Abstract
We have analysed the efficiency of all mitochondrial protein coding genes and six nuclear markers (Adora3, Adrb2, Bdnf, Irbp, Rag2 and Vwf) in reconstructing and statistically supporting known amniote groups (murines, rodents, primates, eutherians, metatherians, therians). The efficiencies of maximum likelihood, Bayesian inference, maximum parsimony, neighbor-joining and UPGMA were also evaluated, by assessing the number of correct and incorrect recovered groupings. In addition, we have compared support values using the conservative bootstrap test and the Bayesian posterior probabilities. First, no correlation was observed between gene size and marker efficiency in recovering or supporting correct nodes. As expected, tree-building methods performed similarly, even UPGMA that, in some cases, outperformed other most extensively used methods. Bayesian posterior probabilities tend to show much higher support values than the conservative bootstrap test, for correct and incorrect nodes. Our results also suggest that nuclear markers do not necessarily show a better performance than mitochondrial genes. The so-called dependency among mitochondrial markers was not observed comparing genome performances. Finally, the amniote groups with lowest recovery rates were therians and rodents, despite the morphological support for their monophyletic status. We suggest that, regardless of the tree-building method, a few carefully selected genes are able to unfold a detailed and robust scenario of phylogenetic hypotheses, particularly if taxon sampling is increased.
Keywords: phylogenetic groups, mitochondrial genes, nuclear genes, tree-building methods, bootstrap tests
Introduction
Different topologies may be obtained for the same set of organisms when different genes, models and methods are used to reconstruct the phylogeny.1,2 Once regarded as a weakness of molecular phylogenetics, the relative independence of molecular markers is now regarded as an important asset, enabling consistency tests on molecular based topologies.3,4
In spite of the positive aspects, however, multiple markers inconsistency does represent a problem with actual data.5–8 The overwhelming and rapidly accumulating amounts of molecular data have just scratched the surface of the problem.9 It is clear that different genes present distinct probabilities of efficiently recovering the correct tree and that a careful marker selection has been shown to be more important than the choice of a tree-building method.1,10 The longer the alignment, for instance, the lower is the sampling error.6,7 Also, a single gene tree may differ from the species tree, which is more likely the result of a multi-marker analysis. On the other hand, systematic errors tend to increase with sequence length9 and evolutionary model selection becomes a far more complex issue7,11,12 in a multi-marker analysis.
While computer simulations have extensively addressed tree-building method reconstruction,13 marker efficiency may only be estimated by the use of known evolutionary trees.1,2,10 In fact, if the amount of fossil, morphological and molecular data that support a particular topology is overwhelming, it becomes possible to assume it to be known. In these cases, efficiency of markers may be estimated for different taxonomic groups and the most efficient markers should be useful to unveil unknown evolutionary relationships in the group.
Previous known-tree studies have already tested the efficiency of genes and tree-building methods,1,14 but most analyses were restricted to either mitochondrial or to nuclear genes, precluding comparisons between genome efficiencies. Nuclear genome is passive of recombination during sexual reproduction and, hence, nuclear markers are truly independent in a way that mitochondrial genes are not.7 Mitochondrial genes, on the other hand, do not present paralogy-related problems and thus comparisons between genomes will be informative for phylogeneticists. A recent analysis included mitochondrial and nuclear genes, but only Bayesian Inference methods were tested.10
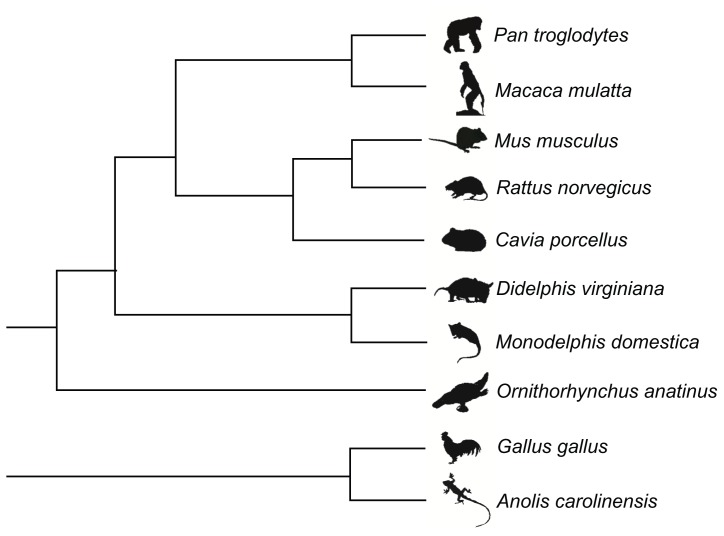
In this work, we have evaluated the efficiency of genes and tree-building methods in recovering a known amniote phylogeny (Fig. 1). We have tested the efficiencies of nucleotide sequences of both nuclear and mitochondrial genes and UPGMA, neighbor-joining, maximum parsimony, Bayesian Inference and maximum likelihood methods of phylogenetic inference.
Figure 1.
Known amniote phylogeny used to test tree-building methods and nuclear and mitochondrial markers to recover this topology.
Materials and Methods
Nucleotide sequences were downloaded for ten vertebrate taxa for which evolutionary relationships were consensual and well established by the fossil record, morphological data and molecular studies. These are Anolis carolinensis, Cavia porcellus, Didelphis virginiana, Gallus gallus, Macaca mulatta, Monodelphis domestica, Mus musculus, Ornithorhynchus anatinus, Pan troglodytes and Rattus norvegicus. Our known phylogeny was established based on multiple and independent lines of evidence. The sister group status, for instance, between squamates (Anolis lineage) and birds (Gallus lineage), and their relationship to mammals are universally supported by paleontological, morphological and molecular data.15–19 Conversely, the relationship between Monotremata (Ornithorhynchus lineage) and the remaining mammals (clade Theria) is also evident by various studies.20–24 Monophyly of didelphid marsupials (Monodelphis and Didelphis) and their relationship to placental mammals are supported by evidence.24,25 The monophyletic status of primates and rodents has been widely sustained by several types of data.21,24,26–29 Even though we acknowledge the fact that uncertainties are always pertinent to any given phylogenetic hypothesis, overwhelming evidences such as those provide a very strong case for the tree topology assumed for this work.
Thirteen mitochondrial (Atp6, Atp8, Cox1, Cox2, Cox3, Nd1, Nd2, Nd3, Nd4, Nd4l, Nd5, Nd6 and Cytb) and six nuclear (Adora3, Adrb2, Bdnf, Irbp, Rag2 and Vwf) genes were used in our analysis. Nuclear genes were selected based on the number of sequences available at GenBank. Other nuclear markers were tested but discarded due to unreliable alignments. All protein coding mitochondrial genes were included in our analyses. Since sequences may have not been available for all species in sequence data banks, not all phylogenetic groups were tested for all markers. For GenBank access numbers, see Supplementary Material 1.
In this paper, we have analysed the efficiency of tree-building methods and mitochondrial and nuclear markers in recovering a known vertebrate phylogeny (Fig. 1). In order to evaluate results in light of evolutionary rates of genes, Table 1 shows number of nucleotides and Rattus vs. Didelphis distance values for all genes evaluated. Among mitochondrial, cytochrome oxidase genes are among the most conservative, whereas Atp8, Nd2 and Nd6 exhibit the highest divergence between Rattus and Didelphis. Considering all markers, the nuclear Irbp is the fastest evolving gene whereas Adora3, also nuclear, is the most conservative.
Table 1.
Nucleotide number of each mitochondrial and nuclear genes and Rattus vs. Didelphis distance values for all genes evaluated.
| Number of base pairs | Rattus x Didelphis | |
|---|---|---|
| Mitochondrial genes | ||
| Atp6 | 684 | 0.29 |
| Atp8 | 211 | 0.42 |
| Cox1 | 1557 | 0.21 |
| Cox2 | 693 | 0.20 |
| Cox3 | 786 | 0.21 |
| Nd1 | 960 | 0.29 |
| Nd2 | 1053 | 0.41 |
| Nd3 | 352 | 0.34 |
| Nd4 | 1385 | 0.31 |
| Nd4l | 297 | 0.37 |
| Nd5 | 1842 | 0.34 |
| Nd6 | 531 | 0.43 |
| Cytb | 1152 | 0.25 |
| Nuclear genes | ||
| Adbr2 | 326 | 0.26 |
| Adora3 | 725 | 0.13 |
| Irbp | 1244 | 0.64 |
| Rag2 | 446 | 0.30 |
| Bndf | 236 | 0.27 |
| Vwf | 1178 | 0.30 |
The decision to analyze nucleotide sequences only was based on the fact that they tend to outperform amino acid sequences due to their three-fold advantage in number.5 Even though they are expected to display a large amount of noise, third codon positions contain informative signal for phylogenetic tree reconstruction30 and were included in our analyses.
Nucleotide sequences were aligned based on translated amino acid sequences using ClustalW.31 After alignment, editing removed poorly aligned flanking regions. In order to evaluate the effect of gene conservativeness, pair-wise proportion of different residues (p-distance) was calculated between all sequence pairs (see Supplemental Material 2).
Phylogenetic trees were reconstructed using the Mega 432 software for UPGMA,33 neighbor-joining (NJ)34 and maximum parsimony (MP),35,36 with default parameters. Both UPGMA and NJ methods are distance based tree-building methods and selected distance model were: proportion of differences, Jukes- Cantor (JC)37 and Kimura 2-parameters (K2P).38
In the case of MP topology search, three different algorithms were used: the (max-mini) branch and bound and two types of heuristic searches. A core tree with three taxa selected for the largest number of steps starts the algorithm. Taxa addition hence continues and, at each step, the number of steps is calculated until it exceeds that of a previously reconstructed tree and all derived trees are ignored. All possible pathways are evaluated. While branch-and-bound algorithms guarantees that all MP tree will be found, this is not the case for heuristic searches. Two heuristics MP algorithms were applied to our dataset: close neighbor interchange (CNI) and min-mini. In the first case, the trees that differed from the provisional tree by 2 and 4 steps are examined, and this search is repeated until there is no remaining tree with a smaller length size. The min-mini algorithm is similar to branch-and-bound in what concerns the initial core tree of three taxa, but the order of taxon addition differs from the former as the taxon chosen for the next step of taxon addition is the one with the minimum of all minimum values. The aim is to reach the MP or a suboptimal MP tree relatively quickly.
Maximum likelihood trees, on the other hand, were computed with the on line version of PhyML.39 In this case, the probability of a certain topology, given alignment and model (ie, likelihood), is computed for each possible tree. The ML tree shows the highest likelihood. ML algorithm is a discrete character method, but, as in UPGMA and NJ, an evolutionary model must also be selected.
In the ML case, BioNJ was chosen as the initial and topology searches were based on Nearest-Neighbor- Interchange and Subtree Prunning and Reconnecting heuristic algorithms. Hasegawa- Kishino-Yano (HKY), Tamura-Nei (TN) and GTR evolutionary models were implemented. It has been shown that more sophisticated models do not necessarily yield better topologies.40 ML methods, however, are not as vulnerable to large variances32 and over- parameterization as distance methods.9,41
Bayesian analyses were performed using Mr. Bayes software42 as estimated by Monte Carlo Markov Chain (MCMC). Analyses were found to be robust, since mean standard deviations were not higher than 0.01 after 100.000 generations. As in ML, more complex substitution models, JC69, HKY and GTR, were selected for analyses. Bayesian posterior probabilities were estimated.
The reliability of each tree was measured using the bootstrap test43 with 100 replicates.44 We have summarized the vast amount of phylogenies reconstructed by calculating the number of nodes that have recovered such partitions. Separate values were estimated for correct and incorrect partitions so that overall efficiencies of markers and genes may be evaluated. Also, since in phylogenetic studies branches with low statistical support are seldom considered, we have included results for correct and incorrect branching patterns with bootstrap values over 90. If sequences were not available for the test, values were removed from the total.
Even though it is a reliable test,44 the phylogenetic bootstrap test does not yield the probability of a given partition being on the true tree (ie, accuracy), but it actually tests the probability of recovering the same partition given an independent data set (ie, repeatability).43,45 It is shown, for instance, that a high bootstrap value itself does not indicate that the grouping is correct due to systematic errors.2,9 Also, bootstrapped data need to be independent and identically distributed (iid) which is probably not the case for sequence data.46 Nonetheless, since test results are usually interpreted this way in literature it is useful to understand properties of the test regarding this issue.
Results
We will discuss our results in light of the efficiency of tree-building methods, of markers and of genomes for recovering particular vertebrate groups. Correct and incorrect recovered partition numbers are displayed separately for each tree-building method and taxonomic group for mitochondrial (Table 2) and nuclear (Table 3) genes. In those tables, correct values correspond to the number of nodes that correctly recovered known amniote partitions. Accordingly, incorrect values are the number of nodes that break the monophyly of known amniote partitions. In both cases, results may be observed for all nodes and, separately, for those branches with high bootstrap values, ie, >90%.
Table 2.
Correct and incorrect recovered partition numbers separately for each tree-building method and taxonomic group for mitochondrial genes.
| Murinae | Rodentia | Primates | Eutheria | Metatheria | Theria | Mammalia | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|
|
|||||||||
| # | #>90 | # | #>90 | # | #>90 | # | #>90 | # | #>90 | # | #>90 | # | #>90 | # | #>90 | |
| Correct | ||||||||||||||||
| Atp6 | 15 | 13 | 11 | 4 | 15 | 12 | 12 | 10 | 15 | 15 | 3 | 0 | 15 | 15 | 86 | 69 |
| Atp8 | 15 | 9 | 15 | 4 | 12 | 0 | 3 | 0 | 15 | 10 | 0 | 0 | 15 | 15 | 75 | 38 |
| Cox1 | 12 | 12 | 0 | 0 | 15 | 15 | 0 | 0 | 14 | 13 | 4 | 1 | 8 | 2 | 53 | 43 |
| Cox2 | 15 | 9 | 3 | 0 | 15 | 15 | 0 | 0 | 15 | 15 | 0 | 0 | 3 | 0 | 51 | 39 |
| Cox3 | 13 | 9 | 0 | 0 | 15 | 12 | 3 | 0 | 13 | 3 | 0 | 0 | 15 | 14 | 59 | 38 |
| Nd1 | 15 | 13 | 0 | 0 | 15 | 15 | 14 | 3 | 15 | 15 | 0 | 0 | 12 | 3 | 71 | 49 |
| Nd2 | 15 | 15 | 0 | 0 | 15 | 15 | 15 | 5 | 15 | 15 | 0 | 0 | 15 | 12 | 75 | 62 |
| Nd3 | 15 | 15 | 12 | 0 | 15 | 14 | 5 | 0 | 13 | 5 | 0 | 0 | 15 | 15 | 75 | 49 |
| Nd4 | 15 | 15 | 4 | 0 | 15 | 15 | 11 | 4 | 15 | 15 | 3 | 0 | 15 | 13 | 78 | 62 |
| Nd4l | 15 | 15 | 15 | 0 | 15 | 15 | 15 | 6 | 15 | 12 | 0 | 0 | 15 | 12 | 90 | 60 |
| Nd5 | 15 | 15 | 15 | 3 | 15 | 15 | 15 | 3 | 15 | 15 | 4 | 2 | 12 | 5 | 91 | 58 |
| Nd6 | 15 | 15 | 8 | 2 | 15 | 15 | 15 | 15 | 15 | 15 | 0 | 0 | 15 | 15 | 83 | 77 |
| Cytb | 15 | 14 | 4 | 0 | 15 | 15 | 3 | 0 | 15 | 15 | 0 | 0 | 4 | 2 | 56 | 46 |
| Average | 14.6 | 13.0 | 6.7 | 1.0 | 14.8 | 13.3 | 8.5 | 3.5 | 14.6 | 12.5 | 1.1 | 0.2 | 12.2 | 9.5 | 72.5 | 53.1 |
| Incorrect | ||||||||||||||||
| Atp6 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 12 | 8 | 0 | 0 | 19 | 8 |
| Atp8 | 0 | 0 | 0 | 0 | 3 | 0 | 12 | 3 | 0 | 0 | 15 | 3 | 0 | 0 | 30 | 6 |
| Cox1 | 3 | 0 | 15 | 3 | 0 | 0 | 15 | 3 | 0 | 0 | 11 | 1 | 7 | 1 | 51 | 8 |
| Cox2 | 0 | 0 | 12 | 3 | 0 | 0 | 15 | 9 | 0 | 0 | 15 | 6 | 12 | 3 | 54 | 21 |
| Cox3 | 2 | 0 | 15 | 3 | 0 | 0 | 12 | 3 | 2 | 0 | 15 | 3 | 0 | 0 | 46 | 9 |
| Nd1 | 0 | 0 | 15 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 15 | 4 | 3 | 0 | 36 | 5 |
| Nd2 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 7 | 0 | 0 | 30 | 7 |
| Nd3 | 0 | 0 | 3 | 0 | 0 | 0 | 10 | 1 | 2 | 0 | 15 | 2 | 0 | 0 | 30 | 3 |
| Nd4 | 0 | 0 | 11 | 3 | 0 | 0 | 4 | 0 | 0 | 0 | 12 | 9 | 0 | 0 | 27 | 12 |
| Nd4l | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 1 | 0 | 0 | 15 | 1 |
| Nd5 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 11 | 0 | 3 | 0 | 15 | 0 |
| Nd6 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 7 | 0 | 0 | 22 | 7 |
| Cytb | 0 | 0 | 11 | 1 | 0 | 0 | 15 | 1 | 0 | 0 | 15 | 5 | 11 | 1 | 52 | 8 |
| Average | 0.4 | 0.0 | 8.3 | 1.1 | 0.2 | 0.0 | 6.9 | 1.5 | 0.3 | 0.0 | 13.9 | 4.3 | 2.8 | 0.4 | 32.8 | 7.3 |
| Correct | ||||||||||||||||
| NJ | 39 | 34 | 20 | 2 | 39 | 36 | 18 | 6 | 38 | 31 | 0 | 0 | 27 | 24 | 181 | 133 |
| UPGMA | 39 | 39 | 21 | 3 | 39 | 30 | 21 | 6 | 39 | 36 | 0 | 0 | 33 | 27 | 192 | 141 |
| MP | 34 | 22 | 20 | 0 | 39 | 32 | 19 | 4 | 37 | 26 | 1 | 0 | 30 | 22 | 180 | 106 |
| ML | 39 | 33 | 9 | 0 | 36 | 36 | 30 | 9 | 37 | 29 | 8 | 0 | 36 | 17 | 195 | 124 |
| Bayesian | 39 | 39 | 17 | 7 | 39 | 36 | 23 | 21 | 39 | 36 | 5 | 2 | 33 | 32 | 195 | 173 |
| Average | 38.0 | 33.4 | 17.4 | 2.4 | 38.4 | 34.0 | 22.2 | 9.2 | 38.0 | 31.6 | 2.8 | 0.4 | 31.8 | 24.4 | 188.6 | 135.4 |
| Incorrect | ||||||||||||||||
| NJ | 0 | 0 | 19 | 0 | 0 | 0 | 24 | 3 | 14 | 0 | 25 | 0 | 12 | 0 | 94 | 3 |
| UPGMA | 0 | 0 | 18 | 0 | 0 | 0 | 18 | 3 | 12 | 0 | 27 | 3 | 6 | 0 | 81 | 6 |
| MP | 5 | 0 | 19 | 0 | 0 | 0 | 20 | 0 | 22 | 0 | 18 | 0 | 9 | 0 | 93 | 0 |
| ML | 0 | 0 | 30 | 0 | 3 | 0 | 12 | 0 | 18 | 0 | 15 | 3 | 3 | 0 | 81 | 3 |
| Bayesian | 0 | 0 | 22 | 27 | 0 | 0 | 16 | 14 | 0 | 0 | 34 | 29 | 6 | 5 | 78 | 75 |
| Average | 1.0 | 0.0 | 21.6 | 5.4 | 0.6 | 0.0 | 18.0 | 4.0 | 13.2 | 0.0 | 23.8 | 7.0 | 7.2 | 1.0 | 85.4 | 17.4 |
Notes: For a particular taxonomic group, results are given by marker (sum of nodes for all among tree-building methods) and by tree-building method (sum of nodes among different markers). On the first column of each taxonomic group is displayed the number of nodes recovered (correctly or incorrectly), whereas on the second column is the number of nodes recovered with a bootstrap value higher than 90. The average value for each taxonomic group is shown at the bottom of each table, as well as the sum (#) of each correct and incorrect node totally and the ones higher than 90 (bootstrap value).
Table 3.
Correct and incorrect recovered partition numbers separately for each tree-building method and taxonomic group for nuclear genes.
| Murinae | Rodentia | Primates | Eutheria | Metatheria | Theria | Mammalia | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|
|
|||||||||
| # | #>90 | # | #>90 | # | #>90 | # | #>90 | # | #>90 | # | #>90 | # | #>90 | # | #>90 | |
| Correct | ||||||||||||||||
| Adrb2 | 15 | 15 | 0 | 0 | 15 | 15 | 0 | 0 | 0 | 0 | 12 | 9 | 12 | 9 | 54 | 48 |
| Adora3 | 15 | 15 | NT | NT | 15 | 15 | 15 | 6 | NT | NT | 15 | 12 | 15 | 12 | 75 | 60 |
| Irbp | 15 | 15 | 9 | 2 | NT | NT | 9 | 2 | NT | NT | NT | NT | NT | NT | 33 | 19 |
| Rag2 | 15 | 15 | NT | NT | 15 | 12 | 15 | 15 | NT | NT | 15 | 15 | NT | NT | 60 | 57 |
| Bndf | 15 | 9 | 7 | 1 | 12 | 7 | 6 | 0 | 15 | 15 | 3 | 0 | 3 | 0 | 61 | 32 |
| Vwf | 15 | 15 | 5 | 0 | 12 | 12 | 9 | 9 | 15 | 15 | NT | NT | 12 | 12 | 68 | 63 |
| Average | 15.0 | 14.0 | 5.3 | 0.8 | 13.8 | 12.2 | 9.0 | 5.3 | 10.0 | 10.0 | 11.3 | 9.0 | 10.5 | 8.3 | 58.5 | 46.5 |
| Incorrect | ||||||||||||||||
| Adrb2 | 0 | 0 | 15 | 6 | 0 | 0 | 15 | 4 | 15 | 9 | 3 | 0 | 3 | 0 | 51 | 19 |
| Adora3 | 0 | 0 | NT | NT | 0 | 0 | 0 | 0 | NT | NT | 0 | 0 | 0 | 0 | 0 | 0 |
| Irbp | 0 | 0 | 6 | 0 | NT | NT | 6 | 0 | NT | NT | NT | NT | NT | NT | 12 | 0 |
| Rag2 | 0 | 0 | NT | NT | 0 | 0 | 0 | 0 | NT | NT | 0 | 0 | NT | NT | 0 | 0 |
| Bndf | 3 | 0 | 6 | 0 | 5 | 2 | 9 | 2 | 0 | 0 | 12 | 0 | 12 | 0 | 47 | 4 |
| Vwf | 0 | 0 | 10 | 3 | 0 | 0 | 6 | 3 | 0 | 0 | NT | NT | 3 | 0 | 19 | 6 |
| Average | 0.5 | 0.0 | 9.3 | 2.3 | 1.0 | 0.4 | 6.0 | 1.5 | 5.0 | 3.0 | 3.8 | 0.0 | 4.5 | 0.0 | 21.5 | 4.8 |
| Correct | ||||||||||||||||
| NJ | 18 | 18 | 5 | 0 | 15 | 13 | 12 | 9 | 6 | 6 | 9 | 9 | 9 | 9 | 74 | 64 |
| UPGMA | 18 | 18 | 0 | 0 | 15 | 15 | 12 | 6 | 6 | 6 | 9 | 3 | 9 | 3 | 69 | 51 |
| MP | 18 | 15 | 3 | 0 | 15 | 12 | 11 | 6 | 6 | 6 | 10 | 9 | 10 | 9 | 73 | 57 |
| ML | 15 | 15 | 9 | 0 | 9 | 6 | 9 | 3 | 6 | 6 | 6 | 6 | 3 | 3 | 57 | 39 |
| Bayesian | 18 | 18 | 4 | 3 | 15 | 15 | 10 | 8 | 6 | 6 | 11 | 9 | 11 | 9 | 75 | 68 |
| Average | 17.4 | 16.8 | 4.2 | 0.6 | 13.8 | 12.2 | 10.8 | 6.4 | 6.0 | 6.0 | 9.0 | 7.2 | 8.4 | 6.6 | 69.6 | 55.8 |
| Incorrect | ||||||||||||||||
| NJ | 0 | 0 | 7 | 0 | 0 | 0 | 6 | 0 | 3 | 3 | 3 | 0 | 3 | 0 | 22 | 3 |
| UPGMA | 0 | 0 | 12 | 3 | 0 | 0 | 6 | 3 | 3 | 3 | 3 | 0 | 3 | 0 | 27 | 9 |
| MP | 0 | 0 | 9 | 0 | 0 | 0 | 7 | 0 | 3 | 0 | 2 | 0 | 2 | 0 | 23 | 0 |
| ML | 3 | 0 | 3 | 0 | 6 | 0 | 9 | 0 | 3 | 0 | 6 | 0 | 9 | 0 | 39 | 0 |
| Bayesian | 0 | 0 | 8 | 8 | 0 | 0 | 8 | 6 | 3 | 3 | 1 | 0 | 1 | 0 | 21 | 17 |
| Average | 0.6 | 0.0 | 7.8 | 2.2 | 1.2 | 0.0 | 7.2 | 1.8 | 3.0 | 1.8 | 3.0 | 0.0 | 3.6 | 0.0 | 26.4 | 5.8 |
Notes: For a particular taxonomic group, results are given by marker (sum of nodes for all among tree-building methods) and by tree-building method (sum of nodes among different markers). On the first column of each taxonomic group is displayed the number of nodes recovered (correctly or incorrectly), whereas on the second column is the number of nodes recovered with a bootstrap value higher than 90. NT shows non tested groups. The average value for each taxonomic group is shown at the bottom of each table, as well as the sum (#) of each correct and incorrect node totally and the ones higher than 90 (bootstrap value).
For a particular taxonomic group, results are also explicit, given by marker (sum of nodes for all among tree-building methods; Tables 2 and 3) and by tree-building method (sum of nodes among different markers; Tables 2 and 3). For each gene, fully represented by all taxonomic groups, the total number of nodes is 15 (five tree-building methods and three substitution models), whereas for each method, the sum is 39 for mitochondrial (13 genes and three models) and 18 for nuclear genes (six genes and three models). Our results may be regarded as empirical tests of the power (number of correct nodes with >90 values) and type one error (number of incorrect nodes with >90 values) associated with the tests.
Tree building methods
When mitochondrial genes are used (Table 2), ML and Bayesian show the overall highest number of correct nodes (195 CB, correct branches), but if analysis is restricted to significant (BP > 90) nodes, Bayesian outperforms other methods (173 SCB, significant correct branches), followed by UPGMA (141 SCB). If incorrect recovered branches, however, are examined, Bayesian again (78 IB, incorrect branches) perform best. ML and UPGMA also show high efficiency (both 81 IB) with poor support for incorrect branches. In the Bayesian analysis, however, incorrect branches are often significantly supported as theoretically expected.47 MP is the only tree-building method that shows no significantly supported incorrect branches. For nuclear markers (Table 3), as observed for mitochondrial genes, Bayesian method seems to surpass others considering the total number of correct branches (75 CB) and the number of significant correct branches (68 SCB). Taking into account the incorrect branches, Bayesian analysis also performs better (21 IB), but again, they were highly supported by significant nodes. Only MP and ML, show no significantly supported incorrect branches for nuclear markers.
Bayesian approaches were introduced into phylogenetics in the mid-1990s, but became very popular during the 2000’s.42 In spite of the high efficiency in recovering and supporting correct branches in this study, the higher values also correspond to incorrect partitions, indicating that the posterior probability are higher than bootstrap values, whether the node is correct or incorrect as suggested by simulations.48
The high efficiency observed of the neglected UPGMA method is somewhat surprising. This tree-building method is hardly ever used in phylogenetic studies due to its high dependence on molecular clock assumptions.51 It has been shown, however, that, when variances are high, UPGMA algorithm does yield a good tree solution reconstruction for microsatellite data.52 It might be considered that, in some cases, the evolutionary time between the two divergent markers is so long that the evolutionary rates have been equalized, creating an artificially constant substitution rate.53
Furthermore, simulations have shown that ML is robust to model violations54 that it tends to outperform NJ and MP on the Felsenstein zone, that is, when long branches are on the opposite sides of an interior node.54 Nevertheless, ML might not perform as well at the anti-Felsenstein zone, ie, where long branches are neighbors.11 Under realistic parameters Bayesian analysis has been shown highly support incorrect clades and to be susceptible to Long-branch attraction.48,49 Such points might explain our results in which differences among methods were very small.1
Markers and genomes
Among mitochondrial markers (Table 2), Nd5 (91 CB), Nd4l (90 CB) and Atp6 (86 CB) perform best considering number of correctly recovered branches whereas Nd6 (77 SCB) and, again, Atp6 (69 SCB) yielded the highest number of significant ones. If incorrect branches are considered, again, Nd4l has the lead, together with Nd5 (both 15 IB). These mitochondrial markers also exhibit no significantly supported incorrect branches.
Comparing nuclear genes (Table 3), Adora3 shows the highest number of correct partitions (75 CB) recovered, but Vwf exhibits the largest number of significant partitions (63 SCB). Adora3 and Rag2 shows no incorrect branch recovered and Adrb2 performed poorly with 19 significant incorrect branches. Mitochondrial and nuclear results are, in fact, comparable since the same number of tree-building methods and models were used in all alignments regardless of the genome.
Discussion
Examining our results, it is clear that nuclear markers do not necessarily show a better performance than mitochondrial genes.55 Also, we would have expected a higher dependence on the efficiency among mitochondrial genes results when compared with efficiencies among nuclear genes, but our results show differently. Also surprising is the lack of correlation between gene size and gene efficiency that has been shown in previous known tree studies.1
When recovered groups are considered, both mitochondrial and nuclear markers were quite efficient recovering murines, primates, metatherians, and mammals. Rodents, eutherians, and therians, on the other hand, presented much lower recovery rates. Our results show that therians (ie, the branching of mammals excluding monotremes) was the most difficult group to recover as a clade.
For mitochondrial markers, ML (8 CB), Bayesian (5 CB) and MP (1 CB) and markers Atp6 (3 CB), Cox1 (4 CB), Nd4 (3 CB) and Nd5 (4 CB) were able to recover it. Topologies for all mitochondrial markers, with the Nd5 exception, included statistically supported branches that broke therian monophyly. Also, in all mitochondrial based phylogenies, therian monophyly was only significantly supported using the Bayesian method.
Conversely, the nuclear markers Adora3 and Rag2 recovered therians as a clade for all tree building methods and models. Only Adrb2 trees presented nonsignificant breakage of therian monophyly. Therian rupture was usually due to the grouping of marsupials and monotremes, known as the Marsupionta hypothesis.
A closer relationship between monotremes and marsupials has been proposed36 and had some morphological support in the past.56,57 Nevertheless, morphological characters recorded to support Marsupionta, such as the columnar stapes and number of thoracolumbar vertebrae, are now thought to be primitive features, found in many early mammals and even in advanced cynodonts.20 Today, there is little doubt that both Metatheria plus Eutheria constitute a monophyletic group, as indicated by a number of shared synapomorphies, including large stylar cusps A, an extensive conular region in the upper molars and hypoconulid from the ultimate lower molar tall and sharply recurved.23 Monophyletism of Theria seems also to be supported by other molecular data.21,22,24
The other group that was hard to recover was rodents. In that case, however, issues were not as drastic since mitochondrial Atp8, Nd4l and Nd5 presented no incorrectly recovered branch for the group. On the other hand, in the trees reconstructed with Cox1, Cox3, Nd1 and Nd2 markers, rodents were non-monophyletic for all tree-building methods.
Rodent monophyly has been the focus of many studies,58–60 but morphological bases of the natural status of Rodentia are very well established.21 Morphological characters that support the monophyly of Rodentia include the presence of a single pair of (upper and lower) enlarged and ever-growing incisors, incisor enamel restricted to the outer surface, absence of canines, P1/p1, and p2, creating a diastema between incisors and cheek teeth, presence of a long and shallow mandibular fossa, relative deep and short horizontal ramus of the mandible, reduced coronoid process and expanded angular process, among others.61 Also Rodentia monophyly is corroborated by recent and more comprehensive molecular studies,26–29 and contradictory results are now largely attributed to a case of long-branch attraction.62 Besides, a putative reason for the non-monophyletism is that Muridae nucleotide sequences evolve at higher rates, with a large number of substitutions due to the short generation time.63 It has been suggested that this problem may be solved with a larger taxon sampling.64
Recent studies tend to use as many markers as possible but taxon sampling and markers are inversely correlated. Also, the variance is small for larger datasets, increasing the odds in having statistically supported contrasting phylogenies among markers.65 Hence, a few carefully selected genes with increased taxon sampling may be better to unfold a detailed and robust phylogenetic scenario.66 Naturally, marker choice must take into account the taxon sampling and intrinsic marker limitations, but nature, ie, mitochondrial or nuclear, seems to bear no significant difference on marker performance.
Supplementary Materials
Supp 1. GeneBank accession numbers of mitochondrial and nuclear genes for each taxonomic group.
Supp 2. Pair-wise proportion of different residues (p-distance) between all sequence pairs.
Footnotes
Author Contributions
Conceived and designed the experiments: CAMR. Analysed the data: JL-F, CAMR, FAP. Wrote the first draft of the manuscript: JL-F, CAMR. Contributed to the writing of the manuscript: JL-F, CAMR, FAP. Agree with manuscript results and conclusions: JL-F, CAMR, FAP. Jointly developed the structure and arguments for the paper: JL-F, CAMR, FAP. Made critical revisions and approved final version: JL-F, CAMR, FAP. All authors reviewed and approved of the final manuscript.
Funding
The authors wish to thank FAPERJ (Rio de Janeiro State Research Foundation) and CNPq (Brazilian National Research Council) for research grants that enabled this study.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
References
- 1.Russo CAM, Takezaki N, Nei M. Efficiencies of different genes and different tree-building methods in recovering a known vertebrate phylogeny. Mol Biol Evol. 1996;13:525–36. doi: 10.1093/oxfordjournals.molbev.a025613. [DOI] [PubMed] [Google Scholar]
- 2.Zardoya R, Meyer A. Phylogenetic performance of mitochondrial protein-coding gene resolving relationships among vertebrates. Mol Biol Evol. 1996;13:933–42. doi: 10.1093/oxfordjournals.molbev.a025661. [DOI] [PubMed] [Google Scholar]
- 3.Chang H, Fuchs M. Limit theorems for patterns in phylogenetic trees. J Math Biol. 2010;60:481–512. doi: 10.1007/s00285-009-0275-6. [DOI] [PubMed] [Google Scholar]
- 4.San Mauro D, Agorreta A. Molecular systematics: a synthesis of the common methods and the state of knowledge. Cell Mol Biol Lett. 2010;15:311–41. doi: 10.2478/s11658-010-0010-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Towsend JP, López-Giráldez F, Friedman R. The phylogenetic informativeness of nucleotide and amino acid sequences for reconstructing the vertebrate tree. J Mol Evol. 2008;67:437–47. doi: 10.1007/s00239-008-9142-0. [DOI] [PubMed] [Google Scholar]
- 6.Shan Y, Li XQ. Maximum gene-support tree. Evol Bioinf. 2008;4:181–91. doi: 10.4137/ebo.s652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spinks PQ, Thomson RC, Lovely GA, Shaffer B. Assessing what is needed to resolve a molecular phylogeny: simulations and empirical data from emydid turtles. BMC Evol Biol. 2009;9:1–17. doi: 10.1186/1471-2148-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shan Y, Gras R. Genome-wide EST data mining approaches to resolving incongruence of molecular phylogenies. Adv Exp Med Biol. 2010;680:237–43. doi: 10.1007/978-1-4419-5913-3_27. [DOI] [PubMed] [Google Scholar]
- 9.Brinkmann H, Philippe H. Animal phylogeny and large-scale sequencing. J Syst Evol. 2008;46:274–86. [Google Scholar]
- 10.Makowsky R, Cox Christian L, Roelke C, et al. Analyzing the relationship between sequence divergence and nodal support using Bayesian phylogenetic analyses. Mol Phyl Evol. 2010;57:485–94. doi: 10.1016/j.ympev.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Bruno WJ, Halpern AL. Topological bias and inconsistency of maximum likelihood using wrong models. Mol Biol Evol. 1999;16:564–6. doi: 10.1093/oxfordjournals.molbev.a026137. [DOI] [PubMed] [Google Scholar]
- 12.Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press; Oxford; 2000. [Google Scholar]
- 13.Som A. ML or NJ-MCL? A comparison between two robust phylogenetic methods. Comput Biol Chem. 2009;33:373–8. doi: 10.1016/j.compbiolchem.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Cummings MP, Otto SP, Wakeley J. Sampling properties of DNA sequence data in phylogenetic analysis. Mol Biol Evol. 1995;12:814–22. doi: 10.1093/oxfordjournals.molbev.a040258. [DOI] [PubMed] [Google Scholar]
- 15.Benton MJ. Classification and phylogeny of the diapsid reptiles. Zool J Linn Soc. 1985;84:97–164. [Google Scholar]
- 16.Laurin M. The osteology of a lower permian eosuchian from texas and a review of diapsid phylogeny. Zool J Linn Soc. 1991;101:59–95. [Google Scholar]
- 17.Cao Y, Sorenson MD, Kumazawa Y, Mindell DP, Hasegawa M. Phylogenetic position of turtles among amniotes: evidence from mitochondrial and nuclear genes. Gene. 2000;259:139–48. doi: 10.1016/s0378-1119(00)00425-x. [DOI] [PubMed] [Google Scholar]
- 18.Cotton JA, Page RDM. Going nuclear: gene family evolution and vertebrate phylogeny reconciled. Proc R Soc Lond Ser B: Biol Sci. 2002;269:1555–61. doi: 10.1098/rspb.2002.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller J. The relationships among diapsid reptiles and the influence of taxon selection. In: Arratia G, Wilson MVH, Cloutier R, editors. Recent Advances in the Origin and Early Radiation of Vertebrates. Verlag Dr. Friedrich Pfeil; Munich: 2004. pp. 379–408. [Google Scholar]
- 20.Luo ZX, Kielan-Jaworowska Z, Cifelli RL. In quest for a phylogeny of Mesozoic mammals. Acta Palaeont Pol. 2002;47:1–78. [Google Scholar]
- 21.Springer MS, Stanhope MJ, Madsen O, Jong WW. Molecules consolidate the placental mammal tree. Trends Ecol Evol. 2004;19:430–8. doi: 10.1016/j.tree.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 22.Prasad AB, Allard MW. Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol. 2008;25:1795–808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wible JR, Rougier GW, Novacek MJ, Asher RJ. The eutherian mammal Maelestes gobiensis from the late cretaceous of mongolia and the phylogeny of cretaceous eutheria. Bull Amer Mus Nat Hist. 2009;327:1–123. [Google Scholar]
- 24.Meredith RW, Janečka JE, Gatesy J, et al. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science. 2011;334:521–4. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 25.Beck RMD. A dated phylogeny of marsupials using a molecular supermatrix and multiple fossil constraints. J Mammal. 2008;89:175–89. [Google Scholar]
- 26.Lin YH, Waddell PJ, Penny D. Pika and vole mitochondrial genomes increase support for both rodent monophyly and glires. Gene. 2002;294:119–29. doi: 10.1016/s0378-1119(02)00695-9. [DOI] [PubMed] [Google Scholar]
- 27.Poux C, Douzery EJP. Primate phylogeny, evolutionary rate variation, and divergence times: a contribution From the Nuclear Gene. IRBP Am J Anthropol. 2004;124:1–16. doi: 10.1002/ajpa.10322. [DOI] [PubMed] [Google Scholar]
- 28.Bininda-Emonds ORP, Cardillo M, Jones KE, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–11. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 29.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–21. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voelker G, Edwards SV. Can weighting improve bushy trees? Models of cytochrome b evolution and the molecular systematics of pipits and wagtails (Aves: Motacillidae) Syst Biol. 1998;47(4):589–603. doi: 10.1080/106351598260608. [DOI] [PubMed] [Google Scholar]
- 31.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 33.Sneath PH, Sokal RR. Numerical Taxonomy: The Principles and Practice of Numerical Classification. Freeman WH; San Francisco CA: 1973. [Google Scholar]
- 34.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35.Hendy MD, Penny D. Branch and bound algorithms to determine minimal evolutionary trees. Math Biosci. 1982;59:277–90. [Google Scholar]
- 36.Kumar S, Tamura K, Nei M. Mega: Molecular Evolutionary Genetic Analysis. Pennsylvania State University; University Park: 1993. [Google Scholar]
- 37.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 38.Kimura M. A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 39.Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 40.Nei M. Phylogenetic analysis in molecular evolutionary genetics. Annu Rev Genet. 1996;30:371–403. doi: 10.1146/annurev.genet.30.1.371. [DOI] [PubMed] [Google Scholar]
- 41.Lemmon AR, Moriarty EC. The importance of proper model assumption in Bayesian Phylogenetics. Syst Biol. 2004;53:265–77. doi: 10.1080/10635150490423520. [DOI] [PubMed] [Google Scholar]
- 42.Ronquist F, Deans AR. Bayesian Phylogenetics and its influence on insect systematics. Annu Rev Entomol. 2010;55:189–206. doi: 10.1146/annurev.ento.54.110807.090529. [DOI] [PubMed] [Google Scholar]
- 43.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 44.Russo CAM. Efficiencies of different statistical tests in supporting a known-vertebrate phylogeny. Mol Biol Evol. 1997;14:1078–80. doi: 10.1093/oxfordjournals.molbev.a025714. [DOI] [PubMed] [Google Scholar]
- 45.Hillis D, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–92. [Google Scholar]
- 46.Ewens WJ, Grant GR. Statistical Methods in Bioinformatics. 2nd ed. New York: 2005. [Google Scholar]
- 47.Misawa K, Nei M. Reanalysis of Murphy et al’s data gives various mammalian phylogenies and suggests overcredibility of Bayesian trees. J Mol Evol. 2003;57(S1):S290–6. doi: 10.1007/s00239-003-0039-7. [DOI] [PubMed] [Google Scholar]
- 48.Misawa K, Nei M. Reanalysis of Murphy et al’s data gives various mammalian phylogenies and suggests overcredibility of bayesian trees. J Mol Evol. 2003;57(1):290–6. doi: 10.1007/s00239-003-0039-7. [DOI] [PubMed] [Google Scholar]
- 49.Kolaczkowski B, Thornton JW. A mixed branch length model of heterotachy improves phylogenetic accuracy. Mol Biol Evol. 2008;25(6):1054–66. doi: 10.1093/molbev/msn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kolaczkowski B, Thornton JW. Long-branch attraction bias and inconsistency in bayesian phylogenetics. PLoS ONE. 2009;4(12):e7891. doi: 10.1371/journal.pone.0007891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pamilo P. Tests of phenograms based on genetic distances. Mol Biol Evol. 1990;12:689–97. doi: 10.1111/j.1558-5646.1990.tb05948.x. [DOI] [PubMed] [Google Scholar]
- 52.Takezaki N, Nei M. Genetic distances and reconstruction of phylogenetic trees from microsateillite DNA. Genetics. 1996;144:389–99. doi: 10.1093/genetics/144.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorne JL, Kishino H, Painter IS. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol. 1998;15:1647–57. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- 54.Katoh K, Kuma T, Miyata T. Genetic algorithm-based maximum- likelihood analysis for molecular phylogeny. J Mol Evol. 2001;53:477–84. doi: 10.1007/s002390010238. [DOI] [PubMed] [Google Scholar]
- 55.Springer MS, DeBry RW, Douady C, et al. Mitochondrial versus nuclear gene sequences is deep-level mammalian phylogeny reconstruction. Mol Biol Evol. 2001;18:132–43. doi: 10.1093/oxfordjournals.molbev.a003787. [DOI] [PubMed] [Google Scholar]
- 56.Kühne WG. The Systematic Position of Monotremes Reconsidered (Mammalia) Z Morph Tiere. 1973;75:59–64. [Google Scholar]
- 57.Gregory WK. The monotremes and the palimpsest theory. Bull Amer Mus Nat Hist. 1947;88:1–52. [Google Scholar]
- 58.Graur D, Hide WA, Li W. Is the guinea-pig a rodent? Nature. 1991;351:649–52. doi: 10.1038/351649a0. [DOI] [PubMed] [Google Scholar]
- 59.D’Erchia AM, Gissi C, Pesole G, Saccone C, Arnason U. The guinea-pig is not a rodent. Nature. 1996;381:597–600. doi: 10.1038/381597a0. [DOI] [PubMed] [Google Scholar]
- 60.Blanga-Kanfi S, Miranda H, Penn O, Pupko T, DeBry RW, Huchon D. Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evol Biol. 2009;9:71. doi: 10.1186/1471-2148-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meng J, Wyss AR. Glires (Lagomorpha, Rodentia) In: Rose KD, Archibald JD, editors. The Rise of Placental Mammals. Baltimore: The John Hopkins University Press; 2001. pp. 145–58. [Google Scholar]
- 62.Bergsten J. A review of long-branch attraction. Cladistics. 2005;21(2):163–93. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- 63.Li WH, Ellsworth DL, Krushkal J, Chang BHJ, Hewett-Emmett D. Rates of nucleotide substitution in primates and rodents and the generation-time effect hypothesis. Mol Phylogenet Evol. 1996;5:182–7. doi: 10.1006/mpev.1996.0012. [DOI] [PubMed] [Google Scholar]
- 64.Huchon D, Madsen O, Sibbald MJJB, et al. Rodent phylogeny and a timescale for the evolution of glires: evidence from an extensive taxon sampling using three nuclear. Genes Mol Biol Evol. 2002;19:1053–65. doi: 10.1093/oxfordjournals.molbev.a004164. [DOI] [PubMed] [Google Scholar]
- 65.Kumar S, Filipski AJ, Battistuzzi FU, Kosakovsky Pond SL, Tamura K. Statistics and truth in phylogenomics. Mol Biol Evol. 2012;29:457–72. doi: 10.1093/molbev/msr202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nishihara H, Okada N, Hasegawa M. The power and pitfalls of phylogenomics. Genome Biol. 2007;8:R199. doi: 10.1186/gb-2007-8-9-r199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp 1. GeneBank accession numbers of mitochondrial and nuclear genes for each taxonomic group.
Supp 2. Pair-wise proportion of different residues (p-distance) between all sequence pairs.