Abstract
Background
The zebrafish is well established as a model organism for the study of vertebrate embryogenesis, but transgenic lines enabling restricted gene expression are still lacking for many tissues.
Results
We first generated the hoxb1a(β–globin):eGFPum8 line that expresses eGFP in hindbrain rhombomere 4 (r4), as well as in facial motor neurons migrating caudally from r4. Second, we generated the hoxb1a(β-globin):Gal4VP16um60 line to express the exogenous Gal4VP16 transcription factor in r4. Lastly, we prepared the UAS(β-actin):hoxa3aum61 line where the hoxa3a gene, which is normally expressed in r5 and r6, is under control of Gal4-regulated UAS elements. Crossing the hoxb1a(β-globin):Gal4VP16um60 line to the UAS(β-actin):hoxa3aum61 line drives robust hoxa3aexpression in r4. We find that transgenic expression of hoxa3a in r4 does not affect hoxb1a expression, but has variable effects on migration of facial motorneurons and formation of Mauthner neurons. While cases of somatic transgene silencing have been reported in zebrafish, we have not observed such silencing to date – possibly because of our efforts to minimize repetitive sequences in the transgenic constructs.
Conclusion
We have generated three transgenic lines that will be useful for future studies by permitting the labeling of r4-derived cells, as well as by enabling r4-specific expression of various transgenes.
Introduction
The zebrafish has become well established as a model system due to its small size, rapid external development of optically transparent embryos and genetic tractability. Much work in zebrafish embryos has relied on the microinjection of mRNA or antisense oligonucleotides to modulate gene function, but it was rapidly realized that the transient nature of such approaches has limited utility for some applications and that methods for direct manipulation of the zebrafish germline are also needed. While the development of such tools initially lagged behind technology development in other areas, it has recently seen significant advances. In particular, the introduction of transgenes, which initially suffered from low rates of integration and high rates of mosaicism, has been significantly improved by the development of transposon-based (Tol2) techniques (Kawakami et al., 2000; Kawakami et al., 2004). Indeed, with the aid of the Tol2 system, it is now feasible to generate transgenic lines to address a multitude of biological questions. Initially, much of this effort was directed at deriving zebrafish reporter lines expressing markers such as GFP in various tissues, but more recently transgenesis efforts have been focused on generating lines permitting conditional gene expression. Conditional gene expression is achieved by the use of promoters that can be activated in response to specific external stimuli (e.g. heat shock), or that are active only in specific tissues. Both of these approaches have been applied successfully in zebrafish (e.g. (Higashijima et al., 1997; Scheer et al., 2002)). Conditional transgene expression is further improved by bi-partite systems (e.g. Cre/Lox and Gal4/UAS) where a tissue specific ‘driver line’ is used to control expression of genes in ‘reporter lines’. These systems permit both temporal and spatial control of gene expression and also enable multiple uses of lines such that one driver line can be used to control several different reporter lines (and vice versa).
The Gal4/UAS system makes use of the yeast Gal4 transcription factor, which activates transcription of genes under control of UAS (upstream activator sequence) promoter elements. The Gal4/UAS system was first implemented in Drosphila (Fischer et al., 1988; Brand and Perrimon, 1993), but is also being used in the mouse (e.g. (Ornitz et al., 1991; Matsumoto et al., 2004; Iyer et al., 2005; Ovchinnikov et al., 2008)). By selective use of promoters, transgenic lines can be generated with Gal4 expressed in tissue-specific patterns. These tissue-specific Gal4 lines are then crossed to transgenic lines where genes of interest have been placed under UAS control. While this approach requires an initial investment in the generation of multiple lines, it has the advantage of subsequently allowing ‘mixing and matching’, such that any Gal4 line can be crossed to any UAS line and the various lines become useful to the entire research community. The first application of the Gal4/UAS system to zebrafish preceded the development of Tol2-based transgenesis and resulted in relatively inefficient expression, as well as infrequent integration events (Scheer and Campos-Ortega, 1999). The efficiency of expression was subsequently improved upon by using Gal4VP16 – a fusion between the Gal4 protein and the strong transcription activation domain from herpes simplex viral protein 16 – as well as by using a UAS construct that contains 14 repeats of the UAS element. While these modifications improved expression in transient assays (Koster and Fraser, 2001), stable lines prepared using Gal4VP16/14xUAS revealed variegated expression, possibly as a result of the incorporation of multiple transgene copies that were subsequently subjected to silencing (e.g. (Sagasti et al., 2005)). In an effort to minimize variegation effects, the Gal4/UAS technology has recently been integrated with the Tol2 transposon system, which increases the frequency of single insertions and may therefore minimize silencing effects. While Tol2-mediated Gal4/UAS transgenesis has now been used successfully by several groups for gene- and enhancer-trapping, as well as for the generation of tissue specific Gal4 driver lines (Davison et al., 2007; Scott et al., 2007; Asakawa et al., 2008; Distel et al., 2009), there have been reports of variegation effects and transgene silencing with this system as well (Goll et al., 2009; Akitake et al., 2011). Nevertheless, we reasoned that sufficient information can be derived from previous efforts at Gal4/UAS transgenesis in zebrafish that it is now feasible to establish Gal4/UAS tools to probe the development of specific tissues in the zebrafish embryo.
Our specific goal is to generate Gal4/UAS tools for the analysis of hindbrain development in zebrafish and here we report transgenic lines specifically for the analysis of rhombomere 4 (r4) development. We first identified a 1.0 kb fragment from the zebrafish hoxb1a promoter and demonstrate that it is sufficient to drive expression of GFP in r4 of stable transgenic Tg(hoxb1a-βglobin:eGFP)um8 embryos, suggesting that this promoter fragment contains all required regulatory elements. We next established a transgenic line where this 1.0 kb fragment drives expression of the Gal4VP16 transcription factor Tg(hoxb1a-βglobin:Gal4VP16)um60. We find that Gal4VP16 expression is restricted to r4 in Tg(hoxb1a-βglobin:Gal4VP16)um60 embryos and we further demonstrate that this r4-restricted Gal4VP16 can transactivate GFP expression from a transiently introduced UAS:GFP plasmid. Lastly, we report the derivation of a UAS:Hoxa3a transgenic line Tg(UAS:hoxa3a)um61 and we demonstrate that Tg(hoxb1a-βglobin:Gal4VP16)/Tg(UAS:hoxa3a)um61 embryos display robust expression of hoxa3a in r4. This ectopic expression disrupts Mauthner neuron formation in r4 and interferes with facial motorneuron migration from r4, but does not affect hoxb1a expression in r4. We have not yet observed any variegation or silencing of these transgenes, in spite of the Tg(hoxb1a-βglobin:eGFP)um8 line having been bred for 6 generations. We speculate that the lack of silencing may stem from our use of a bipartite system where the Gal4VP16 driver and the UAS reporter are established as separate lines, as opposed to co-integrated as has previously been the norm in zebrafish, as well as to our use of fewer UAS elements in the UAS line.
Methods
Generation of transgenic lines
A genomic fragment of the hoxb1a enhancer encompassing position +1 (hoxb1a translation start site as defined by Zv9 Ensembl release) to position -1158 was PCR amplified (using forward oligo 5′-GGACTAGTTTTTGCTTGCCAATTCAA-3′ and reverse oligo 5′-CGGGATCCTCTGGAACTGTCCATACG-3′) and subcloned into the pCR2-TA vector (Invitrogen). The hoxb1a promoter fragment was excised from the pCR2-TA vector by digestion with SpeI (an SpeI site was included in the forward primer used for PCR amplification) and EcoRI (cuts in pCR2-TA polylinker) and cloned into a pBS vector (via SpeI/EcoRI sites in the pBS polylinker) that already contained the eGFP gene in the EcoRV site. The resulting pBS-hoxb1a:eGFP construct was digested with NarI (cuts at an internal site at position -174 of the hoxb1a promoter) and EcoRI (cuts in pBS polylinker), thereby eliminating the hoxb1a translation start site. In place of this sequence was ligated an adapter (generated by hybridization of two oligos: 5′-CGCCGGGCTGGGCATAAAAGTCAGGGCAGAGCCATCTATTGCTTACATTTGC TTCTGG-3′ and 5′-AATTCCAGAAGCAAATGTAAGCAATAGATGGCTCTGCCCTGACTTTTATGCCCAGCCCGG-3′) that contains 53bp from the human basal β-globin promoter. The resulting pBS-hoxb1a(β–globin):eGFP construct was modified by insertion of an oligo containing an XhoI site into the NotI site of the pBS polylinker. The hoxb1a(β–globin):eGFP construct was then released as an XhoI fragment (XhoI now cuts both at the modified NotI site upstream of the hoxb1a promoter and downstream of eGFP in the pBS polylinker) and cloned into the XhoI site of the pTOL2 vector to generate hoxb1a(β–globin):eGFP.
To generate hoxb1a(β–globin):Gal4VP16, Gal4VP16 was amplified from pBSGal4VP16 (using primers 5′-GGAATTCGCCACCATGGATTATAAGGATGATGACGACAAAAAGCTACTGTCT TCTATCGAA-3′ and 5′-CCGAAGCTTTTAAAAAACCTCCCACACCTCCCC-3′), followed by digestion with EcoRI (cuts site in 5′ primer) and HindIII (cuts site in 3′ primer). The eGFP gene was released from pBS-hoxb1a(β–globin):eGFP by digestion with EcoRI and HindIII and the Gal4VP16-containing EcoRI/HindIII fragment was cloned in its place to generate pBS-hoxb1a(β–globin):Gal4VP16. A NotI adapter was inserted into the HindIII site of pBS-hoxb1a(β–globin):Gal4VP16 and the hoxb1a(β–globin):Gal4VP16 fragment was released as a NotI fragment followed by cloning into the PspoM1 site of pTol2 to generate hoxb1a(β–globin):Gal4VP16.
To prepare UAS(β-actin):hoxa3a, HA-tagged hoxa3a was released from the pCS2+ vector with BamHI and NotI, the NotI site was blunted and the resulting fragment was cloned into the BamHI/BssHII site of the pUAS-L vector. A fragment containing 5 UAS elements followed by the carp β-actin promoter and the HA-tagged hoxa3a gene was PCR amplified from the resulting plasmid (using primers 5′-GAAGATCTGTGGATCAGCTTGCATGCCTG-3′ AND 5′-CAAGATCTACCCACACCTCCCCCTGAACC-3′) and cloned into the BglII site of pTol2 via BglII sites included in the primers.
Microinjections
Linearized DNA constructs were injected at the early 1 cell stage together with Tol2 transposase mRNA. Injected embryos were raised to adulthood and genotyped by PCR using oligos 5′-CCGAAGCTTTTAAAAAACCTCCCACACCTCCCC-3′ and 5′-GGAATTCGCCACCATGGATTATAAGGATGATGACGACAAAAAGCTACTGTCTTCTATCGAA-3′ for hoxb1a(β–globin):Gal4VP16um60 and oligos 5′-ATTGCCGAGCCGTCGCAGTA-3′ and 5′-GCTGCAAATAGCAGGAAACG-3′ for UAS(β-actin):hoxa3aum61, or by assaying the offspring for GFP expression for hoxb1a(β–globin):eGFPum8. The lines used herein have been outbred to wild type fish (TL and/or Ekwill) for 6 generations (hoxb1a(β–globin):eGFPum8) or 3 generations (hoxb1a(β–globin):Gal4VP16um60 and UAS(β-actin):hoxa3aum61). In situ hybridizations and immunohistochemistry. In situ hybridizations and immunohistochemistry were carried out as reported previously (Vlachakis et al., 2001; Choe et al., 2002).
Results
A 1.0kbp hoxb1a promoter fragment is sufficient to drive GFP expression in r4 of the hindbrain
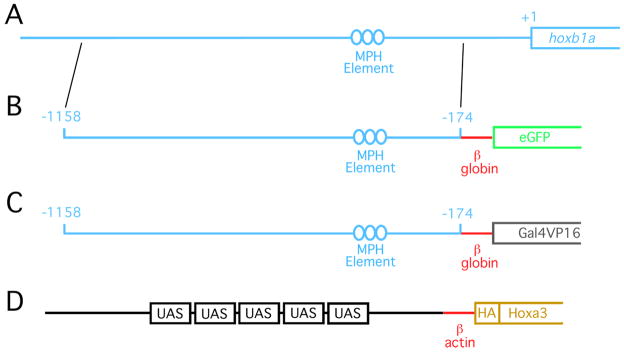
The zebrafish hoxb1a gene is the earliest hox gene to be expressed in a rhombomere-restricted manner. Previous work has identified a series of binding sites for paralog group 1 (PG1) Hox proteins and their Meis and Pbx cofactors as being necessary and sufficient to drive hoxb1 expression in r4 of the murine hindbrain (Popperl et al., 1995; Jacobs et al., 1999). In zebrafish, these elements are located in a region approximately between position -285 and -210 upstream of the hoxb1a translation start site (as defined in the Zv9 Ensembl release). To obtain a genomic region capable of driving gene expression in zebrafish rhombomere 4, we isolated a 1.0kbp fragment from the zebrafish hoxb1a locus by PCR (see methods section for details of cloning) that includes sequence from position -1158 to position -174 upstream of the hoxb1a translation start site. This fragment lacks the hoxb1a translation start site and is also likely to lack the hoxb1a transcription start site (TSS), although the location of the hoxb1a TSS is less well defined. The resulting 1.0kbp fragment and the location of the Hox/Meis/Pbx binding elements are shown in Fig. 1A.
Figure 1.
Diagram of transgenic constructs. (A) Genomic DNA between positions -1158 and -174 (relative to translation start site) was isolated from the zebrafish hoxb1a locus. This sequence was cloned upstream of the murine β–globin promoter and used to drive eGFP (B) or Gal4VP16 (C). (D) Five UAS elements were cloned upstream of the carp β-actin promoter and used to drive HA-tagged hoxa3a. See methods section for specific details of transgene construction. ‘MPH element’ indicates the position of previously defined Meis/Pbx/Hox binding sites required for r4-restricted expression of the hoxb1a gene.
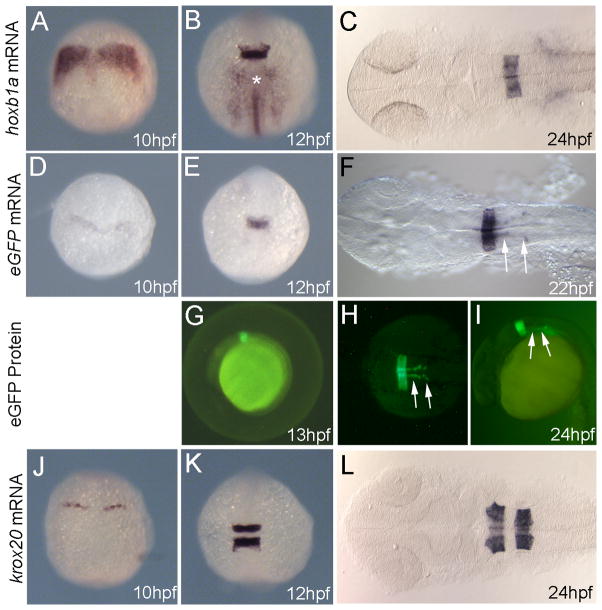
The 1.0kbp fragment was placed upstream of a 53bp sequence encoding the basal promoter of human β-globin followed by the eGFP gene and used to generate stable transgenic lines (Fig. 1B, see methods section for details). The existence of this line has been reported previously (Choe et al., 2009), but it has not been described in detail. Analysis of the hoxb1a(β-globin):eGFPum8 transgenic line demonstrates detectable eGFP mRNA in the zebrafish hindbrain at 10hpf (Fig. 2D). eGFP mRNA is robustly detected by 12hpf and persists at least until 22hpf (Fig. 2E, F). This time course of eGFP mRNA transcription appears somewhat delayed relative to the expression of hoxb1a mRNA, which is detectable by 8hpf and robust by 10hpf ((Prince et al., 1998), Fig 2A, B). We also note that eGFP mRNA is detected primarily in the hindbrain, while endogenous hoxb1a mRNA is detected both in the hindbrain and in adjacent mesoderm (asterisk in Fig. 2B), suggesting that the 1.0kbp fragment contains regulatory elements required for hindbrain expression, but may lack elements required for mesoderm expression.
Figure 2.
hoxb1a(β-globin):eGFPum8 transgenic embryos express GFP in r4, but are otherwise indistinguishable from non-transgenic siblings. Embryos were collected at the indicated stages and assayed for expression of hoxb1a (A–C), eGFP (D–F) or krox20 (J–L) by in situ hybridization as well as for eGFP fluorescence (G–I). All embryos are from an outcross of a heterozygous hoxb1a(β-globin):eGFPum8 carrier. Embryos in D–I are heterozygous for the hoxb1a(β-globin):eGFPum8 transgene. Embryos in A–C and J–L may be either heterozygous or non-transgenic, but there is no difference in expression of hoxb1a and krox20 within the clutch of embryos. Asterisk in B indicates hoxb1a expression in mesoderm. Arrows in F, H, I indicate eGFP expression in migrating nVII facial motor neurons. All embryos are in dorsal view except G and I that are lateral views. Anterior is to the top (A, B, D, E, J, K) or to the left (C, F, G, H, I, L).
eGFP protein is readily detectable by 13hpf (Fig. 2G) and remains highly expressed until 24hpf (Fig. 2H, I). While we occasionally observe the onset of eGFP expression in a salt-and-pepper like pattern, this pattern appears to be transient and resolves into a uniform eGFP expression domain in r4. By 24hpf, both eGFP mRNA and eGFP protein is detectable in a well-defined r4 expression domain as well as in nVII facial motorneurons migrating caudally from r4 (arrows in Figs. 2F, H, I). hoxb1a expression in nVII neurons has been reported previously and hoxb1a is required for proper nVII migration (Cooper et al., 2003).
Examination of several hindbrain genes in hoxb1a(β-globin):eGFPum8 transgenic embryos did not reveal any expression defects. In particular, hoxb1a and krox20 expression in hoxb1a(β-globin):eGFPum8 embryos is indistinguishable from that in non-transgenic control embryos (Fig. 2A–C, J–L). We conclude that the 1.0kbp promoter fragment recapitulates hoxb1a expression, albeit with a somewhat later onset, and does not affect normal hindbrain gene expression.
The 1.0kbp hoxb1a promoter fragment drives Gal4VP16 expression in hindbrain rhombomere 4
We next replaced the eGFP reporter with Gal4VP16 to generate the hoxb1a(β-globin):GAL4VP16um60 line (Fig. 1C). Gal4VP16 is a fusion protein containing the DNA-binding domain of the yeast Gal4 protein fused in-frame to the activation domain from the viral VP16 protein. The Gal4VP16 protein is a potent transcriptional activator that binds yeast UAS (upstream activator sequence) elements. Since there are no reported UAS elements in the zebrafish genome, Gal4VP16 is not expected to affect zebrafish gene expression or embryogenesis. Accordingly, our analysis reveals that expression of hoxb1a (Fig. 3A–C), valentino (Fig. 3E), krox20 (Fig. 3F; detected in red) and hoxa3a (Fig. 3G) in the hoxb1a(β-globin):GAL4VP16um60 line is indistinguishable from control embryos. As expected, in situ hybridization analysis revealed expression of the Gal4VP16 transcript in r4 of the hindbrain (Fig. 3D, F, H). To confirm that the Gal4VP16 protein is functional, we microinjected a plasmid containing 10 UAS elements driving GFP into 1-cell hoxb1a(β-globin):GAL4VP16um60 embryos. At 24hpf, GFP expression is readily detected in r4 of the injected embryos (Fig. 3I), confirming that the Gal4VP16 protein is active in zebrafish embryos.
Figure 3.
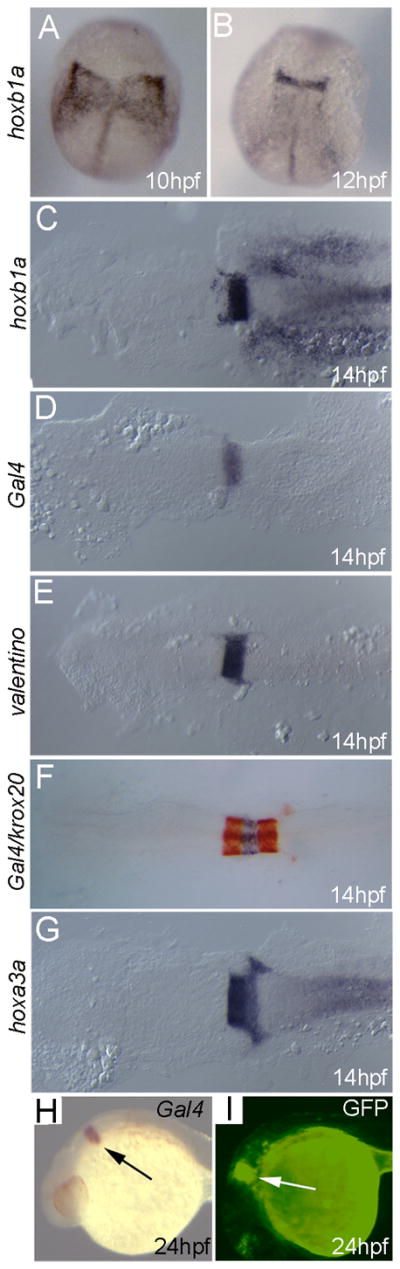
Transgenic embryos from the hoxb1a(β-globin):Gal4VP16um60 line are indistinguishable from their non-transgenic siblings with respect to expression of all hindbrain genes tested. Embryos were collected at the indicated stages and assayed for expression of hoxb1a (A–C), Gal4 (D, F, H), valentino (E), krox20 (F; detected in red) or hoxa3a (G) by in situ hybridization. (I). hoxb1a(β-globin):Gal4VP16um60 embryos were injected with a 10X UAS:GFP plasmid and assayed for GFP fluorescence at 24hpf. All embryos are from an outcross of a heterozygous hoxb1a(β-globin):Gal4VP16um60 carrier. Embryos in D, F, H, I are heterozygous for the hoxb1a(β-globin):Gal4VP16um60 transgene while embryos in A–C, E, G may be either heterozygous or non-transgenic, but there is no difference in expression of hoxb1a, valentine, hoxa3a or krox20 within the clutch of embryos. Arrows in H and I indicate expression in r4. A–G are dorsal views with anterior to the top (A, B) or the left (C–G). H and I are in lateral view with anterior to the left.
hoxb1a(β-globin):GAL4VP16um60 drives expression of a UAS(β-actin):hoxa3aum61 transgene in r4
To further test the function of the hoxb1a(β-globin):GAL4VP16um60 line, we generated an additional stable transgenic line where 5 UAS elements are cloned upstream of the carpβ-actin minimal promoter driving the zebrafish hoxa3a gene (UAS:hoxa3aum61; Fig. 1D). This construct should be inactive in zebrafish, which lack endogenous Gal4-like proteins. Accordingly, UAS(β-actin):hoxa3aum61 embryos were indistinguishable from control non-transgenic embryos with normal hoxa3a expression (Fig. 4D), as well as normal expression of hoxb1a (Fig. 4A), valentino (Fig. 4B) and krox20 (Fig. 4C).
Figure 4.
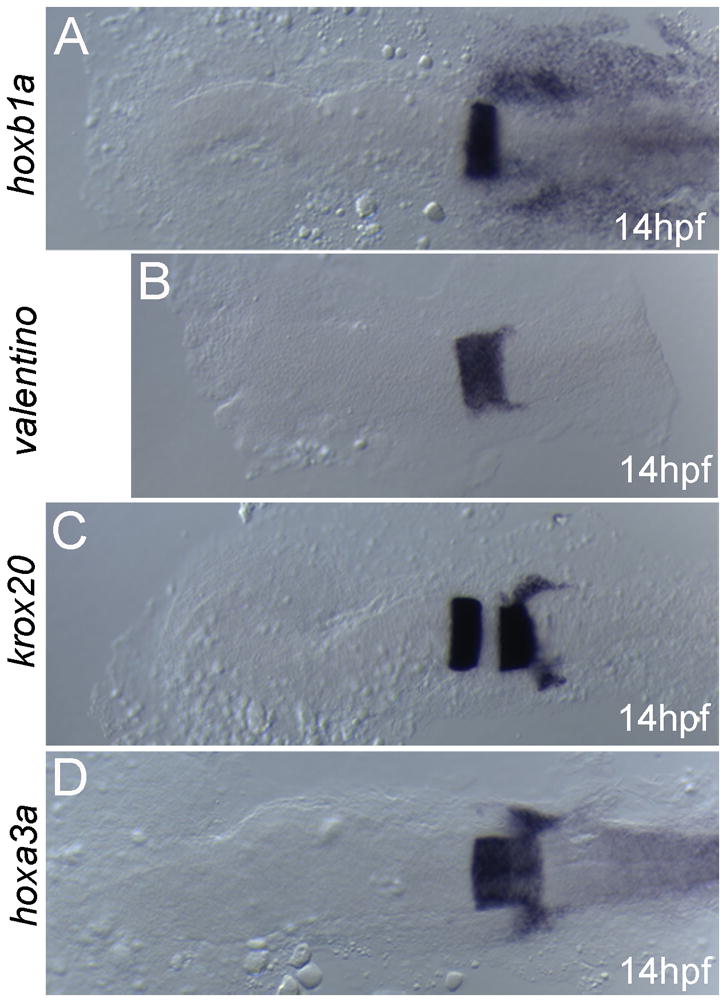
UAS(β-actin):hoxa3aum61 transgenic embryos are indistinguishable from their non-transgenic siblings with respect to expression of all hindbrain genes tested. Embryos were collected at 14hpf and assayed for expression of hoxb1a (A), valentino (B), krox20 (C) or hoxa3a (D). All embryos are from an outcross of a heterozygous UAS(β- actin):hoxa3aum61 carrier and may be either heterozygous or wild type, but there is no difference in expression of hoxb1a, valentine, hoxa3a and krox20 within the clutch of embryos. Embryos are in dorsal view with anterior to the left.
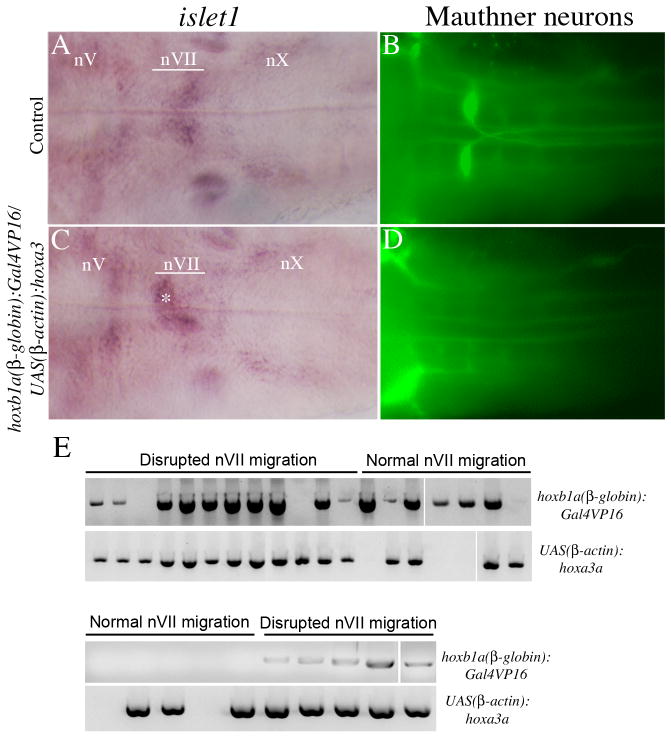
We next crossed UAS(β-actin):hoxa3aum61 to the hoxb1a(β-globin):Gal4VP16um60 fish and examined gene expression in the offspring. We elected to analyze these embryos at 24hpf to ensure that Gal4VP16 had sufficient time to accumulate and drive hoxa3a expression. Data from three separate crosses reveal that ~30% of embryos (31/101) express hoxa3a ectopically in r4 (arrows in Fig. 5B, E). This is close to the expected 25% and suggests that the Gal4VP16 construct efficiently drives expression from the UAS:hoxa3 transgene in r4. Notably, hoxb1a expression is indistinguishable between hoxb1a(β-globin):Gal4VP16um60/UAS(β-actin):hoxa3aum61 embryos and control embryos (Fig. 5C), indicating that hoxa3a expression does not interfere with hoxb1a expression in r4. However, analysis of later stage hoxb1a(β-globin):Gal4VP16um60/UAS(β-actin):hoxa3aum61 embryos revealed disruption of nVII (facial) motorneuron migration from r4 towards the caudal hindbrain (asterisk in Fig. 6C), as well as loss of Mauthner neuron formation in r4 (Fig. 6D), indicating that ectopic expression of hoxa3a may affect later stage r4 function. Genotyping of two separate experiments revealed that the majority of embryos with disrupted nVII migration were doubly transgenic (15/17 in Fig. 6E). However, this phenotype is variable and may not be fully penetrant, as indicated by some phenotypically wild type embryos also carrying both transgenes (3/12 in Fig. 6E).
Figure 5.
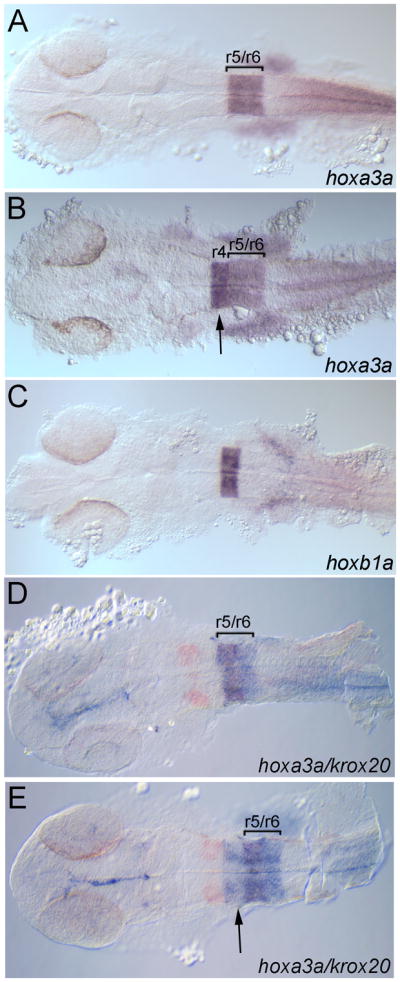
hoxb1a(β-globin):Gal4VP16um60 drives expression of the UAS(β-actin):hoxa3aum61 transgene in rhombomere 4. Embryos were collected at 24 hpf and assayed for expression of hoxa3a (A, B, D, E), hoxb1a (C) or krox20 (D, E; detected in red). All embryos are from a hoxb1a(β-globin):Gal4VP16um60 x UAS(β-actin):hoxa3aum61 cross. Embryos in B, E are double transgenic while embryos in A, C, D may be either single transgenic or wild type. Brackets in A, B, D, E delineate r5/r6. Arrows in B, E point to hoxa3a expression in r4. All embryos are in dorsal view with anterior to the left.
Figure 6.
Characterization of embryos from a hoxb1a(β-globin):Gal4VP16um60/UAS(β-actin):hoxa3aum61 cross. Embryos were collected at 48 hpf and assayed for expression of islet1 to detect motor neurons (A, C) and 3A10 to detect Mauthner neurons (B, D). (E) Embryos with normal or abnormal nVII migration were picked from two representative experiments and genotyped by PCR for presence of the Gal4VP16 and the UAS:Hoxa3a transgene. nV = trigeminal motorneuron, nVII = facial motorneuron, nX = vagal motorneuron. Asterisk in C indicates nVII neurons remaining in r4. All embryos are in dorsal view with anterior to the left.
Discussion
The zebrafish represents a useful model organism that provides experimentally accessible transparent embryos for the study of vertebrate embryogenesis. Recent advances in zebrafish transgenesis now allow for relative straightforward production of transgenic lines, but the number of such lines available for the analysis of any specific tissue is still limited. Here we report the generation of three transgenic lines for analysis of the zebrafish hindbrain. The hoxb1a(β-globin):eGFPum8 line drives expression of eGFP in rhombomere 4 (r4) of the zebrafish hindbrain, as well as in facial motorneurons migrating from r4 into the caudal hindbrain. eGFP mRNA is first detectable in r4 at 10hpf (Fig. 2G) and eGFP fluorescence is observed by 13hpf (Fig. 2J). This onset of expression lags behind that of hoxb1a expression from the endogenous promoter by approximately 2 hours. The hoxb1a(β-globin):Gal4VP16um60 line drives expression of the exogenous Gal4VP16 transcription factor in r4 with approximately the same kinetics as observed for eGFP (Fig. 3D). Lastly, the UAS(β-actin):hoxa3aum61 transgene is not expressed in zebrafish embryos until crossed to the hoxb1a(β-globin):Gal4VP16um60 line, in which case hoxa3a becomes ectopically expressed in r4. These lines will be useful by allowing the marking of r4 cells using the hoxb1a(β–globin):eGFPum8 line and by enabling forced r4-expression of any UAS-controlled transgene by crossing to the hoxb1a(β-globin):Gal4VP16um60. In addition, crossing of the UAS(β-actin):hoxa3aum61 line to other Gal4 lines will drive hoxa3a expression in any number of tissues. We note that a line labeling portions of the murine hindbrain was recently reported (Makki and Capecchi, 2010), but this line uses a different hox promoter (hoxa1, equivalent to zebrafish hoxb1b) and labeling is therefore not restricted to r4. Notably, labeling in the murine line also shows a delay relative to expression of the endogenous promoter, suggesting that this may be a common feature of transgenic reporter lines.
We find that hoxb1a(β-globin):Gal4VP16um60/UAS(β-actin):hoxa3aum61 embryos express hoxa3a ectopically in r4 as expected. Notably, this ectopic expression does not affect hoxb1a expression, but has variable effects on Mauthner neuron formation and nVII facial motorneuron migration. This may be the result of partial re-specification of r4 upon expression of hoxa3a, but this hypothesis would need to be tested by additional experimentation. Our result is distinct from a recent report that drove hoxb3 expression in murine r4 by making use of the r4-specific enhancer from the hoxb2 gene (Wong et al., 2011) and that observed loss of hoxb1 expression, as well as abnormal neurogenesis in r4. While it is possible that the difference between these studies are due to different activities of hoxa3a versus hoxb3, we consider it more likely that differences in the time of onset of transgene expression account for the differing effects on hoxb1a expression. In particular, since our system relies on initial expression of Gal4VP16 that then transactivates hoxa3a, ectopic expression of hoxa3a is not initiated in r4 until after hoxb1a is already transcribed.
It has been reported that Gal4/UAS transgenesis in zebrafish suffers from somatic silencing, apparently mediated by DNA methylation of the transgenic locus (Goll et al., 2009). This silencing manifests itself as loss of transgene expression in subsequent generations and as variable expression in individual embryos. While we have observed ‘salt and pepper’ expression of the hoxb1a(β–globin):eGFPum8 transgene, this appears restricted to the earliest stages of development and is subsequently ‘filled in’ such that consistent expression is observed throughout the eGFP expression domain. We interpret this to represent stochastic onset of eGFP expression rather than silencing and conclude that we do not observed silencing in any of our lines. While we cannot exclude the possibility that silencing will become apparent in subsequent generations, it also is possible that some features of our transgenic constructs make them less susceptible to silencing. In particular, we took several approaches to minimize repetitive sequences in our transgenic lines. First, we made use of the Tol2 system that is more likely to produce single-copy insertions than non-Tol2 approaches, which can lead to insertion of multiple concatamerized transgenes at a single locus. Second, while previous investigators frequently made use of ‘self-reporting’ Gal4/UAS systems where the Gal4VP16-driver and the UAS-reporter are present on the same plasmid, we have separated the two onto different plasmids and used them to raise separate driver and reporter lines. Third, while UAS constructs with 10 or 14 repeated UAS elements have been used previously, we used a UAS reporter containing 5 UAS elements. Similar conclusions were recently drawn by Goll and coworkers in a more systematic study (Akitake et al., 2011). Indeed, by directly comparing silencing of reporter lines containing 4 or 14 UAS elements, these investigators found that fewer UAS elements correlates with lower levels of silencing and lower levels of DNA methylation at the transgenic locus. This supports the general concept that minimization of repetitive elements is important in order to avoid somatic silencing of transgenes in zebrafish.
Bullet points.
A hoxb1a(β–globin):eGFP transgene drives GFP expression in hindbrain rhombomere 4
A hoxb1a(β-globin):Gal4VP16 transgene drives expression of hoxa3a from UAS(β-actin):hoxa3a in r4
Ectopic expression of hoxa3a in r4 disrupts neurogenesis and neuronal migration
Acknowledgments
We are grateful to Matthew Gorgoglione for help with fish care and in situ analysis and Nathan Lawson for the gift of pUAS-L vector. This work was supported by NIH grants NS038183 and HD065081to CGS.
References
- Akitake CM, Macurak M, Halpern ME, Goll MG. Transgenerational analysis of transcriptional silencing in zebrafish. Developmental biology. 2011 doi: 10.1016/j.ydbio.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, Kawakami K. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci U S A. 2008;105:1255–1260. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Choe S-K, Vlachakis N, Sagerström CG. Meis family proteins are required for hindbrain development in the zebrafish. Development. 2002;129:585–595. doi: 10.1242/dev.129.3.585. [DOI] [PubMed] [Google Scholar]
- Choe SK, Lu P, Nakamura M, Lee J, Sagerstrom CG. Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Dev Cell. 2009;17:561–567. doi: 10.1016/j.devcel.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KL, Leisenring WM, Moens CB. Autonomous and nonautonomous functions for Hox/Pbx in branchiomotor neuron development. Developmental biology. 2003;253:200–213. doi: 10.1016/s0012-1606(02)00018-0. [DOI] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Developmental biology. 2007;304:811–824. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel M, Wullimann MF, Koster RW. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13365–13370. doi: 10.1073/pnas.0903060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- Goll MG, Anderson R, Stainier DY, Spradling AC, Halpern ME. Transcriptional silencing and reactivation in transgenic zebrafish. Genetics. 2009;182:747–755. doi: 10.1534/genetics.109.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP inwhole muscles or the whole body by using promoters of zebrafish origin. Developmental biology. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- Iyer M, Salazar FB, Lewis X, Zhang L, Wu L, Carey M, Gambhir SS. Non-invasive imaging of a transgenic mouse model using a prostate-specific two-step transcriptional amplification strategy. Transgenic research. 2005;14:47–55. doi: 10.1007/s11248-004-2836-1. [DOI] [PubMed] [Google Scholar]
- Jacobs Y, Schnabel CA, Cleary ML. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol Cell Biol. 1999;19:5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Koster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–346. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- Makki N, Capecchi MR. Hoxa1 lineage tracing indicates a direct role for Hoxa1 in the development of the inner ear, the heart, and the third rhombomere. Dev Biol. 2010;341:499–509. doi: 10.1016/j.ydbio.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Kiguchi K, Jiang J, Carbajal S, Ruffino L, Beltran L, Wang XJ, Roop DR, DiGiovanni J. Development of transgenic mice that inducibly express an active form of c-Src in the epidermis. Molecular carcinogenesis. 2004;40:189–200. doi: 10.1002/mc.20027. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Moreadith RW, Leder P. Binary system for regulating transgene expression in mice: targeting int-2 gene expression with yeast GAL4/UAS control elements. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:698–702. doi: 10.1073/pnas.88.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov DA, van Zuylen WJ, DeBats CE, Alexander KA, Kellie S, Hume DA. Expression of Gal4-dependent transgenes in cells of the mononuclear phagocyte system labeled with enhanced cyan fluorescent protein using Csf1r-Gal4VP16/UAS-ECFP double-transgenic mice. Journal of leukocyte biology. 2008;83:430–433. doi: 10.1189/jlb.0807585. [DOI] [PubMed] [Google Scholar]
- Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes:expression in the hindbrain region of wild-type and mutants of the segmentation gene valentino. Development. 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Guido MR, Raible DW, Schier AF. Repulsive interactions shape the morphologies and functional arrangement of zebrafish peripheral sensory arbors. Current biology : CB. 2005;15:804–814. doi: 10.1016/j.cub.2005.03.048. [DOI] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mechanisms of development. 1999;80:153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- Scheer N, Riedl I, Warren JT, Kuwada JY, Campos-Ortega JA. A quantitative analysis of the kinetics of Gal4 activator and effector gene expression in the zebrafish. Mech Dev. 2002;112:9–14. doi: 10.1016/s0925-4773(01)00621-9. [DOI] [PubMed] [Google Scholar]
- Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nature methods. 2007;4:323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- Vlachakis N, Choe SK, Sagerstrom CG. Meis3 synergizes with Pbx4 and Hoxb1b in promoting hindbrain fates in the zebrafish. Development. 2001;128:1299–1312. doi: 10.1242/dev.128.8.1299. [DOI] [PubMed] [Google Scholar]
- Wong EY, Wang XA, Mak SS, Sae-Pang JJ, Ling KW, Fritzsch B, Sham MH. Hoxb3 negatively regulates Hoxb1 expression in mouse hindbrain patterning. Developmental biology. 2011;352:382–392. doi: 10.1016/j.ydbio.2011.02.003. [DOI] [PubMed] [Google Scholar]