-to-cell junctions of endothelial and epithelial cells and displays different patterns of homo- and heterophilic adhesion (2, 5, 13, 17, 19). For example, via homotypic trans interactions, JAM-A regulates occlusion of the brain endothelial cell paracellular space, but it also interacts with β2 integrin leukocyte function-associated antigen 1 (LFA-1; integrin aLβ2), which is associated with transendothelial cell migration and recruitment of a variety of circulating leukocytes (i.e., monocytes, neutrophils) (5, 13, 18, 19, 57). An important feature of the involvement of JAM-A in endothelial cell-leukocyte interactions is a contribution to a specific ring-like structure that forms on the endothelial cell surrounding a transmigrating leukocyte, which has already been described for another adhesion molecule, intercellular adhesion molecule 2 (ICAM-2) (22, 33, 41). JAM-A, together with CD99 and CD31, maintains a transient ring structure supporting both paracellular and transcellular leukocyte transmigration (53, 54). Analysis of this phenomenon revealed that JAM-A, unlike other TJ and adherens junction (AdJ) components (e.g., claudin and Ve-cadherin), accumulates at the point of transmigration and is present during and after transmigration is completed (32, 54). The ring-like structures, in conjunction with cytoskeletal components and regulatory molecules, act as transmigration tunnels which also play an active role in spatially and temporally organizing the transmigration machinery for this complex process to occur (4, 53, 54).
JAM-A exhibits a behavior different from that of other TJ and AdJ proteins during inflammation-induced junctional complex remodeling. How JAM-A accumulates at the appropriate place and becomes available on the apical surface for LFA-1-mediated leukocyte binding still awaits clarification. Our present study, therefore, focused on elucidating the mechanisms underlying JAM-A relocalization at the brain endothelial cell barrier under inflammatory conditions in order to enable leukocyte interaction.
MATERIALS AND METHODS
mBMEC culture.
Mouse brain microvascular endothelial cells (mBMECs) were prepared using a modified protocol already described (46, 47). Briefly, brains were collected from 4- to 6-week-old C57BL/6 mice, minced in Hanks balanced salt solution (HBSS; Invitrogen, Carlsbad, CA), and homogenized gently in a Dounce-type homogenizer. Myelin was removed by resuspending homogenates in an 18% dextran suspension (dextran molecular weight, 60,000 to 90,000; USB, Cleveland, OH) and centrifuging. Red blood cells were removed by centrifuging isolated microvessels in a Percoll gradient (Pharmacia, Peapack, NJ) at 2,700 rpm for 11 min. The isolated microvessels were digested in HBSS solution containing 1 μg/ml collagenase/dispase (Roche, Indianapolis, IN), 10 U/ml DNase I (Sigma-Aldrich, St. Louis, MO), and 1 μg/ml Na-p-tosyl-l-lysine chloromethyl ketone (TLCK) for 20 min at 37°C and precipitated with CD31-coated magnet beads (Dynabeads; Invitrogen, Carlsbad, CA). These vessels were further cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% inactivated fetal calf serum, 2.5 μg/ml heparin (Sigma-Aldrich, St. Louis, MO), 20 mM HEPES, 2 mM glutamine, and 1× antibiotic/antimycotic (all from Invitrogen, Carlsbad, CA) plus endothelial cell growth supplement (BD Bioscience, San Jose, CA) and grown in 6-well plates coated with collagen type IV (BD Bioscience, San Jose, CA). This protocol typically produces primary endothelial cell cultures that are approximately 99% pure (determined by immunocytochemistry with an anti-platelet endothelial cellular adhesion molecule 1 [anti-PECAM-1] antibody; BD Bioscience, San Jose, CA).
Mouse brain endothelial cell line (bEnd.3).
Cells of a mouse brain endothelial cell line (bEnd.3) were purchased from the American Type Culture Collection (Manassas, VA) and cultured in medium (DMEM supplemented with 10% fetal bovine serum, 2 mM glutamine, and 1× antibiotic/antimycotic; Invitrogen, Carlsbad, CA) at 37°C in a humidified incubator (10% CO2, 90% air).
GFP-tagged JAM-A.
Mouse JAM-A cDNA was obtained from Open Biosystem (Huntsville, AL). JAM-A was tagged with green fluorescent protein (GFP) in the hinge region between the membrane-proximal C2 domain and the transmembrane part of JAM-A. The cDNA encoding JAM-A was generated by PCR using 5′-GCCCAGATCTGGAGGAGGAGGATAAGGGCACCGAGGGGAA-3′ and 5′-GGGAATTCCCACAGTGAGCGGATG-3′, incorporating BglII and EcoRII sites. All cDNA fragments were then cloned in frame into the pEGFP-C1 plasmid vector using an In Fusion Dry-Down PCR cloning kit (Clontech, Mountain View, CA). Purified plasmids were verified by DNA sequencing. bEnd.3 cells were transfected with the plasmids for 24 h using Lipofectamine 2000. The control plasmid, pEGFP1-C1, was used as a positive control for transfection and expression in bEnd.3 cells. Negative controls were untransfected or mock-transfected cells. At 24 h posttransfection, cells were exposed to medium containing G418 (1 mg/ml; Invitrogen, Carlsbad, CA) for 10 to 14 days to generate a stable cell line. Surviving cells were ring cloned and grown to confluence. The expression of GFP-tagged JAM-A was determined by Western blot analysis using anti-JAM-A antibody (R&D Systems, Minneapolis, MN) and anti-GFP antibody (Invitrogen, Carlsbad, CA). The ratio between endogenous and GFP-tagged proteins was evaluated. The GFP-tagged proteins were also assessed via fluorescence microscopy.
Cell treatment.
Cells were exposed to chemokine (C-C motif) ligand 2 (CCL2; 100 ng/ml; PeproTech, Rocky Hill, NJ), lipopolysaccharide (LPS; 5 μg/ml; Sigma-Aldrich, St. Louis, MO) or medium only (negative control) for 0 to 120 min. Inhibitors were introduced 30 min prior to treatment with CCL2 or LPS. Control cells were exposed to assay medium (DMEM) without inhibitors. The following inhibitors (all from Sigma-Aldrich, St. Louis, MO) were used: 0.4 M sucrose, 1 to 50 μM chlorpromazine (clathrin-dependent internalization), 1 to 10 μM filipin III (caveolin-dependent internalization), 10 to 100 μM 5-(N-ethyl-N-isopropyl)amiloride (EIPA; macropinocytosis), 50 nM bafilomycin A1 (vesicle recycling), 10 μM Y27632 (Rho kinase), 50 μg/ml cycloheximide (protein synthesis), and 1 μM lactacystin (proteasome). Cell viability assays were performed to exclude possible toxic effects of inhibitors, and only cells with 98% viability were used in experiments. The effects of treatment and inhibitors were evaluated by Western blot, immunocytochemistry, or biotinylation assays.
Cell transfection.
Optimal inhibition of Rab4, Rab5, Rab34, caveolin 1, and α-adaptin was achieved after transfection with a cocktail of three selected small interfering RNA (siRNA) oligonucleotides (all from Applied Biosystems, Carlsbad, CA) targeting three different regions of selected genes (siRab4RNA, catalog numbers s72620, s72619, and s201891; siRab5RNA, catalog numbers s114661, s114662, and s114663; siRab34RNA, catalog numbers s72704, s72702, and s201895; sicaveolin-1RNA, catalog numbers s63423, 63424, and s63425; siα-adaptinRNA, catalog numbers s62403, s62402, and s62401). For controls, cells were transfected with siRNA (sicontrol 1; catalog number AM4611). For transient transfection, mBMECs and bEnd.3 cells were transfected with annealed siRNAs using siPortNeoFX siRNA (Applied Biosystems, Carlsbad, CA) transfection agent and were subcultured 24 h later. Cells were used 48 h later for experiments. To specifically inhibit RhoA activity, a dominant-negative mutant, pCMVRhoT19N (Millipore, Billerica, MA), was used. Confluent mBMEC cultures were transiently transfected with plasmid pCMVRhoT19N (1 μg/ml) or empty pCMV vector in Opti-MEM serum-deprived medium supplemented with Lipofectin (10 μg/ml; Invitrogen, Carlsbad, CA) for 6 h. The medium was then replaced with mBMEC growth medium. Experiments were performed 24 h later. Transfection efficiency was evaluated by Western blot analysis.
Time-lapse imaging.
Time-lapse microscopy was performed using a Leica DMIRB inverted microscope (objective, 40×; Leica Microsystems, Germany). The stage was maintained at 37°C by a temperature hood. Time-lapse experiments were conducted for 0 to 2 h. Images were collected every 5 min using an Olympus DP-30 charge-coupled-device camera.
Immunofluorescence.
Samples were fixed in 4% paraformaldehyde and then preincubated in blocking solution containing 5% normal goat serum and 0.05% Tween in phosphate-buffered saline (PBS). Samples were then incubated overnight at 4°C with the following primary antibodies: JAM-A (R&D Systems); claudin-5, occludin, and ZO-1 (Invitrogen); Rab4, Rab5, Ve-cadherin, caveolin-1, and α-adaptin (BD Bioscience); and Rab34 and early endosome antigen 1 (EEA-1) (Abcam, Cambridge, MA). Reactions were visualized by fluorescein-conjugated anti-mouse, anti-rabbit, and/or anti-goat antibodies. All samples were viewed on a confocal laser scanning microscope (LSM 510; Zeiss, Germany). The actin cytoskeleton was visualized with phalloidin-Alexa 568 (Invitrogen) according to the manufacturer's instructions. The cell membrane was labeled with CellLight membrane-CFP Bacmam 2.0 (Invitrogen) according to the manufacturer's instructions.
Cell-based enzyme-linked immunosorbent assay (ELISA).
mBMEC monolayers were treated with CCL2 or LPS for 0 to 60 min, washed with ice-cold PBS, and then washed twice with ice-cold PBS–0.1% bovine serum albumin (BSA). The cells were then incubated with goat anti-mouse JAM-A antibody (R&D Systems) in PBS–0.1% BSA for 1 h at 4°C with occasional shaking and washed with PBS–0.1% BSA, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody for 30 min at room temperature. After washing with PBS–0.1% BSA buffer, cells were fixed in 4% paraformaldehyde (PFA) for 20 min and washed with PBS. For detection, equal parts of substrate reagents hydrogen peroxide and 3,3′,5,5′-tetramethylbenzidine solution (Sigma, Australia) were added to each well and the plates were incubated in the dark at room temperature for 15 min while shaking. The color reaction was stopped by the addition of 1 N HCl. Absorbance was measured at 450 nm using a Tecan microplate reader (58). To adjust for cell number, wells were washed with H2O and stained with 0.08% crystal violet in PBS for 5 min. The nuclear stain was solubilized with 33% acetic acid, and the optical density was measured at 595 nm. To assess potential JAM-A internalization during exposure to anti-JAM-A antibody, mBMECs were treated with primary antibody, fixed with 4% PFA, permeabilized with 0.05% Triton X-100 in PBS, and exposed to HRP-conjugated secondary antibody, followed by visualization with hydrogen peroxide and 3,3′,5,5′-tetramethylbenzidine solution. Samples were compared with those without fixation and permeabilization. There was no evidence for internalization of the JAM-A–anti-JAM-A antibody complex. Time course experiments were performed by stimulating cells with CCL2 or LPS for 0 to 60 min, and maximal surface expression of JAM-A was shown at 10 and 20 min, respectively. Dose-response curves for CCL2 and LPS (0 to 500 ng/ml and 1 to 10 μg/ml, respectively) were also carried out. On the basis of these curves, 100 ng/ml CCL2 and 5 μg/ml LPS were chosen to activate monolayers in later experiments. This concentration was submaximal and allowed the detection of JAM-A expression on the surface of mBMECs. Controls were untreated cells.
Tracer study.
The macropinocytosis marker lysine-fixable Texas Red-dextran (0.5 mg/ml), the caveola internalization marker Alexa 596-cholera toxin and BODIPY-TR ceramide (10 μg/ml and 5 μM, respectively), and the clathrin internalization marker Texas Red-transferrin (5 μg/ml), all from Invitrogen (Carlsbad, CA), were dissolved in ice-cold medium (DMEM without phenol red) and added to the apical side of mBMEC or bEnd.3 cell monolayers with and without CCL2 (100 ng/ml). Monolayers were kept at 4°C for 30 min to allow tracer accumulation on the cell surface and then incubated for 0 to 60 min at 37°C. At this time point, transendothelial electrical resistance (TEER) was simultaneously measured, and cells were subject to time-lapse microscopy analysis.
Evaluation of colocalization.
Images for quantitative florescence analysis were acquired using a Zeiss LSM META 510 laser scanning microscope with sequential mode to avoid interference between channels and saturation. Contrast brightness and the pinhole were held constant. Cells from five independent experiments and three areas per experiment were analyzed. For each area, z stacks of five consecutive optical sections were acquired. To quantify the colocalization of JAM-A and JAM-A–GFP with various vesicle markers, each z-stack optical section was analyzed using the colocalization finder plug-in of ImageJ software (NIH Image, Bethesda MD). The background contribution to colocalization was corrected using the formula corrected colocalization = [(measured colocalization − background colocalization)/(1 − background colocalization)]/100 (24). The colocalization of JAM-A with vesicular markers was estimated by the use of Pearson's correlation coefficient (Rr), determined as
where S1i represents the signal intensity of the pixels in channel 1 and S2i represents signal the signal intensity of the pixels in channel 2; S1over and S2 over reflect the average intensities of the respective channels. The Pearson coefficient ranges from −1 (perfect negative correlation) to +1 (perfect positive correlations between two images). A coefficient of 0 means no correlation between two images (24, 61).
Western blotting.
Western blotting was performed with the following antibodies: mouse anti-JAM-A antibody (R&D Systems, Minneapolis, MN) and anti-Rab4, anti-Rab5, anti-Rab7, and anti-Rab34 antibodies (Abcam, Cambridge, MA). Immunoblots were exposed to secondary anti-mouse or rabbit-HRP conjugated antibody, visualized with a chemiluminescent HRP substrate kit, and analyzed using Image J software.
Fractional analysis of tight junctions.
Fractional analysis of TJ proteins was performed utilizing a ProteoExtract subcellular proteome extraction kit (Calbiochem, La Jolla, CA). Membrane, cytosolic, cytoskeletal, and nuclear fractions were separated. The specificity of the fractions was confirmed using anti-cytochrome P450 reductase (membrane fraction), anticalpain (cytosolic fraction), and antivimentin (actin cytoskeletal fraction) antibodies. For total cell lysate samples, cells were washed in PBS, scraped, and rinsed in 1 ml of the lysis buffer (25 mM Tris-HCl, pH 7.4, with 150 mM NaCl, 0.1% SDS, 1% Triton X-100, and 1% deoxycholate).
Isolation of endosomes.
Isolation of endosomes was performed by isopycnic centrifugation on a continuous density gradient as previously described (14). Briefly, cells were harvested in PBS and resuspended in homogenization buffer containing 0.25 M sucrose, 3 mM imidazole, pH 7.4, 1 mM EGTA, 0.1 mM phenylmethylsulfonyl fluoride, and a 1:100 (vol/vol) cocktail of protease inhibitors. The cell suspension was homogenized in a Dounce-type homogenizer, and the nuclei were removed by centrifugation for 10 min at 1,000 rpm at 4°C. The resultant supernatant was loaded on the top of a linear 10 to 40% sucrose gradient prepared in buffer containing 3 mM imidazole (pH 7.4), 0.15 M NaCl, and 0.01% Triton X-100 and centrifuged for 16 h at 40,000 rpm. After centrifugation, 20 fractions were collected from the bottom of the tube. The protein concentration of each fraction was determined, and the fractions were subjected to electrophoresis and Western blot analysis. In addition, in control experiments, brain endothelial cells were treated with a tracer for fluid-type endocytosis, HRP (1 mg/ml), for 30 min and chased for 40 min at 37°C. After endosome isolation, fractions were treated with 1% Triton in PBS (pH 7.4) for 10 min, and enzymatic HRP activity was evaluated in each collected fraction using a Pierce Immunopure Tmb substrate kit (49). To analyze colocalization of JAM-A with certain Rab-positive (Rab+) vesicles, immunoprecipitation followed by Western blotting was performed. Briefly, vesicular fractions from sucrose gradients were first treated with solubilization buffer (SB; PBS containing 1% Triton X-100) as described previously (28) and then mixed with coated beads (Dynabeads M-500 subcellular; Invitrogen, Carlsbad, CA) conjugated with goat anti-JAM-A antibody (10 μg/107 beads) and incubated for 18 h at 4°C with slow rocking. After incubation, a first fraction was collected and labeled total (fraction that contains coated beads plus endosomal fraction). The incubating mixture was then placed in a magnetic rack (Invitrogen) for 5 min at 18°C. The resulting supernatant was collected and labeled the unbound (UB) fraction, while collected bound beads were labeled the bound (B) fraction. The bound fraction was washed four times to remove loosely bound protein and membrane, while the unbound fraction was spun twice at 100,000 × g for 1 h at 4°C. Western blot analysis was then performed using anti-Rab5, anti-Rab4, anti-Rab7, or anti-Rab34 antibodies. Control beads were conjugated with secondary anti-goat antibody and were subjected to the same procedures in parallel with the experimental groups. No detectable signal was seen in Western blots with these control samples (data not shown).
Biotinylation assay for endocytosis and recycling.
Cells were incubated with 0.5 mg/ml sulfosuccinimidyl-2-(biotin-amido)-ethyl-1,3′-dithioproprionate (sulfo-NHS-SS-biotin; Term Scientific, Rockford, IL) at 0°C, followed by washing with PBS containing 50 mM NH4Cl, 1 mM MgCl2, and 0.1 mM CaCl2 to quench any excess of sulfo-NHS-SS-biotin. Cells were then lysed to quantify surface biotinylated proteins. To determine the total amount of JAM-A within cells using the biotin reagent, the cells were lysed with lysis buffer, 25 mM Tris-HCl (pH 7.4) with 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 1% deoxycholate, protease inhibitor cocktail, and then biotinylated, followed by quenching of excess of sulfo-NHS-SS-biotin by PBS-NH4Cl buffer. Biotin-labeled proteins were quantified using an antibody-capture ELISA. For internalization, after surface biotinylation, cells were incubated with CCL2 at 37°C for 0 to 60 min and then washed with glutathione solution (50 mM glutathione, 75 mM NaCl, 75 mM NaOH, and 1% BSA) at 0°C to release the biotin label from proteins at the cell surface (glutathione stripping). Cells were then suspended in lysis buffer and centrifuged, and the supernatant was incubated with streptavidin beads to collect bound, biotinylated protein. Samples were then subjected to SDS-PAGE, followed by Western blotting. For recycling/degradation assay, after biotinylation of cell membrane surface proteins and internalization in the presence or absence of CCL2, cells were washed with cold glutathione solution and returned to 37°C for various times (0 to 60 min) in the presence or absence of CCL2. At the different time points, cells first underwent a second glutathione stripping to remove any biotin present on the membrane surface and then lysed in lysis buffer and processed further for Western blot analysis (45).
Adhesion and transmigration assay.
Mouse blood (5 ml) was collected by cardiac puncture. Neutrophils were prepared as described previously (60). Briefly, blood was centrifuged to separate white blood cells from erythrocytes (white buffy coat), followed by separation of neutrophils on a Percoll density gradient. For macrophages, we used thioglycolate-elicited macrophages from the peritoneal cavity as a surrogate mononuclear leukocyte source (8). Neutrophil or macrophage purity was evaluated by Diff-Quik staining solution (Fisher Scientific), and viability was tested by trypan blue exclusion. Purity was >98% for neutrophils and >95% for macrophages, and viability was >99%. Isolated macrophages or neutrophils were loaded with the acetomethoxy derivative of calcein (calcein-AM; 4 μM; Invitrogen, Carlsbad, CA) for 30 min at 37°C.
mBMECs were grown to confluence in a Transwell dual-chamber system and then exposed to CCL2 (100 ng/ml) or LPS (5 μg/ml) for 2 and 6 h, respectively. The negative control was cells exposed to assay medium only. Calcein-AM-labeled leukocytes (104) were then added to the upper chamber of the Transwell system and left for 1 h at 37°C. After medium removal and washing of the cells once, the total number of adherent cells on the surface of the mBMECs as well as migrated cells on the bottom of the wells of the Transwell system was evaluated by a florescence reader (Infinity FL200; Tecan Group Ltd., Männedorf, Switzerland), and the number of cells was calculated from a standard curve. To neutralize JAM-A activity, a JAM-A antibody (10 μg/ml; R&D Systems, Minneapolis, MN) or custom-made peptide against JAM-A (1 μg/ml; New England Peptide, Gardner, MA) was added 1 h before and during the adhesion assay. An isotypic control for anti-JAM-A antibodies, IgG2a, and a mismatch control peptide acted as control groups. Cell adhesion was also blocked by neutralizing anti-ICAM-1 antibody (10 μg/ml; R&D Systems) in a control study. In addition, to confirm JAM-A involvement in leukocyte adhesion, adhesion assays were performed on JAM-A-knockout cells prepared after stable transfection of JAM-A short hairpin RNA (shRNA; OriGene, Rockville, MD).
TEER.
TEERs were measured in Endohm TEER measurement chambers equipped with an epithelial voltohmmeter (EVOM) resistance meter (World Precision Instruments, Sarasota, FL). All experiments were carried out in triplicate in five independent experiments (46, 48).
Brain endothelial cell monolayer permeability.
The permeability of brain endothelial cell monolayers to fluorescein isothiocyanate (FITC)-inulin was measured as described in our previous studies (16, 44, 46, 48).
Statistical analysis.
Unpaired Student's t test and one-way analysis of variance (ANOVA) were used to test group-level differences. For post hoc comparisons, Bonferroni's tests were applied.
RESULTS
Redistribution of JAM-1 during the inflammatory remodeling of TJ complex.
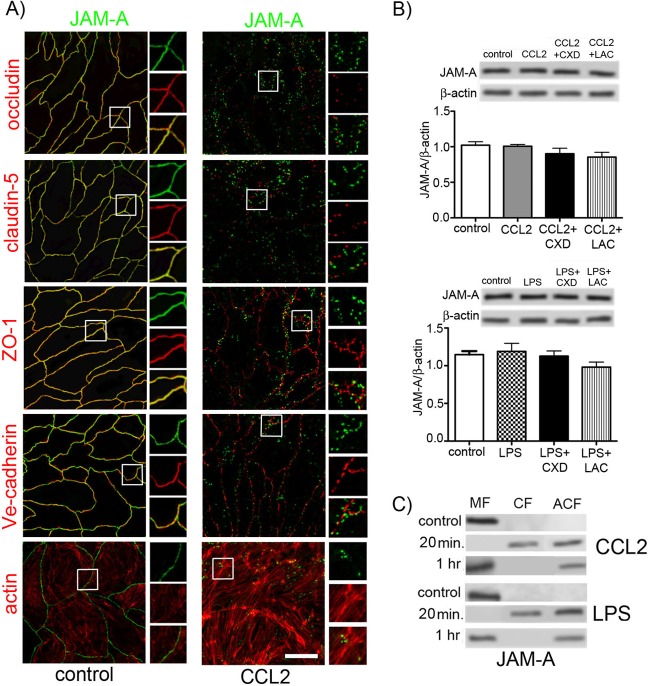
Proinflammatory mediators such as cytokines, chemokines, and oxidative radicals have the ability to remodel the brain endothelial cell barrier, promoting and enhancing the inflammatory response (1, 21, 46, 48). The brain endothelial cell barrier opening is closely associated with significant changes in endothelial cell TJs at the morphological and biochemical level. This includes lost or fragmented staining, particularly for transmembrane TJ proteins, and relocalization of occludin and claudin-5 from membrane to cytosol and then to the actin cytoskeletal fraction (36, 44, 46), which ultimately cause the loss of paracellular occlusion. Besides occludin and claudin-5, JAM-A is denoted a third transmembrane TJ protein, although the contribution of its homotypic trans interaction to paracellular space occlusion is still not clearly defined. Under resting (control) conditions, JAM-A is localized on the lateral membrane of brain endothelial cells, appearing as continuous immunostaining on the interendothelial cell border, and it is colocalized with other TJ proteins, claudin-5, occludin, and ZO-1, but not with an adherens junction (AdJ) protein, Ve-cadherin (Fig. 1A).
Fig 1.
(A) Double immunostaining for JAM-1 and TJ proteins (occludin, claudin-5, and ZO-1), adherens junction protein (Ve-cadherin), and phalloidin-Alexa 596 staining for actin in mBMECs under resting conditions (control, nontreated cells) or with treatment with CCL2 (100 ng/ml) for 30 min. All samples were examined using confocal microscopy. Notice the close association of JAM-A with occludin, claudin-5, and ZO-1 under control conditions, while CCL2 induced relocalization of JAM-A away from the lateral cell border. Boxes indicate locations of high magnification of JAM-A immunostaining with TJ and AdJ proteins as well as the actin cytoskeleton. Bar, 50 μm. (B) During CCL2 or LPS exposure, there were no changes in total JAM-A protein levels. Treatment with an inhibitor of protein synthesis, cycloheximide (CXD; 5 μg/ml), or an inhibitor of protein degradation, lactacystin (LAC; 1 μM), did not affect the total level of protein expression determined by Western blot analysis. Blots represent one of the three independent experiments. Data represent means of 3 independent experiments. (C) mBMECs were exposed to either CCL2 (100 ng/ml) or LPS (5 μg/ml) for 20 or 60 min and then underwent cell fractionation and Western blotting. Under control conditions, JAM-A was found in the membrane fraction (MF; Triton X-100-insoluble fraction). CCL2 and LPS treatment for 20 min resulted in movement of JAM-A to the cytosolic fraction (CF; Triton X-100-soluble fraction) and actin cytoskeletal fraction (ACF; Triton X-100-insoluble fraction). By 60 min, most JAM-A returned to the membrane fraction, while some remained in the actin cytoskeleton fraction. Blots represent one of three successful experiments.
Exposing cells to the inflammatory mediator CCL2 or LPS remodels the TJ complex, inducing JAM-A relocalization, characterized as intense punctate staining away from the lateral membrane. Occludin and claudin-5 also showed relocalization with punctate staining away from the lateral membrane, but there was no association of JAM-A with occludin or claudin-5 after relocalization (Fig. 1A). ZO-1 and Ve-cadherin also had an altered localization in the presence of CCL2 or LPS, characterized as a serrated pattern. This pattern did not overlap that found for JAM-A (Fig. 1A). CCL2- and LPS-induced morphological alterations were examined for time and dose dependence. The presence of CCL2 or LPS at 1 to 200 ng/ml and 1 to 10 μg/ml, respectively, did not show significant differences regarding alterations in the pattern of JAM-A immunostaining (data not shown).
We further investigated whether CCL2 or LPS might affect JAM-A protein expression, thereby altering JAM-A localization. However, Western blot analysis showed that total JAM-A protein expression was not changed during CCL2 or LPS exposure (Fig. 1B). Adding inhibitors of protein synthesis (cycloheximide) and degradation (lactacystin) did not alter total JAM-A protein content during CCL2 or LPS exposure, indicating that degradation and de novo protein synthesis or replacement of JAM-A from an intracellular pool did not play a role in JAM-A alterations under inflammatory stimuli (Fig. 1B). Cell fractionation, however, indicated that upon exposure to inflammatory stimuli (CCL2 and LPS), JAM-A undergoes relocalization from the cell membrane fraction to the cytosolic and actin cytoskeletal fractions in the first 20 min. There was a return to the membrane fraction during further inflammatory stimulus (Fig. 1C).
To visualize the dynamics of JAM-A during CCL2- or LPS-induced increases in brain endothelial cell barrier permeability, we generated stably transfected bEnd.3 cells expressing GFP-tagged JAM-A. The tag was inserted into the hinge region of JAM-A between the membrane-proximal C2 domain and the transmembrane part of JAM-A to keep intact the two extracellular domains, the phosphorylation sites, and the PDZ-binding domain motif in the cytoplasmic tail. In bEnd.3 cells with JAM-A–GFP, the level of exogenously expressed JAM-A was approximately 0.50 (data not shown). Monolayers of bEnd.3 cells with JAM-A–GFP did not differ in the permeability coefficient for FITC-inulin compared to normal bEnd.3 cells, implying that JAM-A–GFP expression does not disrupt brain endothelial cell barrier integrity. In addition, the cell lines had similar increases in brain endothelial cell permeation in the presence of recombinant CCL2. Examination of the localization of JAM-A–GFP by immunocytochemistry, immunoprecipitation, and Western blot analysis showed that JAM-A–GFP displayed a pattern of behavior similar to that of endogenous JAM-A (colocalization to the cell border and functional association with ZO-1) (data not shown). The location of JAM-A–GFP was followed during CCL2 or LPS exposure while simultaneously measuring TEER. JAM-A–GFP started to relocalize from the interendothelial cell border during the first 10 min of treatment as the TEER of the brain endothelial cell barrier dropped ∼75%. By 30 min (TEER level, 10 to 15 Ω · cm2), most JAM-A–GFP appeared away from the interendothelial cell border as dispersed punctate staining (Fig. 2A). Confocal z-section analysis of redistributed JAM-A indicated that during maximal opening of brain endothelial cell barrier (at 60 min of exposure), JAM-A was localized on the apical surface of brain endothelial cells (Fig. 2B). CCL2 and LPS induction of apical localization of JAM-A was confirmed by cell-based ELISA with a polyclonal antibody specific for mouse JAM-A. Both CCL2 and LPS induced a short-term but significant decrease in cell surface expression of JAM-A (first 10 and 20 min, P < 0.001 and P < 0.01, respectively), followed by a significant increase in JAM-A presence on the apical surface of mBMEC monolayers compared to that on nontreated cells (P < 0.05) by 30 to 60 min of CCL2 and LPS exposure (Fig. 2C). At 60 min of CCL2 or LPS exposure, ∼80% of total JAM-A was present on the apical surface.
Fig 2.
(A) (Left) Time-lapse confocal microscopy. bEnd.3 monolayers expressing GFP–JAM-A were exposed to CCL2 (100 ng/ml). Images were obtained every 5 min from 0 to 120 min. Representative images are from some critical time points during CCL2-induced alterations in brain endothelial cell barrier permeability. There is a fragmented pattern of JAM-A staining at the cell-cell border of endothelial cells during CCL2 exposure. (Right) TEER after exposure (0 to 120 min) to CCL2 or LPS. Data represent averages ± SDs for 5 independent experiments. (B) Triple-label immunostaining with CellLight membrane-CFP Bacmam 2.0 (blue), anti-JAM-A antibody (green), and anti-ZO-1 antibody (red) supported by optical z-stack section and three-dimensional analysis clarified that after exposure to CCL2 for 30 min, JAM-A redistributed onto the brain endothelial cell apical surface, where it may play a role as a leukocyte adhesion molecule. Bar, 50 μm. (C) Cell-based ELISA of JAM-A surface expression upon exposure to CCL2 (100 ng/ml) or LPS (5 μg/ml). Cells were treated for different time points (0 to 60 min), and every sample was then incubated with anti-JAM-A antibody, followed by HRP-conjugated secondary antibody and fixation. Notice that over a short time (10 to 20 min) JAM-A disappears from the cell surface, with return and expression on the apical membrane from 30 to 60 min. Data represent averages ± SDs for 3 independent experiments. *, P < 0.05 compared with control nontreated cells; **, P < 0.01 compared with control nontreated cells; ***, P < 0.001 compared with control nontreated cells. ODS 450, optical density at 450 nm.
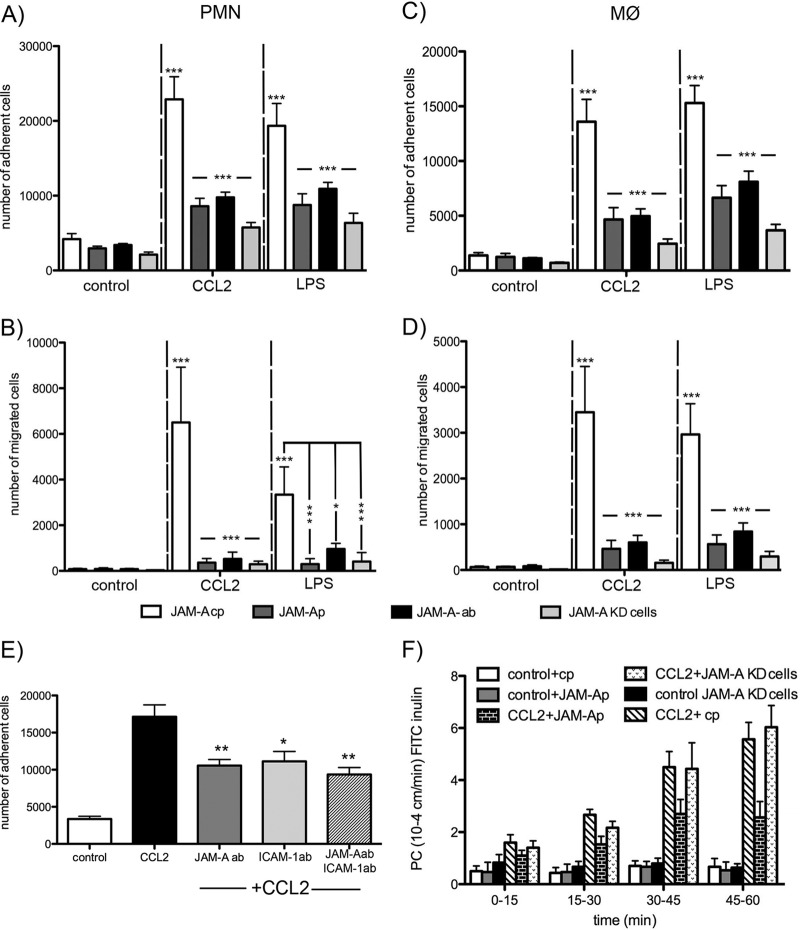
This unique pattern of JAM redistribution in the presence of CCL2 or LPS was associated with increased adhesion and transmigration of monocytes and neutrophils at the brain endothelial cell surface (Fig. 3). Monolayers treated with CCL2 or LPS for 60 min, when the complete redistribution of JAM-A occurred, showed increase adhesion and migration of both neutrophils and macrophages compared to control untreated cells (P < 0.001 and P < 0.01, respectively). The contribution of redistributed JAM-A to leukocyte adhesion and transmigration was shown in experiments (i) with a JAM-A-neutralizing antibody that specifically recognized the extracellular domain of the JAM-A (BV11 clone), (ii) with a peptide antagonist specific for the LFA domain on JAM-A, and (iii) under conditions where JAM-A was depleted by transfection with JAM-A shRNA (JAM-A-knockdown [KD] cells). Under all these experimental conditions, there was a marked reduction in the adhesion and migration of neutrophils (P < 0.001) and macrophages (P < 0.001) compared to controls (cells treated with isotypic IgG, control peptide, or mock transfection; Fig. 3). Blocking JAM-A function with neutralizing antibody or antagonist peptide did not affect the redistribution of other TJ proteins like occludin and claudin-5 or the permeability of the brain endothelial cell barrier (Fig. 3F). However, the complete absence of JAM-A in JAM-A-KD cells affected TJ protein expression and localization and increased the permeability of bEnd.3 monolayers (Fig. 3F and data not shown). Deprivation of JAM-A function did not affect the adhesion property or expression of another leukocyte adhesion molecule, ICAM-1, or the ICAM-1–leukocyte interaction (Fig. 3E).
Fig 3.
Freshly prepared neutrophils (polymorphonuclear leukocytes [PMNs]) (A and B) and macrophages (MØ) (C and D) were labeled with calcein-AM and layered on top of mBMEC monolayers previously treated with CCL2, LPS, or vehicle in the presence of JAM-A-inhibitory peptide (JAM-Ap; 1 μg/ml), control JAM-A peptide (JAM-A-cp; 1 μg/ml), or neutralizing anti-JAM-A antibody (JAM-A-Ab; 10 μg/ml). As a control, we also used JAM-A-KD cells, generated by stable transfection of bEnd.3 cells with JAM-A shRNA. Cells were incubated for 2 h, and then the medium with nonadherent cells was removed and the sample was washed and fixed. The fluorescence was read on a fluorescent reader. Both CCL2 and LPS increased the number of adherent (A and C) and migrated (B and D) neutrophils and macrophages, and that was blocked by treatment with JAM-A-inhibitory peptide or JAM-A antibody or if JAM-A-KD cells were used. Data represent averages ± SDs for 3 independent experiments. *, P < 0.05; ***, P < 0.001. (E) Adhesion assay for neutrophils (polymorphonuclear leukocytes) under conditions of exposure of brain endothelial cell monolayer to CCL2. The adhesion was blocked by adding either neutralizing anti-JAM-A (10 μg/ml; R&D Systems), neutralizing anti-ICAM-1 (10 μg/ml; R&D Systems), or a cocktail of anti-JAM-A and anti-ICAM-A antibodies (both at a concentration of 10 μg/ml) (pretreated and treated for 1 h). Notice the significant reduction in polymorphonuclear leukocyte adhesion if JAM-A or ICAM-1 is blocked. However, there was no amplifying effect if two neutralizing antibodies were applied. Data represent averages ± SDs for 3 independent experiments. *, P < 0.05 compared with cells treated with CCL2 only; **, P < 0.01 compared with cells treated with CCL2 only. ab, antibody. (F) Recombinant murine CCL2 was applied at the top and bottom of a Transwell system. The permeability coefficient (PC) for FITC-inulin was evaluated from 0 to 60 min. CCL2 induced a time-dependent increase in permeability. Adding JAM-A-neutralizing peptide showed a partial protection of this opening of the brain endothelial cell barrier, but CCL2 still increased permeability in JAM-A-KD cells. Data represent averages ± SDs for 5 independent experiments.
Thus, while the loss of JAM-A from the interendothelial cell junctions during inflammatory remodeling is similar to that of other transmembrane TJ proteins (occludin and claudin-5), JAM-A differs in that it is directed to the endothelial cell apical surface and gains a new function as a leukocyte adhesion molecule.
Mechanism of JAM-A redistribution.
Our recent study indicated that inflammation-induced remodeling of the TJ complex is associated with endocytosis of occludin and claudin-5 (caveola-dependent internalization/recycling process), by which they become redistributed from the lateral membrane. Taking this into consideration, we examined whether internalization is the mechanism underlying redistribution of JAM-A by CCL2 or LPS and which internalization pathways are involved. JAM-A and JAM-A–GFP internalization was first analyzed by surface biotinylation assays. Biotin-labeled JAM-A undergoes internalization in a time-dependent manner in the presence of CCL2 or LPS. In the first 10 min of exposure to CCL2 or LPS, approximately 30% of JAM-A was internalized, while 80 to 90% was present intracellularly by 10 to 20 min. However, by 30 to 40 min of CCL2 or LPS treatment, the intracellular level of JAM-A was depleted (∼25% of JAM-A), while at 60 min JAM-A was not present in the cytosol (Fig. 4A and B). Treating cells with cycloheximide (5 μg/ml) during CCL2 or LPS exposure did not affect the JAM-A intracellular content, pinpointing that internalization and not de novo synthesis is the source of the short-term intracellular pool.
Fig 4.
(A) Internalization of surface-biotinylated JAM-A protein. Confluent mBMECs treated with and without cycloheximide (CXD) were surface biotinylated at 0°C and then exposed to CCL2 for 60 min at 37°C to allow internalization. Any membrane-bound biotin was removed by glutathione solution (glutathione stripping [gs]). Lanes 1 and 2, biotinylated JAM-A at the cell surface; lane 3, glutathione stripping of surface biotin; lanes 4 and 5, total cell lysate; lane 6 and 7, portion of biotinylated-internalized proteins. Adjusted blot showing the time course (0 to 60 min) of internalized biotinylated JAM-A during mBMEC exposure to CCL2. (B) Quantification of internalized JAM-A during the exposure to CCL2 and opening of the brain endothelial cell barrier. The percent internalized protein was estimated as the percentage of the total biotinylated JAM-A. GADPH (glyceraldehyde-3-phosphate dehydrogenase) represents an internal loading control. Data represent averages ± SDs of 5 independent experiments.
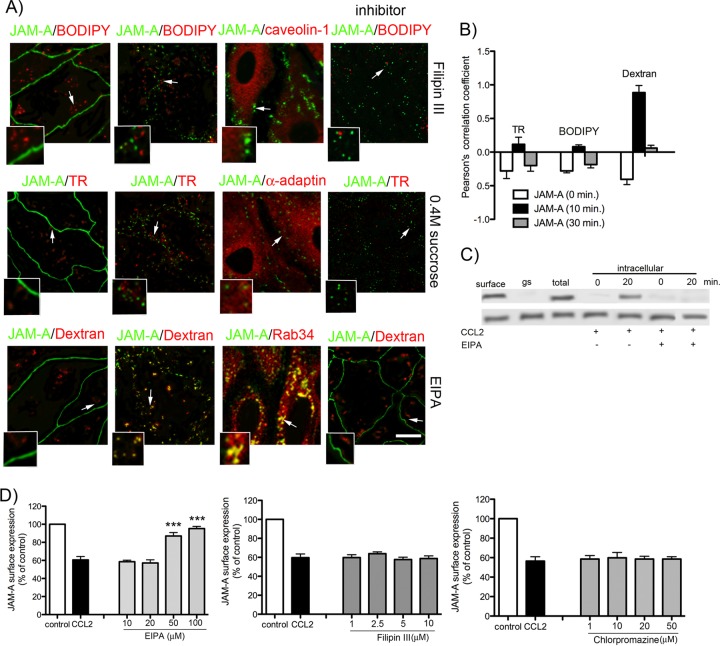
Further analysis was focused on the internalization pathways for JAM-A. Three classic pathways of internalization, the clathrin-dependent, caveola-dependent, and macropinocytosis pathways, were investigated using the specific tracers transferrin-Texas Red, BODIPY-TR ceramide, and dextran-Texas Red, respectively, as was immunocytochemistry. The trafficking of JAM-A and JAM-A–GFP during exposure to CCL2 (100 ng/ml) at 10, 20, 30, and 60 min was observed, when the permeability coefficient increased 1- to 9-fold from control values. As shown in Fig. 5A, JAM-A was colocalized exclusively with the tracer dextran-Texas Red or was associated with Rab34+ vesicles (macropinocytotic vesicles). Quantification of colocalization by calculating Pearson's coefficient for JAM-A and tracers showed the correlation of JAM-A and dextran colocalization at a level of 0.95 (on a scale of from −1 to 1) in the first 10 min of CCL2 treatment, indicating that macropinocytosis is predominantly involved in the redistribution of JAM-A (Fig. 5B). Furthermore, inhibition of the JAM-A internalization with 0.4% sucrose, chlorpromazine (clathrin-dependent pathway), filipin III (caveola-dependent pathway) or EIPA, an inhibitor of macropinocytosis, showed that only EIPA treatment completely prevented JAM-A and JAM-A–GFP vesicular accumulation. Inhibition of caveola- or clathrin-dependent internalization had no effect (Fig. 5A). In addition, EIPA completely prevented JAM-A intracellular localization in a biotinylation assay (Fig. 5C), and this effect was dose dependent (Fig. 5D). Filipin III or chlorpromazine had no effect on JAM-A internalization during the first 10 min of CCL2 exposure even at an increased concentration, excluding the possibility of the involvement of these two pathways in JAM-A internalization (Fig. 5D).
Fig 5.
(A) Morphological analysis of JAM-A–GFP internalization. Briefly, bEnd.3 cell monolayers were first exposed to the indicated concentration of the tracer BODIPY-TR-ceramide (BODIPY; 5 μM; caveola pathway), Texas Red-transferrin conjugate (TR; 10 μg/ml; clathrin pathway), and dextran (10 kDa)-Texas Red conjugate (dextran; 0.5 mg/ml; macropinocytosis) for 30 min at 4°C, and then CCL2 was added at a concentration of 100 ng/ml and samples were placed in an incubator at 37°C for 10 min. Cells were then fixed. In separate experiments, bEnd.3 cells stably transfected with GFP–JAM-A were exposed to CCL2 for 0 to 60 min, fixed, and processed for immunocytochemistry using mouse anti-caveolin-1, anti-α-adaptin, or anti-Rab34 antibodies. Both the tracer study and immunocytochemistry indicated a close association of JAM-A with dextran and Rab34 immunostaining (small boxes). Arrows, magnified colocalization of JAM-A with tracers and antibody-labeled internalization pathways. In the inhibition study (indicated in column inhibitors), cells were preincubated with a certain inhibitor, either filipin III (caveola-dependent internalization), 0.4 M sucrose (clathrin-dependent pathway), or EIPA (100 mM; macropinocytotic uptake), followed by incubation with CCL2 for 10 min. Only the inhibition of macropinocytosis prevented JAM-A movement from the cell border. Bar, 50 μm. (B) Quantification of colocalization of JAM-A with internalization vesicles based on Pearson's correlation coefficient of GFP–JAM-A/BODIPY-TR ceramide, JAM-A–GFP/transferrin-Texas Red, and JAM-A–GFP/dextran-Texas Red. There was a high degree of correlation between total JAM-A and dextran. Other tracers did not show any colocalization pattern. Error bars indicate means ± SDs. (C) The internalization pathway via macropinocytosis was also confirmed in a biotin internalization assay. In confluent mBMECs, JAM-A was biotinylated at 0°C and then exposed to either CCL2 for 20 min at 37°C or CCL2 and the inhibitor of macropinocytosis, EIPA. Surface, biotinylated JAM-A at the cell surface; gs, glutathione stripping (amount of membrane-bound biotin which was removed by glutathione solution); total, total cell lysate; intracellular, the amount of JAM-A internalized during the period from 0 to 20 min in the presence of CCL2 or CCL2 and EIPA. The macropinocytosis inhibitor prevented JAM-A internalization during CCL2 exposure. The blot represents one of three experiments performed. (D) Dose-dependent inhibition of CCL2-induced internalization of JAM-A after exposure to CCL2 for 10 min and in the presence of selected inhibitors: EIPA (10 to 100 μM), filipin III (1 to 10 μM), and chlorpromazine (1 to 50 μM, clathrin-dependent internalization). JAM-A surface expression under these conditions was evaluated by cell-based ELISA, and it is presented as the percentage of the JAM-A under control conditions. Again, only EIPA at a dose of 50 to 100 mM was able to block internalization of JAM-A. Data represent averages ± SDs for 3 independent experiments. ***, P < 0.001 compared with control nontreated cells.
Once internalized, proteins are directed to the cell endosomal system. Their further trafficking may be dependent on the type of internalization stimulus. For example, TJ and AdJ proteins could be directed to early endosomes (Rab5+ vesicles) and then redirected to recycling endosomes (Rab4+, Rab11+ vesicles) and returned to the cell surface, or they could be directed to late endosomes (Rab7+ vesicles) and lysosomes (LAMP-1 positive) (23, 26, 30, 47, 52). Analyzing the fate of internalized JAM-A by immunofluorescence, we found that at early time points (up to 10 min of CCL2 exposure), these proteins were present only in dextran-positive, Rab5+, and Rab34+ isolated vesicles (macropinosomes) (Fig. 5A and B and 6C). At between 20 and 30 min, the majority of JAM-A was localized in Rab4+ vesicles. There was no localization of these proteins in Rab7+ vesicles (late endosomes).
Fig 6.
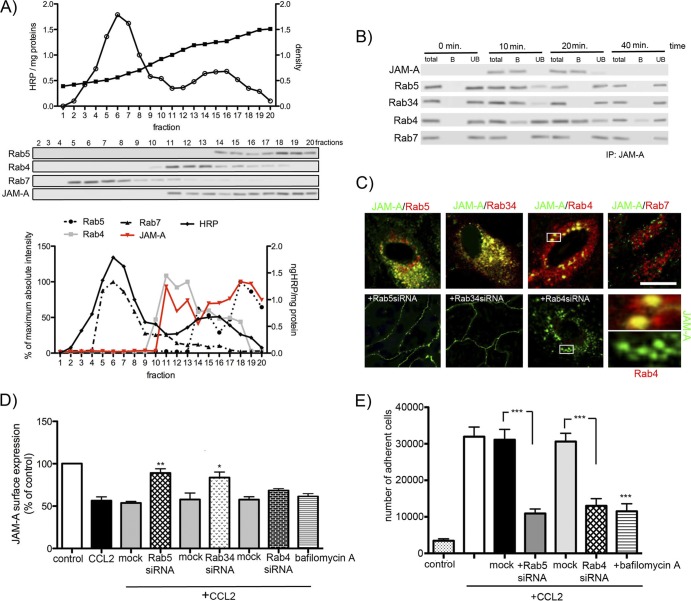
(A) After exposure of mBMEC to CCL2 (100 ng/ml) for 20 min, endosome-rich fractions were prepared using a continuous sucrose gradient (15 to 40%). Twenty fractions were collected and analyzed by Western blotting. Fraction 1 is the top of the gradient and has the lowest density of sucrose. The fraction and its endosomal content were also labeled with an HRP (1 mg/ml) pulse, incubated for 5 min, and chased for 40 min. Fractions 14 to 20 mostly contain Rab5+ vesicles (early endosomes) and fractions 11 to 16 contain Rab4+ vesicles (recycling endosomes), while fractions 5 to 8 contain Rab7+ vesicles (late endosomes). The Western blot represents one of three successful experiments. Densitometric analysis of Rab5, Rab4, Rab7, and JAM-A expression in collected fractions indicated the correlation of JAM-A presence with Rab5+ and Rab4+ endosomes. The graph is a summary of the three independent experiments. (B) Isolated endosomes were pulled down using JAM-A antibody-coated beads. Western blot assays for Rab5, Rab34, Rab4, and Rab7 were performed to assess the JAM-A presence in certain Rab+ vesicles. Total, whole collected fraction; B, endosomal fraction bound to JAM-A antibody-coated magnetic Dynabeads; UB, unbound fraction not absorbed to JAM-A antibody-coated Dynabeads. The blot represents one of three successful experiments. IP, immunoprecipitation. (C) Immunocytochemistry analysis of JAM-A–GFP localization with Rab5+, Rab34+, Rab4+, and Rab7+ vesicles during CCL2 exposure (20 min). Rab5 or Rab34 siRNA prevented JAM-A internalization. Rab4 siRNA did not prevent internalization, but it did prevent JAM-A from appearing on the apical membrane (zoom confocal images). Bar, 20 μm. (D and E) Preventing JAM-A internalization via macropinocytosis (Rab5 siRNA and Rab34 siRNA), preventing JAM-A recycling by depleting Rab4 vesicles (Rab4 siRNA), or treatment with an inhibitor of emptying of the recycling endosomes (bafilomycin A1, 50 nM) reduced the internalization of JAM-A (D) after 30 min of treatment as well as neutrophil adhesion (P < 0.001). Notice that Rab4 inhibition did not affect the internalization of JAM-A but via inhibition of JAM-A recycling affected the adhesion of neutrophils on the brain endothelial cell surface. All data represent averages ± SDs for 3 independent experiments. *, P < 0.05 compared with control or mock-transfected cells; **, P < 0.01 compared with control or mock-transfected cells; ***, P < 0.001 compared with control or mock-transfected cells.
This finding was confirmed by analysis of isolated endosomes. To examine fractions enriched in early (Rab5+ and Rab34+), recycling (Rab4+), and late (Rab7+) endosomes, we performed subcellular fractionation of brain endothelial cells treated with CCL2, followed by Western blot analysis. Rab5, Rab4, and Rab7 were broadly distributed from fractions 5 to 20, with a higher accumulation of Rab5 in fractions 18 to 20, Rab4 in fractions 11 to 13, and Rab7 in fractions 5 to 8. JAM-A was distributed from fractions 11 to 20, which correlated with early and recycling endosomes (Fig. 6A). Further analysis of the content of isolated endosomes was performed using magnetic beads coated with antibodies against JAM-A, Rab5, Rab34, Rab4, and Rab7 incubated with selected fractions (from samples collected after 0 to 40 min of CCL2 exposure), collected, and washed. Unbound material, as well as an equal portion of starting material, was centrifuged, and the compositions of the immunoadsorbed (B), unbound (UB), and starting material (total) were analyzed by Western blotting (Fig. 5B). The Rab5+ and Rab34+ vesicles (from fraction 18) predominantly contained JAM-A in the first 10 min (approximately 90% of the internalized JAM-A). At 20 min of exposure to CCL2, JAM-A is mostly accumulated in Rab4+ vesicles (fraction 11; 75% of the total internalized JAM-A), and it is still present at lower levels (only 25%) at 40 min. Internalized JAM-A disappears from Rab4+ vesicles after 60 min of CCL2 exposure (Fig. 6B). There was no JAM-A in Rab7+ vesicles (fraction 5) at any analyzed time point (Fig. 6B).
Beyond 40 min of CCL2 exposure, JAM-A was not localized in any of the examined vesicles. Indeed, there were only trace amounts of JAM-A present intracellularly (<10%) (Fig. 6B). Correlating these data with those presented in Fig. 2 to 4, by 40 min of CCL2 exposure, most JAM-A is present on the apical membrane of the brain endothelial cells, gaining a new role as a leukocyte adhesion molecule. The importance of this vesicular pathway for this new function was further confirmed by an inhibition study where either Rab5 vesicles were depleted using siRab5RNA, Rab34+ vesicles were depleted by siRNA, Rab4+ vesicles were depleted by siRab4RNA, or recycling was blocked by the pharmacological inhibitor bafilomycin A1 (Fig. 6C to E; data not shown). Blocking JAM-A internalization and sorting by transient transfection with siRab5RNA or siRab34RNA prevented JAM-A internalization after CCL2 treatment and left JAM-A mostly localized on the lateral membrane (Fig. 6C and D). This localization of JAM-A was incompatible with a role of JAM-A as a leukocyte adhesion molecule. Thus, blocking Rab5 and Rab34 reduced neutrophil adhesion to brain endothelial cells (Fig. 6E) and transmigration (P < 0.01; data not shown). In contrast, siRab4RNA and bafilomycin A1 did not inhibit JAM-A internalization but reduced JAM-A movement to the apical membrane. Thus, immunofluorescence analysis showed typical JAM-A punctate staining away from the lateral membrane (Fig. 6C) but reduced cell surface expression at 30 min, with JAM-A being mostly present intracellularly (Fig. 6D). Consistent with the role of apical JAM-A in leukocyte trafficking, siRab4RNA and bafilomycin A1 both reduced neutrophil adhesion to brain endothelial cells after CCL2 treatment (Fig. 6E).
On the basis of the presented data, our suggestion is that after internalization JAM-A is sorted into macropinosomes/early endosomes (Rab34+ and Rab5+), and due to the presence in Rab5+, vesicles get sorted into recycling vesicles (Rab4+) which are involved in returning JAM-A to the membrane surface, but now at the apical membrane rather than the lateral membrane.
Signaling pathways involved in JAM-A redistribution.
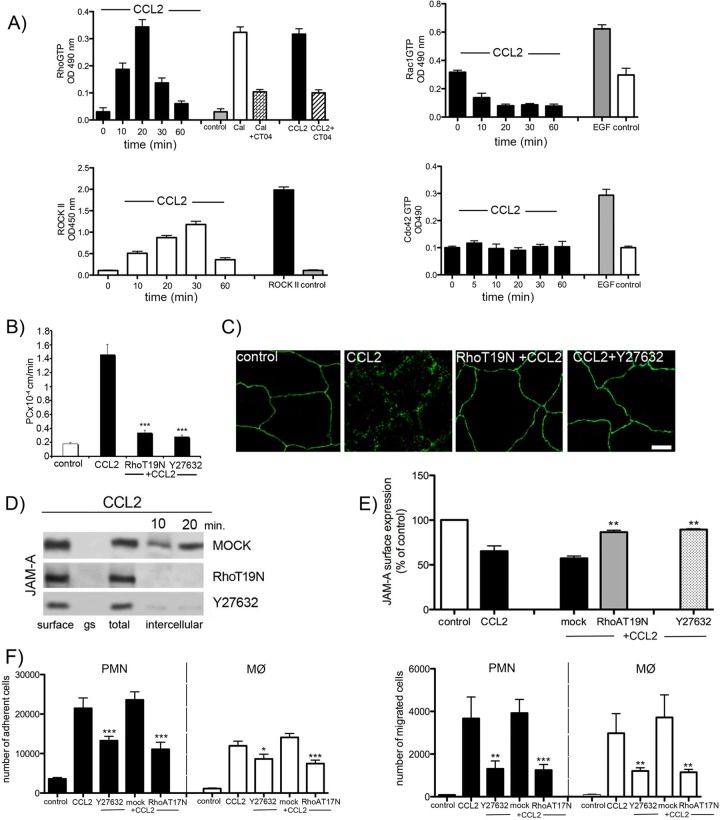
The dynamic behavior of the TJ complex is intimately connected to fundamental cell regulatory mechanisms, including vesicle trafficking and cytoskeletal reorganization (7–9). Thus, the actin cytoskeleton plays a prominent role during inflammation, where there is extensive cytoskeletal remodeling with robust stress fiber formation and accumulation of phosphorylated regulatory myosin light chains on centrally positioned stress fibers, leading to actomyosin contraction, cell retraction, and disruption of endothelial cell monolayer integrity. In addition, junctional plasticity may be directly regulated via actin-binding components of TJ or indirectly by organizing intracellular vesicular trafficking and other cytoskeletal structures. The master regulators of these events are members of the Rho GTPase family, RhoA, Rac1, and Cdc42, and Rho kinase (43). We analyzed the activation of three Rho GTPases, RhoA, Rac1, and Cdc42, as well as Rho kinase (ROCK I), during CCL2 exposure. Of the three Rho GTPases, only RhoA showed short-term activation in brain endothelial cells during CCL2 exposure (Fig. 7A). The downstream target of RhoA, Rho kinase, had very similar activation kinetics (Fig. 7A). Our previous studies already indicated that both RhoA and Rho kinase play a critical role in modulating brain endothelial cell TJ complexes during CCL2 exposure by inducing occludin and claudin-5 redistribution, which in turn leads to increased brain endothelial cell barrier permeability (36, 44, 46). Inhibition of RhoA, via dominant-negative mutant RhoT19N, or inhibition of the Rho kinase activity, using Y27632, diminished the increased brain endothelial cell barrier permeability induced by CCL2 (Fig. 7B). As RhoA and Rho kinase are involved in the redistribution of occludin and claudin-5, we also examined whether the absence of RhoA and Rho kinase would affect JAM-A redistribution. Knockdown of RhoA by transient transfection with RhoT19N and inhibition of Rho kinase activity with Y27632 diminished the redistribution of JAM-A during 30 min of CCL2 exposure. It also affected JAM-A internalization, preventing JAM-A expression on the apical membrane (Fig. 7C to E). As a result, both RhoA and Rho kinase blockage affected JAM-A function as a leukocyte adhesion molecule, reducing neutrophil and macrophage adhesion and migration in the presence of CCL2 (P < 0.001) (Fig. 7F).
Fig 7.
Role of RhoA and Rho kinase in JAM-A internalization. (A) Time course of RhoA Rac1, Cdc42, and ROCK II activation during exposure to CCL2 in brain endothelial cells. Rho GTP, Rac1, and Cdc42 levels and ROCK II activity were measured with RhoA–, Rac1–, or Cdc42–small G-protein activation assay (G-lisa) or ROCK activation assay using MYPT1 as a substrate. The positive controls for RhoA GTP levels and for Rac1 GTP and Cdc42 GTP levels were Swiss 3T3 cells treated with calpeptin (Cal; 0.1 mg/ml) and epidermal growth factor (EGF; 10 ng/ml), respectively. The specificity of the RhoA activation was determined under conditions of blocking the RhoA activation in Swiss 3T3 cells (calpeptin induced) and brain endothelial cells (CCL2 induced) by CT04 inhibitor (1 μg/ml; cytoskeleton). Data represent averages ± SDs for 3 independent experiments. OD 490 nm, optical density at 490 nm. (B) Permeability coefficient for FITC-inulin of brain endothelial cell monolayers exposed to CCL2 for 30 min. The brain endothelial cell monolayer was transfected with pCMVRhoT19N or pretreated with Y27632 to block RhoA or Rho kinase and treated with CCL2 for 30 min. The absence of RhoA or Rho kinase activation significantly diminished the increased permeation for FITC-inulin at the brain endothelial cell monolayer. Data represent averages ± SDs for 3 independent experiments. ***, P < 0.001. (C) mBMEC monolayers were mock transfected or transfected with pCMVRhoT19N or pretreated with Rho kinase inhibitor Y27632. Cells were then treated with CCL2 (100 ng/ml). Inhibiting either Rho or Rho kinase diminished CCL2-induced relocalization of JAM-A (absence of the punctate pattern of immunostaining). Bar, 10 μm. (D) Inhibition of RhoA or Rho kinase activity in mBMECs also prevented CCL2-induced internalization of biotin-labeled JAM-A, also evaluated by cell-based ELISA for JAM-A (E). Data represent averages ± SDs for 3 independent experiments. **, P < 0.01 compared with cells treated only with CCL2. (F) Adhesion and migration assay for neutrophils (polymorphonuclear leukocytes [PMN]) and macrophages (MØ) under conditions of exposure of brain endothelial cell monolayer to CCL2, transient depletion of RhoA activity via RhoT19N dominant inactive mutant, or deprivation of Rho kinase activity via Rho kinase inhibitor Y27632 (10 μM). Notice the significant reduction in polymorphonuclear leukocyte and MØ adhesion and migration if RhoA and Rho kinase activity is blocked. Data represent averages ± SDs for 3 independent experiments. *, P < 0.05 compared with cells treated only with CCL2; **, P < 0.01 compared with cells treated only with CCL2; ***, P < 0.001 compared with cells treated only with CCL2.
In addition, we investigated whether RhoA affects JAM-A phosphorylation during inflammation-induced remodeling of the brain endothelial cell barrier, as RhoA controls initial phosphorylation at the Ser residue during the redistribution of occludin and claudin-5 (44). However, we did not find additional phosphorylation of JAM-A on Ser, Thr, or Typ residues during redistribution (data not shown). Thus, our observation is that RhoA and Rho kinase are mostly involved in regulating macropinocytotic uptake of JAM-A and relocalization of JAM-A to the apical membrane of brain endothelial cells.
DISCUSSION
Opening of the blood-brain barrier during inflammatory leukocyte recruitment is still a poorly understood event. Most studies have focused on the role of the endothelial cell apical region in mediating leukocyte-endothelial cell interactions (12, 20). There are very limited data regarding leukocyte movement between adjacent endothelial cells and the role of interendothelial cell junctional complexes in that process (22, 39, 59). The objective of the present study was, therefore, to investigate the effects of monocyte chemoattractant protein 1/CCL2 (MCP-1/CCL2) and LPS on one TJ complex component, JAM-A, which is pivotal in leukocyte migration. We found that (i) JAM-A, like other TJ proteins, undergoes redistribution from the brain endothelial cell lateral membrane during inflammatory remodeling of the TJ complex; (ii) redistribution of JAM-A is via internalization involving macropinocytosis; (iii) internalized JAM-A is transiently stored in recycling endosomes and then recruited to the apical side of the endothelial cell; (iv) inhibition of JAM-A or its redistribution significantly reduces the ability of CCL2 to increase the adhesion and transmigration of monocytes and neutrophils at the brain endothelium; and (v) Rho and Rho kinase signaling and actin cytoskeleton reorganization play a critical role in JAM-A internalization during inflammatory opening of the brain endothelial cell barrier.
Inflammatory stimuli cause marked alterations in brain endothelial cells. This complex process changes the endothelial cell surface, manifested as an upregulation in expression of adhesion molecules such as ICAM-1, vascular cell adhesion molecule 1 (VCAM-1), and selectins (12, 20, 30, 50, 53). It also results in intensive TJ complex remodeling directed toward forming a paracellular route and facilitating leukocyte entry into brain parenchyma (29, 36, 48). This TJ complex remodeling is closely associated with a relocalization of occludin, claudin-5, and ZO-1 from TJ strands and loosening of TJ protein adhesion, which ultimately leads to loss of occlusion and gap formation between brain endothelial cells (44, 46, 47). During this process, transmembrane TJ proteins (occludin and claudin-5) are redistributed from the membrane fraction to the cytosolic and actin cytoskeletal fractions (44, 46, 47). JAM-A, as part of the TJ complex, also undergoes redistribution to those fractions early during inflammatory remodeling (25, 35). However, for claudin-5 and occludin, redistribution back to the plasma membrane occurs only when the inflammatory stimulus is removed, but relocalization of JAM-A to the plasma membrane fraction occurs in the presence of the inflammatory stimulus and during maximal opening of the brain endothelial cell barrier (47). In addition, there is a new localization of JAM-A on the apical plasma membrane (not a return to the TJ), and this localization enables JAM-A to interact with leukocytes (monocytes and neutrophils), as indicated by others and in our study (leukocyte adhesion and migration assays).
Our study pinpoints one potential mechanism underlying this JAM-A relocalization, internalization/endocytosis. That process is critical for TJ transmembrane protein redistribution during inflammation (6, 23, 26, 27, 47, 51, 52). For example, internalization of transmembrane TJ proteins (i.e., claudin-5 and occludin) via caveolae is proposed to transiently reduce the lateral adhesive property of brain endothelial cells (disturbance of trans interactions of occludin and claudin-5) and increase paracellular permeability (47). JAM-A, as part of the TJ complex, also forms homophilic trans interactions on the extracellular domain and contributes to occlusion of the paracellular space. Thus, in order to form a paracellular route, trans JAM-A interactions need to be broken and JAM-A also needs to be removed from the lateral membrane via internalization. However, although there is a close temporal correlation in JAM-A internalization with occludin and claudin-5, JAM-A was not found to be associated with either of these TJ proteins after uptake and did not use the caveola- or clathrin-internalizing vesicles found for occludin and claudin-5. JAM-A showed a separate internalization mechanism via macropinocytosis. This finding differs from those from recent studies by Utech et al. (51) and Bruewer et al. (6), who described so-called block internalization of JAM-A, occludin, and claudin-1 via macropinocytosis in epithelial cells exposed to tumor necrosis factor alpha and gamma interferon (6, 51). Although there are similarities between the epithelial and brain endothelial cell barriers, they also have unique structural properties, which could affect barrier responses. For example, occludin and claudin-5 are localized in the lipid raft microdomain in brain endothelial cells, and it is not surprising that caveolae play an important role in function and endocytotic turnover of these TJ proteins (23, 27, 31). In contrast, JAM-A is not localized in the lipid raft microdomain, leaving an option that a caveola-dependent process may not influence the internalization pathway.
After internalization, vesicular sorting determines molecular fate. In this study, JAM-A (uptake by dextran-positive and Rab5/Rab34+ vesicles, sorted toward Rab4+ vesicles) was removed from the lateral membrane in order to unseal the TJ complex but sorted to Rab4+ vesicles for return to the membrane. Both occludin and claudin-5 also have the ability to be recycled back to the lateral membrane after internalization under inflammatory stimuli (47), but the pattern for JAM-A was different. JAM-A stayed in Rab4+ vesicles for a relatively short time, and already after 30 min of CCL2 treatment, most of the JAM-A was away from Rab4+ endosomes and recycled back to the plasma membrane. There was no sorting to late endosomes and lysosomes, confirming that degradation, the process mostly described as being associated with morphological alteration of TJ proteins, does not take place. JAM-A recycling to the plasma membrane is different from that for claudin-5 and occludin. The recycling is not associated with recovery of the brain endothelial cell barrier, and it occurs during exposure to the inflammatory stimuli (47). As breaking JAM-A trans interactions has a role in unmasking of the JAM-A LFA domain, JAM-A internalization and recycling to the apical membrane could be associated with transformation of JAM-A into a leukocyte adhesion molecule regulating leukocyte transmigration. Thus, blocking JAM-A macropinocytotic uptake, intracellular sorting, or delivery to the apical membrane can affect the leukocyte-endothelial cell interaction and leukocyte transmigration.
Rho GTPases are critical signaling molecules involved in regulating the endothelial cell barrier opening during inflammation and leukocyte transmigration (7, 36, 40, 46, 56). The major target of Rho GTPase is the actin cytoskeleton, and both RhoA and Rho kinase were critically involved in regulating macropinocytotic internalization of JAM-A. Their inhibition prevented JAM-A internalization and appearance as a leukocyte adhesion molecule. Our previous studies have also shown that Rho and Rho kinase can directly and indirectly affect TJ integrity by regulating TJ protein phosphorylation (36, 44, 46), but we did not find any participation of RhoA or Rho kinase in JAM-A phosphorylation.
It is important to address the potential implications of our findings. In the tightly regulated process of leukocyte rolling, firm adhesion, and diapedesis, it is clear that JAM-A plays a prominent role, as shown by the effects on leukocyte adhesion and transmigration of blocking JAM-A and its relocalization (9, 11). However, the importance of JAM-A relocation on the apical surface during brain endothelial cell barrier opening may be closely associated with its contribution in directing/triggering leukocyte migration through the paracellular space in concert with other adhesion molecules (PECAM-1 and CD99) (37). Macropinocytotic uptake of JAM-A from the lateral side of brain endothelial cells and transfer to the apical membrane are fundamental in JAM-A obtaining a new role as a leukocyte adhesion molecule. Thus, understanding of JAM-A relocalization during inflammatory events at the blood-brain barrier sheds light on this molecule as an important target for preventing leukocyte infiltration during CNS inflammation.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant NS-062853 from the National Institute of Neurological Disorders and Stroke (to A.V.A.).
The confocal microscopy work was performed in the Microscopy and Image-Analysis Laboratory (MIL) at the University of Michigan, Department of Cell and Developmental Biology.
Footnotes
Published ahead of print 25 June 2012
REFERENCES
- 1. Abbott NJ. 2000. Inflammatory mediators and modulation of blood-brain barrier permeability. Cell. Mol. Neurobiol. 20: 131– 147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Babinska A, et al. 2007. The F11 receptor (F11R/JAM-A) in atherothrombosis: overexpression of F11R in atherosclerotic plaques. Thromb. Haemost. 97: 272– 281 [PubMed] [Google Scholar]
- 3. Bazzoni G. 2011. Pathobiology of junctional adhesion molecules. Antioxid. Redox Signal. 15: 1221– 1234 [DOI] [PubMed] [Google Scholar]
- 4. Bazzoni G, Dejana E. 2004. Endothelial cell-to-cell junctions: molecular organization and role in vascular homeostasis. Physiol. Rev. 84: 869– 901 [DOI] [PubMed] [Google Scholar]
- 5. Bazzoni G, et al. 2000. Homophilic interaction of junctional adhesion molecule. J. Biol. Chem. 275: 30970– 30976 [DOI] [PubMed] [Google Scholar]
- 6. Bruewer M, et al. 2005. Interferon-gamma induces internalization of epithelial tight junction proteins via a macropinocytosis-like process. FASEB J. 19: 923– 933 [DOI] [PubMed] [Google Scholar]
- 7. Carbajal JM, Schaeffer RC., Jr 1999. RhoA inactivation enhances endothelial barrier function. Am. J. Physiol. 277: C955– C964 [DOI] [PubMed] [Google Scholar]
- 8. Cohn ZA. 1974. The isolation and cultivation of mononuclear phagocytes. Methods Enzymol. 32: 758– 765 [DOI] [PubMed] [Google Scholar]
- 9. Corada M, et al. 2005. Junctional adhesion molecule-A-deficient polymorphonuclear cells show reduced diapedesis in peritonitis and heart ischemia-reperfusion injury. Proc. Natl. Acad. Sci. U. S. A. 102: 10634– 10639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crane IJ, Liversidge J. 2008. Mechanisms of leukocyte migration across the blood-retina barrier. Semin. Immunopathol. 30: 165– 177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Del Maschio A, et al. 1999. Leukocyte recruitment in the cerebrospinal fluid of mice with experimental meningitis is inhibited by an antibody to junctional adhesion molecule (JAM). J. Exp. Med. 190: 1351– 1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Diacovo TG, Roth SJ, Buccola JM, Bainton DF, Springer TA. 1996. Neutrophil rolling, arrest, and transmigration across activated, surface-adherent platelets via sequential action of P-selectin and the beta 2-integrin CD11b/CD18. Blood 88: 146– 157 [PubMed] [Google Scholar]
- 13. Fraemohs L, Koenen RR, Ostermann G, Heinemann B, Weber C. 2004. The functional interaction of the beta 2 integrin lymphocyte function-associated antigen-1 with junctional adhesion molecule-A is mediated by the I domain. J. Immunol. 173: 6259– 6264 [DOI] [PubMed] [Google Scholar]
- 14. Griffith GR, Consigli RA. 1984. Isolation and characterization of monopinocytotic vesicles containing polyomavirus from the cytoplasm of infected mouse kidney cells. J. Virol. 50: 77– 85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Itoh M, et al. 2001. Junctional adhesion molecule (JAM) binds to PAR-3: a possible mechanism for the recruitment of PAR-3 to tight junctions. J. Cell Biol. 154: 491– 497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kazakoff PW, McGuire TR, Hoie EB, Cano M, Iversen PL. 1995. An in vitro model for endothelial permeability: assessment of monolayer integrity. In Vitro Cell. Dev. Biol. Anim. 31: 846– 852 [DOI] [PubMed] [Google Scholar]
- 17. Keiper T, Santoso S, Nawroth PP, Orlova V, Chavakis T. 2005. The role of junctional adhesion molecules in cell-cell interactions. Histol. Histopathol. 20: 197– 203 [DOI] [PubMed] [Google Scholar]
- 18. Kostrewa D, et al. 2001. X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif. EMBO J. 20: 4391– 4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lamagna C, et al. 2005. Dual interaction of JAM-C with JAM-B and alpha(M)beta2 integrin: function in junctional complexes and leukocyte adhesion. Mol. Biol. Cell 16: 4992– 5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ley K, Laudanna C, Cybulsky MI, Nourshargh S. 2007. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7: 678– 689 [DOI] [PubMed] [Google Scholar]
- 21. Lochhead JJ, et al. 2010. Oxidative stress increases blood-brain barrier permeability and induces alterations in occludin during hypoxia-reoxygenation. J. Cereb. Blood Flow Metab. 30: 1625– 1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mamdouh Z, Mikhailov A, Muller WA. 2009. Transcellular migration of leukocytes is mediated by the endothelial lateral border recycling compartment. J. Exp. Med. 206: 2795– 2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marchiando AM, et al. 2010. Caveolin-1-dependent occludin endocytosis is required for TNF-induced tight junction regulation in vivo. J. Cell Biol. 189: 111– 126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martineau M, Galli T, Baux G, Mothet JP. 2008. Confocal imaging and tracking of the exocytotic routes for d-serine-mediated gliotransmission. Glia 56: 1271– 1284 [DOI] [PubMed] [Google Scholar]
- 25. Martin-Padura I, et al. 1998. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 142: 117– 127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsuda M, Kubo A, Furuse M, Tsukita S. 2004. A peculiar internalization of claudins, tight junction-specific adhesion molecules, during the intercellular movement of epithelial cells. J. Cell Sci. 117: 1247– 1257 [DOI] [PubMed] [Google Scholar]
- 27. McCaffrey G, et al. 2007. Tight junctions contain oligomeric protein assembly critical for maintaining blood-brain barrier integrity in vivo. J. Neurochem. 103: 2540– 2555 [DOI] [PubMed] [Google Scholar]
- 28. Mukherjee K, et al. 2000. Live Salmonella recruits N-ethylmaleimide-sensitive fusion protein on phagosomal membrane and promotes fusion with early endosome. J. Cell Biol. 148: 741– 753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Muller WA. 2003. Leukocyte-endothelial-cell interactions in leukocyte transmigration and the inflammatory response. Trends Immunol. 24: 327– 334 [DOI] [PubMed] [Google Scholar]
- 30. Muro S, et al. 2003. A novel endocytic pathway induced by clustering endothelial ICAM-1 or PECAM-1. J. Cell Sci. 116: 1599– 1609 [DOI] [PubMed] [Google Scholar]
- 31. Nusrat A, et al. 2000. Tight junctions are membrane microdomains. J. Cell Sci. 113(Pt 10): 1771– 1781 [DOI] [PubMed] [Google Scholar]
- 32. Ostermann G, et al. 2005. Involvement of JAM-A in mononuclear cell recruitment on inflamed or atherosclerotic endothelium: inhibition by soluble JAM-A. Arterioscler. Thromb. Vasc. Biol. 25: 729– 735 [DOI] [PubMed] [Google Scholar]
- 33. Ostermann G, Weber KS, Zernecke A, Schroder A, Weber C. 2002. JAM-1 is a ligand of the beta(2) integrin LFA-1 involved in transendothelial migration of leukocytes. Nat. Immunol. 3: 151– 158 [DOI] [PubMed] [Google Scholar]
- 34. Ozaki H, et al. 2000. Junctional adhesion molecule (JAM) is phosphorylated by protein kinase C upon platelet activation. Biochem. Biophys. Res. Commun. 276: 873– 878 [DOI] [PubMed] [Google Scholar]
- 35. Ozaki H, et al. 1999. Cutting edge: combined treatment of TNF-alpha and IFN-gamma causes redistribution of junctional adhesion molecule in human endothelial cells. J. Immunol. 163: 553– 557 [PubMed] [Google Scholar]
- 36. Persidsky Y, et al. 2006. Rho-mediated regulation of tight junctions during monocyte migration across the blood-brain barrier in HIV-1 encephalitis (HIVE). Blood 107: 4770– 4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petri B, Bixel MG. 2006. Molecular events during leukocyte diapedesis. FEBS J. 273: 4399– 4407 [DOI] [PubMed] [Google Scholar]
- 38. Ransohoff RM, Liu L, Cardona AE. 2007. Chemokines and chemokine receptors: multipurpose players in neuroinflammation. Int. Rev. Neurobiol. 82: 187– 204 [DOI] [PubMed] [Google Scholar]
- 39. Roberts TK, Buckner CM, Berman JW. 2010. Leukocyte transmigration across the blood-brain barrier: perspectives on neuroAIDS. Front. Biosci. 15: 478– 536 [DOI] [PubMed] [Google Scholar]
- 40. Sakakibara T, Nemoto Y, Nukiwa T, Takeshima H. 2004. Identification and characterization of a novel Rho GTPase activating protein implicated in receptor-mediated endocytosis. FEBS Lett. 566: 294– 300 [DOI] [PubMed] [Google Scholar]
- 41. Shaw SK, et al. 2004. Coordinated redistribution of leukocyte LFA-1 and endothelial cell ICAM-1 accompany neutrophil transmigration. J. Exp. Med. 200: 1571– 1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sobocki T, et al. 2006. Genomic structure, organization and promoter analysis of the human F11R/F11 receptor/junctional adhesion molecule-1/JAM-A. Gene 366: 128– 144 [DOI] [PubMed] [Google Scholar]
- 43. Spindler V, Schlegel N, Waschke J. 2010. Role of GTPases in control of microvascular permeability. Cardiovasc. Res. 87: 243– 253 [DOI] [PubMed] [Google Scholar]
- 44. Stamatovic SM, Dimitrijevic OB, Keep RF, Andjelkovic AV. 2006. Protein kinase Calpha-RhoA cross-talk in CCL2-induced alterations in brain endothelial permeability. J. Biol. Chem. 281: 8379– 8388 [DOI] [PubMed] [Google Scholar]
- 45. Stamatovic SM, Keep RF, Andjelkovic AV. 2011. Tracing the endocytosis of claudin-5 in brain endothelial cells. Methods Mol. Biol. 762: 303– 320 [DOI] [PubMed] [Google Scholar]
- 46. Stamatovic SM, Keep RF, Kunkel SL, Andjelkovic AV. 2003. Potential role of MCP-1 in endothelial cell tight junction ‘opening’: signaling via Rho and Rho kinase. J. Cell Sci. 116: 4615– 4628 [DOI] [PubMed] [Google Scholar]
- 47. Stamatovic SM, Keep RF, Wang MM, Jankovic I, Andjelkovic AV. 2009. Caveolae-mediated internalization of occludin and claudin-5 during CCL2-induced tight junction remodeling in brain endothelial cells. J. Biol. Chem. 284: 19053– 19066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stamatovic SM, et al. 2005. Monocyte chemoattractant protein-1 regulation of blood-brain barrier permeability. J. Cereb. Blood Flow Metab. 25: 593– 606 [DOI] [PubMed] [Google Scholar]
- 49. Tjelle TE, Brech A, Juvet LK, Griffiths G, Berg T. 1996. Isolation and characterization of early endosomes, late endosomes and terminal lysosomes: their role in protein degradation. J. Cell Sci. 109(Pt 12): 2905– 2914 [DOI] [PubMed] [Google Scholar]
- 50. Turowski P, Adamson P, Greenwood J. 2005. Pharmacological targeting of ICAM-1 signaling in brain endothelial cells: potential for treating neuroinflammation. Cell. Mol. Neurobiol. 25: 153– 170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Utech M, et al. 2005. Mechanism of IFN-gamma-induced endocytosis of tight junction proteins: myosin II-dependent vacuolarization of the apical plasma membrane. Mol. Biol. Cell 16: 5040– 5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Utech M, Mennigen R, Bruewer M. 2010. Endocytosis and recycling of tight junction proteins in inflammation. J. Biomed. Biotechnol. 2010: 484987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weber C. 2003. Novel mechanistic concepts for the control of leukocyte transmigration: specialization of integrins, chemokines, and junctional molecules. J. Mol. Med. (Berl.) 81: 4– 19 [DOI] [PubMed] [Google Scholar]
- 54. Weber C, Fraemohs L, Dejana E. 2007. The role of junctional adhesion molecules in vascular inflammation. Nat. Rev. Immunol. 7: 467– 477 [DOI] [PubMed] [Google Scholar]
- 55. Williams LA, Martin-Padura I, Dejana E, Hogg N, Simmons DL. 1999. Identification and characterisation of human junctional adhesion molecule (JAM). Mol. Immunol. 36: 1175– 1188 [DOI] [PubMed] [Google Scholar]
- 56. Wojciak-Stothard B, Entwistle A, Garg R, Ridley AJ. 1998. Regulation of TNF-alpha-induced reorganization of the actin cytoskeleton and cell-cell junctions by Rho, Rac, and Cdc42 in human endothelial cells. J. Cell. Physiol. 176: 150– 165 [DOI] [PubMed] [Google Scholar]
- 57. Wojcikiewicz EP, et al. 2009. LFA-1 binding destabilizes the JAM-A homophilic interaction during leukocyte transmigration. Biophys. J. 96: 285– 293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wyman TH, et al. 2002. A two-insult in vitro model of PMN-mediated pulmonary endothelial damage: requirements for adherence and chemokine release. Am. J. Physiol. Cell Physiol. 283: C1592– C1603 [DOI] [PubMed] [Google Scholar]
- 59. Xu H, Dawson R, Crane IJ, Liversidge J. 2005. Leukocyte diapedesis in vivo induces transient loss of tight junction protein at the blood-retina barrier. Invest. Ophthalmol. Vis. Sci. 46: 2487– 2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yuan Y, Fleming BP. 1990. A method for isolation and fluorescent labeling of rat neutrophils for intravital microvascular studies. Microvasc. Res. 40: 218– 229 [DOI] [PubMed] [Google Scholar]
- 61. Zinchuk V, Zinchuk O, Okada T. 2007. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta Histochem. Cytochem. 40: 101– 111 [DOI] [PMC free article] [PubMed] [Google Scholar]