Abstract
Phosphorylation is one of the key mechanisms that regulate centrosome biogenesis, spindle assembly, and cell cycle progression. However, little is known about centrosome-specific phosphorylation sites and their functional relevance. Here, we identified phosphoproteins of intact Drosophila melanogaster centrosomes and found previously unknown phosphorylation sites in known and unexpected centrosomal components. We functionally characterized phosphoproteins and integrated them into regulatory signaling networks with the 3 important mitotic kinases, cdc2, polo, and aur, as well as the kinase CkIIβ. Using a combinatorial RNA interference (RNAi) strategy, we demonstrated novel functions for P granule, nuclear envelope (NE), and nuclear proteins in centrosome duplication, maturation, and separation. Peptide microarrays confirmed phosphorylation of identified residues by centrosome-associated kinases. For a subset of phosphoproteins, we identified previously unknown centrosome and/or spindle localization via expression of tagged fusion proteins in Drosophila SL2 cells. Among those was otefin (Ote), an NE protein that we found to localize to centrosomes. Furthermore, we provide evidence that it is phosphorylated in vitro at threonine 63 (T63) through Aurora-A kinase. We propose that phosphorylation of this site plays a dual role in controlling mitotic exit when phosphorylated while dephosphorylation promotes G2/M transition in Drosophila SL2 cells.
INTRODUCTION
In most eukaryotic cells, the centrosome is composed of a pair of centrioles surrounded by an amorphous protein matrix, the pericentriolar material (PCM). The PCM contains proteins required for microtubule nucleation, like γ-tubulin (γ-Tub) ring complex (γ-TuRC) components; anchoring proteins that bind to different enzymes and their targets; scaffolding proteins, which other complexes bind to; and regulatory kinases, phosphatases, and signaling molecules (15, 35). In proliferating cells, the centrosome is duplicated once per cell cycle such that at the onset of mitosis, a cell carries two centrosomes, serving as mitotic spindle poles.
Distinct steps of centrosome biogenesis occur in close coordination with cell cycle progression (60). Centrosome duplication is initiated at the G1/S transition and proceeds throughout S phase. At the G2/M transition, centrosomes recruit additional PCM components required for microtubule nucleation, a process termed maturation (52), and eventually separate and move to opposite poles of the mitotic spindle. Phosphorylation through protein kinases is one of the key mechanisms that control centrosome functions during the cell cycle. Examples are (i) cyclin-dependent kinase 1 (CDK1; Dmel/cdc2), which contributes to the separation of centrosomes in late G2 (11, 17); (ii) Polo-like kinase 1 (PLK1; Dmel/polo), which is involved in recruiting γ-TuRCs and is thus required for centrosome maturation (34); (iii) SAK/PLK4, another member of the polo kinase family, which is a major regulator of centriole duplication (29); and (iv) Aurora-A, which is involved in centrosome maturation (7). Furthermore, Aurora-A has been implicated in centrosome duplication and separation (4). Another kinase that might be involved in the regulation of centrosome functions is the ubiquitously expressed casein kinase II (CK2; Dmel/CkII), which is implicated in a variety of cellular processes, including cell cycle progression. It colocalizes with mitotic spindles and centrosomes in mammalian cells (25), and moreover, an RNA interference (RNAi) screen conducted in Drosophila melanogaster cells revealed a possible centrosome-related function, as knockdown of CkIIα or its regulatory subunit CkIIβ led to mild centrosome abnormalities (8). In addition to its main function as a microtubule organizing center (MTOC), the centrosome also contributes to cell cycle progression at the G1/S and G2/M transitions and is required for efficient asymmetric cell division and cytokinesis (9, 48). Centrosomes are, furthermore, involved in stress response pathways and cell cycle checkpoint control, and aberrant centrosome numbers cause genomic instability and consequently tumor formation (5). Our knowledge regarding the molecular composition of the centrosome has substantially increased during the past several years: direct proteomic analyses (33, 69), protein correlation profiling (2), genome-wide RNAi screens (20, 28), comparative genomics (37), and numerous studies of individual proteins have led to the identification of more than 300 candidate centriole and PCM components, many of which are highly conserved in different species. However, the molecular inventory of the centrosome and knowledge of the mechanisms controlling its functions are still incomplete. Therefore, a detailed exploration of the modulation of centrosomal proteins by kinases would further our understanding of the role of centrosomal phosphoproteins in the context of cellular signaling.
Several large-scale phosphoproteomic data sets have recently been published. Using Drosophila as a model system, Bodenmiller and colleagues (14) determined the phosphoproteome of Kc167 cells and Zhai et al. (70) identified phosphorylation sites derived from Drosophila embryos. In the present study, we identified phosphoproteins of the centrosome of Drosophila embryos. We functionally characterized the identified proteins for their role in centrosome replication and maturation, cell cycle regulation, and chromosome segregation. Using a combinatorial RNAi screening approach, we also identified functional interactions of these proteins with 4 selected kinases (polo, aur, cdc2, and CkIIβ) to integrate the centrosome phosphoproteins into signaling networks.
MATERIALS AND METHODS
Preparation of centrosome samples.
Centrosomes were isolated from Drosophila melanogaster preblastoderm-stage (0- to 3-h) embryos through two sucrose gradient centrifugations and subsequently affinity purified on magnetic beads (Dynabeads Protein G; Invitrogen) as described by Lehmann et al. (36) with the following modifications: all purification steps were carried out in buffers containing phosphatase inhibitors (1 mM sodium fluoride, 2 mM sodium orthovanadate, 25 mM β-glycerophosphate) in order to avoid dephosphorylation of centrosomal proteins during the procedure.
The enrichment of centrosome proteins during consecutive isolation steps was monitored by Western blotting with mouse monoclonal anti-γ-tubulin (anti-γ-Tub) antibody GTU-88 (Sigma) and rabbit polyclonal anti-dGrip84 WD (Y. Zheng, Carnegie Institution of Washington, Washington, DC). In parallel, immunofluorescence microscopy of centrosomes from all isolation steps using anti-γ-Tub antibody was performed as described before (36).
Immunopurified centrosomes were eluted from magnetic beads by treating the samples with 0.1% RapiGestSF protein solubilization reagent (Waters) for 30 min at 9°C while shaking them at 800 rpm in a Thermomixer. The elution buffer was prepared as follows. One vial of RapiGest (1 mg) was dissolved in 100 μl of 50 mM ammonium hydrogen carbonate, producing a 1% stock solution to which 900 μl of a 20 mM HEPES buffer, pH 7.5, containing 100 mM NaCl2, 10 mM EGTA, and 2 μM β-mercaptoethanol was added (final concentration of 0.1% RapiGest). The supernatant containing the RapiGest-eluted proteins was further processed for enrichment by titanium dioxide (TiO2) and subsequently analyzed by mass spectrometry (MS). Magnetic beads cross-linked with rabbit preimmune serum and incubated with centrosomes were treated as described above and served as a negative control in all experiments. The antibody-based purification of centrosomes has been carried out for several biological replicates.
Phosphopeptide enrichment and MS analysis.
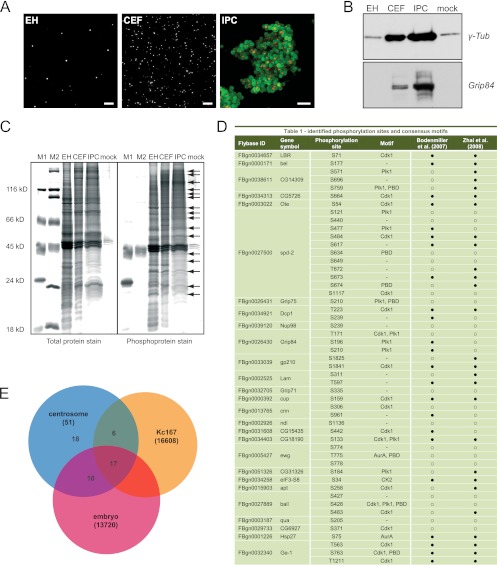
Following tryptic digestions of the RapiGestSF-eluted centrosome samples, phosphopeptides were enriched using a method based on TiO2 affinity (65). Liquid chromatography–matrix-assisted laser desorption ionization mass spectrometry (LC-MALDI MS) was performed using a 1100 Series Nanoflow LC system (Agilent Technologies) and an Ultraflex II LIFT MALDI-time of flight (TOF)/TOF mass spectrometer (Bruker Daltonics) as described previously (43). The tandem mass spectrometry (MS/MS) data were searched against the FlyBase sequence database (http://flybase.bio.indiana.edu/) using the Mascot software (Matrix Science, London, United Kingdom). The following settings were used for the searches: mass error tolerance for the precursor ions, 30 ppm; mass error tolerance for the fragment ions, 0.5 Da; fixed modification, carbamidomethylation; variable modification, methionine oxidation; number of missed cleavage sites, 1; type of instrument, MALDI-TOF-postsource decay (PSD). The Mascot ion score of 26, corresponding to a probability of P = 0.05, was used as a guideline for considering peptides identified or not; however, each matching spectrum was evaluated visually. Matches with Mascot ion scores of >26 were discarded if strong signals in the spectra were unassigned. Matches with ion scores of ≤26 were included if matched signals in the fragment ion spectrum followed expected sequence-specific fragmentation characteristics: predominant cleavage N terminally of proline, resulting in a corresponding strong b- or y-ion, or predominant cleavage C terminally of aspartic acid or glutamic acid, resulting in a corresponding strong y-ion for arginine-terminated peptides (62). A total of 51 utilized phosphorylation sites in 27 different proteins were identified (Fig. 1) (see also Table S1 in the supplemental material).
Fig 1.
Phosphoproteome analysis of the Drosophila centrosome. (A) Immunofluorescence microscopy with anti-cnn antibody shows enrichment of centrosomes (white) at different isolation stages: embryo homogenate (EH), centrosome-enriched fraction (CEF) after velocity sedimentation, and immunopurified centrosomes (red) (IPC) bound to magnetic beads (green). Bar, 10 μm. (B) Western blotting with antibodies against centrosomal proteins γ-Tub and Grip84 confirms enrichment of centrosomes throughout the isolation procedure. (C) To detect the abundance of phosphoproteins in centrosome samples, a negative control (mock) and two molecular mass standards containing two (M1) and one (M2) phosphorylated protein band, respectively, were separated on a polyacrylamide gel and stained with ProQ-Diamond phosphoprotein gel stain and SYPRO Ruby total protein gel stain. The total protein and the phosphoprotein patterns of the embryo homogenate significantly differed from the ones after centrosome enrichment and affinity purification. An enrichment of at least 20 phosphorylated protein bands (arrows) was observed in the immunopurified sample, with the most prominent band at around 130 kDa. (D) Identified phosphorylation sites and their respective consensus kinase motifs. A comparison with two whole-phosphoproteome analyses in Drosophila Kc167 cells and Drosophila early embryos is given in the last two columns, respectively. ● indicates that a site was identified and ○ indicates that a site was not identified in the respective study. (E) The Venn diagram illustrates the overlap of the 51 phosphorylation sites identified with our approach and the two whole-phosphoproteome studies conducted in Drosophila. The total number of identified sites in each study is given in parentheses.
In-gel detection of phosphorylated proteins.
Embryo homogenate (EH), centrosome-enriched fraction (CEF), and immunopurified centrosomes (IPCs) were incubated in sodium dodecyl sulfate (SDS) loading buffer for 10 min at 95°C. Protein samples were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Pro-Q Diamond phosphoprotein gel stain (Invitrogen), which recognizes phosphate groups attached to serine, threonine, and tyrosine residues, followed by SYPRO Ruby protein gel stain (Invitrogen), according to the manufacturer's instructions. Pro-Q Diamond- and SYPRO Ruby-stained proteins were detected at excitation wavelengths of 532 nm (green filter) and 473 nm (blue filter), respectively, through the Fuji FLA-5100 laser scanner. The images were further processed with the AIDA image analyzer software and subsequently annotated with Adobe Photoshop and Adobe Illustrator.
RNA interference.
Long double-stranded RNA (dsRNA) for RNAi experiments was generated by PCR amplifying ∼500-bp-long fragments using genomic DNA as the template. The amplicons contained T7 promoter sites and were amplified using primers designed with the E-RNAi tool from DKFZ (http://rnai.dkfz.de). In vitro transcription was performed using the T7 RiboMAX Express large-scale RNA production system (Promega) according to the manufacturer's instructions, followed by DNase I digestion and ethanol precipitation to purify the RNA. Both PCR-amplified DNA and precipitated dsRNA were subjected to gel electrophoresis and photometric measurements for quality control and quantification. Primer and amplicon sequence information are shown in Table S3 in the supplemental material. We investigated potential off-target effects both by bioinformatics analysis as calculated through the E-RNAi tool (see above) and by carrying out a second round of independent RNAi experiments. We calculated gene specificity, transcript specificity, and RNAi efficiency for all target genes listed in Table S3.
SL2 cells were cultured in Schneider's Drosophila medium (Invitrogen) with 10% fetal bovine serum (FBS; Invitrogen) at 25°C. dsRNA treatment was performed essentially according to the method of Clemens et al. (16) with the following modifications: 1 × 106 cells were incubated with 10 μg dsRNA for 1 h at room temperature in serum-free medium. After 72 h, a second dsRNA treatment was carried out to ensure optimal depletion of protein levels. In the case of simultaneous knockdowns, 10 μg of dsRNA targeting a kinase and 10 μg of dsRNA targeting a phosphoprotein were added to 1 × 106 cells and this treatment was repeated after 72 h. All knockdowns were carried out in biological replicates. After 4 days, cells were harvested and analyzed via automated immunofluorescence microscopy and fluorescence-activated cell sorting (FACS).
Automated immunofluorescence microscopy HCS.
Following RNAi treatment, cells were transferred to concanavalin A (ConA)-coated (Sigma) glass-bottomed 96-well plates (Greiner) and allowed to adhere for 1.5 h before fixation in 4% paraformaldehyde (Applichem). Cells were incubated with rabbit polyclonal anticentrosomin (anti-cnn) (Pineda) and mouse monoclonal anti-phospho-histone H3 (Abcam) followed by labeling with secondary antibodies and high-content screening (HCS) CellMask Blue cytoplasmic/nuclear stain (Invitrogen). Plates were automatically imaged with the ArrayScan VTI HCS reader (Thermo Scientific), using a 40×, 0.75-numerical-aperture (NA) PlanNeoFluar objective lens to obtain high-resolution images. Images were acquired in 3 channels (Hoechst stain, fluorescein isothiocyanate [FITC], and Texas Red) until 100 mitotic cells per well were obtained (depending on the mitotic index of the sample, approximately 500 fields/well). At least 200 mitotic cells per knockdown from two biological replicates were counted and analyzed. High-content analysis was performed using the Morphology Explorer Bioapplication. Cells were identified by segmenting the Hoechst channel based on size, shape, and fluorescence intensity of objects. Mitotic cells were selected in the FITC channel based on the intensity of nuclear anti-phospho-H3 staining. An average of n = 20,000 cells per knockdown was imaged to determine the mitotic index. Amounts and sizes of centrosomes were determined within each mitotic cell based on the anti-cnn fluorescent signal in the Texas Red channel. Algorithm parameters for object segmentation, object selection (gating), and segmentation of intracellular objects (spots) were manually optimized for each individual plate using control cells. Following scan completion, all relevant reported data, including percentage of selected objects/well (mitotic index), spot count, and mean spot area per selected object (number and size of centrosomes per mitotic cell), were exported to an Excel spreadsheet using the Cellomics vHCS View software and analyzed statistically.
Data analysis.
Mitotic indices in each well were automatically determined as the ratio of the total number of cells segmented in the 4′,6-diamidino-2-phenylindole (DAPI) channel to the number of phosphorylated histone H3-positive cells selected in the FITC channel. To obtain a robust estimate of the mitotic index per knockdown, the median mitotic index of the four corresponding replicate wells (2 biological by 2 technical replicates) was calculated. The corresponding (total) numbers of mitotic and nonmitotic cells were then used to compare each single knockdown to the enhanced green fluorescent protein (EGFP) control by means of Fisher's exact test. The resulting P values were adjusted for multiple testing using the Bonferroni-Holm method, and adjusted P values smaller than 0.05 were considered indicative for a significantly altered mitotic index. For the double knockdowns, Fisher's exact test was used to compare each knockdown to both the EGFP control and the single knockdown of the respective kinase. This step was used to identify enhanced or reduced penetrance of phenotypes. Here, the Bonferroni-Holm adjustment was carried out based on the combined set of the resulting P values to ensure a controlled family-wise error rate within each type of double knockdown, and as for the single knockdowns, a significance threshold of 0.05 was used.
Significant phenotypes regarding centrosome number were determined as follows. An average of 200 mitotic cells per knockdown was analyzed, and each cell was assigned to one of four categories: 0, 1, 2, or >2 spot counts/object (centrosomes/mitotic cell). The resulting vectors of membership counts of the two independent experiments per knockdown were averaged and compared to the class membership proportions of the controls (EGFP for single knockdowns and both EGFP and kinase for double knockdowns) by means of a chi-square test. The Bonferroni-Holm adjustment of the resulting P values was carried out as described above, and a significance threshold of 0.01 was used to determine knockdowns causing significantly altered class membership distributions. These knockdowns were assigned to a centrosome number phenotype depending on which category of cells was most increased compared to the control.
To score phenotypes regarding centrosome size, we first computed the mean spot area, μarea, and standard deviation, σarea, over all control cells. Based on these values, three size categories were set: large (spot area is bigger than μarea + σarea), small (spot area is smaller than μarea − σarea), and normal (spot area is within the range of μarea ± σarea). Each sample mitotic cell was then assigned to one of the categories, and significantly altered class membership distributions were determined as described above using a chi-square test. Knockdowns causing a significantly altered size distribution were assigned to large- or small-centrosome phenotypes.
For the identification of chromosome segregation defects, 100 mitotic cells per knockdown were visually inspected and the numbers of normal and misaligned chromosomes were recorded. Significant alignment phenotypes were determined using a chi-square test as described above.
Table S2 in the supplemental material lists all phenotypes identified in single and double knockdowns. Adjusted P values are listed in Table S3.
Cell cycle analysis.
For FACS analysis, SL2 cells were fixed in 70% ice-cold ethanol and stained with propidium iodide (50 μg/ml pancreatic RNase A [Sigma] and 50 μg/ml propidium iodide [Calbiochem] in phosphate-buffered saline [PBS]) for 1.5 h at 37°C. The DNA content of an average of 20,000 cells per knockdown (from two biological replicates) was quantified on a flow cytometer (FACSCalibur; BD) and analyzed using FlowJo software (Treestar). The cell populations were gated, and the proportions of cells that fall into the different cell cycle phases were recorded and averaged over the two replicates. To compare the cell cycle distribution of a knockdown to that of the control, z-scores for each cell cycle phase were calculated, which are given by the absolute difference of the average sample proportion and the average control proportion divided by the standard deviation of the control proportions for the respective phase. Cell cycle phenotypes were considered to be statistically significant when the z-score was found to be ≥3.8.
Generation of SL2 cell lines stably expressing fusion proteins.
For localization studies and immunoprecipitation experiments, we generated SL2 cell lines stably expressing proteins fused to a GFP, FLAG, or tandem affinity purification (TAP) tag. FLAG and TAP vectors carrying Ote, ball, and Dcp1 target genes were obtained from the BDGP Expression clone collection. All other expression vectors used in this study were generated by PCR amplifying the target genes using DGRC clones as the templates. The Gateway cloning technology (Invitrogen) was applied to shuttle genes into EGFP expression vectors pAGW and pAWG, which were obtained from the DGRC Drosophila Gateway vector collection, and into the TAP expression vectors pDEST NTAP and pDEST CTAP. Cells were cotransfected with vectors carrying the tagged target genes and pCoBlast (Invitrogen), a vector carrying a blasticidin resistance gene, by calcium phosphate transfection (Invitrogen) according to the manufacturer's instructions. After 3 weeks of selection for blasticidin-resistant cells, stable expression of target genes was confirmed by immunofluorescence microscopy and Western blotting with the corresponding tag-specific antibodies. Cloning primer sequences and plasmids used in this study are listed in Table S3 in the supplemental material.
Generation of Ote phosphomutant constructs.
Site-directed mutagenesis was performed by SINA Science Services GmbH to introduce the T63E and T63A mutations into the BDGP otefin (Ote) TAP vector using a 2-step PCR method and type IIs restriction enzymes according to the method of Engler et al. (23, 24). The mutations were confirmed by DNA sequencing.
Immunofluorescence microscopy of fixed SL2 cells.
SL2 cells were allowed to settle on ConA-coated coverslips and fixed with methanol for at least 5 min at −20°C. TAP fusion proteins were labeled with rabbit polyclonal anti-calmodulin binding site (CBS) (Eurogentec), FLAG fusion proteins with mouse monoclonal anti-FLAG M2 (Sigma), and EGFP fusion proteins with rabbit polyclonal anti-GFP antibody (Clontech). Lam and Ote were visualized with mouse monoclonal anti-ADL67.10 lamin Dm0 (Hybridoma Bank) and rabbit antiotefin (D. Chen, Beijing, China), respectively. Centrosomes were visualized with either mouse monoclonal anti-γ-Tub GTU-88 (Sigma) or rabbit anti-cnn. Microtubules were labeled with mouse monoclonal anti-α-Tub (Sigma). Following primary antibody labeling, cells were incubated with the appropriate fluorescence-conjugated secondary antibodies (Invitrogen) and DNA was labeled with DAPI. For image acquisition, we used an Axio Imager Z1 fluorescence microscope (Zeiss) with an MRM charge-coupled device (CCD) camera and AxioVision software. Image processing and annotation were done using the Adobe Photoshop and Adobe Illustrator software.
IP.
FLAG fusion proteins were immunoprecipitated from SL2 cell lysates using EZview Red anti-FLAG M2 affinity gel (Sigma-Aldrich). For aur immunoprecipitations (aur IPs), magnetic beads (Dynabeads Protein G; Invitrogen) were cross-linked with anti-aur antibody and incubated with embryo homogenate. Beads with bound protein were washed 5 times for 10 min, and elution was performed at 50°C with SDS-PAGE sample buffer lacking β-mercaptoethanol to avoid elution of the antibody. Immunoprecipitated complexes were resolved by SDS-PAGE followed by Western blotting and probed with antibodies against bait protein (anti-FLAG/anti-aur) and potential complex partners (anti-γ-Tub, α-Tub, aur [rabbit anti-Aurora-A {DM}; Knoblich], Lam, and Ote). Cell lysates from untransfected SL2 cells served as the negative control for FLAG IPs. Embryo homogenate incubated with beads that were cross-linked with rabbit preimmune serum served as a control for the aur IP.
Kinase profiling on peptide microarrays.
To determine whether MS-identified in vivo phosphorylation sites in the proteins Ote, spd-2, cnn, Grip75, Nup98, Grip84, Grip71, ewg, ball, qua, and CG6927 are targeted by the 4 kinases of interest, kinase profiling on customized peptide microarrays (JPT Peptide Technologies GmbH) was employed.
The microarray used in this study was composed of 124 peptides, of which 104 overlapping 15-amino-acid-long peptides covered the substrate protein Ote in an 11/4 format. Two negative-control peptides, in which serine was replaced by alanine, as well as four positive-control peptides for each kinase were present. Additionally, 14 15-mer peptides encompassing the newly identified phosphorylation sites in 10 different substrates were included. Peptides were synthesized on cellulose membranes using SPOT technology (55, 68), deposited onto activated glass slides, and covalently immobilized to the glass slide surface. Each peptide was present in triplicate on the chip, and the microarray was printed in three identical subarrays to enable intrachip reproducibility tests.
Microarrays were incubated with recombinant active kinases Aurora-A, PLK1, CDK1/cyclin B (Invitrogen), and CK2 holoenzyme (Biaffin GmbH) in the presence of radioisotopically labeled ATP (Hartmann Analytic GmbH) in assay buffer (for Aurora-A, 20 mM Tris, pH 7.5, 2 mM dithiothreitol [DTT], 0.5 mM EDTA, 0.01% Triton X-100, 5% glycerol; for PLK1, 20 mM Tris, pH 7.5, 1 mM DTT, 5% glycerol, 1 mg/ml bovine serum albumin [BSA]; for CDK1, 20 mM Tris, pH 7.5, 5% glycerol, 0.1 mg/ml BSA, 1 mM DTT, 0.05% Triton X-100; for CK2, 50 mM Tris, pH 7.5, 10 mM NaCl2, 10 mM MgCl2, 10 mM DTT) for 3 h at 37°C. Subsequent to incubation with kinase–[γ-33P]ATP solution, the microarrays were washed three times with 2% phosphoric acid followed by intense washing steps with dimethylformamide (DMF), dichloromethane (DCM), and double-distilled (dd) water. Finally, the microarrays were washed with methanol and dried using a nitrogen stream. Phosphorylated peptides were detected following phosphorimaging on a Fuji FLA 3000 scanner. SPOT recognition software ArrayPro 4.0 (Media Cybernetics) was used for data analysis. The mean of signal intensities for pixels around recognized spots (background) was subtracted from the mean of signal intensities for pixels within recognized spots (signal), resulting in corrected median values (signal minus background). Mean values of corrected means of signal intensities from 3 identical subarrays on each microarray image were used for data evaluation. Peptide sequences and measured mean signal intensities for each kinase are listed in Table S3 in the supplemental material.
In vitro Aurora-A phosphorylation of Ote and MS identification of phosphorylated residues.
His-tagged Ote was purified from Escherichia coli using magnetic nickel-nitrilotriacetic acid (Ni-NTA) beads (Sigma). To confirm phosphorylation of Ote by Aurora-A, an in vitro kinase assay and subsequent detection of phosphoresidues by MS were conducted. After washing in assay buffer (25 mM Tris, pH 7.5, 10 mM MgCl2, 0.5 mM EGTA, 0.5 mM Na3VO4, 5 mM β-glycerophosphate, 2.5 mM DTT, 0.01% Triton X-100, 100 μM ATP), His-tagged Ote bound to beads was incubated with Aurora-A kinase (Invitrogen) for 1 h at 30°C, boiled in SDS-PAGE sample buffer for 10 min, and separated by SDS-PAGE. The negative control was treated identically without addition of Aurora-A kinase. The resulting bands covering the respective mass range of Ote (45 kDa) in the Aurora-A-treated sample and the control sample were excised, reduced, alkylated, in-gel trypsin digested as described earlier (13), and subjected to MS. LC-MS/MS experiments were performed using a nanoAcquity ultrahigh-pressure liquid chromatography (UPLC; Waters Corp., Milford, MA) system and an LTQ Orbitrap Velos hybrid ion trap mass spectrometer (Thermo Scientific). Separation of peptides was performed by reverse-phase chromatography using a Waters reverse-phase nanocolumn (BEH C18, 75-μm inside diameter [i.d.] by 250 mm, 1.7-μm particle size) at a flow rate of 300 nl/min. Peptides were initially loaded onto a precolumn (Waters UPLC Trap Symmetry C18; 180-μm i.d. by 20 mm, 5-μm particle size) from the nanoAcquity sample manager with 0.1% formic acid for 3 min at a flow rate of 10 μl/min. After this period, the column valve was switched to allow the elution of peptides from the precolumn onto the analytical column. Solvent A was water plus 0.1% formic acid, and solvent B was acetonitrile plus 0.1% formic acid. The linear gradient employed was 5 to 50% solvent B in 60 min. The LC eluant was sprayed into the mass spectrometer by means of a New Objective nanospray source. All m/z values of eluting ions were measured in the Orbitrap mass analyzer, set at a resolution of 30,000. Data-dependent scans (top 20) were employed to automatically isolate and generate fragment ions by collision-induced dissociation in the linear ion trap, resulting in the generation of MS/MS spectra. Ions with charge states of 2+ and above were selected for fragmentation. Postrun, the data were processed using Protein Discoverer (version 1.2; Thermo Scientific). Briefly, all MS/MS data were converted to mgf files and these files were then submitted to the Mascot search algorithm (Matrix Science) and searched against a custom Drosophila melanogaster otefin database, using a fixed modification of carbamidomethyl (C) and variable modifications of oxidation (M) and phosphorylation (S, T, and Y).
RESULTS
Identification of centrosomal phosphoproteins.
We isolated centrosomes from preblastoderm Drosophila embryos (45) and affinity purified them with an antibody to centrosomin (cnn), one of the core components of the pericentriolar material, followed by enrichment of phosphopeptides to identify substrates of regulatory kinases and phosphatases that are critical for centrosome maturation, duplication, and separation. Using this system as starting material has several advantages: multigram quantities of embryos can be obtained in a few hours; during rapid nuclear divisions before cellularization, centrosomes are less likely to be attached to the nuclear membrane; and furthermore, the highly mitotic embryos contain centrosomes with large amounts of PCM compared to mammalian centrosomes that are primarily derived from interphase cells. The enrichment of centrosomes in consecutive isolation stages was validated by immunofluorescence microscopy (Fig. 1A) and immunoblotting with antibodies against centrosomal proteins Grip84 and γ-Tub (Fig. 1B). To minimize dephosphorylation during the isolation procedure, all steps were carried out in the presence of phosphatase inhibitors. Immunopurified centrosome samples showed a high abundance of phosphorylated proteins determined by in-gel labeling of phosphoserine, -threonine, and -tyrosine residues. We detected at least 20 phosphorylated protein bands that were enriched in the immunopurified fraction compared to the embryo homogenate (Fig. 1C). The immunopurified fraction was subjected to tryptic digestion followed by enrichment of phosphopeptides by titanium dioxide (TiO2) affinity chromatography (65). The subsequent LC-MALDI MS analysis resulted in the identification of 51 phosphorylation sites in 27 proteins, shown in Fig. 1D. Among the identified proteins, we found known PCM components such as cnn; the γ-TuRC proteins Grip71, Grip75, and Grip84; and the centriolar protein spd-2, validating the enrichment of centrosomes in our samples. We also identified a number of proteins associated with the nuclear envelope (NE) (LBR, Ote, Nup98, gp210, Lam, and cup) and nuclear proteins (ewg, apt, and ball). Ote, Lam, and ball have previously been shown to localize to the mitotic spindle and/or function in mitotic spindle organization (Uniprot annotation [18, 28]). Additionally, we found proteins involved in translational initiation (eIF3-S8), RNA-mediated gene silencing (P granule proteins Dcp1 and bel), and stress response (Hsp27), as well as proteins of unknown function (CG15435, CG14309, CG5726, CG15435, CG18190, and CG6927). Out of the 27 proteins, 17 were also identified in a previous global proteome analysis of the Drosophila centrosome (47). Hence, the identification of 10 new centrosome candidates in this study was possible because selective enrichment facilitates detection of low-abundance proteins in complex mixtures. With the exception of ewg, all proteins identified in this study have previously been shown to be phosphorylated at different residues in two large-scale phosphoproteome studies conducted in Drosophila embryos (70) and Kc167 cells (14), respectively. However, we identified 18 previously unknown phosphorylated residues (Fig. 1E). For example, we found 11 phosphorylation sites in the centriolar protein spd-2, 5 of which were identified neither in the whole embryo nor in the Drosophila cell phosphoproteome analysis. We manually searched our data set for consensus motifs for the mitotic kinases Cdk1, Plk1, Aurora-A, and CK2 and the polo box domain (PBD) binding motif (see Table S1 in the supplemental material) and found that more than half of the identified sites are predicted to be recognized by these centrosome-associated kinases.
Phosphorylated proteins function in pathways regulating the centrosome cycle, cell division, and chromosome segregation.
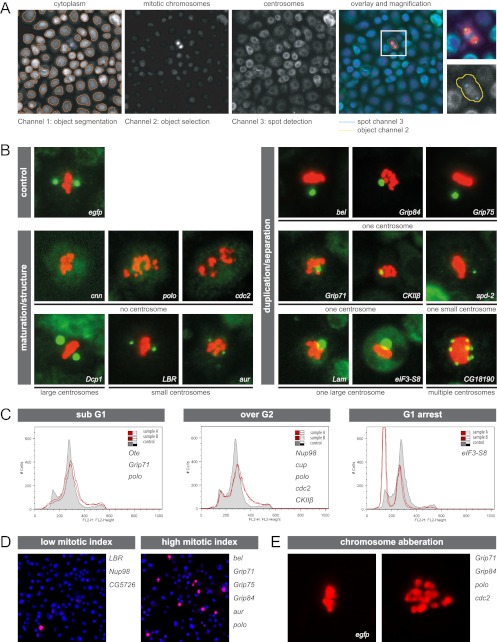
To investigate the function of MS-identified phosphoproteins, we conducted a cell-based RNAi screen followed by automated high-content immunofluorescence microscopy and FACS analysis. We first analyzed the effects on centrosome structure/maturation, centrosome duplication/separation, and chromosome segregation as well as cell cycle progression upon downregulation of 25 phosphoproteins and the 4 kinases polo, aur, cdc2, and CkIIβ. In order to minimize potential off-target effects, we repeated, in addition to bioinformatics analysis, the knockdown of the new centrosomal candidates that developed a centrosome- or cell cycle-dependent phenotype upon depletion using alternative dsRNAs (see Table S3 in the supplemental material). We developed a combinatorial strategy, in which we incubated cells with dsRNA targeting one of the 4 kinases in parallel with dsRNA of the phosphoprotein set to reveal functional interdependencies. We chose the 4 cell cycle and centrosome cycle regulatory kinases based on the assumption that the identified phosphoproteins are probable substrates. RNAi-mediated phenotypes were analyzed with respect to the effect on number and size of centrosomes in mitotic cells, mitotic index, and mitotic chromosome alignment. By using a microscopy system for automated image capture, processing, and analysis, we could efficiently screen large numbers of cells (Fig. 2A). In the initial single knockdown experiments, we found that depletion of 15 out of 29 proteins produced centrosome number and/or size aberrations, depletion of 9 proteins led to a significantly altered number of mitotic cells, and depletion of 4 proteins induced chromosome missegregation. The following 7 different centrosome defects were observed and subsequently classified into two main categories: centrosome maturation/structure and centrosome duplication/separation (Fig. 2B). (i) An increased proportion of mitotic cells without centrosomes occurred after downregulation of cnn, polo, and cdc2, confirming the well-established function of cnn and polo in centrosome maturation and structure maintenance (40, 63). (ii) Downregulation of aur and LBR induced cells with 2 small centrosomes. (iii) Dcp1 induced cells with 2 large centrosomes. The latter two centrosome size aberrations were also classified as centrosome maturation defects, although we cannot rule out the possibility that large centrosomes are a consequence of centriole overduplication. While aur's role in the regulation of maturation has been described before (7), it has not previously been shown that LBR and Dcp1 are implicated in this process. (iv to vi) Inhibition of 8 out of 29 proteins led to an increased number of mitotic cells with only one centrosome, which appeared either normal (iv), large (v), or small (vi). The last three phenotypes indicate a role for the proteins in centrosome duplication and separation, respectively. These results support previous findings for the γ-TuRC components Grip71, Grip75, and Grip84 as well as spd-2 and Lam (28). bel, eIF3-S8, and the kinase CkIIβ had not previously been shown to have functions in centrosome duplication or separation. (vii) Supernumerary centrosomes were observed after depletion of CG18190, an uncharacterized protein that has recently been identified as a component of the microtubule-associated complex (30). Knockdown of 9 proteins induced significant deviations in the proportion of mitotic cells, indicating a regulatory role for these proteins in progression through the cell cycle (Fig. 2D). Low mitotic indices were caused by downregulating LBR, Nup98, and CG5726, while depletion of Grip71, Grip75, Grip84, polo, and aur induced an accumulation of cells in mitosis, which was expected as these proteins are implicated in the regulation of mitotic progression (8, 10, 46, 67). Additionally, a function of phosphoprotein bel in cell cycle regulation was identified, as inhibition also resulted in an increase of mitotic cells. Defects regarding chromosome segregation were observed after depleting Grip71, Grip84, polo, and cdc2, confirming previous results (28, 46, 61). To further dissect specific cell cycle stage distributions following dsRNA treatment, we carried out FACS analysis and found that a fraction of the analyzed proteins (5/29) function in cytokinesis, as their depletion led to accumulation of cells with more than 2N DNA content (Fig. 2C). Depletion of eIF3-S8 resulted in G1-phase arrest, indicating that loss of this translation initiation factor affects G1-to-S transition. Taken together, our RNAi analysis revealed previously unknown functions for the phosphoproteins bel, eIF3-S8, LBR, Dcp1, and CG18190 and the kinase CkIIβ in centrosome cycle regulatory pathways.
Fig 2.
Functional characterization of phosphoproteins and regulatory kinases by RNAi, automated immunofluorescence microscopy, and FACS. (A) The image processing and segmentation approach, which was applied to assign identified phosphoproteins and kinases to regulatory pathways, is illustrated. The effects on number and size of mitotic centrosomes (labeled with anti-cnn, green) in RNAi-treated SL2 cells were analyzed using an algorithm that reports number, intensity, and morphology of intracellular objects. Cell cycle effects were monitored via calculating the ratio of total number of cells (labeled with cytoplasmic stain, blue) segmented in channel 1 to the number of mitotic cells (labeled with anti-phospho-histone H3, red) selected in channel 2. (B) Examples of mitotic cells (centrosomes in green and chromosomes in red) reflecting the 7 aberrant centrosome phenotypes observed after RNAi-mediated protein depletion: one centrosome, one large centrosome, one small centrosome, multiple centrosomes (duplication/separation); no centrosome (structure maintenance); and small and large centrosomes (maturation). Knockdown of EGFP served as a negative control. The corresponding RNAi target proteins are indicated within each image. (C) FACS analysis of SL2 cells incubated with dsRNA reveals genes involved in the regulation of cell cycle progression. Three types of aberrant cell cycle distribution profiles were identified: increased sub-G1 DNA content, increased number of polyploid cells, and accumulation of cells in G0/G1 phase. Each profile shown contains a control histogram (gray; cells treated with dsRNA targeting EGFP) and an aberrant histogram representative for its phenotypic class (red line; names of all target genes exhibiting similar profiles upon depletion are given in the corresponding panel). (D) Two fluorescence microscopy images (superimposition of DAPI [blue] and mitotic chromosomes [red]) are shown representative of cells displaying low or high mitotic indices after dsRNA treatment. Proteins whose depletion induced an aberrant proportion of mitotic cells are given on the right of the corresponding image. (E) Cells were manually scored for chromosome segregation defects after depletion of target proteins. A control cell treated with EGFP dsRNA with normal chromosome (red) alignment in metaphase and an example of abnormally distributed mitotic chromosomes are shown. The proteins inducing this phenotype are given on the right side of the panel.
RNAi screening in kinase-depleted backgrounds identifies functions of phosphoproteins in relation to polo, aur, cdc2, and CkIIβ.
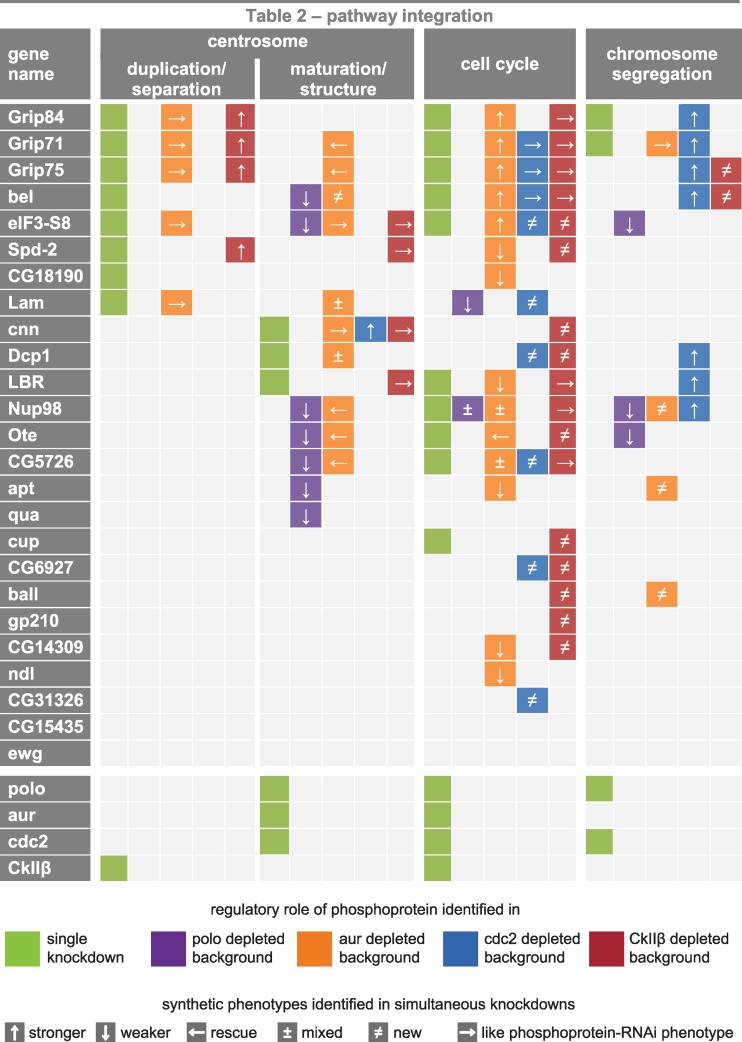
To gain further insight into the relationships between kinases and potential kinase substrates and to identify functionally redundant genes, we used RNAi to inhibit two genes simultaneously in 100 combinations. Twenty-five phosphoproteins were downregulated in 4 different kinase-depleted backgrounds, and synthetic phenotypes were analyzed again with respect to centrosome number and size, mitotic index, and chromosome aberrations (Fig. 3). By suppressing gene activity in the 4 sensitized backgrounds, we integrated 14 out of 25 analyzed phosphoproteins into signaling networks controlling centrosome maturation/structure while the single knockdown approach identified only 3 proteins as being implicated in these pathways. Phenotype analysis of double knockdowns revealed functional implications of 22/25 phosphoproteins in pathways regulating progression through the cell cycle, 12 of which were not detectable upon depletion of the phosphoprotein alone. Surprisingly, 11 phosphoproteins were shown to be integrated in chromosome segregation pathways. Most of these were identified in the cdc2-depleted background. In contrast, only two phosphoproteins (Grip71 and Grip84) were identified to play a role in chromosome segregation in the single knockdown experiments.
Fig 3.
Integration of phosphoproteins in pathways regulating the centrosome cycle, cell cycle, and chromosome segregation. Functional implications of 25 phosphoproteins and 4 kinases (bottom rows) in signaling pathways controlling centrosome duplication/segregation and centrosome maturation/structure as well as cell cycle progression and chromosome segregation were identified by RNAi phenotype scoring in SL2 cells. Protein functions identified in single knockdown experiments are depicted in olive-colored panels. Protein functions identified by the occurrence of synthetic phenotypes resulting from simultaneous downregulation of a phosphoprotein and any of the 4 kinases are color coded as follows: polo in purple, aur in orange, cdc2 in blue, and CkIIβ in red. ↑ and ↓ indicate that simultaneous depletion of the phosphoprotein leads to increased or decreased strength of the kinase RNAi phenotype, respectively. ← indicates that the phenotype observed after kinase inhibition is reverted when additionally depleting a phosphoprotein. ± indicates a mixed phenotype after simultaneous knockdown compared to single knockdown phenotypes of the corresponding phosphoprotein and kinase. ≠ indicates that a phenotype is induced only by simultaneous downregulation of phosphoprotein and kinase while depletion of one component does not lead to aberrations in the respective category. → indicates a dominant phosphoprotein RNAi phenotype.
By analyzing the deviation of synthetic phenotypes from single knockdown phenotypes of the 4 kinases, we determined functional relationships of phosphoproteins with aur, cdc2, polo, and CkIIβ. Probable regulatory mechanisms were assigned to each of the 5 types of deviated synthetic phenotypes that we found in this study. (i) For instance, a weaker double knockdown phenotype suggests negative regulation of the phosphoprotein Nup98 downstream of the kinase polo or in an independent parallel pathway that is required for the maintenance of centrosome structure. (ii) Other examples are aur and Dcp1, which differentially regulate centrosome maturation; hence, opposite effects on centrosome size were observed in single knockdowns, which compensate each other when the two proteins are inhibited simultaneously. (iii) ball is redundantly implicated in the regulation of chromosome segregation, since silencing in an aur-depleted background results in a significantly increased number of cells with chromosome aberrations while single knockdown of ball has no apparent effect on chromosome segregation. (iv) A rescue phenotype indicates that kinase and phosphoprotein function in the same pathway in a mutually dependent mechanism. For instance, negative regulation of CG5726 downstream of aur could be required for effective centrosome maturation. Another possibility is that these genes function in independent pathways. (v) A phosphoprotein is likely to be implicated in a process independent or upstream of the kinase when the phenotype of the phosphoprotein RNAi is dominant over the kinase RNAi phenotype in the simultaneous knockdown of the two. For example, Grip84 is most probably required for centrosome duplication/separation through a mechanism that is independent of aur activity. A summary of all identified phosphoprotein functions arising from combinatorial RNAi screening analysis and functional relations between kinases and phosphoproteins is shown in Fig. 3. In conclusion, our data demonstrate that virtually all identified centrosomal phosphoproteins are implicated in signaling pathways related to centrosome biology or cell cycle regulation, either directly or through functional interaction with the relevant kinases.
Phosphorylated proteins with centrosome-related functions localize to mitotic spindles and centrosomes in SL2 cells.
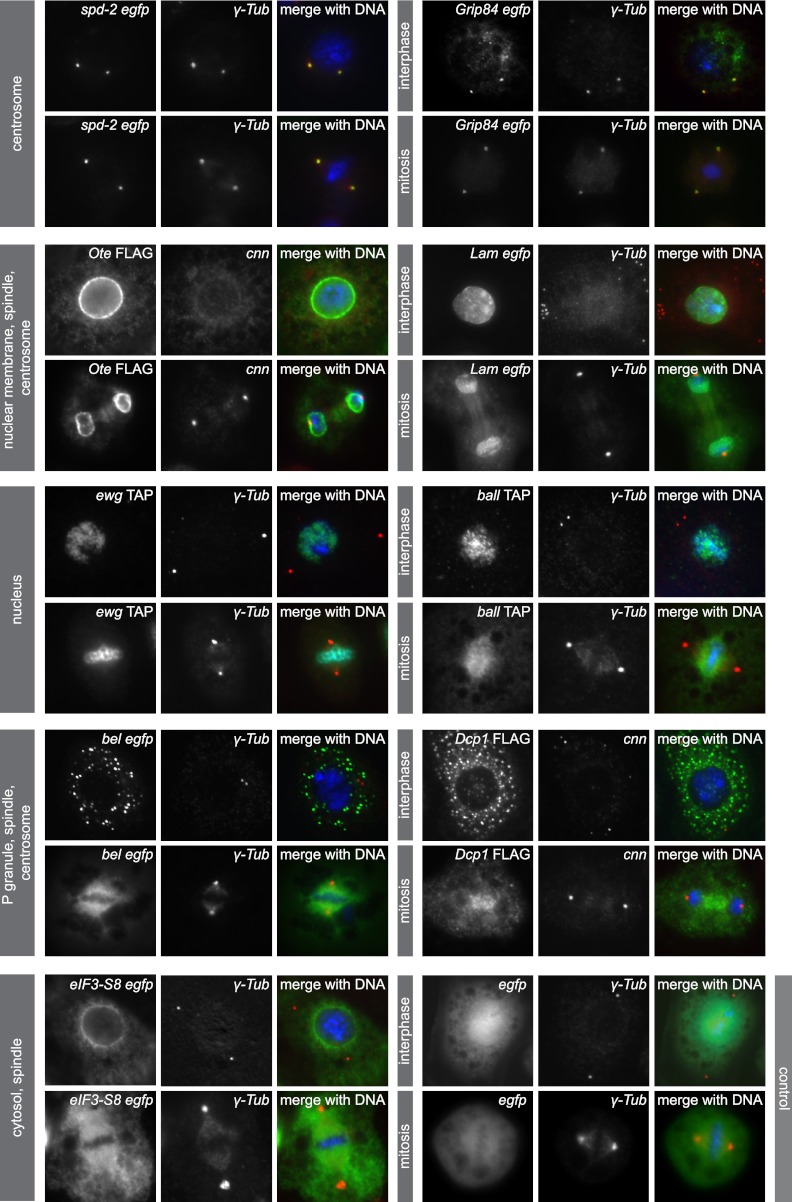
To determine the subcellular localization of phosphoproteins, we generated SL2 cell lines stably expressing GFP, TAP, or FLAG fusion proteins and monitored their expression throughout the cell cycle. We chose 9 proteins from 5 different subcellular compartments based on their previously annotated localizations: centrosome, P granule, nuclear membrane, nucleus, and cytosol (Fig. 4). As expected, GFP-tagged spd-2 and Grip84 were found to localize to centrosomes throughout the cell cycle, validating the specificity of the tagging and overexpression approach. Phosphoproteins Ote and Lam fused to a FLAG and GFP tag, respectively, were primarily associated with the NE but also colocalized with centrosomes and the spindle from metaphase until anaphase. FLAG-tagged Ote was additionally associated with the midbody in telophase and localized to interphase centrosomes (Fig. 5B). Transcriptional regulator ewg fused to a TAP tag localized exclusively to chromatin and neither to spindles nor to centrosomes, consistent with our RNAi data that revealed no centrosome-related function of this protein in any of the sensitized backgrounds. Another TAP fusion protein, ball, which is a nucleosomal histone kinase (1), localized to chromatin, but its expression also partially overlapped with the mitotic spindle, supporting previous findings that this kinase is implicated in sister chromatid segregation and mitotic spindle organization (18). bel and Dcp1, two proteins implicated in RNA-mediated gene silencing, localized to cytoplasmic P granules in interphase cells, as was expected. Interestingly, in mitosis, GFP-bel colocalized with centrosomes and the mitotic spindle in agreement with our functional data, demonstrating this protein to be required for centrosome duplication/separation. Dcp1, a protein which was identified as a negative regulator of centrosome maturation in our study, also associated with metaphase and anaphase spindles when expressed as a FLAG fusion protein in SL2 cells. The expression of the translation initiation factor complex component eIF3-S8 was restricted to the cytosol in interphase and concentrated adjacent to the nuclear membrane. In mitosis, the peak fraction of GFP–eIF3-S8 was localized to the mitotic spindle. Our overexpression studies confirmed known subcellular locations of 9 centrosome candidate proteins and revealed previously unknown associations with mitotic spindles for 3 proteins (bel, Dcp1, and eIF3-S8). Moreover, we describe for the first time a colocalization with Drosophila centrosomes for the phosphoproteins bel and Ote. These findings are consistent with the functional analysis of the candidate centrosome proteins, which identified 7 of the 9 overexpressed fusion proteins to be implicated in centrosome cycle-associated signaling pathways.
Fig 4.
Expression of MS-identified phosphoproteins in SL2 cells confirms known localizations and identifies new spindle and centrosome localizations. Immunofluorescence microscopy of SL2 cells expressing GFP, FLAG, or TAP fusion proteins labeled with anti-GFP, -FLAG, or -CBS (green), a centrosome marker (anti-γ-Tub or anti-cnn, red), and DAPI (blue) confirms known localization to the centrosome (spd-2 and Grip84), the nuclear membrane (Ote and Lam), the nucleus/chromatin (ewg and ball), P granules (bel and Dcp1), and the cytosol (eIF3-S8). GFP control cells show uniform distribution of the tag. Localization was monitored throughout the cell cycle, and representative images for interphase and mitotic localization of each fusion protein are shown. Previously unknown localization to the anaphase spindle was identified for Ote, Lam, and Dcp1. eIF3-S8 and bel localize to the metaphase spindle, and additionally, bel and Ote colocalize with centrosomes.
Fig 5.
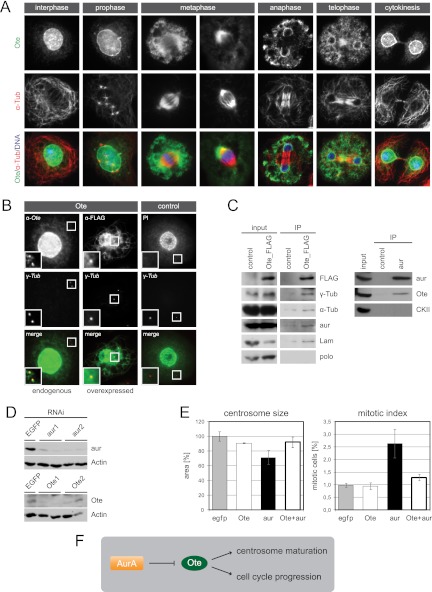
Nuclear membrane protein Ote localizes to interphase centrosomes and functions downstream of aur in centrosome maturation. (A) Localization of endogenous Ote is shown throughout the cell cycle. SL2 cells were labeled with anti-Ote (green) and anti-α-Tub (red). Superimposition with DAPI in blue. (B) Endogenous Ote and overexpressed FLAG-tagged Ote localize to centrosomes in interphase. A fluorescence microscopy image of a cell labeled with rabbit anti-Ote (green) and anti-γ-Tub (red) is shown in the left panel. Cells stably expressing Ote fused to a FLAG tag were labeled with rabbit anti-FLAG (green) and anti-γ-Tub (red) (middle panel). The right panel shows a cell expressing TAP-tagged nuclear protein scra as a negative control, in which the tag was labeled with rabbit immunoglobulin (IgG, green) and the centrosome was labeled with anti-γ-Tub (red). Superimpositions of the two channels are shown in the bottom row, and magnifications of the area around the interphase centrosomes are given in each image. Fluorescent images in the right panel serve as a control for the specificity of the anti-Ote antibody and the specificity of fusion protein localization. Brightness and contrast of endogenous Ote staining were enhanced 3-fold in the magnified section to clarify localization of a minor portion of the protein to the centrosome while the majority of the protein localizes to the nuclear membrane. (C) Extracts from SL2 cells expressing FLAG-tagged Ote were analyzed by immunoprecipitation with anti-FLAG antibody. Interacting proteins were determined by Western blotting with antibodies to α-Tub, γ-Tub, aur, Lam, and polo. The known interaction of Lam and Ote was verified, and additionally, α-Tub, γ-Tub, and aur were found to copurify with FLAG-Ote while polo is not present in the purified complex. Immunoprecipitation of endogenous aur from Drosophila embryo homogenate verifies the interaction with Ote, which was detected in the aur precipitate but not in the control IP. Nontransfected cells served as control for the FLAG IP. (D) Western blots illustrating the efficiency of RNAi experiments. SL2 cells treated with two independent dsRNAs for each target gene show strongly decreased levels of aur and Ote, respectively, compared to control cells treated with dsRNA targeting EGFP. Actin is shown as a loading control. (E) Ote functionally interacts with aur. Phenotypes of single and double knockdowns regarding centrosome size and mitotic index are shown in the graphs. EGFP RNAi served as control. (F) The schematic illustrates the possible regulatory mechanism. Negative regulation of Ote by the kinase aur is indicated by ⊣.
Dynamic localization of NE phosphoprotein Ote throughout the cell cycle.
To shed more light on the role of NE proteins regarding centrosome regulatory pathways, we followed endogenous localization of the protein Ote in SL2 cells at different stages of the cell cycle (Fig. 5A). As expected, interphase cells labeled with an anti-Ote specific antibody exhibited prominent staining of the nuclear periphery. At the onset of mitosis prior to nuclear envelope breakdown (NEBD), we observed invaginations in the nuclear membrane in close proximity to centrosomes. In Drosophila early embryos, complete nuclear lamina breakdown does not occur until after metaphase (51). Instead, Lam persists in an envelope-like structure enclosing the entire mitotic apparatus, including centrosomes. Ote has also been proposed to be a component of this spindle envelope (31). Likewise, ultrastructural studies in cultured Drosophila cells have revealed the persistence of nuclear membranes enveloping the mitotic spindle until anaphase (19, 41). In support of these findings, we observed such a structure in fixed SL2 cells when following Lam distribution at prometaphase until metaphase (data not shown), but this was not seen in cells labeled with anti-Ote, suggesting that Ote leaves the membrane before complete breakdown. Following NEBD, Ote is dispersed in the cytoplasm and concentrated at spindle poles. In anaphase, when the NE starts to reassemble, Ote is recruited to chromosomes. Interestingly, we found that Ote accumulates first at regions adjacent to centrosomes and at peripheral sites of chromosomes but is excluded from the region where midspindle microtubules attach. Only later in telophase, Ote forms a continuous rim around chromatin. At the same time, the signal becomes stronger in the midspindle area. During cytokinesis, Ote returns to the nuclear periphery, but a minor fraction of the protein remains dispersed in the cytoplasm and is also found at the midbody. Despite the colocalization of Ote with spindle poles during metaphase, we also found that a minor portion of the protein localizes to interphase centrosomes, as was demonstrated by immunofluorescence microscopy with antibodies detecting the endogenous protein and the overexpressed FLAG fusion protein. Labeling at the centrosome was specific, as demonstrated by several control experiments (Fig. 5B).
Ote is a component of centrosomes and functionally interacts with aur in maturation.
We conducted immunoprecipitation experiments on FLAG-tagged Ote with the aim of confirming suspected interaction partners from our RNAi analysis results. We found that γ-Tub copurified with Ote (Fig. 5C), supporting the hypothesis that Ote is not only localized to the inner nuclear membrane but indeed a genuine component of Drosophila centrosomes. We also confirmed the well-established binding of Lam and Ote in these experiments (27). Furthermore, Ote coprecipitates with α-Tub. Although it had previously been shown that the protein localizes to or around the mitotic spindle, we hereby for the first time provide biochemical evidence for an association with microtubules. Lastly, hints of a functional interaction between the centrosome-related kinase aur and Ote coming from our RNAi analysis were supported by the fact that aur was copurified in Ote pulldowns. This interaction could be confirmed via reverse IP, in which we purified endogenous aur from embryo homogenate and detected Ote as a binding partner (Fig. 5C). Based on the phenotype analysis of single and double knockdowns of aur and Ote, we had discovered that the two proteins functionally interact in centrosome maturation as well as cell cycle regulation. Knockdown efficiency was validated by Western blotting showing that Ote and aur protein levels are significantly reduced after treatment with two independent dsRNAs for each gene (Fig. 5D). While inhibition of aur induces mitotic cells with small centrosomes, this phenotype is rescued when simultaneously depleting Ote. Similarly, loss of aur function induces accumulation of cells in mitosis, yet additional removal of Ote leads to populations of cells with mitotic indices close to control level (Fig. 5E). Taken together, these data led us to propose a model in which negative regulation of Ote downstream of aur is required for obtaining mature centrosomes and for progression through mitosis (Fig. 5F).
Ote is an in vitro Aurora-A substrate.
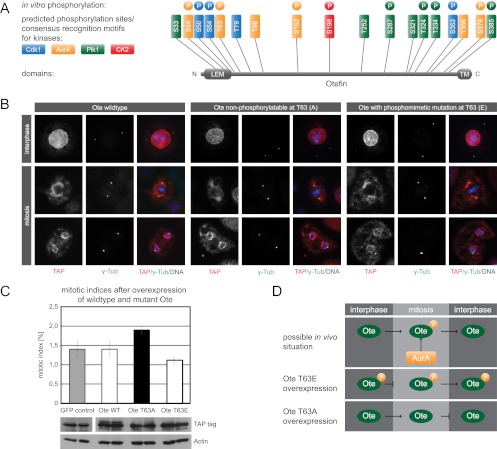
To elucidate the nature of the interaction between aur and Ote, we first wanted to test whether Ote is a substrate of Aurora-A kinase. In silico analysis predicted 6 phosphorylation residues conforming to the Aurora-A consensus motif. In addition, we found 7 Plk1 and 4 Cdk1 consensus sites and one CK2 consensus site within the Ote coding sequence (Fig. 6A). We employed peptide microarrays to investigate whether these sites are phosphorylated by the respective kinases in vitro. Peptides encompassing residues S44, T63, S152, and S378 incorporated radioisotopically labeled phosphate after incubation with recombinant active Aurora-A kinase, indicating that Ote is a substrate for aur. Furthermore, 4 out of 7 potential Plk1 sites were phosphorylated upon incubation with Plk1. CK2 phosphorylated only peptides containing S198, the single consensus site present in the Ote sequence. Serine 54 had been identified as an in vivo phosphorylation site in our and other phosphoproteomic studies (14, 70) and has previously been shown to be phosphorylated by Cdk1 in vitro (3). In our peptide microarray analysis, S54 was phosphorylated by Cdk1, supporting these results. Additionally, S50 and S352 were shown to be phosphorylated by Cdk1 (Fig. 6A; see also Table S4 in the supplemental material). To further confirm a direct phosphorylation of Ote by Aurora-A, we conducted an in vitro kinase assay with recombinant His-tagged Ote purified from E. coli. Subsequent LC-MS/MS analysis confirmed phosphorylation of Ote at threonine 63 by Aurora-A (see Table S4c).
Fig 6.
Phosphorylation of Ote at threonine 63 is critical for cell cycle progression. (A) Schematic drawing of Ote indicating the N-terminal LEM domain, the C-terminal transmembrane domain (TM), and all potential phosphorylatable residues conforming to the consensus motifs of Cdk1 (blue), Aurora-A (orange), Plk1 (green), and CK2 (red). Colored circles designate sites that were phosphorylated in vitro on peptide microarrays by the 4 respective recombinant kinases. (B) Immunofluorescence microscopy images showing SL2 cells stably expressing TAP-tagged wild-type Ote, a nonphosphorylatable mutant (Ote T63A), and a phosphomimetic mutant (Ote T63E) in interphase and mitosis. Ote fusion protein localization was visualized with anti-CBS (red), centrosomes were visualized with anti-γ-Tub (green), and DNA was visualized with DAPI (blue). (C) Effects on cell cycle progression in SL2 cells overexpressing wild-type and mutant Ote are shown in the graph. While wild-type Ote overexpression has no effect on the mitotic index, overexpression of the nonphosphorylatable T63A mutant leads to an increase and overexpression of the phosphomimetic T63E mutant leads to a decrease of the proportion of mitotic cells, respectively, compared to control cells overexpressing TAP-tagged EGFP. Equal expression levels of wild-type Ote, the T63A mutant, and the T63E mutant are demonstrated by Western blotting. (D) Hypothetical model demonstrating the effect of Ote phosphorylation by Aurora-A on cell cycle progression in vivo, which is derived from the results of phosphomutant studies depicted in the bottom panels.
Phosphorylation of Ote at T63 is critical for mitotic progression.
In order to characterize the function of Ote phosphorylation, we generated phosphomimetic and nonphosphorylatable mutants for two sites, serine 54 and threonine 63. We studied the localization of the mutant proteins in SL2 cells and their influence on cell cycle progression and centrosome morphology. Overexpression of Ote mutated at the Cdk1 site serine 54 did not alter its localization, nor did it affect centrosome morphology and cell cycle progression (data not shown). Next, we wanted to analyze a phosphorylation site that is targeted by Aurora-A kinase because our RNAi results suggested a functional interaction of aur and Ote in centrosome maturation and cell cycle progression. We chose threonine 63 since this site had been identified in vivo (14) and was phosphorylated by Aurora-A in vitro in our study. Overexpression of the phosphomimetic Ote mutant led to a decreased number of mitotic cells compared to overexpression of wild-type Ote or GFP, which served as a control for this experiment. In contrast, overexpression of the nonphosphorylatable Ote mutant increased the mitotic index, thereby phenocopying the aur knockdown (Fig. 6C). These results indicate that phosphorylation of Ote at threonine 63 promotes mitotic exit, whereas the presence of this phosphosite in interphase blocks mitotic entry. As aur is most likely responsible for T63 phosphorylation, we propose a model for the interdependency of aur and Ote in which progression through the cell cycle depends on phosphorylation of Ote by aur in mitosis and dephosphorylation of this residue by a yet-unknown phosphatase in interphase (Fig. 6D). Since localization of the protein does not seem to be affected by T63 phosphorylation, as shown in Fig. 6B, we argue for an altered functionality of Ote. Alternatively, this phosphorylation might facilitate the interaction with a certain binding partner, thus allowing cells to complete mitosis and enter the next cell cycle.
DISCUSSION
We identified 51 phosphorylation sites, 18 of which have not been described before, in 27 proteins from purified centrosomes by phosphopeptide enrichment followed by MS-based peptide identification. Among the identified proteins were 6 known centrosome components, but the majority of proteins have so far been linked only to non-centrosome-associated cellular localizations and processes, including NE assembly, transcriptional and translational regulation, and proteolysis. Importantly, however, this work revealed redundant and nonredundant functions of phosphoproteins in centrosome maturation, duplication, or separation; cell cycle regulation; and chromosome segregation and demonstrated their integration into signaling pathways of 4 major protein kinases. For a subset of 4 phosphoproteins, we identified previously unknown centrosome and/or spindle localization.
Proteomic analysis of Drosophila centrosomes identifies novel centrosome components and phosphorylation sites.
A major goal of this study was to determine substrates of centrosome-associated regulatory enzymes (e.g., cdc2, polo, aur, Nek2, and PP1) and in particular centrosome-specific phosphorylation sites. While a number of high-throughput data sets describing the global phosphorylation status of Drosophila proteins (14, 70) as well as phosphoproteome analyses of the human mitotic spindle (49, 59) and the yeast spindle pole body (32) have recently become available, our study provides the first inventory of centrosome phosphoproteins and their phosphorylation sites in Drosophila. By combining an affinity purification method for the isolation of centrosomes, phosphopeptide enrichment, and MS-based proteomics, we identified both unknown centrosome components and unknown phosphorylation sites. These findings imply that some of the sites identified by this approach are likely to be specific to the subcellular localization of the respective protein. Interestingly, more than 50% of the identified residues in this study are predicted consensus sites for the known centrosome-associated kinases polo, aur, and cdc2. Phosphorylation of 6 out of 8 tested consensus sites by the predicted kinases was confirmed in vitro via phosphorylation assays using peptide microarrays (see Table S4 in the supplemental material). These results strongly support the hypothesis that the proteins are targeted by centrosome kinases for phosphorylation-dependent centrosome localization and/or activation of a centrosome-associated function (50).
Cell cycle-dependent localization and function of phosphoproteins.
We used two complementary approaches to confirm an association with centrosomes for the proteins not previously known or expected to be centrosomal. (i) We cloned 9 candidate genes, expressed them in Drosophila SL2 cells, and examined their subcellular localization in interphase and mitosis. In addition to two known centrosome components, spd-2 and Grip84, which served as positive controls, we found colocalization with spindles and/or centrosomes for a further 6 candidate proteins in mitosis. Previous reports have demonstrated that components of the translation machinery associate with spindle microtubules to regulate translation of mRNAs that are required for mitotic progression and spindle assembly (12, 22, 38, 64). In support of this model, 3 proteins involved in RNA processing/translation initiation (bel, Dcp1, and eIF3-S8) were shown to be localized at mitotic spindles in our study. It has been proposed that the structure of the mitotic spindle apparatus might be stabilized by a so-called spindle matrix, a macromolecular complex constituted by several nuclear components in Drosophila (54). Lamin B was reported to be a structural component of this matrix in Xenopus and humans (66), consistent with our findings that Lam colocalizes with spindles; moreover, it was purified in a complex with α-Tub in our study.
(ii) In a second approach aimed at identifying relevant centrosome components, we conducted a cell-based functional RNAi analysis and screened for phenotypes affecting centrosome biogenesis, cell cycle progression, and chromosome segregation. Remarkably, we found that out of the 25 analyzed proteins, 16 appear to be involved in regulating the centrosome cycle, 22 play a role in cell cycle progression, and 11 play a role in chromosome segregation pathways. It is important to note that most of the functions became apparent only by analyzing synthetic phenotypes, in which a phosphoprotein and one of the four centrosome-associated kinases were downregulated concomitantly. While silencing of one gene may be sufficient to identify it as an essential component of a signaling network, such an approach often fails to detect redundant protein functions, most likely due to the existence of alternative pathways which compensate for the loss of activity of only one component of the respective signaling network. Identification of redundant regulators of a certain biological process therefore requires removal of a second element in the respective pathway. In addition to revealing many unsuspected new functions, our combinatorial RNAi approach also allowed us to identify functional interdependencies between kinases and phosphoproteins for previously unknown regulatory mechanisms controlling centrosome and cell cycle events.
The centrosome and NE components.
A striking observation from our results was that 6 out of the 27 MS-identified candidate proteins (22%) were components of the NE. These were not simply contaminants of the centrosome preparations, as we could assign centrosome cycle-related functions to 4 of them, either directly or in kinase-depleted backgrounds. In addition, localization studies of FLAG/GFP-tagged Ote and Lam in SL2 cells support the notion that these proteins have cell cycle-dependent functions for the centrosome and spindle despite their main role in assembling the nuclear membrane. There is accumulating evidence for an interaction between centrosomal and NE components from various studies. For example, it has been shown that nuclear pore subcomplexes relocate to kinetochores upon NEBD, where they interact with the γ-TuRC and promote mitotic spindle assembly (44). Caenorhabditis elegans ZYG-12 localizes to both centrosomes and the NE and is essential for their attachment (42). Centrin 2, a core component of the centriole, also associates with nuclear pore complexes in Xenopus laevis and human cells (56). A microtubule-independent role for the centrosome and Aurora-A for NEBD was also demonstrated previously (53). In this study, several lines of evidence indicate that the nuclear inner membrane protein Ote is also a genuine component of centrosomes. It binds to Lam and is found in a complex with both γ- and α-Tub, suggesting that it may facilitate bridging of the centrosome to the NE in interphase via microtubules. At the onset of mitosis prior to NEBD, we observed invaginations in the nuclear membrane in close proximity to centrosomes. In mammalian cells, it has been shown that such invaginations are generated by dynein-mediated microtubule-dependent forces, which create mechanical tension in the nuclear membrane and thereby trigger NEBD (6, 58). In support of this model, the minus-end-directed microtubule motor dynein is required for nuclear attachment of centrosomes during mitosis in Drosophila (57). However, an interaction partner for dynein at the NE has so far been elusive. Interestingly, Ote has been identified as an in vitro binding partner of dynein light chain Dlc90F in a two-hybrid study (26), a finding that may provide the missing link for centrosome-NE attachment and tearing of the NE in Drosophila. Ote was also shown to be involved in centrosome maturation and cell cycle progression downstream of aur. The results of this study strongly suggest Ote as a substrate for Aurora-A in vitro. Whether Ote is an in vivo substrate of aur remains to be elucidated. However, Ote phosphomutant analysis revealed that the Aurora-A consensus site threonine 63 is critical for progression through mitosis, supporting the results of our combinatorial RNAi study. A functional interdependency was also observed between Nup98 and polo. While depletion of polo leads to severe centrosome aberrations, a simultaneous knockdown of Nup98 significantly weakened the polo-induced phenotype, indicating that Nup98 is a downstream target in a pathway that maintains centrosome structure. Yet another interdependency of the centrosome kinase polo and the NE component Lam was revealed in our functional analysis. While polo is known to be required for mitotic exit and, hence, depletion leads to mitotic arrest, a role for Lam in a polo-dependent pathway of mitotic progression is not described. Based on the observation that parallel inhibition of polo and Lam partially rescues the polo-induced phenotype while depletion of Lam alone has no significant effect on mitotic progression, we suggest that negative regulation of Lam in a parallel signaling pathway downstream of polo is required for mitotic exit. The connection of polo and NE proteins is consistent with previous studies identifying several nuclear pore components as well as lamins as Plk1 binding partners and potential substrates, respectively (39, 59).
In conclusion, the findings of the present study support the notion that the centrosome functions as a signaling platform (21) and is integrated into a number of major cellular signaling pathways (35). Many components transiently associate with the centrosome to fulfill unexpected tasks that differ from their established functions in other cellular compartments (31). Our description of the in vivo phosphorylation status of centrosome-associated proteins provides a basis for future research aimed at understanding the molecular mechanisms controlling centrosome and cell cycle regulatory pathways in health and disease.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to D. Chen, Y. Zheng, and J. Knoblich for providing antibodies for this study; M. Neuenschwander and B. Lukaszewska-McGreal for technical assistance; T. Kessler and S.-H. Jang for critical reading of the manuscript; and M. Cooper from Thermo Fisher Scientific for support in high-content imaging.
This work was supported by NGFN Plus Mutanom, IG Cell, IG Neuronet, and GABI Centroplanta (B.M.H.L.) and Technologiestiftung Berlin funding for the automated microscopes (ESFRI) (J.P.V.K.). M.R. is a Wellcome Trust research career development and Beit-Prize fellow.
Footnotes
Published ahead of print 2 July 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Aihara H, et al. 2004. Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo. Genes Dev. 18:877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andersen JS, et al. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426:570–574 [DOI] [PubMed] [Google Scholar]
- 3. Ashery-Padan R, et al. 1997. Localization and posttranslational modifications of otefin, a protein required for vesicle attachment to chromatin, during Drosophila melanogaster development. Mol. Cell. Biol. 17:4114–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barr AR, Gergely F. 2007. Aurora-A: the maker and breaker of spindle poles. J. Cell Sci. 120:2987–2996 [DOI] [PubMed] [Google Scholar]
- 5. Basto R, et al. 2008. Centrosome amplification can initiate tumorigenesis in flies. Cell 133:1032–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. 2002. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108:83–96 [DOI] [PubMed] [Google Scholar]
- 7. Berdnik D, Knoblich JA. 2002. Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr. Biol. 12:640–647 [DOI] [PubMed] [Google Scholar]
- 8. Bettencourt-Dias M, et al. 2004. Genome-wide survey of protein kinases required for cell cycle progression. Nature 432:980–987 [DOI] [PubMed] [Google Scholar]
- 9. Bettencourt-Dias M, Glover DM. 2007. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8:451–463 [DOI] [PubMed] [Google Scholar]
- 10. Bjorklund M, et al. 2006. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature 439:1009–1013 [DOI] [PubMed] [Google Scholar]
- 11. Blangy A, Arnaud L, Nigg EA. 1997. Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150. J. Biol. Chem. 272:19418–19424 [DOI] [PubMed] [Google Scholar]
- 12. Blower MD, Feric E, Weis K, Heald R. 2007. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J. Cell Biol. 179:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bluemlein K, Ralser M. 2011. Monitoring protein expression in whole-cell extracts by targeted label- and standard-free LC-MS/MS. Nat. Protoc. 6:859–869 [DOI] [PubMed] [Google Scholar]
- 14. Bodenmiller B, et al. 2007. PhosphoPep—a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol. Syst. Biol. 3:139 doi:10.1038/msb4100182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bornens M. 2002. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14:25–34 [DOI] [PubMed] [Google Scholar]
- 16. Clemens JC, et al. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. U. S. A. 97:6499–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crasta K, Huang P, Morgan G, Winey M, Surana U. 2006. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J. 25:2551–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cullen CF, Brittle AL, Ito T, Ohkura H. 2005. The conserved kinase NHK-1 is essential for mitotic progression and unifying acentrosomal meiotic spindles in Drosophila melanogaster. J. Cell Biol. 171:593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Debec A, Marcaillou C. 1997. Structural alterations of the mitotic apparatus induced by the heat shock response in Drosophila cells. Biol. Cell 89:67–78 [DOI] [PubMed] [Google Scholar]
- 20. Dobbelaere J, et al. 2008. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6:e224 doi:10.1371/journal.pbio.0060224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Doxsey S, McCollum D, Theurkauf W. 2005. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21:411–434 [DOI] [PubMed] [Google Scholar]
- 22. Eliscovich C, Peset I, Vernos I, Mendez R. 2008. Spindle-localized CPE-mediated translation controls meiotic chromosome segregation. Nat. Cell Biol. 10:858–865 [DOI] [PubMed] [Google Scholar]
- 23. Engler C, Gruetzner R, Kandzia R, Marillonnet S. 2009. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4:e5553 doi:10.1371/journal.pone.0005553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Engler C, Kandzia R, Marillonnet S. 2008. A one pot, one step, precision cloning method with high throughput capability. PLoS One 3:e3647 doi:10.1371/journal.pone.0003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Faust M, Montenarh M. 2000. Subcellular localization of protein kinase CK2. A key to its function? Cell Tissue Res. 301:329–340 [DOI] [PubMed] [Google Scholar]
- 26. Giot L, et al. 2003. A protein interaction map of Drosophila melanogaster. Science 302:1727–1736 [DOI] [PubMed] [Google Scholar]
- 27. Goldberg M, et al. 1998. Interactions among Drosophila nuclear envelope proteins lamin, otefin, and YA. Mol. Cell. Biol. 18:4315–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goshima G, et al. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316:417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146 [DOI] [PubMed] [Google Scholar]
- 30. Hughes JR, et al. 2008. A microtubule interactome: complexes with roles in cell cycle and mitosis. PLoS Biol. 6:e98 doi:10.1371/journal.pbio.0060098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kalt A, Schliwa M. 1993. Molecular components of the centrosome. Trends Cell Biol. 3:118–128 [DOI] [PubMed] [Google Scholar]
- 32. Keck JM, et al. 2011. A cell cycle phosphoproteome of the yeast centrosome. Science 332:1557–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keller LC, Romijn EP, Zamora I, Yates JR, III, Marshall WF. 2005. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15:1090–1098 [DOI] [PubMed] [Google Scholar]
- 34. Lane HA, Nigg EA. 1996. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 135:1701–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lange BM. 2002. Integration of the centrosome in cell cycle control, stress response and signal transduction pathways. Curr. Opin. Cell Biol. 14:35–43 [DOI] [PubMed] [Google Scholar]
- 36. Lehmann V, Müller H, Lange BM. 2006. Immunoisolation of centrosomes from Drosophila melanogaster. Curr. Protoc. Cell Biol. 3.17.1–3.17.13 doi:10.1002/0471143030.cb0317s29 [DOI] [PubMed] [Google Scholar]
- 37. Li JB, et al. 2004. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117:541–552 [DOI] [PubMed] [Google Scholar]
- 38. Liska AJ, et al. 2004. Homology-based functional proteomics by mass spectrometry: application to the Xenopus microtubule-associated proteome. Proteomics 4:2707–2721 [DOI] [PubMed] [Google Scholar]
- 39. Lowery DM, et al. 2007. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J. 26:2262–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lucas EP, Raff JW. 2007. Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J. Cell Biol. 178:725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maiato H, et al. 2006. The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells. Chromosoma 115:469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Malone CJ, et al. 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115:825–836 [DOI] [PubMed] [Google Scholar]
- 43. Mirgorodskaya E, Braeuer C, Fucini P, Lehrach H, Gobom J. 2005. Nanoflow liquid chromatography coupled to matrix-assisted laser desorption/ionization mass spectrometry: sample preparation, data analysis, and application to the analysis of complex peptide mixtures. Proteomics 5:399–408 [DOI] [PubMed] [Google Scholar]
- 44. Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM, Dasso M. 2010. The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat. Cell Biol. 12:164–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moritz M, Alberts BM. 1999. Isolation of centrosomes from Drosophila embryos. Methods Cell Biol. 61:1–12 [DOI] [PubMed] [Google Scholar]
- 46. Müller H, Fogeron ML, Lehmann V, Lehrach H, Lange BM. 2006. A centrosome-independent role for gamma-TuRC proteins in the spindle assembly checkpoint. Science 314:654–657 [DOI] [PubMed] [Google Scholar]
- 47. Müller H, et al. 2010. Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 29:3344–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nigg EA, Raff JW. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell 139:663–678 [DOI] [PubMed] [Google Scholar]
- 49. Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. 2006. Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. U. S. A. 103:5391–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Okuda M. 2002. The role of nucleophosmin in centrosome duplication. Oncogene 21:6170–6174 [DOI] [PubMed] [Google Scholar]
- 51. Paddy MR, Saumweber H, Agard DA, Sedat JW. 1996. Time-resolved, in vivo studies of mitotic spindle formation and nuclear lamina breakdown in Drosophila early embryos. J. Cell Sci. 109:591–607 [DOI] [PubMed] [Google Scholar]
- 52. Palazzo RE, Vogel JM, Schnackenberg BJ, Hull DR, Wu X. 2000. Centrosome maturation. Curr. Top. Dev. Biol. 49:449–470 [DOI] [PubMed] [Google Scholar]
- 53. Portier N, et al. 2007. A microtubule-independent role for centrosomes and aurora a in nuclear envelope breakdown. Dev. Cell 12:515–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Qi H, et al. 2004. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol. Biol. Cell 15:4854–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reineke U, Volkmer-Engert R, Schneider-Mergener J. 2001. Applications of peptide arrays prepared by the SPOT-technology. Curr. Opin. Biotechnol. 12:59–64 [DOI] [PubMed] [Google Scholar]
- 56. Resendes KK, Rasala BA, Forbes DJ. 2008. Centrin 2 localizes to the vertebrate nuclear pore and plays a role in mRNA and protein export. Mol. Cell. Biol. 28:1755–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Robinson JT, Wojcik EJ, Sanders MA, McGrail M, Hays TS. 1999. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol. 146:597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Salina D, et al. 2002. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108:97–107 [DOI] [PubMed] [Google Scholar]
- 59. Santamaria A, et al. 2011. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol. Cell. Proteomics 10:M110.004457 doi:10.1074/mcp.M110.004457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sluder G. 2005. Two-way traffic: centrosomes and the cell cycle. Nat. Rev. Mol. Cell Biol. 6:743–748 [DOI] [PubMed] [Google Scholar]
- 61. Somma MP, et al. 2008. Identification of Drosophila mitotic genes by combining co-expression analysis and RNA interference. PLoS Genet. 4:e1000126 doi:10.1371/journal.pgen.1000126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Steen H, Mann M. 2004. The ABC's (and XYZ's) of peptide sequencing. Nat. Rev. Mol. Cell Biol. 5:699–711 [DOI] [PubMed] [Google Scholar]
- 63. Sunkel CE, Glover DM. 1988. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 89:25–38 [DOI] [PubMed] [Google Scholar]
- 64. Suprenant KA. 1993. Microtubules, ribosomes, and RNA: evidence for cytoplasmic localization and translational regulation. Cell Motil. Cytoskeleton 25:1–9 [DOI] [PubMed] [Google Scholar]
- 65. Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. 2006. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat. Protoc. 1:1929–1935 [DOI] [PubMed] [Google Scholar]
- 66. Tsai MY, et al. 2006. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science 311:1887–1893 [DOI] [PubMed] [Google Scholar]
- 67. Verollet C, et al. 2006. Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J. Cell Biol. 172:517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wenschuh H, et al. 2000. Coherent membrane supports for parallel microsynthesis and screening of bioactive peptides. Biopolymers 55:188–206 [DOI] [PubMed] [Google Scholar]
- 69. Wigge PA, et al. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141:967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhai B, Villen J, Beausoleil SA, Mintseris J, Gygi SP. 2008. Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res. 7:1675–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.