Abstract
Objective:
Head and neck squamous cell carcinoma, including oral squamous cell carcinoma (OSCC) is the sixth most common cancer in the human population. Despite significant efforts committed in treatment of OSCC the overall survival rate of OSCC has not improved significantly. Activating mutations in the fibroblast growth factor receptor 3 (FGFR3) genes are responsible for some human cancers, including bladder and cervical carcinoma. Despite a high frequency in some benign skin disorders, FGFR3 mutations have not been reported in cutaneous malignancies. Therefore, FGFR3 gene may play a role in epithelial biology and mutations of FGFR3 gene may contribute to the development of OSCC.
Materials and Methods:
In this cross-sectional study, DNA was extracted and purified from snap frozen tissue biopsy sections of 20 OSCC cases. Exons 7 and 15 were amplified by polymerase chain reaction (PCR) and sequenced in both directions.
Results:
In three cases silent mutations were identified in exon 7 (882 T to C) which may be introduced as Single Nucleotide Polymorphism (SNP) and no mutation was identified in exon 15.
Conclusion:
FGFR3 gene mutation in exon 7 and 15 has no significant role in the development and progression of OSCC. Analyzing other exons or considering other advanced gene mutation assessment techniques may clarify the role of this receptor mutation in OSCC pathogenesis.
Keywords: FGFR3, Oral Squamous Cell Carcinoma, PCR, Mutation
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC), including oral squamous cell carcinoma (OSCC), is the sixth most common malignancy worldwide and the third most common cancer in developing countries [1].
In Iran, although registered statistical reports do not include HNSCC in the ten most common cancers among Iranian population but consider it as a frequent malignancy in the head and neck region [2].
The survival rate of patients with HNSCC has not improved significantly despite the improvement of treatment modalities.
Therefore, recent studies have focused on molecular target therapies instead of conventional combination of surgery and radiation therapy [3]. Fibroblast growth factor receptor 3 (FGFR3) is a well-known member of growth factor receptors with tyrosine kinase activity which binds to its specific ligands with high affinity [4].
Like other receptors with tyrosine kinase activity, it is composed of an extracellular domain that binds to the ligands; a transmembrane domain, placed in the cytoplasmic membrane and an intracellular domain which is responsible for tyrosine kinase activity of the receptor [5].
The extracellular region of FGFR contains three immunoglobulin-like (Ig-like) domains. To produce various extracellular regions, FGFR3 associated mRNA undergoes different splicing in the posttranscriptional level [6].
Specific germline mutations, mainly localized in exons 7, 10, 12, 15 and 19, considered as the main etiologic factor in some kinds of human skeletal disorders [7]. These disorders include craniosynostoses and chondrodysplasias, such as hypochondroplasia, achondroplasia, severe achondroplasia with developmental delay and acanthosis nigricans (SADDAN), and thanatophoric dysplasia (TD) [8]. Similar but somatic FGFR3 mutations are observed in various benign skin tumors such as seborrheic keratoses and epidermal nevi and also in cancers like multiple myeloma and epithelial malignancies (bladder and cervix carcinomas) [9]. Recently, the functional roles of FGFR3 mutations in the development of actinic cheilitis and cutaneous squamous cell carcinoma have been proposed [4]. Although recently in addition to numerical and structural chromosomal disorders [10] FGFR3 point mutations have been reported in 62% of OSCC cases [11], it seems that complementary studies are needed to clarify the role of FGFR3 mutations in the pathogenesis of OSCC. In the present study, we investigated the presence of FGFR3 mutations in exons 7 and 15 of oral squamous cell carcinoma samples.
MATERIALS AND METHODS
Tissue specimens and clinical data:
Snap frozen tissue biopsies of OSCC (n = 20) cases were retrieved from Iran National Tumor Bank which is funded by the Cancer Institute of Tehran University.
All cases were assessed for suitability of DNA analysis. Clinical data, including patient demographics, grading and staging (based on Neville et al. [14] classification) were retrieved from the institute (Table I).
Table I.
Clinical data of the cases
| Case | Sex | Age | Site | Smoking Status |
|---|---|---|---|---|
| 1 | F | 23 | T | No |
| 2 | M | 49 | T | Yes |
| 3 | M | 49 | T | - |
| 4 | M | 35 | T | No |
| 5 | M | 63 | T | Yes |
| 6 | M | 64 | T | No |
| 7 | M | 53 | G | No |
| 8 | M | 79 | T | No |
| 9 | M | 64 | T | No |
| 10 | M | 72 | T | No |
| 11 | F | 76 | G | Yes |
| 12 | M | 25 | T | No |
| 13 | M | 73 | T | - |
| 14 | M | 72 | B | No |
| 15 | F | 32 | T | No |
| 16 | M | 54 | HP | No |
| 17 | M | 52 | G | Yes |
| 18 | M | 75 | B | Yes |
| 19 | M | 79 | FL | Yes |
| 20 | F | 64 | T | No |
T: Tongue; G: Gingiva; B: Buccal mucosa; HP: Hard Palate; FL: Floor of the mouth
DNA extraction & PCR:
DNA was extracted using the Accu®Prep Genomic DNA Extraction kit (Bioneer Corp., Seoul, South Korea) by the protocol supplied by the manufacturer.
For the PCR procedure, primers were designed to amplify exons 7 and 15 of the FGFR3 gene, regions known to harbor mutations in other cancers.
Primers were as follows:
Exon 7: 5′-AGTGGCGGTGGTGGTGAGGGAG - 3′ and 5′-CTGCAAGGTGTGTACAGTGACGCACA -3′;
Exon 15: 5′-GTGACCGAGGACAACGTGATG -3′ and 5′-GGTGAGTGTAGACTCGGTCAAA - 3′.
Primer sequences were created using previous study data [12]. PCR was performed using Premix Taq TM Hot Start Version master mix (Takara Bio. Inc., Japan). PCR was carried out with the following cycling conditions:
- Exon 7: One cycle at 95°C for 2 minutes and 30 cycles at 95°C for 30 seconds (denaturation), 58°C for 30 seconds (annealing) and 72°C for 50 seconds (extension) followed by one cycle at 72°C for 5 minutes.
- Exon 15: One cycle at 95°C for 2 minutes and 30 cycles at 95°C for 30 seconds (denaturation), 61°C for 30 seconds (annealing) and 72°C for 50 seconds (extension) followed by one cycle at 72°C for 5 minutes.
The PCR products were visualized after electrophoresis in 2% agarose to isolate and confirm fragments of the expected size.
DNA sequencing:
Using the Accu®Prep Genomic DNA Extraction kit, the PCR products were isolated and then sent to Millegene DNA Sequencing Service (Toulouse, France) for sequencing.
Sequencing was performed in both directions using the same PCR primers.
Each exon analysis was performed using Ex-PAPy-UniProt Knowledge base: Swiss-Prot and TrEMBL.
RESULTS
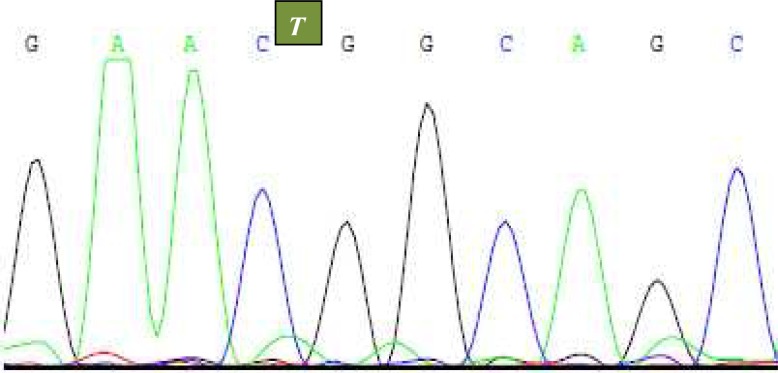
Exons 7 and 15 of the FGFR3 gene were successfully extracted from OSCC cases. In three cases (3 of 20) silent mutations were identified in exon 7. None resulted in aminoacid changes. All these mutations contained T to C substitution at nucleotide 882 and amino acid position 294 (Fig I). No mutation was identified in exon 15 (Table II).
Fig. 1.
Case 5, 10 and 14, T-to-C substitution at nucleotide 882 (exon 7).
Table II :
Fibroblast growth factor receptor 3 (FGFR3) mutational status in Oral Squamous Cell Carcinoma (OSCC)
| Case | Sex | Age | Stage | Exon | Mutation | AA change |
|---|---|---|---|---|---|---|
| 1 | F | 23 | II | - | - | - |
| 2 | M | 49 | III | - | - | - |
| 3 | M | 49 | I | - | - | - |
| 4 | M | 35 | IV | - | - | |
| 5 | M | 63 | III | 7 | 882 T to C | 294(Si) |
| 6 | M | 64 | II | - | - | - |
| 7 | M | 53 | IV | - | - | - |
| 8 | M | 79 | III | - | - | - |
| 9 | M | 64 | III | - | - | - |
| 10 | M | 72 | I | 7 | 882 T to C | 294(Si) |
| 11 | F | 76 | IV | - | - | - |
| 12 | M | 25 | IV | - | - | - |
| 13 | M | 73 | IV | - | - | - |
| 14 | M | 72 | II | 7 | 882 T to C | 294(Si) |
| 15 | F | 32 | IV | - | - | - |
| 16 | M | 54 | II | - | - | - |
| 17 | M | 52 | IV | - | - | - |
| 18 | M | 75 | IV | - | - | - |
| 19 | M | 79 | I | - | - | - |
| 20 | F | 64 | I | - | - | - |
AA, Amino acid; Si, Silent
DISCUSSION
Demographic studies have shown that oral and oropharyngeal squamous cell carcinomas represent 3% of cancers in men and 2% of cancers in women. In addition, these kind of cancers represent approximately 2% of the total death in men and 1% in women [13].
Although OSCC treatment strategies vary based on the stage and location of the cancer, wide surgical excision followed by radiotherapy is still considered as the main core of treatment. Based on the tumor stage, the 5-year disease-free survival rate for intraoral carcinoma will be 76%, if metastasis does not occur by the time of diagnosis (stage I and II), 41% when the cervical nodes are involved (stage III) and only 9% when metastasis below the clavicle is present (stage IV) [14].
Because of high mortality and morbidity associated with conventional treatment of OSCC, new therapeutic strategies are focused on molecular targeted therapies, especially against epidermal growth factor receptors (EGFRs). So, it is recommended to consider monoclonal antibodies and tyrosine kinase inhibitors (TKIs) in the chemotherapeutic regimen to improve treatment effectiveness and reduce post-surgical morbidity [15, 16]. Recent studies have introduced another receptor with tyrosine kinase activity, Fibroblast Growth Factor Receptor (FGFR), which may be considered as a target for molecular therapy. Although somatic mutations of FGFRs are reported in several other benign and malignant neoplasms including multiple myeloma, bladder carcinoma, cervix carcinoma, non-small cell lung cancer and benign skin epidermal tumors [9, 17–19], only in urothelial cell carcinomas FGFRs, especially FGFR3, have been introduced as a biomarker and potential therapeutic target [17].
In the present study, we analyzed exons 7 and 15 of FGFR3 gene to identify novel mutations in OSCC lesions which would lead to structural change and constitutive activation of the receptor. These exons were selected because previous studies have introduced exons 7, 10, 12, 15 and 19 as common mutation sites in related lesions [7].
Exon 7 encodes for the linker region between Ig`-like domains II–III in the extracellular ligand-binding domain; whereas, exon 15 is involved in the synthesis of the intracellular tyrosine kinase domain II [5].
We observed T-to-C transition of nucleotide 882 in three cases (3 out of 20) localized in exon 7. Both wild type and mutational codons encode the same amino acid (aspargine) and consequently this mutation has no significant effect on the receptor structure or function, so we may call it as a “silent mutation”.
One recent study [4] has reported the same mutation in SCC arising in actinic cheilitis (AC) and one additional missense mutation (753 C-to-T) in another SCC case. All these mutations are localized in exon 7. Although they additionally reported one missense and two silent mutations in AC cases, they did not interpret them without considering mutations observed in exons 15 and 17. Besides, Zhang et al. [11] did not report any mutation in exon 7 of OSCC cases using PCR-SSCP analysis method. We did not observe mutations reported by Chou et al. [4] in exon 15. They showed one missense mutation in SCC arising in AC and three mutations, one missense and two silent, in AC cases. The authors declared that the higher frequency of missense mutations in the early stage of SCC arising in AC may help in the early detection of malignancy transition potential of premalignant lesions and this issue would be relevant to possible target therapy.
In addition, in another viewpoint, the different mutational status observed between our results and their results may describe the different pathogenesis of OSCC and carcinoma of the lip, considering activating FGFR3 mutations as a receptor with tyrosine kinase activity. Similar to this conclusion has been made in other malignancies. For example Ware et al. [19] divided non-small cell lung cancers (NSCLCs) into two separate categories; the lesions in the first category respond to treatment with EGFR tyrosine kinase inhibitors (TKIs) such as imatinib, but the lesions in the second category develop resistance against TKIs and relapse; so the authors suggested combination therapy of EGFR and FGFR TKIs as a more efficient treatment for patients with NSCLCs. In the present study we did not analyze exon 17, but Zhang et al. showed a constitutive activating mutation of FGFR3 in 62% of oral squamous cell carcinoma cases.
They observed cases containing G697C mutation with enhanced tyrosine kinase activity compared to non-mutated cases and introduced this somatic mutation as a diagnostic or prognostic factor [11].
Despite the previous study, Chou et al. found only one missense mutation in AC without any dysplasia and one silent mutation in SCC arising in AC. Furthermore, considering mutations in exons 7, 15 and 17, they suggest activating FGFR3 mutations as a target for molecular therapy with small molecular tyrosine kinase inhibitors such as PD173074, SU5402 and PKC41230 which are in phase II trial studies [4].
Such controversial results have been demonstrated in other malignancies. For example, although Wu et al. introduced somatic mutations of FGFR3 in cervical carcinomas, they emphasized that it may not be as common as initial studies reported [20].
Zuivarloon et al. did not confirm the presence of FGFR3 mutations in prostate tumors which was reported in the previous studies. They suggested a minor role of these mutations to select patients for conservative management [21]. However, the process began initially with identifying FGFR3 germline mutations in skeletal disorders [22, 23] and was continued until the recognition of somatic mutation at the same location in various cancers, especially urothelial carcinomas and multiple myeloma [24, 25]. This is not the end of progression. In summary, although we did not find any activating FGFR3 mutation in exons 7 and 15 in snap frozen tissue biopsies of OSCC cases, it seems analyzing other exons or considering other advanced gene mutation assessment techniques could be helpful to clarify the role of these receptor mutations in OSCC pathogenesis.
Acknowledgments
This paper was supported financially by a grant of Dental Research Center of Tehran University of Medical Sciences (#89-03-70-10778). We would like to thank Dr. Fereidooni, Dr. Kamali, Dr. Hosseini and Mr. Joulaei (Iran Naional Tumor Bank) for kindly providing the specimens.
REFERENCES
- 1.Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ, Tsai TH, et al. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res. 2008 Jul 1;14(13):4085–95. doi: 10.1158/1078-0432.CCR-07-4404. [DOI] [PubMed] [Google Scholar]
- 2.The Diseases Control Center of Ministry of Health, Treatment and Medical Education of Islamic Republic of Iran. The registered statistical report of cancers. 2005.
- 3.Han J, Kioi M, Chu WS, Kasperbauer JL, Strome SE, Puri RK. Identification of potential therapeutic targets in human head & neck squamous cell carcinoma. Head Neck Oncol. 2009 Jul 14;1:27. doi: 10.1186/1758-3284-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou A, Dekker N, Jordan RC. Identification of novel fibroblast growth factor receptor 3 gene mutations in actinic cheilitis and squamous cell carcinoma of the lip. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009 Apr;107(4):535–41. doi: 10.1016/j.tripleo.2008.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005 Apr;16(2):139–49. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, et al. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996 Jun 21;271(25):15292–7. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 7.Hafner C, Vogt T, Hartmann A. FGFR3 mutations in benign skin tumors. Cell Cycle. 2006 Dec;5(23):2723–8. doi: 10.4161/cc.5.23.3509. [DOI] [PubMed] [Google Scholar]
- 8.Bernard-Pierrot I, Brams A, Dunois-Larde C, Caillault A, Diez de Medina SG, Cappellen D, et al. Oncogenic properties of the mutated forms of fibroblast growth factor receptor 3b. Carcinogenesis. 2006 Apr;27(4):740–7. doi: 10.1093/carcin/bgi290. [DOI] [PubMed] [Google Scholar]
- 9.Logie A, Dunois-Larde C, Rosty C, Levrel O, Blanche M, Ribeiro A, et al. Activating mutations of the tyrosine kinase receptor FGFR3 are associated with benign skin tumors in mice and humans. Hum Mol Genet. 2005 May;14(9):1153–60. doi: 10.1093/hmg/ddi127. [DOI] [PubMed] [Google Scholar]
- 10.Agha-Hosseini F, Khazabb M, Parvaneroo A. Evaluation of chromosomal disorders in Tissueand Blood Samples in Patients with oral squamous cell carcinoma. J Dent TUMS. 2004;1(4):25–30. [Google Scholar]
- 11.Zhang Y, Hiraishi Y, Wang H, Sumi KS, Hayashido Y, Toratani S, et al. Constitutive activating mutation of the FGFR3b in oral squamous cell carcinomas. Int J Cancer. 2005 Oct;117(1):166–8. doi: 10.1002/ijc.21145. [DOI] [PubMed] [Google Scholar]
- 12.Billerey C, Chopin D, Aubriot-Lorton MH, Ricol D, Gil Diez de Medina S, Van Rhijn B, et al. Frequent FGFR3 mutations in papillary non-invasive bladder (pTa) tumors. Am J Pathol. 2001 Jun;158(6):1955–9. doi: 10.1016/S0002-9440(10)64665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Regezi JA, Sciubba JJ, Jordan RC. 5th ed. Missouri: Saunders; 2008. Oral pathology; Clinical pathologic correlations; pp. 48–52. [Google Scholar]
- 14.Neville BW, Damm DD, Allen CM, Bouqout JE. 3rd ed. Missouri: Saunders; 2009. Oral and maxillofacial pathology; pp. 409–12. [Google Scholar]
- 15.Bozec A, Formento P, Fischel JL, Etienne-Grimaldi MC, Milano G. Tapered dose versus constant drug exposure to anti-EGFR drugs on head-and-neck cancer xenografts. A comparison between cetuximab and gefitinib. Oral Oncol. 2009 Mar;46(3):172–7. doi: 10.1016/j.oraloncology.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Molinolo AA, Amornphimoltham P, Squarize CH, Castilho RM, Patel V, Gutkind JS. Dysregulated molecular networks in head and neck carcinogenesis. Oral Oncol. 2009 Apr-May;45(4–5):324–34. doi: 10.1016/j.oraloncology.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Rhijn BW, Zuiverloon TC, Vis AN, Radvanyi F, van Leenders GJ, Ooms BC, et al. Molecular grade (FGFR3/MIB-1) and EORTC risk scores are predictive in primary non-muscle-invasive bladder cancer. Eur Urol. 2010 Sep;58(3):433–41. doi: 10.1016/j.eururo.2010.05.043. [DOI] [PubMed] [Google Scholar]
- 18.Vatsveen TK, Brenne AT, Dai HY, Waage A, Sundan A, Børset M. FGFR3 is expressed and is important for survival in INA-6, a human myeloma cell line without a t (4;14) Eur J Haematol. 2009 Nov;83(5):471–6. doi: 10.1111/j.1600-0609.2009.01312.x. [DOI] [PubMed] [Google Scholar]
- 19.Ware KE, Marshall ME, Heasley LR, Marek L, Hinz TK, Hercule P, et al. Rapidly acquired resistance to EGFR tyrosine kinase in hibitors in NSCLC cell lines through de-repression of FGFR2 and FGFR3 expression. PLoS One. 2010 Nov;5(11):e14117. doi: 10.1371/journal.pone.0014117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu R, Connolly D, Ngelangel C, Bosch FX, Munoz N, Cho KR. Somatic mutations of fibroblast growth factor receptor 3 (FGFR3) are uncommon in carcinomas of the uterine cervix. Oncogene. 2000 Nov;19(48):5543–6. doi: 10.1038/sj.onc.1203934. [DOI] [PubMed] [Google Scholar]
- 21.Zuiverloon TC, Boormans JL, Trapman J, van Leenders GJ, Zwarthoff EC. No evidence of FGFR3 mutations in prostate cancer. Prostate. 2011 May;71(6):637–41. doi: 10.1002/pros.21279. [DOI] [PubMed] [Google Scholar]
- 22.Webster MK, Donoghue DJ. FGFR activation in skeletal disorders: too much of a good thing. Trends Genet. 1997 May;13(5):178–82. doi: 10.1016/s0168-9525(97)01131-1. [DOI] [PubMed] [Google Scholar]
- 23.Passos-Bueno MR, Wilcox WR, Jabs EW, Sertie AL, Alonso LG, Kitoh H. Clinical spectrum of fibroblast growth factor receptor mutations. Hum Mutat. 1999;14(2):115–25. doi: 10.1002/(SICI)1098-1004(1999)14:2<115::AID-HUMU3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Dodurga Y, Tataroglu C, Kesen Z, Satiroglu-Tufan NL. Incidence of fibroblast growth factor receptor 3 gene (FGFR3) A248C, S249C, G372C, and T375C mutations in bladder cancer. Genet Mol Res. 2011 Jan;10(1):86–95. doi: 10.4238/vol10-1gmr923. [DOI] [PubMed] [Google Scholar]
- 25.Qing J, Du X, Chen Y, Chan P, Li H, Wu P, et al. Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice. J Clin Invest. 2009 May;119(5):1216–29. doi: 10.1172/JCI38017. [DOI] [PMC free article] [PubMed] [Google Scholar]