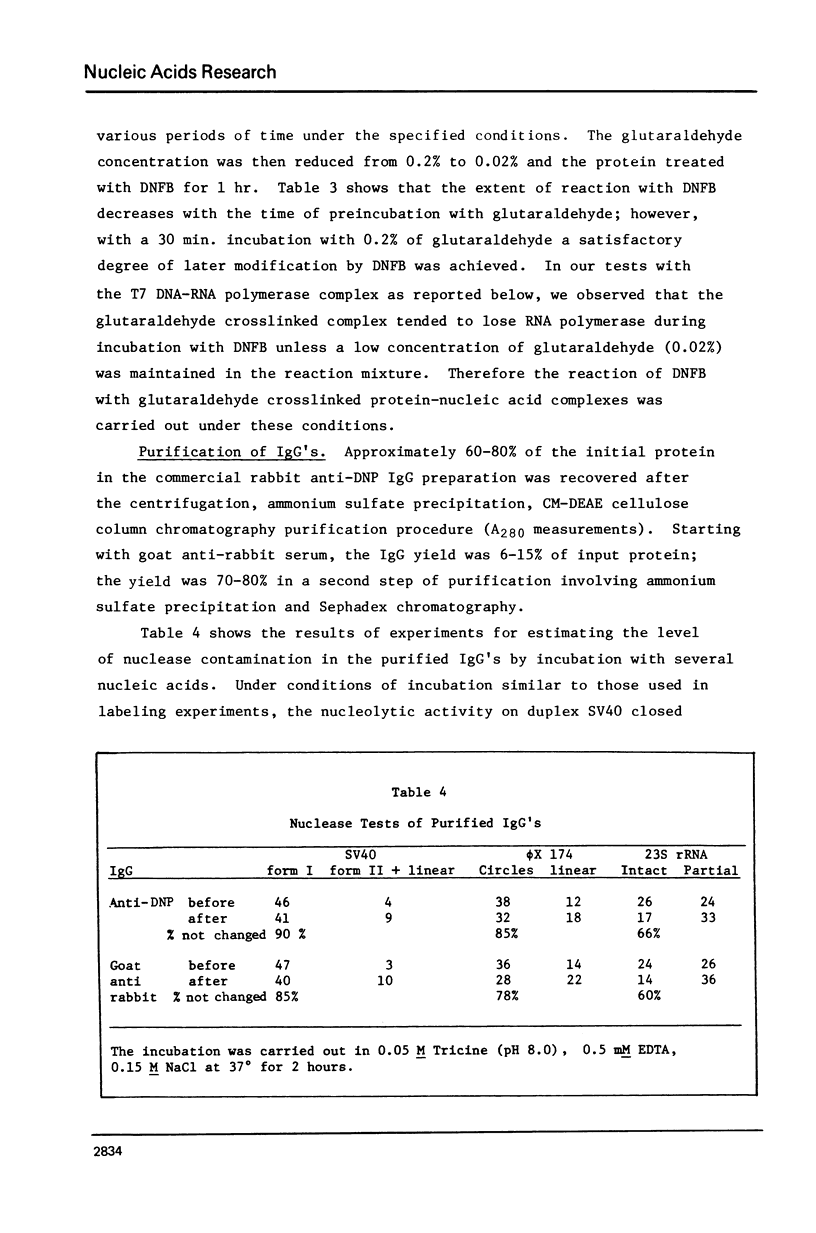
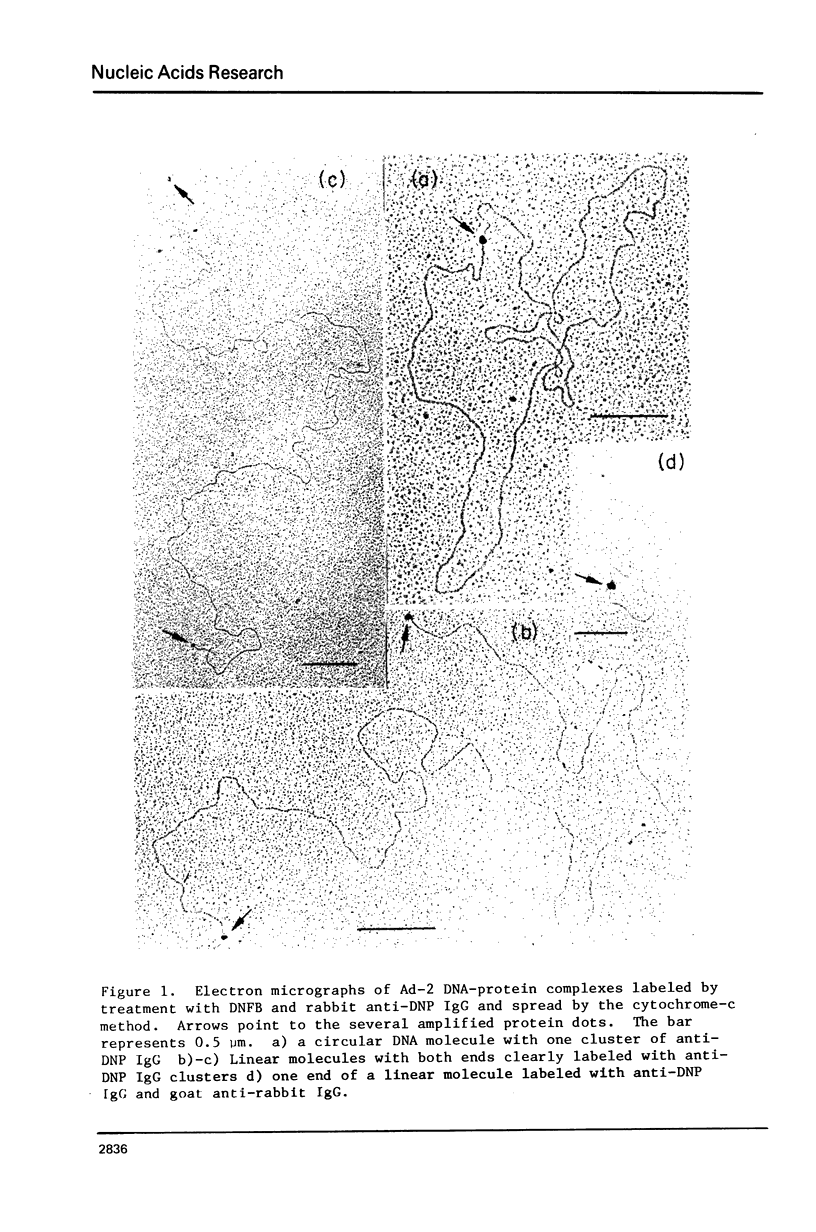
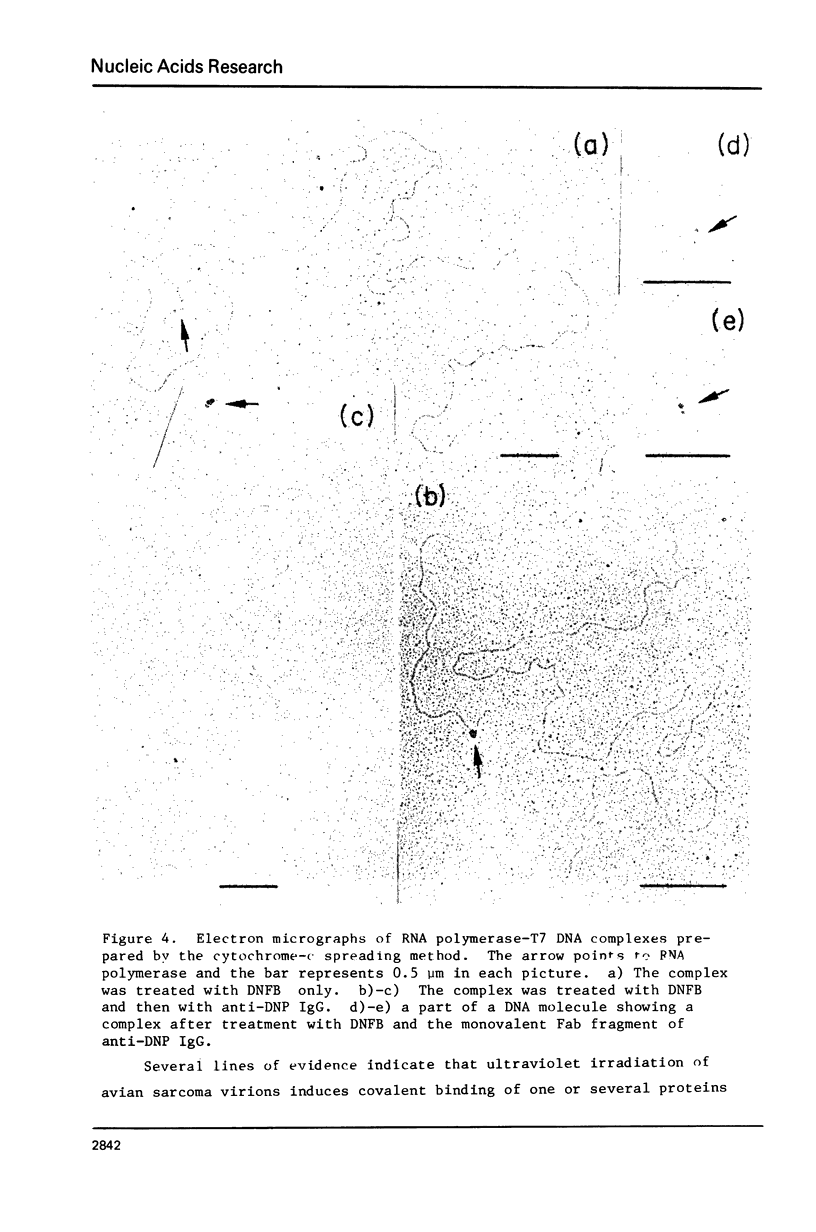
Abstract
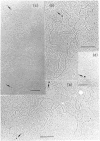
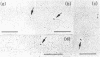
An electron microscopic method for demonstrating the presence of and mapping the positions of proteins specifically bound to nucleic acids is described. The nucleic acid-protein complex is treated with dinitrofluorobenzene under conditions such that dinitrophenyl (DNP) groups are attached to nucleophilic groups on the protein, with only a low level of random attachment to the nuclei acid. This product is treated with rabbit anti-DNP IgG. The position of the protein-(DNP)n(IgG)m complex on the nucleic acid strand can be observed by electron microscopy by protein free spreading methods and, in many cases, by cytochrome-c spreading. If necessary for visualization by the latter method, the size of the labeled region can be increased by treatment with goat anti-rabbit IgG. High efficiency of electron microscopic labeling is achieved. Examples studied are: the adenovirus-2 DNA terminal protein, a protein covalently bound to SV40 DNA, DNA polymerase I bound to DNA, E. coli RNA polymerase bound to T7 DNA, and proteins UV crosslinked to avian sarcoma virus RNA.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albring M., Griffith J., Attardi G. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1348–1352. doi: 10.1073/pnas.74.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W., Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell. 1976 Apr;7(4):595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- Bister K., Varmus H. E., Stavnezer E., Hunter E., Vogt P. K. Biological and biochemical studies on the inactivation of avian oncoviruses by ultraviolet irradiation. Virology. 1977 Apr;77(2):689–704. doi: 10.1016/0042-6822(77)90492-5. [DOI] [PubMed] [Google Scholar]
- Bordier C., Dubochet J. Electron microscopic localization of the binding sites of Escherichia coli RNA polymerase in the early promoter region of T7 DNA. Eur J Biochem. 1974 May 15;44(2):617–624. doi: 10.1111/j.1432-1033.1974.tb03519.x. [DOI] [PubMed] [Google Scholar]
- Brack C., Eberle H., Bickle T. A., Yuan R. Mapping of recognition sites for the restriction endonuclease from Escherichia coli K12 on bacteriophage PM2 DNA. J Mol Biol. 1976 Dec 15;108(3):583–593. doi: 10.1016/s0022-2836(76)80138-6. [DOI] [PubMed] [Google Scholar]
- Bunnett J. F., Hermann D. H. Kinetics of dinitrophenylation of amino acids. Biochemistry. 1970 Feb 17;9(4):816–822. doi: 10.1021/bi00806a014. [DOI] [PubMed] [Google Scholar]
- Griffith J., Huberman J. A., Kornberg A. Electron microscopy of DNA polymerase bound to DNA. J Mol Biol. 1971 Jan 28;55(2):209–214. doi: 10.1016/0022-2836(71)90192-6. [DOI] [PubMed] [Google Scholar]
- Hinkle D. C., Chamberlin M. J. Studies of the binding of Escherichia coli RNA polymerase to DNA. I. The role of sigma subunit in site selection. J Mol Biol. 1972 Sep 28;70(2):157–185. doi: 10.1016/0022-2836(72)90531-1. [DOI] [PubMed] [Google Scholar]
- Hirsh J., Schleif R. High resolution electron microscopic studies of genetic regulation. J Mol Biol. 1976 Dec;108(2):471–490. doi: 10.1016/s0022-2836(76)80131-3. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Kung H. J., Davidson N. An electron microscope study of Sindbis virus RNA. Cold Spring Harb Symp Quant Biol. 1974;38:943–950. doi: 10.1101/sqb.1974.038.01.096. [DOI] [PubMed] [Google Scholar]
- Kasamatsu H., Wu M. Protein-SV40 DNA complex stable in high salt and sodium dodecyl sulfate. Biochem Biophys Res Commun. 1976 Feb 9;68(3):927–936. doi: 10.1016/0006-291x(76)91234-1. [DOI] [PubMed] [Google Scholar]
- Kasamatsu H., Wu M. Structure of a nicked DNA-protein complex isolated from simian virus 40: covalent attachment of the protein to DNA and nick specificity. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1945–1949. doi: 10.1073/pnas.73.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller T., Sogo J. M., Bujard H. An electron microscopic method for studying nucleic acid-protein complexes. Visualization of RNA polymerase bound to the DNA of bacteriophages T7 and T3. Biopolymers. 1974 May;13(5):995–1009. doi: 10.1002/bip.1974.360130514. [DOI] [PubMed] [Google Scholar]
- Kung H. J., Bailey J. M., Davidson N., Vogt P. K., Nicolson M. O., McAllister R. M. Electron microscope studies of tumor virus RNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):827–834. doi: 10.1101/sqb.1974.039.01.096. [DOI] [PubMed] [Google Scholar]
- NISONOFF A., WISSLER F. C., LIPMAN L. N., WOERNLEY D. L. Separation of univalent fragments from the bivalent rabbit antibody molecule by reduction of disulfide bonds. Arch Biochem Biophys. 1960 Aug;89:230–244. doi: 10.1016/0003-9861(60)90049-7. [DOI] [PubMed] [Google Scholar]
- Palacios R., Palmiter R. D., Schimke R. T. Identification and isolation of ovalbumin-synthesizing polysomes. I. Specific binding of 125 I-anti-ovalbumin to polysomes. J Biol Chem. 1972 Apr 25;247(8):2316–2321. [PubMed] [Google Scholar]
- Reed S. I., Ferguson J., Davis R. W., Stark G. R. T antigen binds to simian virus 40 DNA at the origin of replication. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1605–1609. doi: 10.1073/pnas.72.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. J., Bellett J. D. A circular DNA-protein complex adenoviruses and its possible role in DNA replication. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):523–531. doi: 10.1101/sqb.1974.039.01.064. [DOI] [PubMed] [Google Scholar]
- Robinson A. J., Younghusband H. B., Bellett A. J. A circula DNA-protein complex from adenoviruses. Virology. 1973 Nov;56(1):54–69. doi: 10.1016/0042-6822(73)90287-0. [DOI] [PubMed] [Google Scholar]
- Sanger F. The free amino groups of insulin. Biochem J. 1945;39(5):507–515. doi: 10.1042/bj0390507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A., Todaro G. J. The genome-associated, specific RNA binding proteins of avian and mammalian type C viruses. Cell. 1977 Jan;10(1):91–99. doi: 10.1016/0092-8674(77)90143-x. [DOI] [PubMed] [Google Scholar]
- Stillman B. W., Bellett A. J., Robinson A. J. Replication of linear adenovirus DNA is not hairpin-primed. Nature. 1977 Oct 20;269(5630):723–725. doi: 10.1038/269723a0. [DOI] [PubMed] [Google Scholar]
- Tabak H. F., Griffith J., Geider K., Schaller H., Kornberg A. Initiation of deoxyribonucleic acid synthesis. VII. A unique location of the gap in the M13 replicative duplex synthesized in vitro. J Biol Chem. 1974 May 25;249(10):3049–3054. [PubMed] [Google Scholar]
- Vollenweider H. J., Koller T. Physical mapping of Qbeta replicase binding sites on Qbeta RNA. J Mol Biol. 1976 Mar 5;101(3):367–377. doi: 10.1016/0022-2836(76)90153-4. [DOI] [PubMed] [Google Scholar]
- Vollenweider H. J., Sogo J. M., Koller T. A routine method for protein-free spreading of double- and single-stranded nucleic acid molecules. Proc Natl Acad Sci U S A. 1975 Jan;72(1):83–87. doi: 10.1073/pnas.72.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland B. C., Szybalski W., Ris H. Mapping of deletions and substitutions in heteroduplex DNA molecules of bacteriophage lambda by electron microscopy. Science. 1969 Mar 21;163(3873):1343–1348. doi: 10.1126/science.163.3873.1343. [DOI] [PubMed] [Google Scholar]
- Williams R. C. Use of polylysine for adsorption of nuclei acids and enzymes to electron microscope specimen films. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2311–2315. doi: 10.1073/pnas.74.6.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]