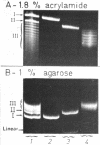
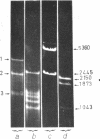
Abstract
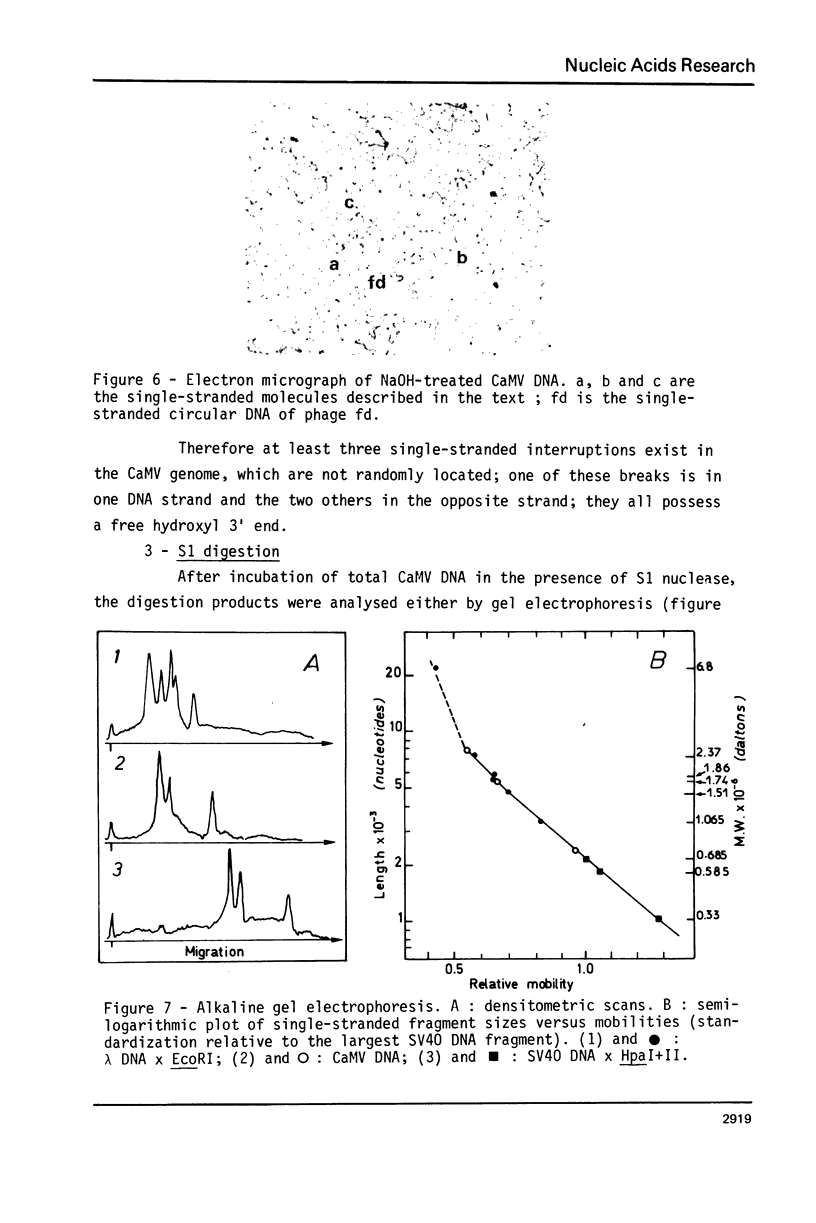
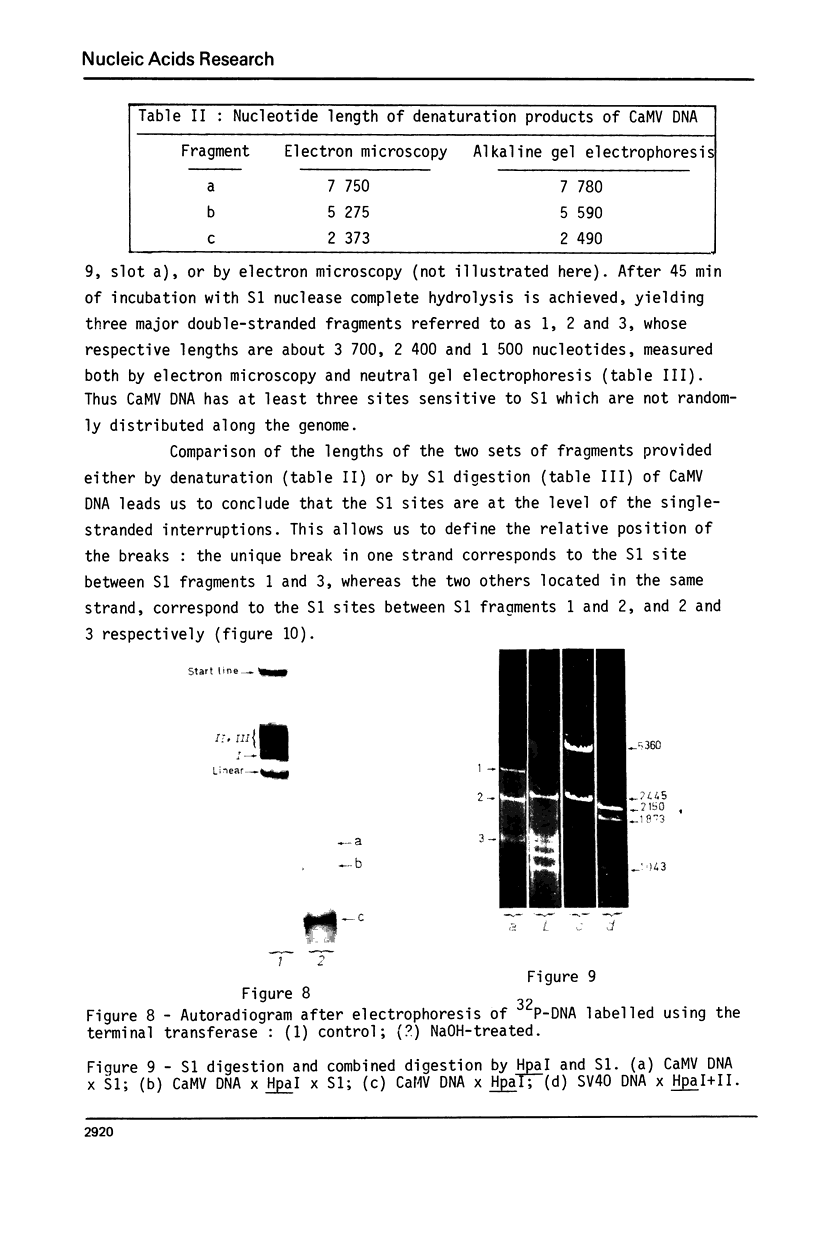
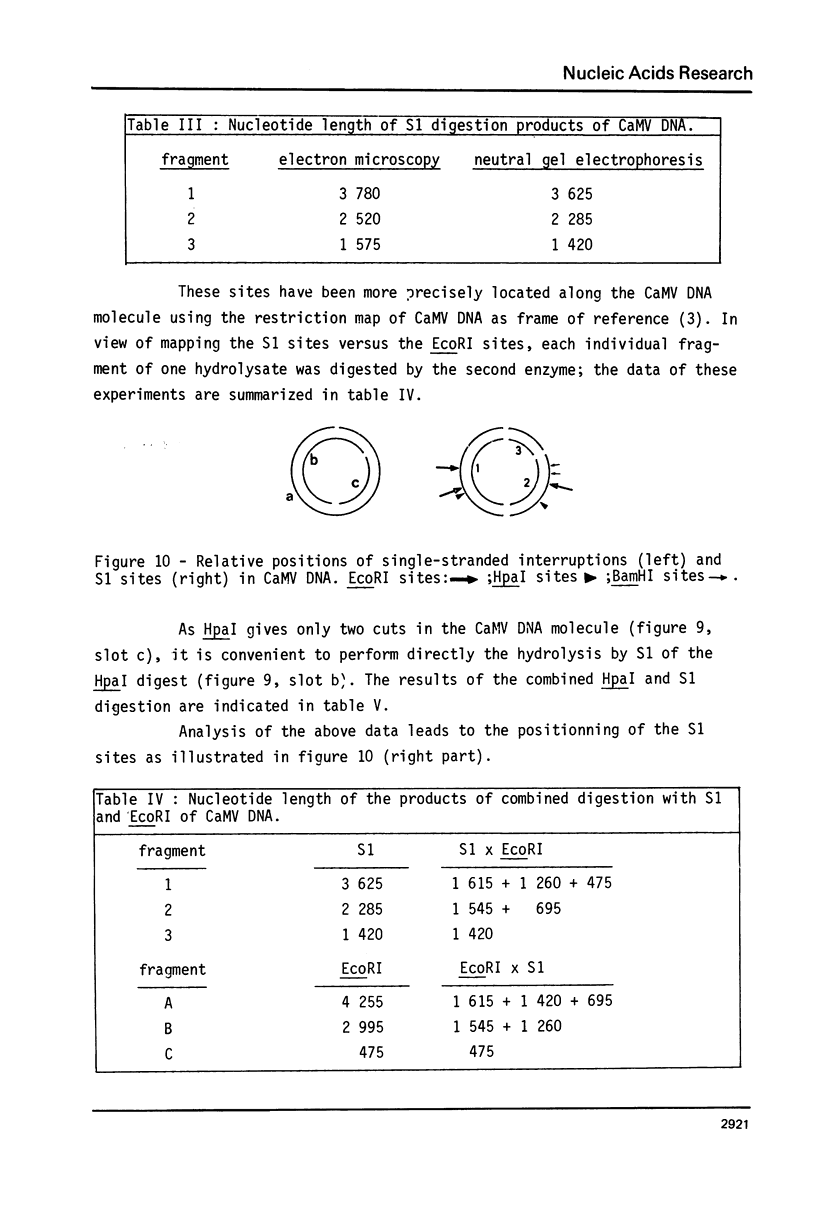
The Cauliflower Mosaic Virus (CaMV) genome is a double-stranded DNA molecule of about 5 million daltons. Native DNA molecules appear heterogeneous when analysed by gel electrophoresis. We have examined the nature of this apparent heterogeneity. Besides, this genome is shown here to contain three single-stranded breaks, as revealed by different denaturation experiments: heating at 75 degrees C, treatment with NaOH or dimethyl sulfoxide (DMSO). Labelling with terminal transferase proves that the 3' ends at these interruptions all have free hydroxyl groups. Electron microscopy and alkaline gel electrophoresis indicate that these three discontinuities are shared by both strands, and that they are not randomly located. S1 nuclease is active on CaMV DNA and generates three fragments. The comparison between the sizes of these fragments and of the products of denaturation leads us to consider that S1 acts at the level of the interruptions. We have determined that two of them, distant by one third genome unit, are in the same strand; the other is in the opposite strand, distant by one sixth genome unit from the nearest other one. The combined use of restriction enzymes and S1 nuclease has enabled us to locate these three discontinuities on the restriction map of the CaMV genome that we have otherwise established.
Full text
PDF


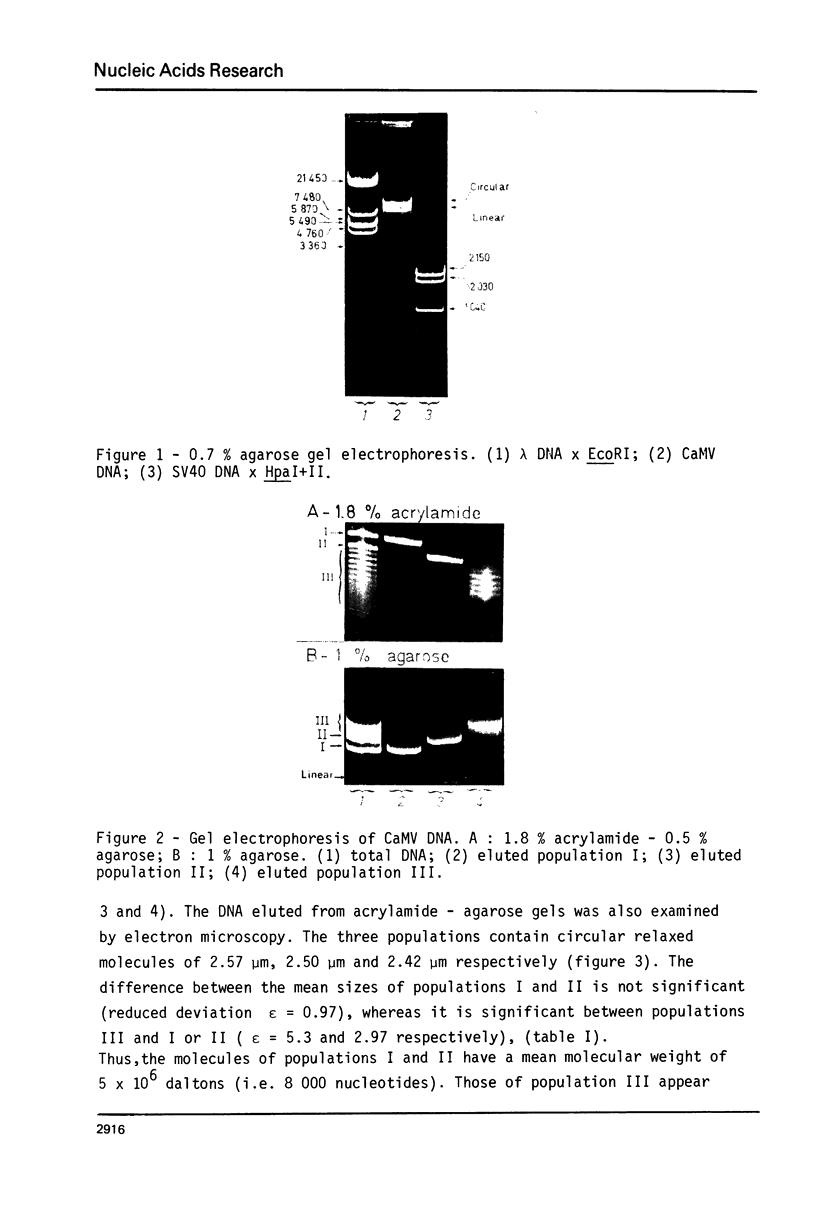
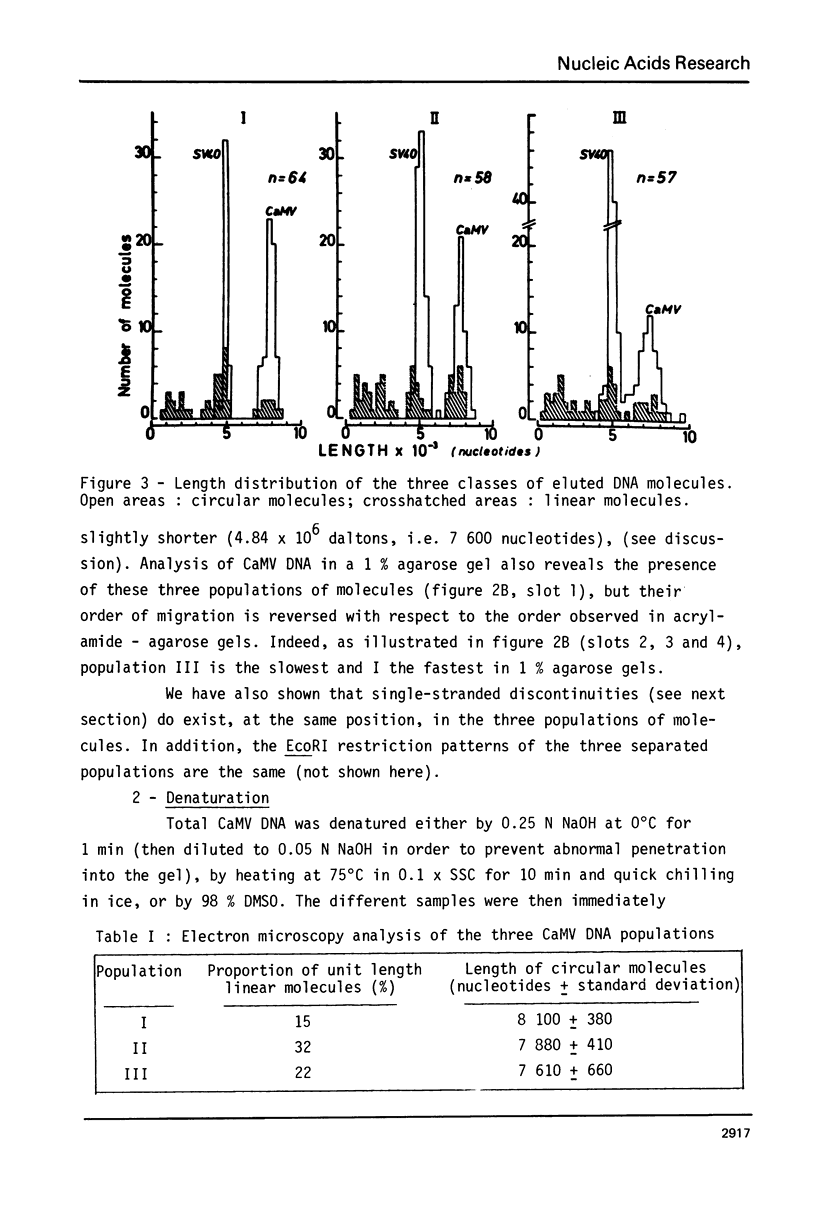





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brack C., Eberle H., Bickle T. A., Yuan R. A map of the sites on bacteriophage PM2 DNA for the restriction endonucleases HindIII and HpaII. J Mol Biol. 1976 Jun 14;104(1):305–309. doi: 10.1016/0022-2836(76)90016-4. [DOI] [PubMed] [Google Scholar]
- Danna K. J., Sack G. H., Jr, Nathans D. Studies of simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol. 1973 Aug 5;78(2):363–376. doi: 10.1016/0022-2836(73)90122-8. [DOI] [PubMed] [Google Scholar]
- Hull R., Shepherd R. J. The structure of cauliflower mosaic virus genome. Virology. 1977 Jun 1;79(1):216–230. doi: 10.1016/0042-6822(77)90346-4. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Meagher R. B., Shepherd R. J., Boyer H. W. The structure of cauliflower mosaic virus. I. A restriction endonuclease map of cauliflower mosaic virus DNA. Virology. 1977 Jul 15;80(2):362–375. doi: 10.1016/s0042-6822(77)80012-3. [DOI] [PubMed] [Google Scholar]
- PIRONE T. P., POUND G. S., SHEPHERD R. J. Purification and properties of cauliflower mosaic virus. Nature. 1960 May 21;186:656–657. doi: 10.1038/186656b0. [DOI] [PubMed] [Google Scholar]
- Roychoudhury R., Jay E., Wu R. Terminal labeling and addition of homopolymer tracts to duplex DNA fragments by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 1976 Jan;3(1):101–116. doi: 10.1093/nar/3.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell G. J., Follett E. A., Subak-Sharpe J. H., Harrison B. D. The double-stranded DNA of cauliflower mosaic virus. J Gen Virol. 1971 Jun;11(3):129–138. doi: 10.1099/0022-1317-11-3-129. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Shepherd R. J., Bruening G. E., Wakeman R. J. Double-stranded DNA from cauliflower mosaic virus. Virology. 1970 Jun;41(2):339–347. doi: 10.1016/0042-6822(70)90086-3. [DOI] [PubMed] [Google Scholar]
- Shepherd R. J. DNA viruses of higher plants. Adv Virus Res. 1976;20:305–339. doi: 10.1016/s0065-3527(08)60508-4. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Nathans D. Letter: A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J Mol Biol. 1973 Dec 15;81(3):419–423. doi: 10.1016/0022-2836(73)90152-6. [DOI] [PubMed] [Google Scholar]
- Studies on bacteriophage fd DNA I. A cleavage map of the fd genome. J Mol Biol. 1975 Jun 15;95(1):21–31. [PubMed] [Google Scholar]
- Subramanian K. N., Zain B. S., Roberts R. J., Weissman S. M. Mapping of the HhaI and HinfI cleavage sites on simian virus 40 DNA. J Mol Biol. 1977 Feb 25;110(2):297–317. doi: 10.1016/s0022-2836(77)80074-0. [DOI] [PubMed] [Google Scholar]
- Summers J., O'Connell A., Millman I. Genome of hepatitis B virus: restriction enzyme cleavage and structure of DNA extracted from Dane particles. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4597–4601. doi: 10.1073/pnas.72.11.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Wilson G. A., Young F. E. Isolation of a sequence-specific endonuclease (BamI) from Bacillus amyloliquefaciens H. J Mol Biol. 1975 Sep 5;97(1):123–125. doi: 10.1016/s0022-2836(75)80028-3. [DOI] [PubMed] [Google Scholar]