Abstract
Background
MicroRNAs (miRNAs) are a group of short (~22 nt) noncoding RNAs that specifically regulate gene expression at the post-transcriptional level. miRNA precursors (pre-miRNAs), which are imperfect stem loop structures of ~70 nt, are processed into mature miRNAs by cellular RNases III. To date, thousands of miRNAs have been identified in different organisms. Several viruses have been reported to encode miRNAs.
Findings
Here, we extended the analysis of miRNA-encoding potential to the Anatid herpesvirus 1 (AHV-1). Using computational approaches, we found that AHV-1 putatively encodes 12 mature miRNAs. We then compared the 12 mature miRNAs candidates with the all known miRNAs of the herpesvirus family. Interestingly, the “seed sequences” (nt 2 to 8) of 2 miRNAs were predicted to have the high conservation in position and/or sequence with the 2 miRNAs of Marek’s disease virus type 1 (MDV-1). Additionally, we searched the targets from viral mRNAs.
Conclusions
Using computational approaches, we found that AHV-1 putatively encodes 12 mature miRNAs and 2 miRNAs have the high conservation with the 2 miRNAs of MDV-1. The result suggested that AHV-1 and MDV-1 should have closed evolutionary relation, which provides a valuable evidence of classification of AHV-1. Additionally, seven viral gene targets were found, which suggested that AHV-1 miRNAs could affect its own gene expression.
Keywords: Anatid herpesvirus 1, microRNA, Conservation, Computational analyses
Findings
MicroRNAs (miRNAs) are noncoding small RNA molecules with important regulatory functions in expression of target genes [1,2]. The miRNAs are about 19 to 25 nucleotides (nt) long. They are firstly transcribed as long primary miRNAs, which are processed into 60–70 nt miRNA precursor (pre-miRNA) by nuclear RNase III Drosha [3]. Pre-miRNA is then exported to the cytoplasm by the export factor Exportin 5 and further cleaved into ∼ 22 nt duplexes [4]. Mature miRNAs regulate protein-coding gene expression via the RNA silencing machinery, typically by forming imperfect duplexes with target messenger RNAs (mRNAs).
To date, thousands of miRNAs have been identified in different organisms [5]. The discovery of miRNAs encoded by DNA viruses suggests that viruses have evolved to exploit RNA silencing for regulation of the expression of their own genes, host genes, or both [6]. Most viral miRNAs (vmiRNAs) have been identified by cDNA cloning of small RNAs from virus-infected cells [7-10], whereas others have been identified following computational prediction and hybridization analysis [10-12]. Experimental screening of vmiRNAs via high-throughput sequencing of large numbers of cDNA clones from infected cells is technically challenging, time consuming and could be incomplete, given that viral gene expression can have highly constrained tissue-, time-, and replication state-specific patterns [12]. Of the vmiRNAs identified so far, most are encoded by viruses in the herpesvirus family, containing α [13-16], β [17], γ subfamily [8,9,18]. AHV-1, an unassigned virus in the family Herpesviridae, can induce duck viral enteritis in waterfowl of the family Anatidae. To query whether this strategy is also employed by AHV-1 or not, we have analyzed putative miRNA-encoding capacity of AHV-1.
The AHV-1 miRNA prediction was performed using the complete genome sequence of AHV-1 strain CHv (JQ647509) [19]. The genome size is 162,175 nt. Figure 1 shows a flowchart of the computational prediction process. Briefly, the viral genome was scanned for hairpin-structured miRNA precursors using VMir Analyzer program [20,21]. 197 sequences with potential hairpin-like structures were extracted as candidate miRNA precursors. Then candidates within or antisense to protein-coding regions were removed according to the NCBI genome annotations. 50 precursors were further identified using MiPred program (http://www.bioinf.seu.edu.cn/miRNA/) and the sequences with lower minimum free energy (equivalent or below −25 kcal.mol-1) were remained, subsequently, the remained 24 real pre-microRNA sequences were conducted BLASTn searching against itself to remove repeated sequences. Finally, 16 sequences were selected as miRNA precursors candidates. At the last step, the mature sequences were predicted by Bayes-SVM-MiRNA web server v1.0 (http://wotan.wistar.upenn. edu/BayesSVMmiRNAfind/). After that, 12 mature sequences were predicted with 21nt in length (Table 1) and the secondary structure of pre-miRNAs were shown in Figure 2.
Figure 1.
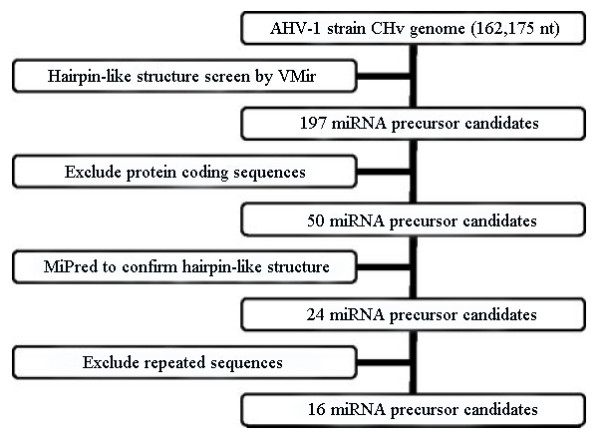
Flowchart of the AHV-1 miRNA prediction procedure.
Table 1.
Sequences and genomic positions of putative AHV-1 miRNAs
| No. |
Predicted mature miRNA sequence (5’ to 3’) |
Position, orientation |
Location |
|---|---|---|---|
| 1 |
CUCCCUUGCUUUGACAUGUCC |
26325-26345, - |
Within intergenic region between UL44 and UL45 |
| 2 |
UCGUUGGGCGGUUUCUUCGUG |
72302-72322, - |
Within intergenic region between UL26 and UL27 |
| 3 |
UUCAAACGGAGGCGUUGUGCG |
72512-72532, + |
Within intergenic region between UL26 and UL27 |
| 4 |
UUUCUGGGACCUCACCGCGGA |
79262-79282, + |
Within intergenic region between UL22 and UL23 |
| 5 |
UAAGAACUGCUGGUACCUUGC |
112559-112579, + |
Within intergenic region between UL4 and UL5 |
| 6 |
CAACGGAUGAACGUCGGCGCG |
112720-112740, - |
Within intergenic region between UL4 and UL5 |
| 7 |
CAUGGGAACAUUUAACACCCC |
123128-123148, + |
Within intergenic region between ICP22 and LORF2 |
| 8 |
UGGAUGGUUUGGAGACAGCUG |
125173-125193, + |
Within intergenic region between ICP4 and ICP22 |
| 9 |
UAUGUUUUGCCCGGGCAAAUG |
132907-132927, + |
Within intergenic region between US1 and ICP4 |
| 10 |
AAAUCUGGCGUUCGCACUCUG |
134522-134542, - |
Within intergenic region between US1 and ICP4 |
| 11 |
AUUUCGGAGUGCGAAUAUGUG |
134586-134606, - |
Within intergenic region between US1 and ICP4 |
| 12 | GGUAGGUUGUUUGGAGAUUGC | 160321-160341, + | Within unique short terminal repeat region |
Figure 2.
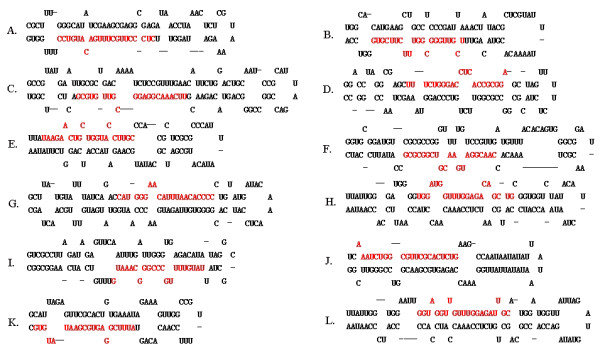
Secondary structure predictions of AHV-1 pre-miRNAs. The putative mature miRNAs sequence were shown in red. A-L in order named AHV-1-pre-miR-1 to AHV-1-pre-miR-12.
In order to investigate whether the AHV-1 miRNAs predicted are conserved in other herpesviruses, each of the 12 putative mature AHV-1 miRNA candidates was compared with the all known miRNAs of the herpesvirus family independently in database (http://www.sanger.ac.uk/Software/Rfam/mirna/). Interestingly, the “seed sequences” (nt 2 to 8) of 2 miRNAs were predicted to have the high conservation with the 2 miRNAs of MDV-1 (Table 2). The genomic positions of the 2 miRNAs encoded by MDV-1 are proximal to the latencyassociated transcript region. The vmiRNAs have generally been reported to lack sequence conservation across different viral species [10], with the exception of the primate polyomaviruses [11]. But among closed viral species, they could showed conservation in position and/or sequence. Jurak et al identified 16 and 17 miRNAs expressed by herpes simplex viruses 1 and 2 (HSV-1 and −2), respectively. The genomic positions of most miRNAs encoded by these two viruses are within or proximal to the latency associated transcript region. Nine miRNAs are conserved in position and/or sequence, particularly in the seed region, between these two viruses [16]. Additional, Waidner et al reported the genome locations, but not microRNA sequences, are conserved among four avian herpesviruses, infectious laryngotracheitis virus (ILTV) and herpesvirus of turkeys (HVT), as well as Marek's disease viruses (MDV-1 and MDV-2). Most are clustered in the repeats flanking the unique long region (I/TRL), except in ILTV which lacks these repeats [14]. So the result suggested that AHV-1 and MDV-1 should have closed evolutionary relation, which provides a valuable evidence of classification of AHV-1. Meanwhile, the prevalence of microRNAs in the genomic repeat regions suggests that the latent infection in herpesviruses could be relevant to function of microRNA.
Table 2.
miRNA homologs expressed by AHV-1 and MDV-1
| Name | miRNA sequencea |
No. of identical nt/total in seed region (nt positions 2–8) |
|---|---|---|
| AHV-1-pre-miR-7 |
-CAUGGGAACAUU-UAACACCCC GCAUGGAAACGUCCUGGGAAA- - * * * * * †* * * † * † * † † * |
5/7 |
| MDV-1-miR-M13 |
|
|
| AHV-1-pre-miR-9 |
UAUGUUUUGCCCGGGCAAAUG-UCUGUUGUUCCGUAGUGUUCUC * * * * * * * * † * † † † |
5/7 |
| MDV-1-miR-M6-5p |
a Sequences of orthologous miRNAs expressed by AHV-1 and MDV-1 aligned using ClustalW2. The seed sequences are shown in boldface. Identical nucleotides and nucleotides with conserved target binding potential are indicated with * and †, respectively.
What is the function of the vmiRNA? In order to know whether the vmiRNA could modulate its own genes expression, we checked the 3’UTR of viral mRNAs that could perfectly complement with the “seed sequence” of vmiRNA. AHV-1-pre-miR-4 was predicted to target UL29 gene (DNA replication-recombination; binds single-stranded DNA) and US5 gene (unknown function). AHV-1-pre-miR-7 was predicted to target UL16 gene (capsid maturase). AHV-1-pre-miR-9 was predicted to target UL15B gene (DNA cleavage-encapsidation), UL45 (tegument/envelope protein), and US1 gene (immediate-early and late transrepressor protein). AHV-1-pre-miR-12 was predicted to target UL45 gene and US7 gene (cell-cell spread). However, none of gene targets were found for the other vmiRNAs. Additionally, we wonder whether the putative vmiRNA could be used by AHV-1 to modulate host cell genes expression profiles. But so far genome of duck is in the process of being annotated and there is not available 3’UTR database of duck genes, so prediction can not be carried on.
Here, we introduced a concept that the AHV-1 genome could reasonably encode candidate pre-miRNAs. Studies are in progress to experimentally identify the putative vmiRNAs during AHV-1 infection.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JX carried out most of the data collection, data analysis and drafted the manuscript. ACC, MSW, SCZ, DKZ, RYJ, SC, YZ, XYW and XYC helped draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jun Xiang, Email: mypersonalbest_07927@126.com.
Anchun Cheng, Email: chenganchun@vip.163.com.
Mingshu Wang, Email: mshwang@163.com.
Shunchuan Zhang, Email: zsc060606@gmail.com.
Dekang Zhu, Email: zhudk24@gmail.com.
Renyong Jia, Email: cqrcjry@yahoo.com.cn.
Shun Chen, Email: sophia_cs@163.com.
Yi Zhou, Email: ZhouYi196111@163.com.
Xiaoyu Wang, Email: renyupeng1986@163.com.
Xiaoyue Chen, Email: Anchun@sicau.edu.cn.
Acknowledgements
The research was supported by China 973 program(2011CB111606), National Natural Science Foundation of China (31072157), China Agricultural Research System (CARS-43-8), the Changjiang Scholars and Innovative Research Team of Sichuan Agricultural University (PCSIRT0848), and National science and technology support program for agriculture(2011BAD34B03). Anchun Cheng and Mingshu Wang are the corresponding authors at: Institute of Preventive Veterinary Medicine, Sichuan Agricultural University & Key Laboratory of Animal Disease and Human Health of Sichuan Province, Sichuan Agricultural University, Wenjiang, Chengdu city, Sichuan, 611130, P.R. China & Avian Disease Research Center, College of Veterinary Medicine of Sichuan Agricultural University, 46# Xinkang Road, Yucheng district, Yaan 625014, P.R.China. Tel.: +86 835 2885774; fax: +86 835 2885774. E-mail address: chenganchun@vip.163.com. (A. Cheng); mshwang@163.com (M. Wang).
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Structural requirements for pre-microRNAbinding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Ganem D. MicroRNAs and viral infection. Mol Cell. 2005;20:3–7. doi: 10.1016/j.molcel.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Gra¨sser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci USA. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Gra¨sser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- Cui C, Griffiths A, Li GL, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. Prediction and identification of Herpes Simplex Virus 1-encoded microRNAs. J Virol. 2006;80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R, Anderson A, Bernberg E, Kamboj S, Huang E, Lagasse G, Isaacs G, Parcells M, Meyers BC, Green PJ, Burnside J. Sequence conservation and differential expression of Marek's disease virus microRNAs. J Virol. 2008;82:12213–12220. doi: 10.1128/JVI.01722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidner LA, Morgan RW, Anderson AS, Bernberg EL, Kamboj S, Garcia M, Riblet SM, Ouyang M, Isaacs GK, Markis M, Meyers BC, Green PJ, Burnside J. MicroRNAs of Gallid and Meleagrid herpesviruses show generally conserved genomic locations and are virus-specific. Virology. 2009;388:128–136. doi: 10.1016/j.virol.2009.02.043. [DOI] [PubMed] [Google Scholar]
- Glazov EA, Horwood PF, Assavalapsakul W, Kongsuwan K, Mitchell RW, Mitter N, Mahony TJ. Characterization of microRNAs encoded by the bovine herpesvirus 1 genome. J Gen Virol. 2010;91:32–41. doi: 10.1099/vir.0.014290-0. [DOI] [PubMed] [Google Scholar]
- Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, Coen DM. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J Virol. 2010;84:4659–4672. doi: 10.1128/JVI.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W, Trang P, Zhong Q, Yang E, Belle CV, Liu FY. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. 2005;7:1684–1695. doi: 10.1111/j.1462-5822.2005.00598.x. [DOI] [PubMed] [Google Scholar]
- Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Wu, Anchun Cheng, Mingshu Wang, Qiao Yang, Dekang Zhu, Renyong Jia, Shun Chen, Yi Zhou, Xiaoyu Wang, Xiaoyue Chen. Complete genomic sequence of chinese virulent duck enteritis virus. J Virol. 2012;86:5965. doi: 10.1128/JVI.00529-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullivan CS, Grundhoff A. Identification of Viral MicroRNAs. Methods Enzymol. 2007;427:1–23. doi: 10.1016/S0076-6879(07)27001-6. [DOI] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]


