Abstract
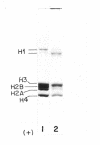
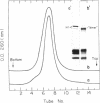
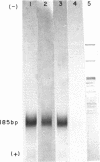
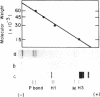
Ultraviolet (UV)-induced cross-linking was utilized in order to identify histone-DNA interacting regions in the chromatin repeating unit. Fractionated mononucleosomes which contained 185 base pairs of DNA and a full complement of the histones, including histone H1, were irradiated with light of lambda greater than 290nm in the presence of a photosensitizer. Equimolar amounts of histones H2A and H2B were found, by two independent labeling experiments, to be cross-linked to the DNA. Based on previous finding that the UV irradiation specifically cross-links residues which are in close proximity, irrespective of the nature of the amino acid side chain or the nucleotide involved, our results indicate that the four core histones are not positioned equivalently with respect to the DNA. This arrangement allows histones H2A and H2B to preferentially cross-link to the DNA. A water soluble covalent complex of DNA and histones was isolated. This complex was partially resistant to mild nuclease digestion, it exhibited a CD spectrum similar to that of chromatin, and was found to contain histone H1. These results are compatible with a model which suggests that histone H1, though anchored to the linker, is bound to the DNA at additional sites. By doing so it spans the whole length of the nucleosome and clamps together the DNA fold around the histone core.
Full text
PDF




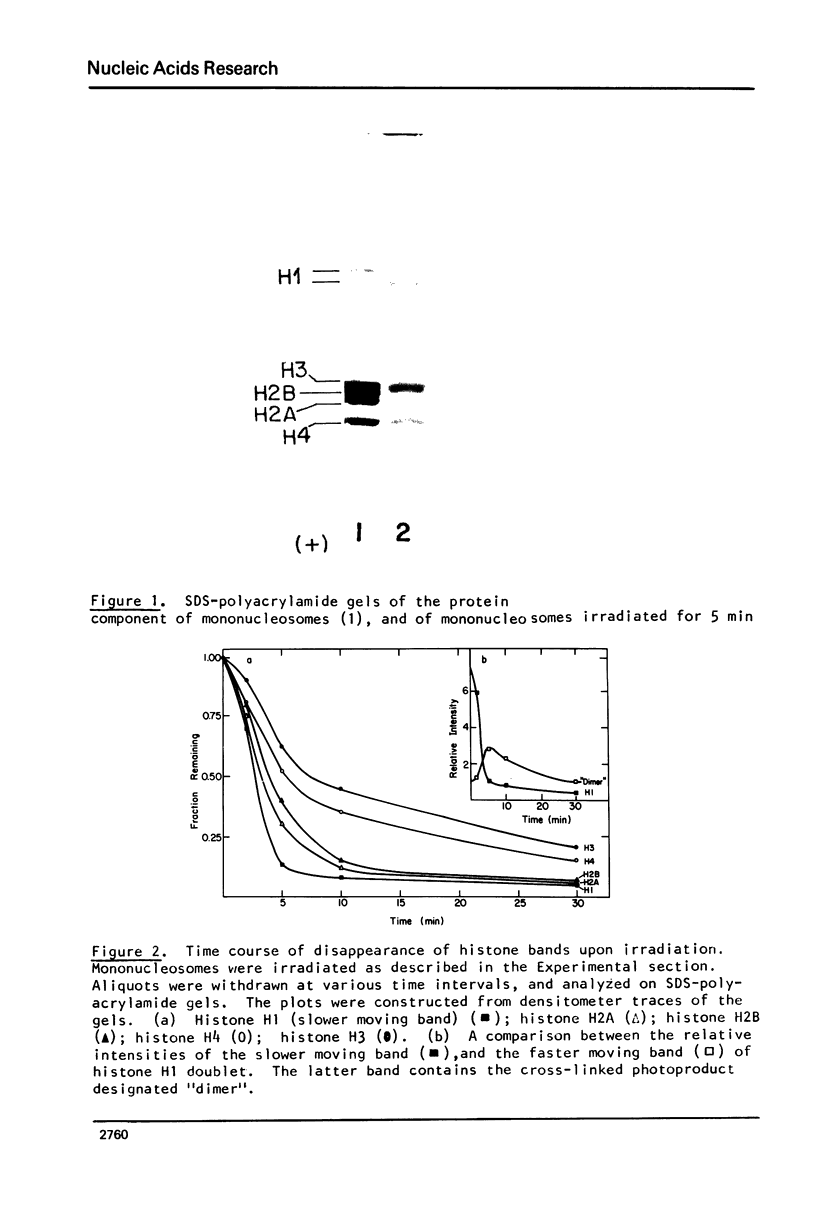
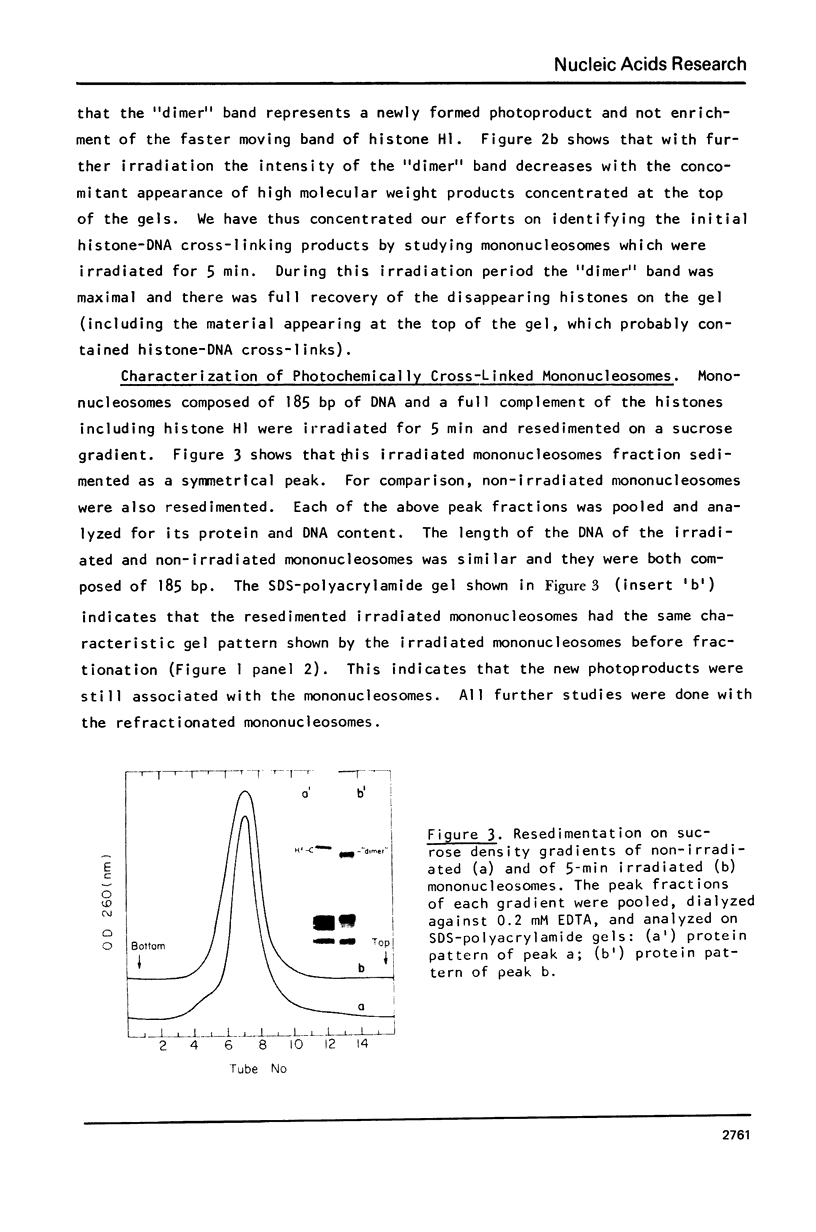


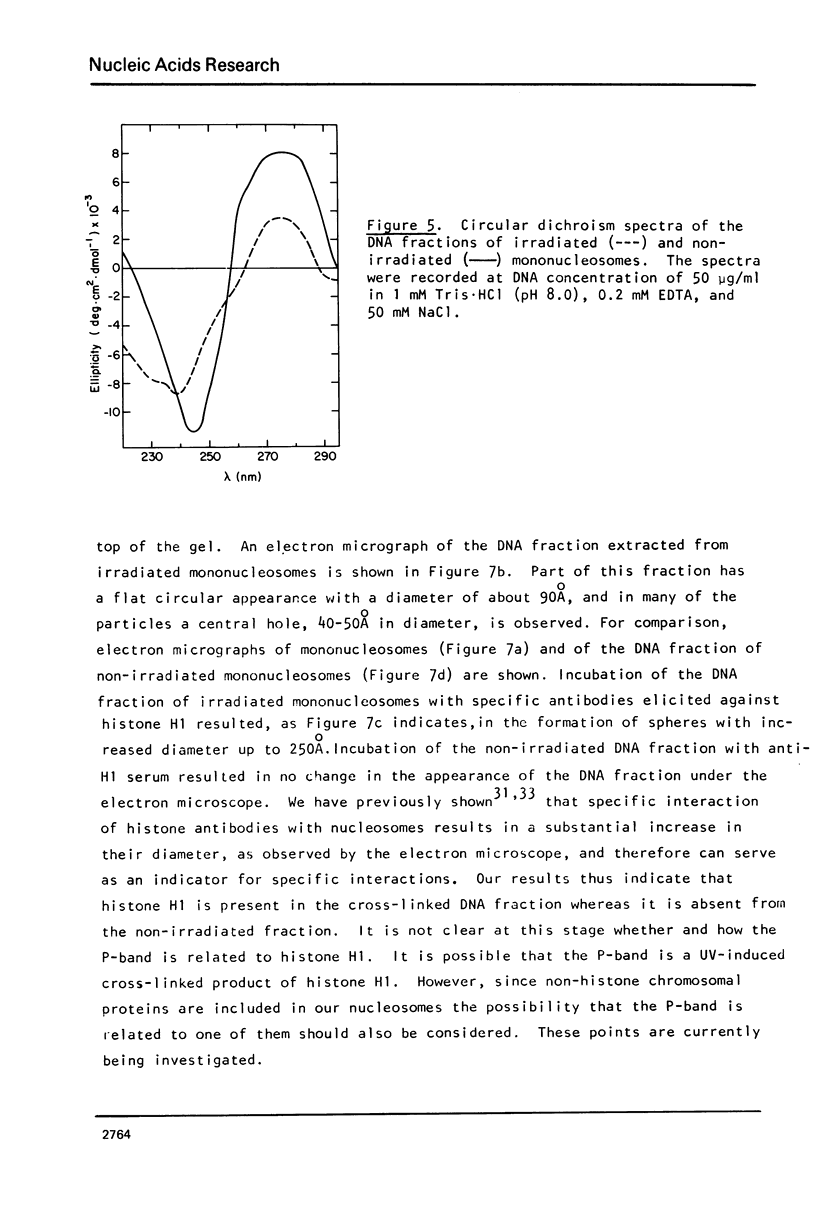
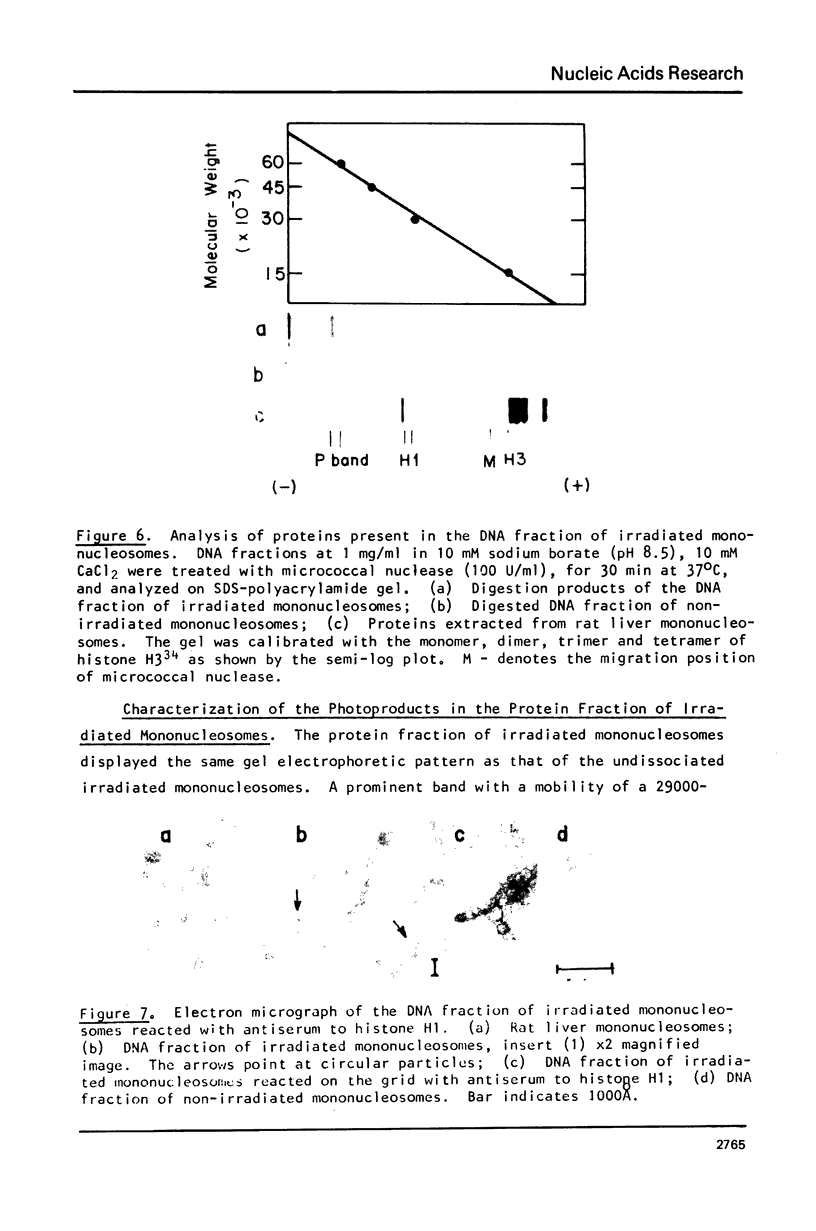
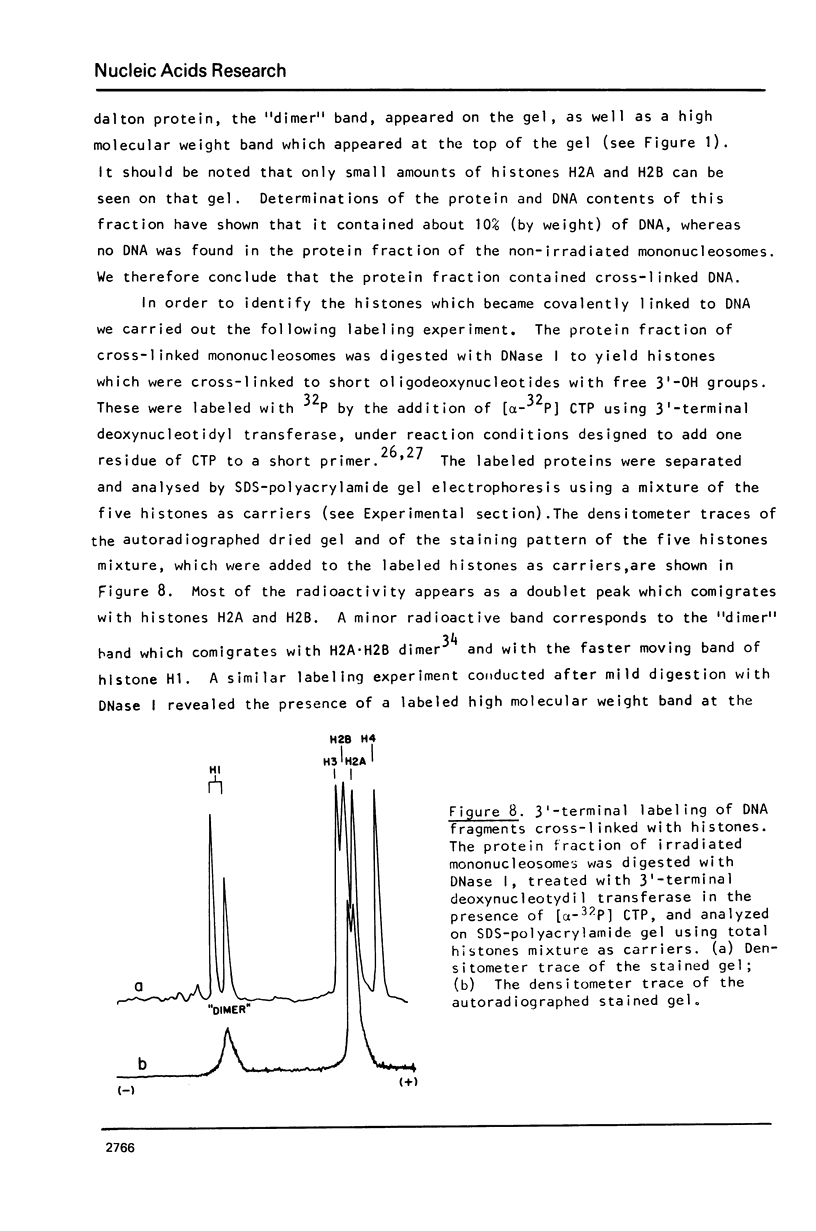

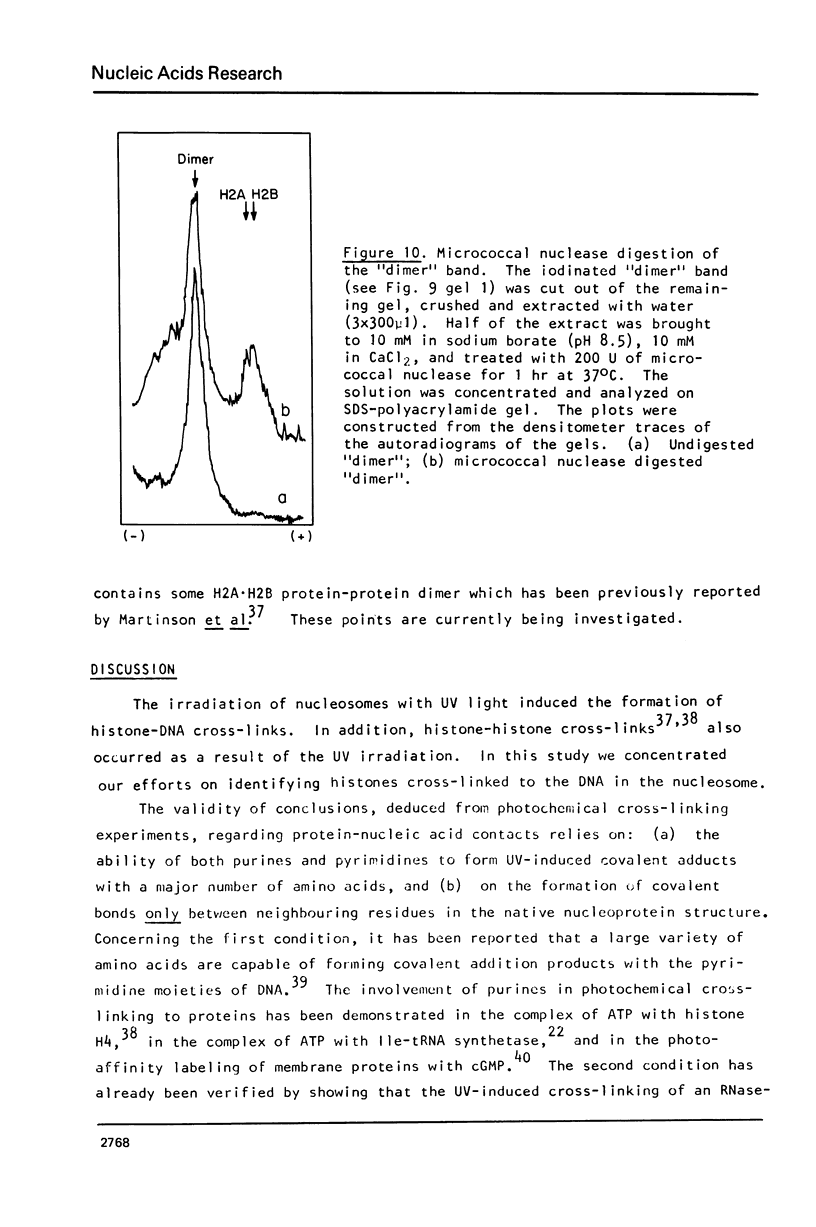





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antonoff R. S., Ferguson J. J., Jr, Idelkope G. Direct photo-affinity labeling of cyclic nucleotide binding proteins with guanosine-3',5'-monophosphate. Photochem Photobiol. 1976 May;23(5):327–329. doi: 10.1111/j.1751-1097.1976.tb07256.x. [DOI] [PubMed] [Google Scholar]
- Axel R. Cleavage of DNA in nuclei and chromatin with staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2921–2925. doi: 10.1021/bi00684a020. [DOI] [PubMed] [Google Scholar]
- Bakayev V. V., Melnickov A. A., Osicka V. D., Varshausky A. J. Studies on chromatin. II. Isolation and characterization of chromatin subunits. Nucleic Acids Res. 1975 Aug;2(8):1401–1419. doi: 10.1093/nar/2.8.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin J. P., Boseley P. G., Bradbury E. M., Ibel K. The subunit structure of the eukaryotic chromosome. Nature. 1975 Jan 24;253(5489):245–249. doi: 10.1038/253245a0. [DOI] [PubMed] [Google Scholar]
- Biroc S. L., Reeder R. H. Iodination of Xenopus laevis histone F2a1 in chromatin. Biochemistry. 1976 Apr 6;15(7):1440–1448. doi: 10.1021/bi00652a014. [DOI] [PubMed] [Google Scholar]
- Boseley P. G., Bradbury E. M., Butler-Browne G. S., Carpenter B. G., Stephens R. M. Physical studies of chromatin. The recombination of histones with DNA. Eur J Biochem. 1976 Feb 2;62(1):21–31. doi: 10.1111/j.1432-1033.1976.tb10093.x. [DOI] [PubMed] [Google Scholar]
- Burton D. R., Hyde J. E., Walker I. O. Histones F2a1 and F3 interact reversibly and cooperatively with DNA to form an equimolar complex in chromatin. FEBS Lett. 1975 Jul 15;55(1):77–80. doi: 10.1016/0014-5793(75)80962-8. [DOI] [PubMed] [Google Scholar]
- Bustin M., Goldblatt D., Sperling R. Chromatin structure visualization by immunoelectron microscopy. Cell. 1976 Feb;7(2):297–304. doi: 10.1016/0092-8674(76)90029-5. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Felsenfeld G. Supercoiling energy and nucleosome formation: the role of the arginine-rich histone kernel. Nucleic Acids Res. 1977;4(5):1159–1181. doi: 10.1093/nar/4.5.1159-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Danna K. J., Nathans D. Bidirectional replication of Simian Virus 40 DNA. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3097–3100. doi: 10.1073/pnas.69.11.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Goldblatt D., Bustin M. Exposure of histone antigenic determinants in chromatin. Biochemistry. 1975 Apr 22;14(8):1689–1695. doi: 10.1021/bi00679a022. [DOI] [PubMed] [Google Scholar]
- Havron A., Sperling J. Specificity of photochemical cross-linking in protein-nucleic acid complexes: identification of the interacting residues in RNase- pyrimidine nucleotide complex. Biochemistry. 1977 Dec 13;16(25):5631–5635. doi: 10.1021/bi00644a038. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Kössel H., Roychoudhury R. Synthetic polynucleotides. Ther terminal addition of riboadenylic acid to deoxyoligonucleotides by terminal deoxynucleotidyl transferase as a tool for the specific labelling of deoxyoligonucleotides at the 3'-ends. Eur J Biochem. 1971 Sep 24;22(2):271–276. doi: 10.1111/j.1432-1033.1971.tb01541.x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Martinson H. G., Shetlar M. D., McCarthy B. J. Histone-histone interactions within chromatin. Crosslinking studies using ultraviolet light. Biochemistry. 1976 May 4;15(9):2002–2007. doi: 10.1021/bi00654a030. [DOI] [PubMed] [Google Scholar]
- McConnell B., von Hippel P. H. Hydrogen exchange as a probe of the dynamic structure of DNA. I. General acid-base catalysis. J Mol Biol. 1970 Jun 14;50(2):297–316. doi: 10.1016/0022-2836(70)90194-4. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Rill R., Van Holde K. E. Properties of nuclease-resistant fragments of calf thymus chromatin. J Biol Chem. 1973 Feb 10;248(3):1080–1083. [PubMed] [Google Scholar]
- Roychoudhury R., Jay E., Wu R. Terminal labeling and addition of homopolymer tracts to duplex DNA fragments by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 1976 Apr;3(4):863–877. doi: 10.1093/nar/3.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih T. Y., Fasman G. D. Conformation of deoxyribonucleic acid in chromatin: a circular dichroism study. J Mol Biol. 1970 Aug 28;52(1):125–129. doi: 10.1016/0022-2836(70)90182-8. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Histones H3 and H4 interact with the ends of nucleosome DNA. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4400–4404. doi: 10.1073/pnas.73.12.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P. Mapping DNAase l-susceptible sites in nucleosomes labeled at the 5' ends. Cell. 1976 Oct;9(2):347–353. doi: 10.1016/0092-8674(76)90124-0. [DOI] [PubMed] [Google Scholar]
- Singer D. S., Singer M. F. Studies on the interaction of H1 histone with superhelical DNA: characterization of the recognition and binding regions of H1 histones. Nucleic Acids Res. 1976 Oct;3(10):2531–2547. doi: 10.1093/nar/3.10.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Camerini-Otero R. D., Felsenfeld G. Chromatin structure as probed by nucleases and proteases: evidence for the central role of histones H3 and H4. Cell. 1976 Sep;9(1):179–193. doi: 10.1016/0092-8674(76)90063-5. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Sperling J., Havron A. Photochemical cross-linking of neighboring residues in protein-nucleic acid complexes: rnase and pyrimidine nucleotide inhibitors. Biochemistry. 1976 Apr 6;15(7):1489–1495. doi: 10.1021/bi00652a020. [DOI] [PubMed] [Google Scholar]
- Sperling J. Photochemical crosslinking of ATP to histone H4. Photochem Photobiol. 1976 May;23(5):323–326. doi: 10.1111/j.1751-1097.1976.tb07255.x. [DOI] [PubMed] [Google Scholar]
- Sperling R., Amos L. A. Arrangement of subunits in assembled histone H4 fibers. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3772–3776. doi: 10.1073/pnas.74.9.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling R., Bustin M. Dynamic equilibrium in histone assembly: self-assembly of single histones and histone pairs. Biochemistry. 1975 Jul 29;14(15):3322–3331. doi: 10.1021/bi00686a006. [DOI] [PubMed] [Google Scholar]
- Sperling R., Bustin M. Self assembly of histone F2a1. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4625–4629. doi: 10.1073/pnas.71.11.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A., Bina-Stein M., Simpson R. T. Crosslinked histone octamer as a model of the nucleosome core. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2780–2784. doi: 10.1073/pnas.74.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strniste G. F., Rall S. C. Induction of stable protein-deoxyribonucleic acid adducts in Chinese hamster cell chromatin by ultraviolet light. Biochemistry. 1976 Apr 20;15(8):1712–1719. doi: 10.1021/bi00653a019. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Georgiev G. P. Heterogeneity of chromatin subunits in vitro and location of histone H1. Nucleic Acids Res. 1976 Feb;3(2):477–492. doi: 10.1093/nar/3.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T., Singer M. The interaction of histones with simian virus 40 supercoiled circular deoxyribonucleic acid in vitro. J Biol Chem. 1975 Jan 25;250(2):796–798. [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Removal of histone H1 exposes a fifty base pair DNA segment between nucleosomes. Biochemistry. 1976 Jul 27;15(15):3307–3314. doi: 10.1021/bi00660a022. [DOI] [PubMed] [Google Scholar]
- Wilhelm F. X., Wilhelm M. L., Erard M., Duane M. P. Reconstitution of chromatin: assembly of the nucleosome. Nucleic Acids Res. 1978 Feb;5(2):505–521. doi: 10.1093/nar/5.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue V. T., Schimmel P. R. Direct and specific photochemical cross-linking of adenosine 5'-triphosphate to an aminoacyl-tRNA synthetase. Biochemistry. 1977 Oct 18;16(21):4678–4684. doi: 10.1021/bi00640a023. [DOI] [PubMed] [Google Scholar]