The widespread availability of peptides and oligonucleotides synthesized by solid-phase methods has had a profound impact upon biology and medicine, with a myriad of important uses in research, diagnostics, and therapeutics. A limitation of current technologies is the relatively short length of the molecules that can be synthesized, as determined by the stepwise reaction yield, and thus peptides and oligonucleotides are usually restricted to lengths below approximately 50 amino acids or 100 nucleotides (nt), respectively. This synthetic limitation has driven interest in the development of alternative approaches for the production of full-length genes and proteins. The most common strategy has been to splice together shorter segments into a full-length, functional assembly; for example, the Staudinger ligation reaction permits full-length proteins to be constructed from a series of peptides,[1] and full-length genes can be obtained from multiple short single strands in a series of sequential ligation steps[2] or by polymerase cycling assembly (PCA).[3] However, the assembly based strategies for gene synthesis reported to date remain laborious, expensive, and time-consuming, and thus have not yet provided the level of accessibility needed for widespread utility.
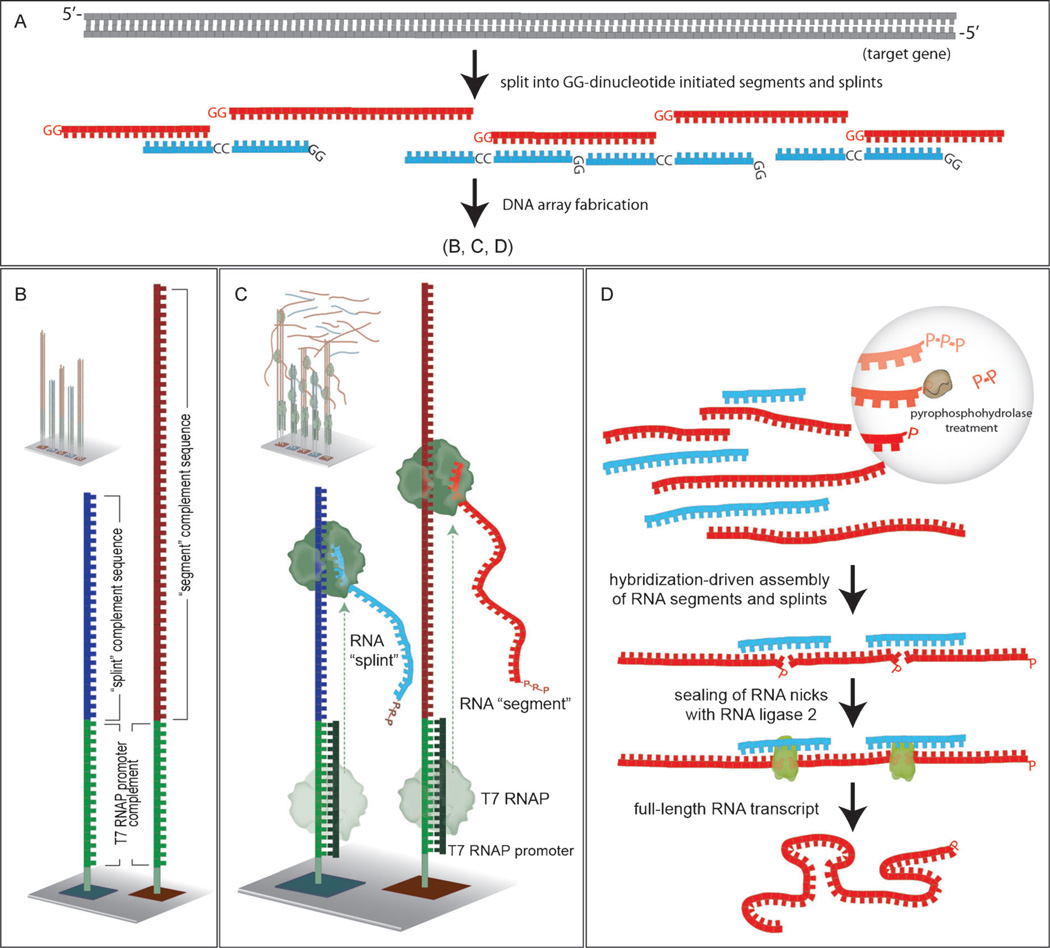
We present herein a strategy for the assembly of full-length RNA transcripts from DNA array elements (Figure 1). In this approach, each element of the DNA array includes a T7 RNA polymerase promoter sequence at the 5′ end. Transcription from these surface-bound promoters yields many RNA copies of the oligonucleotide elements encoded in the array. These amplified RNA molecules self-assemble to yield the desired full-length transcript. The transcript, once synthesized, is readily copied by reverse transcription polymerase chain reaction (RT-PCR) to yield the corresponding gene.
Figure 1.
Illustration of the RNA-mediated gene assembly process. A) Design of segment (red) and splint (blue) sequences to be employed. B) Segment (dark red) and splint (dark blue) complement sequences as synthesized on the DNA array with the complement of the T7 RNAP promoter sequence (green) at their 3′-termini. C) Hybridization of an oligonucleotide encoding the T7 RNAP promoter sequence yields the necessary double-stranded promoter, and addition of RNA polymerase causes transcription to occur. D) RNA segments and splints have their terminal triphosphate units trimmed to monophosphates, assembly occurs by RNA:RNA hybridization, and nicks are sealed to yield the desired full-length RNA.
We designed an oligonucleotide array with the sequences necessary to produce a full-length transcript for the fluorescent protein ZsGreen1. We chose ZsGreen1 for a proof-of-principle demonstration for several reasons: a) the protein is relatively small in size, consisting of 231 amino acids; b) it has been shown to fold correctly under in vitro translation conditions; and c) it is fluorescent and thus its translation is easily monitored. A full-length RNA transcript, comprising the 696 nt that encode ZsGreen1 and an additional 10 nt corresponding to the Kozak consensus sequence (5′-GGT CGC CAC C-3′, added to the 5′ end of the RNA transcript to enhance eukaryotic in vitro translation efficiency[4]), was assembled from RNAs produced from photolithographically fabricated oligonucleotide arrays. The 706 nt RNA molecule was divided into 18 segment sequences ranging in length from 18 to 58 nt, and 17 splints of 32 nt each (Supporting Information).
Figure 1 depicts the process of generating RNA sequences from a DNA microarray, and their subsequent assembly and ligation to produce the desired full-length RNA molecule. The process consists of six successive steps, as follows: a) design the oligonucleotide array; b) fabricate the array; c) produce many RNA copies of each array element (“splints” and “segments”, see Figure 1 and the text below) using T7 RNA polymerase; d) remove pyrophosphate from 5′ terminal triphosphates on the splints and segments with RNA 5′ pyrophosphohydrolase; e) allow self-assembly of the splints and segments into the desired full-length construct by RNA:RNA hybridization; and f) seal the nicks with T4 RNA ligase 2. This final RNA product may then be converted into a DNA copy by reverse transcription, whereupon it may be either cloned, or employed directly to produce more full-length RNAs for in vitro translation or other purposes.
Oligonucleotide arrays were designed to encode “segment sequences”, which are the sections of the desired full-length RNA transcript, and “splint sequences”, which are complementary RNAs that serve as templates to direct the correct assembly of the RNA segments (Figure 1A). Two parameters determined the choice of segment and splint sequences. First, each segment had to be at least 30 nt in length, to provide at least two 15 nt stretches of sequence for hybridization during assembly (the last segment however, is not subject to this limitation, and was only 18 nt in length, Supporting Information). Second, it was required that the 5′ end of each RNA transcript corresponded to a GG dinucleotide, based upon the higher efficiency of transcription exhibited by T7 RNA polymerase (T7 RNAP) when multiple guanine nucleotides are present at the 5′ terminus of the transcript being synthesized (see Figure 1A).[5] GGG trinucleotide sequences at the 5′ terminus were avoided however, as they have been shown to give rise to a ladder of poly G transcripts in which the number of G residues can range from 1–3, attributed to “slippage” of the enzyme during coupling of GTP.[6]
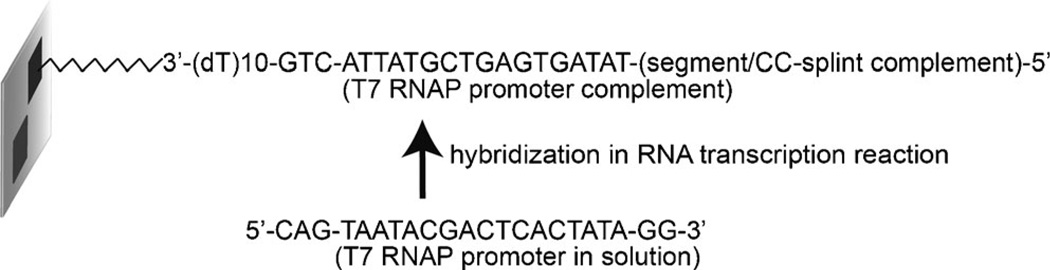
These design criteria yielded 18 segment sequences to encompass the desired 706 nt transcript. Each of the 17 splint sequences had a length of 32 nt, corresponding to two 15 nt regions complementary to the segments that it was to join, and an additional 5′ GG dinucleotide to enhance transcription efficiency. Each surface-bound oligonucleotide also included at the 3′ end a ten base dT spacer sequence,[7] and the three base sequence CTG to improve the hybridization stability of the T7 RNA polymerase complement (see below). The overall design of the surface-bound oligonucleotides is illustrated in Figure 2, and thus consists of five different sequences; a 3′-(dT)10 spacer, a CTG trinucleotide, the 17 mer T7 promoter sequence, a CC dinucleotide, and finally the desired segment or splint sequence. To make the necessary double-stranded DNA T7 RNA polymerase promoter, the 22 nt complementary strand (consisting of a 5′-CAG, the 17 nt T7 promoter complement, and two 3′ guanines) is included in the T7 RNA transcription reaction. The addition of RNA polymerase results in the synthesis of multiple copies of each RNA segment from each oligonucleotide sequence (Figure 1C).
Figure 2.
Design details of the surface-bound oligonucleotides, along with their complements from solution.
The DNA arrays used in this case were synthesized in situ, in a base-by-base manner, using maskless array synthesizer (MAS) technology.[8] The arrays were synthesized on either glass or amorphous carbon substrates with similar results. Silanized glass substrates are the industry standard for DNA microarrays, whereas we have found that DNA arrays fabricated on amorphous carbon substrates are more stable than their glass analogues to prolonged incubations at elevated temperatures and repeated hybridization cycles.[9]
The fidelity of the oligonucleotide sequences on the microarray is of critical importance for the correct assembly of a full-length RNA transcript. The light-directed synthesis methods used in this work were thoroughly optimized to maximize sequence fidelity and to reduce the number of errors that occur during array fabrication. Synthesis errors (which can result in truncates, incorrect sequences, etc.) are not detrimental to hybridization-based assays, but can have adverse consequences in the production of useful gene and protein products. The Supporting Information contains the methods employed in the present work, and highlights the differences from previously published methods.[8b, 9b]
Milligan et al. have shown that T7 RNA polymerase will produce RNAs from single-stranded synthetic DNA templates having a duplex DNA promoter, producing hundreds to thousands of RNA transcripts per template molecule.[5, 10] This amplification capability is central to the approach described herein, as the increased concentrations of segment and splint strands drive the hybridization-based assembly process, obviating the need for further PCR amplification prior to the PCA employed in all other gene assembly strategies reported to date.[11]
The assembly of the RNA segment sequences into the full-length RNA transcript includes ligation with T4 RNA ligase 2. However, the transcripts generated by T7 RNA polymerase are triphosphorylated and therefore must be “trimmed” to their monophosphorylated analogues before ligation. This trimming is accomplished by treatment of the transcript pool with RNA 5′ pyrophosphohydrolase (Figure 1D), removing a pyrophosphate group from the 5′ end of each RNA. The assembled RNA segments are then ligated with T4 RNA ligase 2 to produce the desired full-length transcript. The pyrophosphate removal and ligation steps utilize a compatible buffer, which permits them to be performed successively, in a single tube, without intervening buffer-exchange steps and thereby simplifies the overall assembly process. T4 RNA ligase 2 with ATP is thus added directly into the RNA 5′ pyrophosphohydrolase-treated reaction mixture, which contains the RNA segments and splints from the oligonucleotide array. The RNA product was reverse transcribed and PCR amplified using forward and reverse primers for the ZsGreen1 gene. The reverse primer included a sequence encoding six histidine residues to enable His-tag purification of the protein product.[12]
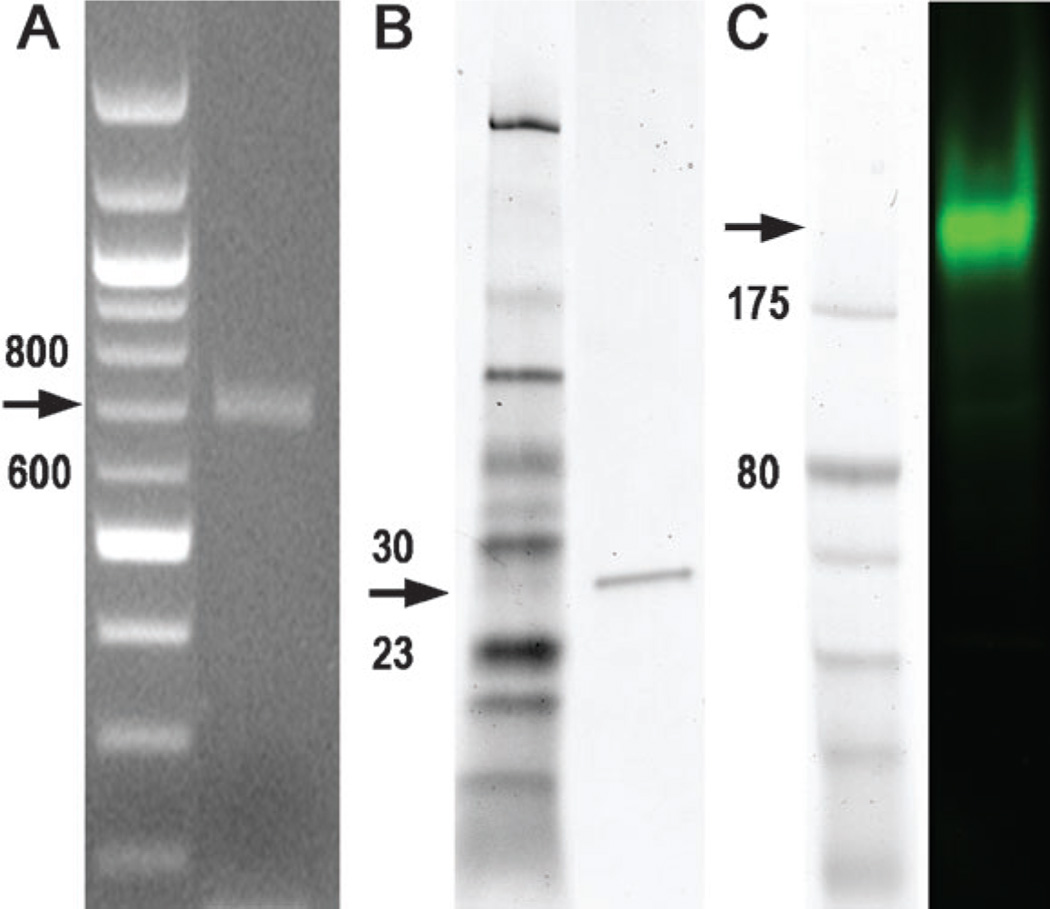
The fidelity of the assembly process was monitored in four ways. First, the RT-PCR product was analyzed by agarose gel electrophoresis. Figure 3A shows that a single DNA band of the expected size (714 bp) is obtained. It is likely that a variety of incomplete products also form during the assembly process, but as the RT-PCR step uses primers from the ends of the desired final construct, such incomplete products are not amplified and therefore are not visible on the gel. Second, the RT-PCR product was subjected to in vitro translation and the resultant protein product was analyzed by reducing sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE). Figure 3B shows that only a single band of the expected molecular weight (26 950 daltons) is visible by Coomassie Blue staining. Third, the same protein product was analyzed by nonreducing SDS-PAGE and detected by fluorescence imaging. Figure 3C shows that only a single fluorescent protein is observed under these nonreducing electrophoretic conditions. Finally, we cloned the PCR product directly (without enzymatic error corrections), and subjected 51 randomly chosen colonies to Sanger sequencing. 22 of the clone sequences were a perfect match to the desired target sequence; in total 33711 bases of DNA sequence were obtained and 49 transitions, three transversions, two deletions, and one insertion were identified (1.63 errors/kb; see Supporting Information). This high rate of generation of the correct gene sequence (22/51 ≈ 40%) is invaluable for practical applications of gene-synthesis technology.
Figure 3.
Gel electrophoresis of ZsGreen1 gene and protein synthesized from the RNA assemblies. A) Agarose gel electrophoresis of the product of RT-PCR amplification of the assembled ZsGreen1 transcript with a ZsGreen1 forward primer and a His-tag-appended ZsGreen1 reverse primer, along with a 100 bp DNA ladder marker (left). The expected size is 714 bp. No RT-PCR product was detected in control experiments without template. B) Electrophoretic analysis in a reducing SDS polyacrylamide gel of the ZsGreen1 protein product obtained in an E. coli cell-free expression system from the gene shown in (A). The expected size is 26.9 kDa. C) Electrophoretic analysis in a nonreducing SDS polyacrylamide gel of the same protein product and standard marker set, as shown in (B). ZsGreen1 exists under nonreducing conditions as a tetramer of theoretical molecular weight 107.6 kDa. It does not migrate true to the expected molecular weight under these nonreducing gel conditions.
Gene assembly from DNA arrays was first described in 2004,[11a] and has since been the subject of several other reports.[11b,d–f, 13] Its allure lies in the potential to make complete genes as rapidly and inexpensively as single oligonucleotides are made today, enabled by the ability of DNA arrays to easily provide many thousands of oligonucleotides for assembly. However, gene assembly has remained a costly and laborious endeavor. Reasons for this include: a) the oligonucleotides that are synthesized on DNA arrays must be cleaved from the surface prior to use and are impure, containing many truncated or chemically modified sequences and thus necessitating various labor- and time-intensive purification or error correction procedures;[11a,b,d–f, 13b] b) only minute amounts of oligonucleotide are made per array feature, necessitating complicated amplification strategies that include adaptor ligation and several other steps;[11a,b,d–f,13b] c) virtually all strategies reported to date are based upon PCA,[11, 13] which although widely used, is complex, laborious, and prone to error.[14]
Previous work on gene assembly from oligonucleotide arrays has employed the DNA sequences themselves, rather than assembling RNA intermediates as in this work. The generation of an RNA intermediate has several advantages: a) approximately 100 to 1000 copies of the RNA are produced by transcription from each DNA strand present on the array;[10] this obviates the need for complex PCR-based oligonucleotide amplification[15] prior to gene assembly;[11e,f] b) parallel gene assembly of the RNA segment and splint sequences, directly from the oligonucleotide array, eliminates a number of laborious steps (e.g., cleavage of the oligonucleotides from the array, amplification of the oligonucleotide pool, and purification of the oligonucleotide pool); c) the sequencing results obtained in the present study show that the full-length RNA transcripts produced have a high sequence fidelity (i.e., a low number of incorrect sequences), whereas the individual oligonucleotides produced during in situ syntheses may include a variety of defects owing to side reactions and incomplete nucleotide-coupling reactions.[16] Sequence errors that are present on the array are presumably copied into the RNA transcripts; however, these deleterious sequences may be incorporated less often into the full-length RNA transcripts owing to the additional sequence fidelity constraints innate to the hybridization/ligation assembly procedure; d) the assembled product is an RNA transcript that is readily copied into DNA for cloning or for production of more RNA copies by in vitro transcription. The RNA-mediated assembly process described herein is also considerably simpler and more rapid than previously described multi-step and multi-day strategies,[11e,f] involving only four successive enzymatic procedures that are readily performed in a few hours (Supporting Information, Table S1).
There are several interesting directions in which to pursue the present work. First, although we provide here a proof-of-principle demonstration of the feasibility of RNA-mediated gene assembly, it will be necessary to undertake the synthesis of many different genes to ascertain what, if any, limitations exist with respect to the universality of the approach. To this end, we have begun to improve and generalize the design principles employed, developing algorithms to select segment and splint sequences that hybridize with similar thermodynamic stabilities, avoiding the use of RNA sequences that fold into excessively stable secondary structures,[17] and exploring the effects of relaxing the requirement of a 5′ terminal GG dinucleotide to a single G residue. Second, we need to explore the length limits of the strategy and determine how long a gene construct it will be possible to assemble in a robust manner. Third, it will be interesting to explore the ability to assemble multiple genes in parallel, which may then them-selves be assembled into larger final constructs. For example, it would be advantageous to be able to assemble ten constructs of 1 kb each and to then stitch them together into a final construct of 10 kb, perhaps using conventional overlapping PCR. Even more ambitious goals are readily imagined, such as the assembly, in a step-wise manner, of large gene clusters, chromosomes, or even genomes.
In summary, we have described a strategy for the RNA-mediated assembly of genes from DNA arrays. Proof-of-principle was demonstrated in the assembly of a small gene encoding the green fluorescent protein, ZsGreen1 and its in vitro translation to yield a functional protein. Sequence analysis of cloned constructs indicated a yield of correct constructs of approximately 40%.
Supplementary Material
Footnotes
This work was supported by the Wisconsin Center of Excellence in Genomics Science (USA), through NIH/NHGRI grant 1P50HG004952. We gratefully acknowledge Gloria M. Kreitinger for assistance with Figure preparation. We thank Yi-Chun Shih for helping with sequence designs. The ZsGreen1 encoding plasmid was a gift from Ya-Fang Chiu.
Supporting information for this article (experimental details) is available on the WWW under http://dx.doi.org/10.1002/anie.201109058.
References
- 1.Nilsson BL, Soellner MB, Raines RT. Annu. Rev. Biophys. Biomol. Struct. 2005;34:91–118. doi: 10.1146/annurev.biophys.34.040204.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itakura K, Hirose T, Crea R, Riggs AD, Heyneker HL, Bolivar F, Boyer HW. Science. 1977;198:1056–1063. doi: 10.1126/science.412251. [DOI] [PubMed] [Google Scholar]
- 3.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 4.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin CT, Muller DK, Coleman JE. Biochemistry. 1988;27:3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- 7.Guo Z, Guilfoyle RA, Thiel AJ, Wang RF, Smith LM. Nucleic Acids Res. 1994;22:5456–5465. doi: 10.1093/nar/22.24.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Singh-Gasson S, Green RD, Yue YJ, Nelson C, Blattner F, Sussman MR, Cerrina F. Nat. Biotechnol. 1999;17:974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]; b) Phillips MF, Lockett MR, Rodesch MJ, Shortreed MR, Cerrina F, Smith LM. Nucleic Acids Res. 2008;36:e7. doi: 10.1093/nar/gkm1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Lockett MR, Smith LM. Anal. Chem. 2009;81:6429–6437. doi: 10.1021/ac900807q. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lockett MR, Weibel SC, Phillips MF, Shortreed MR, Sun B, Corn RM, Hamers RJ, Cerrina F, Smith LM. J. Am. Chem. Soc. 2008;130:8611–8613. doi: 10.1021/ja802454c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milligan JF, Uhlenbeck OC. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 11.a) Richmond KE, Li MH, Rodesch MJ, Patel M, Lowe AM, Kim C, Chu LL, Venkataramaian N, Flickinger SF, Kaysen J, Belshaw PJ, Sussman MR, Cerrina F. Nucleic Acids Res. 2004;32:5011–5018. doi: 10.1093/nar/gkh793. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tian JD, Gong H, Sheng NJ, Zhou XC, Gulari E, Gao XL, Church G. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]; c) Xiong AS, Yao QH, Peng RH, Li X, Fan HQ, Cheng ZM, Li Y. Nucleic Acids Res. 2004;32:e98. doi: 10.1093/nar/gnh094. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kim C, Kaysen J, Richmond K, Rodesch M, Binkowski B, Chu L, Li M, Heinrich K, Blair S, Belshaw P, Sussman M, Cerrina F. Microelectron. Eng. 2006;83:1613–1616. [Google Scholar]; e) Kosuri S, Eroshenko N, LeProust EM, Super M, Way J, Li JB, Church GM. Nat. Biotechnol. 2010;28:1295–1299. doi: 10.1038/nbt.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Matzas M, Stahler PF, Kefer N, Siebelt N, Boisguerin V, Leonard JT, Keller A, Stahler CF, Haberle P, Gharizadeh B, Babrzadeh F, Church GM. Nat. Biotechnol. 2010;28:1291–1294. doi: 10.1038/nbt.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hochuli E, Bannwarth W, Dobeli H, Gentz R, Stuber D. Bio/Technology. 1988;6:1321–1325. [Google Scholar]
- 13.a) Quan JY, Saaem I, Tang N, Ma SM, Negre N, Gong H, White KP, Tian JD. Nat. Biotechnol. 2011;29:449–452. doi: 10.1038/nbt.1847. [DOI] [PubMed] [Google Scholar]; b) Borovkov AY, Loskutov AV, Robida MD, Day KM, Cano JA, Le Olson T, Patel H, Brown K, Hunter PD, Sykes KF. Nucleic Acids Res. 2010;38:e180. doi: 10.1093/nar/gkq677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong AS, Peng RH, Zhuang J, Liu JG, Gao F, Chen JM, Cheng ZM, Yao QH. Biotechnol. Adv. 2008;26:121–134. doi: 10.1016/j.biotechadv.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Cleary MA, Kilian K, Wang YQ, Bradshaw J, Cavet G, Ge W, Kulkarni A, Paddison PJ, Chang K, Sheth N, Leproust E, Coffey EM, Burchard J, McCombie WR, Linsley P, Hannon GJ. Nat. Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- 16.a) Gao X, Gaffney BL, Senior M, Riddle RR, Jones RA. Nucleic Acids Res. 1985;13:573–584. doi: 10.1093/nar/13.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pon RT, Damha MJ, Ogilvie KK. Nucleic Acids Res. 1985;13:6447–6465. doi: 10.1093/nar/13.18.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Pon RT, Usman N, Damha MJ, Ogilvie KK. Nucleic Acids Res. 1986;14:6453–6470. doi: 10.1093/nar/14.16.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Crippa S, Digennaro P, Lucini R, Orlandi M, Rindone B. Gazz. Chim. Ital. 1993;123:197–203. [Google Scholar]
- 17.All of the RNA segments employed for the assembly of ZsGreen1 have higher Gibbs free energy (less stable) for secondary structure folding than for hybridization to their complementary splints.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.