Abstract
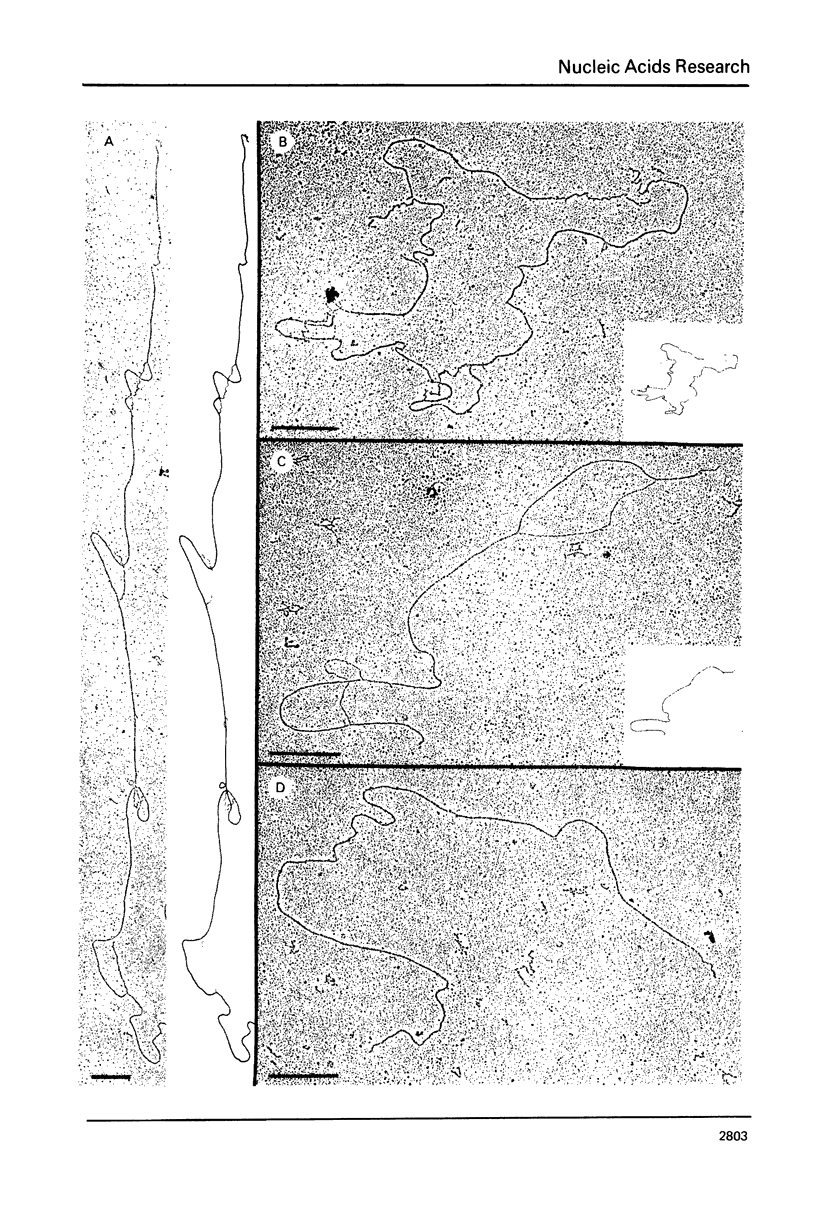
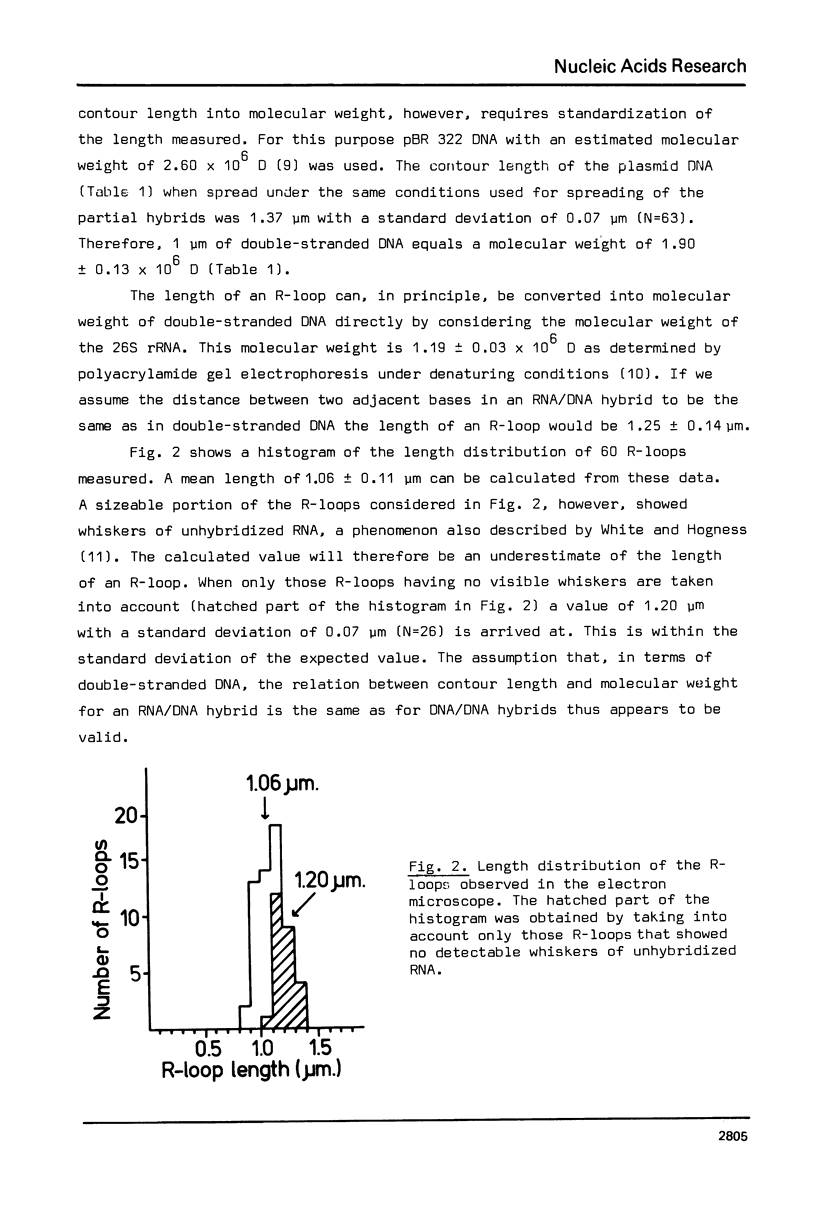
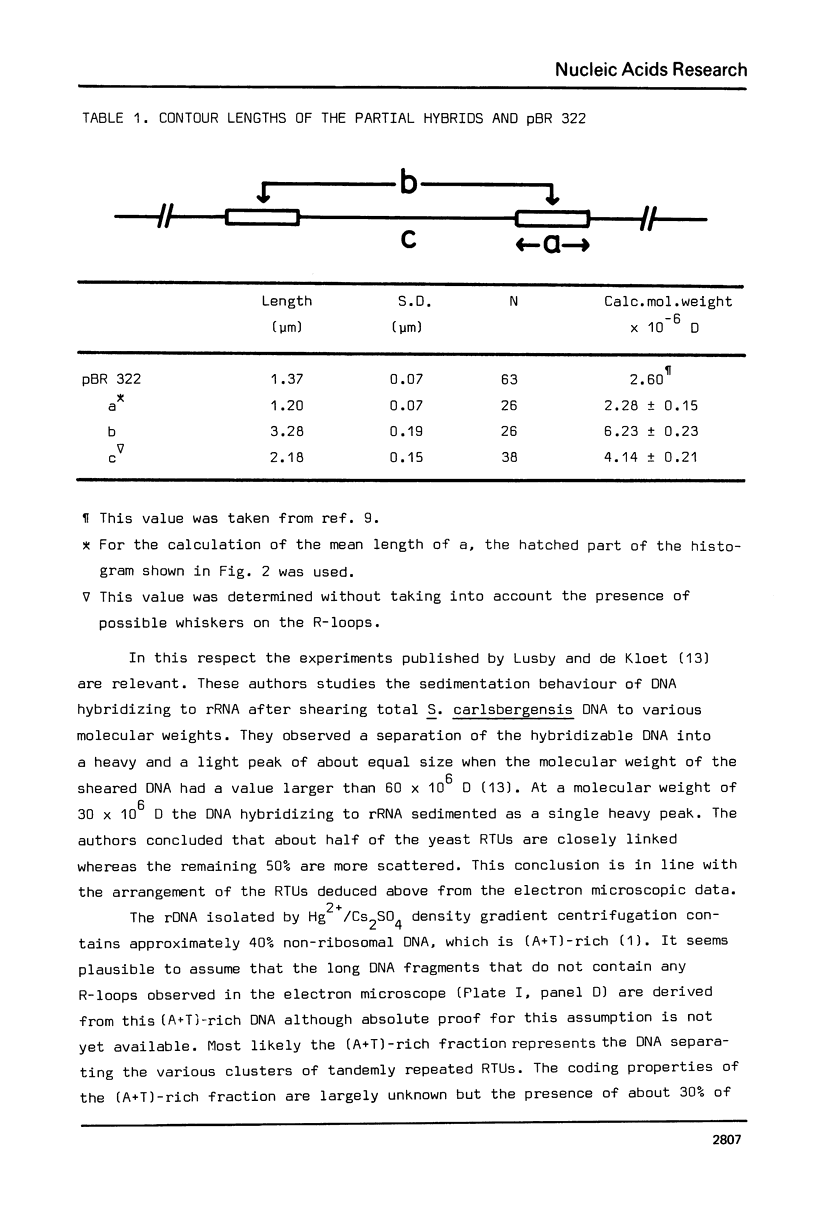
The genetic organization of the multiple ribosomal transcription units (RTUs) on the genome of the yeast Saccharomyces carlsbergensis was studied by electron microscopy of purified ribosomal DNA hybridized to 26S rRNA using the R-loop technique (Thomas, M., White, R.L. and Davis, R.W. (1973) Proc. Natl. Acad. Sci. U.S. 73, 2294-2298). Plasmid pBR 322, the molecular weight of which is known, was used as a standard for converting contour length of double-stranded DNA into molecular weight. The 140 yeast RTUs were found to be arrayed in tandem repeats, each repeat containing at most 0.4 X 10(6) D (about 6% of the length of the RTU) of non-transcribed spacer DNA. The repeats, in turn, are arranged in a number of clusters separated by much longer stretches of non-ribosomal DNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brand R. C., Planta R. J. The molecular weights of yeast ribosomal precursor RNAs. Mol Biol Rep. 1975 Dec;2(4):321–325. doi: 10.1007/BF00357019. [DOI] [PubMed] [Google Scholar]
- Cramer J. H., Farrelly F. W., Rownd R. H. Restriction endonuclease analysis of ribosomal DNA from Saccharomyces cerevisiae. Mol Gen Genet. 1976 Nov 17;148(3):233–241. doi: 10.1007/BF00332897. [DOI] [PubMed] [Google Scholar]
- Goebel W., Bonewald R. Class of small multicopy plasmids originating from the mutant antibiotic resistance factor R1 drd-19B2. J Bacteriol. 1975 Aug;123(2):658–665. doi: 10.1128/jb.123.2.658-665.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusby E. W., Jr, de Kloet S. R. The heavy DNA satellite of yeast and its relationship to the ribosomal RNA genes. Arch Biochem Biophys. 1976 May;174(1):187–191. doi: 10.1016/0003-9861(76)90337-4. [DOI] [PubMed] [Google Scholar]
- Meyerink J. H., Retèl J., Planta R. J., Heidekamp F. Topographical analysis of yeast ribosomal DNA by cleavage with restriction endonucleases. Mol Biol Rep. 1976 Apr;2(5):393–400. doi: 10.1007/BF00366261. [DOI] [PubMed] [Google Scholar]
- Meyerink J. H., Retèl J. Topographical analysis of yeast ribosomal DNA by cleavage with restriction endonucleases. II. The physical map of EcoRI fragments. Nucleic Acids Res. 1976 Oct;3(10):2697–2707. doi: 10.1093/nar/3.10.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder J. C., Staynov D. Z., Gratzer W. B. Properties of RNA in formamide. Biochemistry. 1974 Dec 17;13(26):5367–5373. doi: 10.1021/bi00723a018. [DOI] [PubMed] [Google Scholar]
- Retèl J., Planta R. J. Nuclear satellite DNAs of yeast. Biochim Biophys Acta. 1972 Oct 27;281(3):299–309. doi: 10.1016/0005-2787(72)90442-x. [DOI] [PubMed] [Google Scholar]
- Retèl J., Planta R. J. On the mechanism of the biosynthesis of ribosomal RNA in yeast. Biochim Biophys Acta. 1970 Dec 14;224(2):458–469. doi: 10.1016/0005-2787(70)90578-2. [DOI] [PubMed] [Google Scholar]
- Retèl J., Van Keulen H. Characterization of yeast ribosomal DNA. Eur J Biochem. 1975 Oct 1;58(1):51–57. doi: 10.1111/j.1432-1033.1975.tb02347.x. [DOI] [PubMed] [Google Scholar]
- Thomas M., White R. L., Davis R. W. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. The structural organization of ribosomal DNA in Drosophila melanogaster. Cell. 1977 Feb;10(2):193–212. doi: 10.1016/0092-8674(77)90214-8. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Reeder R. H., Dawid I. B., Brown D. D. Arrangement of length heterogeneity in repeating units of amplified and chromosomal ribosomal DNA from Xenopus laevis. J Mol Biol. 1976 Aug 25;105(4):487–505. doi: 10.1016/0022-2836(76)90230-8. [DOI] [PubMed] [Google Scholar]
- White R. L., Hogness D. S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):177–192. doi: 10.1016/0092-8674(77)90213-6. [DOI] [PubMed] [Google Scholar]
- van den Bos R. C., Retèl J., Planta R. J. The size and the location of the ribosomal RNA segments in ribosomal precursor RNA of yeast. Biochim Biophys Acta. 1971 Mar 25;232(3):494–508. doi: 10.1016/0005-2787(71)90603-4. [DOI] [PubMed] [Google Scholar]



